Abstract
In addition to phosphatidylglycerol (PG), cardiolipin (CL), and phosphatidylethanolamine (PE), Sinorhizobium meliloti also possesses phosphatidylcholine (PC) as a major membrane lipid. The biosynthesis of PC in S. meliloti can occur via two different routes, either via the phospholipid N-methylation pathway, in which PE is methylated three times in order to obtain PC, or via the phosphatidylcholine synthase (Pcs) pathway, in which choline is condensed with CDP-diacylglycerol to obtain PC directly. Therefore, for S. meliloti, PC biosynthesis can occur via PE as an intermediate or via a pathway that is independent of PE, offering the opportunity to uncouple PC biosynthesis from PE biosynthesis. In this study, we investigated the first step of PE biosynthesis in S. meliloti catalyzed by phosphatidylserine synthase (PssA). A sinorhizobial mutant lacking PE was complemented with an S. meliloti gene bank, and the complementing DNA was sequenced. The gene coding for the sinorhizobial phosphatidylserine synthase was identified, and it belongs to the type II phosphatidylserine synthases. Inactivation of the sinorhizobial pssA gene leads to the inability to form PE, and such a mutant shows a greater requirement for bivalent cations than the wild type. A sinorhizobial PssA-deficient mutant possesses only PG, CL, and PC as major membrane lipids after growth on complex medium, but it grows nearly as well as the wild type under such conditions. On minimal medium, however, the PE-deficient mutant shows a drastic growth phenotype that can only partly be rescued by choline supplementation. Therefore, although choline permits Pcs-dependent PC formation in the mutant, it does not restore wild-type-like growth in minimal medium, suggesting that it is not only the lack of PC that leads to this drastic growth phenotype.
Rhizobia are soil bacteria that are able to form a symbiosis with legume plants that leads to the formation of nitrogen-fixing root nodules. The formation and functioning of this symbiosis are based on specific recognition of signal molecules, which are produced by both the bacterium and the plant partner. Recognition factors of the bacterial endosymbiont include lipochitin oligosaccharides or nodulation (Nod) factors, extracellular polysaccharides, lipopolysaccharides, K-antigens, and cyclic glucans (55), and these factors are required for nodule formation, the infection process, and the colonization of the root nodule. Recently it was demonstrated that adequate levels of certain bacterial membrane lipids, i.e., phosphatidylcholine (PC), also are required in order to allow the formation of a fully functional symbiosis between Bradyrhizobium japonicum and its host plant soybean (38). Under conditions of phosphate limitation, phosphorus-free membrane lipids (sulfolipids, ornithine-containing lipids, and diacylglyceryl-N,N,N-trimethylhomoserine lipids) are formed in rhizobia (22). Rhizobial mutants lacking the ability to form any one of these phosphorus-free membrane lipids form effective nitrogen-fixing root nodules (31), demonstrating that not all major bacterial membrane lipids are required for the onset of a successful symbiosis.
Escherichia coli is the prokaryote with the best-studied membrane lipid biosynthesis. For E. coli three major membrane phospholipids, phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL), are present. Certain functions have been defined for specific membrane phospholipids in E. coli. Anionic phospholipids (PG and CL) are involved in the initiation of DNA replication (64) and in the translocation of outer membrane precursor proteins (28). The zwitterionic PE is essential for proper functioning of the electron transfer chain (36), for the assembly and function of lactose permease (4, 6), and for motility and chemotaxis (51).
The membrane lipid composition in Sinorhizobium meliloti is more complex. In addition to PE, PG, and CL, S. meliloti membranes possess PC as a major membrane lipid, as well as monomethyl PE (MMPE) and dimethyl PE (DMPE) as minor membrane lipids. Two pathways for PC biosynthesis exist for S. meliloti. PC can be synthesized by the phospholipid N-methylation (Pmt) pathway from PE (12), or it can be synthesized by the PC synthase (Pcs) pathway directly from free choline and CDP-diacylglycerol (13, 52). Therefore, PC biosynthesis in this organism can happen via PE as an intermediate or in the alternative Pcs pathway in a PE-independent manner, offering the opportunity to uncouple PC biosynthesis from PE biosynthesis. Previously we described S. meliloti mutants deficient in the Pmt pathway as well as the Pcs pathway and therefore completely lacking PC (12). These mutants were not only strongly affected in their vegetative functions but also lacked the ability to form nitrogen-fixing root nodules on their host plant alfalfa (12, 53).
The first committed step in the biosynthesis of PE is catalyzed by phosphatidylserine synthase (Pss). A gene coding for the Pss enzyme (pssA) has been found and cloned in prokaryotes (10, 21, 41), lower eukaryotes like Saccharomyces cerevisiae (29, 40), and plants (11). Pss is responsible for the formation of phosphatidylserine from CDP-diacylglycerol and serine (EC 2.7.8.8). In a subsequent step, phosphatidylserine is decarboxylated by phosphatidylserine decarboxylase (Psd) to yield PE.
Genetic and biochemical studies have revealed the existence of two different types of Pss. Subclass I Pss, the prototype of which is the Pss from E. coli, was thought to occur primarily in gram-negative bacteria (32). Members of the type I subclass are soluble enzymes that seem to be tightly associated with the ribosomal fraction (44). Subclass I Pss are part of a protein superfamily that furthermore includes CL synthases, poxvirus envelope proteins, phospholipases D, and nucleases (26). In contrast, subclass II Pss, like the Bacillus subtilis Pss or the S. cerevisiae Pss, are not related on a sequence level to type I Pss and are predicted to be integral membrane proteins. Subclass II Pss are related to phosphatidylinositol synthases, phosphatidylglycerolphosphate synthases, and Pcs (53). So far, pssA genes have been cloned or characterized from E. coli, B. subtilis, S. cerevisiae, Helicobacter pylori, Agrobacterium sp. strain ATCC 31749, and Tritium aestivum (wheat). However, a detailed mutant characterization with respect to lipid pattern and growth has been performed only with the mutant of type I Pss from E. coli.
Here we describe the functional complementation of a PE-deficient chemical mutant of S. meliloti with a sinorhizobial gene bank and the isolation of the sinorhizobial gene (pssA) coding for a type II Pss. A mutant in which the sinorhizobial pssA gene is replaced with an antibiotic marker was constructed, and we show that PE is not essential for growth of S. meliloti in complex medium. However, on minimal media, PE-deficient mutants of S. meliloti show severe growth defects that can only partially be rescued by a functional PC biosynthesis.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used and their relevant characteristics are shown in Table 1. S. meliloti strains were grown at 30°C either in complex tryptone yeast extract (TY) medium containing 4.5 mM CaCl2 (2) or in morpholinepropanesulfonic acid (MOPS) minimal medium using glucose (15 mM) as a carbon source (1). Choline chloride (Sigma) was added to the minimal medium in the amounts indicated. For determination of growth rates, strains were first cultivated on TY medium containing 4.5 mM CaCl2 before being resuspended at cell densities of 9 × 107 cells ml−1 in TY medium containing CaCl2 concentrations varying between 0 and 50 mM and grown for more than three generations per subcultivation. The growth rates of strains were determined during the third subcultivation in TY medium with the respective calcium concentration.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| S. meliloti 1021 | SU47 str-21 | 34 |
| Sm 1021 derivatives | ||
| KDR309 | PmtA-deficient chemical mutant | 14 |
| KDR310 | PE-deficient chemical mutant | 14; this work |
| CS111 | pssA gene replaced with a gentamicin resistance cassette | This work |
| E. coli | ||
| DH5α | recA1 φ80 lacZΔM15 | 24 |
| BL21(DE3)/pLysS | Expression strain | 57 |
| Plasmids | ||
| pLAFR3 | Cosmid vector, tetracycline resistant | 56 |
| pRK2013 | Helper plasmid, kanamycin resistant | 20 |
| pRK404 | Broad-host-range vector, tetracycline resistant | 15 |
| pBBR1MCS-3 | Broad-host-range vector, tetracycline resistant | 27 |
| pACΩ-Gm | Broad-host-range vector containing gentamicin resistance cassette | 49 |
| pET3a | Expression vector, carbenicillin resistant | 57 |
| pET9a | Expression vector, kanamycin resistant | 57 |
| pK18mobsacB | Suicide vector, kanamycin resistant | 47 |
| pMP3510 | Broad-host-range vector, tetracycline resistant | 54 |
| pUC18/pUC19 | Cloning vectors, carbenicillin resistant | 65 |
| pCOS9.4 | pssA-containing DNA of S. meliloti in pLAFR3 | This work |
| pTB2009 | pssA-containing 6.6-kb BamHI/BamHI insert in pRK404 | This work |
| pTB2036 | pssA-containing 2.1-kb BamHI/HindIII insert in pRK404 | This work |
| pTB2086 | psd-containing HindIII/BclI insert in pRK404 | This work |
| pTB2104 | pssA-containing EcoRI/BamHI-insert in pMP3510 | This work |
| pTB2112 | pssA gene of S. meliloti in pMP3510 | This work |
| pTB2559 | pcs gene of S. meliloti in pET9a | 52 |
| pTB2919 | pssA gene of S. meliloti in pET9a | This work |
E. coli strains were cultured on Luria-Bertani medium at 37°C. Antibiotics were added to the media at the following final concentrations when required (in micrograms per milliliter): 500 for streptomycin, 200 for neomycin, 70 for gentamicin, 20 for piperacillin, 4 for tetracycline for S. meliloti, and 100 for carbenicillin, 10 for gentamicin, and 20 for tetracycline for E. coli.
Cosmids, pRK404, pMP3510, and pK18mobsacB derivatives were mobilized into S. meliloti strains by triparental mating using the mobilizing plasmid pRK2013 as described previously (45).
DNA manipulations.
Recombinant DNA techniques were performed according to standard protocols (46). DNA was sequenced by the dideoxy-mediated chain termination method using an SQ3 sequencer (Hoefer) and pUC19 derivatives.
Functional complementation of PE-deficient mutant S. meliloti KDR310.
Cosmids of a sinorhizobial gene bank (52) were mobilized into mutant KDR310 (14), and transconjugants were selected for wild-type-like growth on minimal medium. From colonies able to grow like the wild type on minimal medium, cosmids were isolated and transformed in E. coli DH5α, and the respective DNA inserts were analyzed.
In vivo labeling of S. meliloti with [14C]acetate and quantitative analysis of lipid extracts.
The lipid compositions of S. meliloti 1021 wild-type and mutant strains were determined following labeling with [1-14C]acetate (Amersham). Cultures (1 ml) of wild-type and mutant strains in TY medium were inoculated from precultures grown in the same medium. After addition of 0.4 μCi of [14C]acetate (60 nCi/nmol) to each culture, the cultures were incubated for either 4 or 16 h. The cells were harvested by centrifugation, washed with 500 μl of water, and resuspended in 100 μl of water. The lipids were extracted according to the method of Bligh and Dyer (3). The chloroform phase was used for lipid analysis on thin-layer chromatography (TLC) plates (high-performance TLC aluminum sheets, silica gel 60; Merck), and after one-dimensional or two-dimensional separation using the solvent systems described previously (14), the individual lipids were quantified as described previously (14) or by using a PhosphorImager (Storm 820; Molecular Dynamics).
Cloning of the Pss gene (pssA) of S. meliloti for complementation and expression studies.
The oligonucleotide primers oTBL235 (5′-gAg gAA TTC ATA Tgg AAg AgC CCC g-3′) and oTBL236 (5′-AgA AgC TTg gAT CCT ATT CgC TAT CAC CCg-3′), introducing EcoRI/NdeI sites and HindIII/BamHI sites (underlined), respectively, were used in the PCR to amplify the sinorhizobial pssA gene using the plasmid pTB2036 as a template. After digestion with EcoRI and HindIII, the PCR product was cloned into pUC19 to yield the plasmid pTB2071. The latter was digested with NdeI and BamHI, and the pssA-containing fragment was recloned into pET3a to yield pTB2081. The plasmid pTB2081 was digested with XbaI and BamHI, and the pssA gene was recloned into the XbaI/BamHI sites of pMP3510 to yield pTB2112, a plasmid used for complementation studies with S. meliloti. Alternatively, pTB2081 was digested with NdeI and BamHI, and the pssA gene was recloned into the NdeI/BamHI sites of pET9a to obtain pTB2919, a plasmid used for expression of sinorhizobial PssA in E. coli.
Exchange of the sinorhizobial pssA gene with a gentamicin resistance cassette.
The oligonucleotide primers opss5 (5′-ACg TTC TAg ATg gAg gCg AAg CgC TTC gAC AAg TCg ATg gAg C-3′) and opss6 (5′-ACg TgC CCg ggC CTT gTC gTg CgT gCC ggT ggC g-3′) were used in the PCR (XL-PCR kit; Applied Biosystems) to amplify about 1.8 kb of genomic DNA upstream of the putative pssA gene from S. meliloti, introducing XbaI and SmaI sites (underlined) into the PCR product. Similarly, the primers opss7 (5′-ACg TCC Cgg gAg ACg ATg gTC gTg ACT gCg gTg gCC TAT C-3′) and opss8 (5′-ACg TCT gCA ggA AAg Cgg CAT CCC CTT CgA gCT Cgg CAT C-3′) were used to amplify about 1.8 kb of genomic DNA downstream of the pssA gene, introducing SmaI and PstI sites (underlined) into this PCR product. After digestion with the respective enzymes, the PCR products were cloned as XbaI/SmaI or SmaI/PstI fragments into pUC18 to yield the plasmids pCCS01 and pCCS02, respectively. Then, the XbaI/SmaI fragment from pCCS01 was recloned into pBBR1MCS-3 (27) to yield pCCS03. The plasmid pCCS02, linearized with SmaI, was cloned into the SmaI site of pCCS03. The plasmid containing the two sinorhizobial genomic fragments adjacent was named pCCS04. pCCS04 was digested with PstI, the pUC18 backbone was cut out, and the rest of pCCS04 was religated to yield pCCS04a. The plasmid pCCS04a was digested with SmaI, and the gentamicin resistance cassette derived from pACΩ-Gm (49) was cloned as a SmaI fragment into pCCS04a to yield pCCS04b. The plasmid pCCS04b was digested with PstI and XbaI to reclone the regions usually flanking the sinorhizobial pssA gene and the gentamicin resistance gene located in between those regions as a PstI/XbaI fragment into the suicide vector pK18mobsacB (47) to yield pCCS05. Via triparental mating using pRK2013 as a helper plasmid, pCCS05 was introduced into the wild-type strain S. meliloti 1021. Transconjugants were selected on TY medium containing neomycin and gentamicin to select for single recombinants in a first step. pK18mobsacB contains the sacB gene (50), which confers sucrose sensitivity to many bacteria. Growth of the single recombinants on high levels of sucrose will therefore select for a further recombination and the loss of the vector backbone of pK18mobsacB from the bacterial genome. Single recombinants were grown under nonselective conditions in complex medium for 1 day before being plated on TY medium containing 10% (wt/vol) sucrose and gentamicin. Several large and small colonies grew after 5 days, and the membrane lipids of 16 candidates were analyzed by in vivo labeling during growth on complex medium with [14C]acetate and subsequent TLC (data not shown). Six clones lacking PE were identified. Southern blot analysis confirmed that the PE-deficient strains were indeed double recombinants in which the pssA gene was replaced with a gentamicin resistance cassette (data not shown). Using PCR, the orientation of the gentamicin resistance cassette was determined, and in CS111 the cassette has the same orientation as the replaced pssA gene.
Determination of phospholipid methyltransferase activity in the presence or absence of added PE.
Cultures (30 ml) of mutant or wild-type strains were grown to an optical density at 620 nm of 0.6. Cells were harvested by centrifugation for 10 min at 5,000 × g, and the pellets obtained were resuspended in 3 ml (total volume) of ice-cold reaction buffer (100 mM Tris-HCl [pH 9.5]). The cell suspension was passed three times through a French pressure cell at 20,000 lb/in2. Unbroken cells and cell debris were removed by centrifugation at 4,000 × g for 4 min to obtain the cell extract. The protein concentration was determined by the method of Dulley and Grieve (17). The phospholipid methyltransferase activity was determined by quantifying the transfer of labeled methyl groups from S-adenosyl-l-methionine to chloroform-soluble material. Under the conditions chosen, methylated derivatives of PE were the only chloroform-soluble substances formed (14). When the effect of added PE was studied, first, per reaction tube, 100 μg of PE (l-α-phosphatidylethanolamine, dipalmitoyl; Sigma) in 10 μl of chloroform-methanol (1:1 [vol/vol]) was mixed with 3.6 μl of 1% Triton X-100 (wt/vol) and dried under vacuum before other components were added. Therefore, in a total of 180 μl in Eppendorf reaction tubes, the reaction mixture contained 100 μg of PE or no PE, 0.02% Triton X-100, 200 μg of sinorhizobial protein, 100 μg of bovine serum albumin, and 125 nCi of S-adenosyl-l-[methyl-3H]methionine (100 nCi/nmol) in 100 mM Tris-HCl (pH 9.5). The mixtures were incubated for 15 min in a 30°C water bath. The reactions were stopped by addition of 180 μl of ice-cold 20% (wt/vol) trichloroacetic acid, and precipitations were completed by a 10-min incubation on ice. The precipitates were pelleted by centrifugation (5 min, 14,000 × g), dissolved by the addition of 60 μl of 1 M Tris base, and neutralized by adding 40 μl of 20% (wt/vol) trichloroacetic acid, respectively. Lipids were extracted according to the method of Bligh and Dyer (3). The chloroform phases were dried, and aliquots of the redissolved lipids were counted in scintillation vials.
Determination of Pcs and Pss activities.
E. coli BL21(DE3)/pLysS strains harboring one of the plasmids pTB2919, pTB2559, or pET9a were grown in Luria-Bertani medium containing the appropriate antibiotics. At a cell density of 5 × 108 cells/ml, isopropyl-β-d-thiogalactoside was added to a final concentration of 100 μM. After 4 h of induction, cells were harvested and stored at −20°C. The cell pellets were resuspended in 50 mM Tris-HCl, pH 8.0, and cell extracts were prepared as described above.
The optimized standard assay to determine Pcs activity (13) contained, in a total volume of 50 μl in Eppendorf tubes, 50 μg of protein, 50 mM Tris-HCl, pH 8, 10 mM MnCl2, 20 μM CDP-DAG (cytidine 5′-diphospho-sn-glycerol, 1,2-dipalmitoyl; Sigma), 0.2% (wt/vol) Triton X-100, and 50 μM [methyl-14C]choline (55 mCi/mmol; Amersham Biosciences). The mixture was incubated for 30 min at 30°C in a water bath, and the reaction was stopped by mixing with 188 μl of methanol-chloroform (2:1 [vol/vol]). Addition of 63 μl of chloroform and 63 μl of water led to phase separation, and after washing of the chloroform phase with another 100 μl of water, it was dried and an aliquot was analyzed using one-dimensional TLC. The Pss assay was performed in an identical way, except that [14C]choline was replaced with equimolar amounts of [U-14C]serine (151 mCi/mmol; Amersham Biosciences). In some Pss assays, 10 mM hydroxylamine, an inhibitor of Psd (7, 18, 61), was present in the reaction mixture.
Nucleotide sequence accession number.
The nucleotide sequence (2,183 bp) of the HindIII-BamHI fragment complementing the PE-deficient mutant KDR310 has been deposited in GenBank under accession number AF247564.
RESULTS
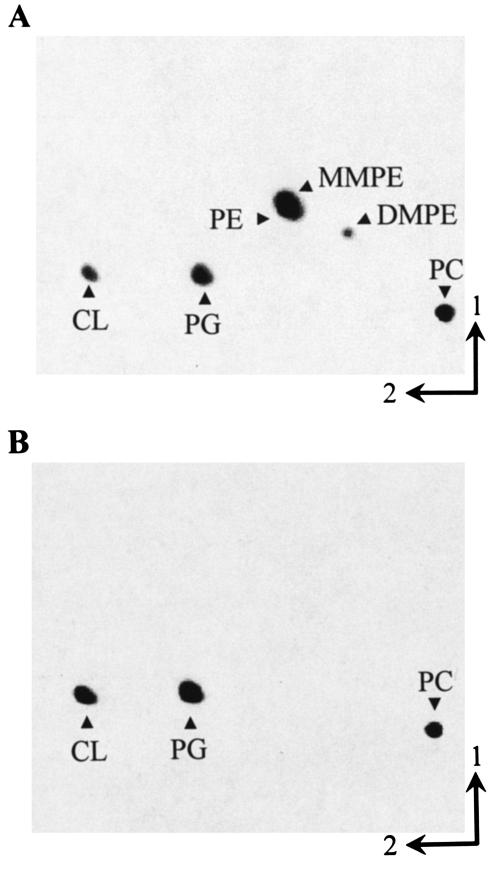
Identification of a PE-deficient mutant of S. meliloti. During our search for phospholipid N-methyltransferase-deficient mutants of S. meliloti using a colony autoradiography method (42), we had identified six chemically generated mutants that showed no phospholipid N-methyltransferase (Pmt) activity when the respective cell-free crude extracts were assayed (14). One-dimensional TLC of mutant lipid extracts demonstrated that none of the mutants was able to form MMPE or DMPE after growth on complex medium (data not shown). However, in one of the mutant extracts (KDR310), even another lipid seemed to be absent. To further compare the phospholipid patterns of the wild type and mutant KDR310, two-dimensional TLC analysis of the lipids was performed (Fig. 1). The S. meliloti wild type possesses PG, CL, PE, MMPE, DMPE, and PC as membrane phospholipids (Fig. 1A). Mutant KDR310, however, has only PG, CL, and PC and lacks MMPE, DMPE, and PE (Fig. 1B). Staining of TLC plates with ninhydrin confirmed that in the case of the mutant KDR310, no ninhydrin-positive compound could be detected at the position where PE was expected (data not shown). The relative amounts of the individual phospholipids from the S. meliloti 1021 wild type and mutant KDR310 are shown in Table 2. In mutant KDR310, the relative amounts of all three remaining phospholipids are elevated, with a large increase in the relative quantities of the anionic phospholipids PG and CL (from 19 to 60%) and a smaller increase in the relative amount of PC (from 27 to 36%).
FIG. 1.
Separation of [14C]acetate-labeled lipids from S. meliloti 1021 wild type (A) and PE-deficient mutant KDR310 (B) by two-dimensional TLC. Phospholipids are indicated.
TABLE 2.
Phospholipid composition of S. meliloti 1021 (wild type) and PE-deficient mutant strain KDR310 after growth in complex medium
| Lipid | Lipid composition (% of total 14C)
|
|
|---|---|---|
| Wild type | KDR310 | |
| PE + MMPE | 44.6 | 0.3 |
| DMPE | 3.1 | 0.0 |
| PC | 26.9 | 35.8 |
| PG | 13.4 | 36.7 |
| CL | 5.5 | 23.0 |
| Other | 6.5 | 4.2 |
PE restores phospholipid N-methyltransferase activity in mutant KDR310.
The absence of PE in membranes of mutant KDR310 and therefore the lack of one substrate could explain why we did not detect any Pmt activity in our usual assay that did not contain externally added PE (14). The effect of PE on Pmt activities of different sinorhizobial cell extracts was studied in the presence of the detergent Triton X-100 to ensure equal accessibility of added lipid (Table 3). When PE was added to S. meliloti wild-type extracts, the methyl groups transferred to lipid increased by a factor of 33.7. Addition of PE to the Pmt-deficient mutant KDR309 increased the amount of radiolabeled methyl groups incorporated into the lipid phase by a factor of 6.0. As no Pmt activity is present in KDR309, the stimulation of methyl group incorporation into the lipid phase might be due to cyclopropane synthases acting on the fatty acyl groups of PE (23). In the case of mutant KDR310, however, the stimulation of Pmt activity by PE was 27.1-fold (Table 3), suggesting that, as in the wild type, stimulation of methyl group incorporation into lipids by KDR310 extracts was much higher than by KDR309 extracts and that Pmt activity was present in KDR310. However, in KDR310, Pmt activity can only be detected when external PE is added.
TABLE 3.
Stimulation of in vitro phospholipid N-methyltransferase activity by added PEa
| Strain | Incorporation of methyl groups (pmol/mg of protein/min)
|
||
|---|---|---|---|
| Without PE | With PE | Stimulation (x-fold) | |
| Wild type | 0.71 | 23.91 | 33.7 |
| KDR309 | 0.13 | 0.78 | 6.0 |
| KDR310 | 0.23 | 6.24 | 27.1 |
The assay for phospholipid N-methyltransferase was done with cell-free extracts of S. meliloti wild-type 1021, Pmt-deficient mutant KDR309, and PE-deficient mutant KDR310 after growth in complex medium.
PE-deficient mutant KDR310 is defective in Pss.
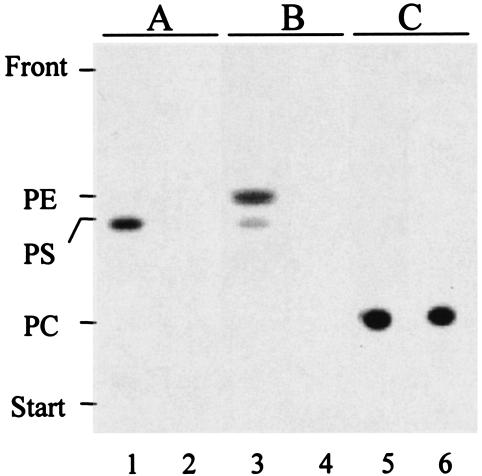
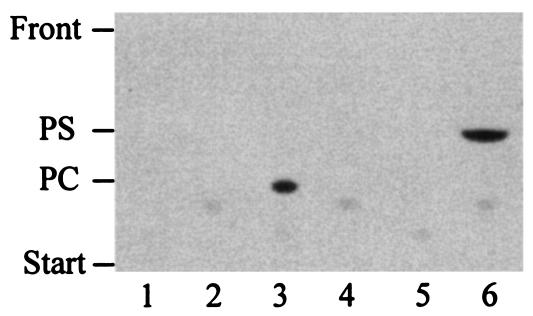
Formation of phosphatidylserine (PS) and PE by cell extracts of S. meliloti wild type and PE-deficient mutant KDR310 was studied by incorporation of [14C]serine into lipid products (Fig. 2). With wild-type extracts, only PS was formed when hydroxylamine, an inhibitor of Psd activity, was present in the reaction mixture, demonstrating the presence of Pss activity in the S. meliloti wild type (lane 1). When hydroxylamine was omitted in the Pss assay, both PS and PE were formed in the crude cell extracts of the wild type (lane 3). For mutant KDR310, neither PS nor PE was formed under such conditions, demonstrating that Pss activity was absent in this mutant (lanes 2 and 4). As a control to verify that other activities were not impaired, we investigated the recently discovered Pcs activity (13, 52) and could show that both wild-type (lane 5) and mutant KDR310 (lane 6) extracts were able to incorporate radiolabeled choline into PC.
FIG. 2.
Mutant KDR310 is defective in Pss activity. Lipid products obtained by in vitro activity tests for Pss in the presence of hydroxylamine (A) or without hydroxylamine (B) and for Pcs (C) were separated by one-dimensional TLC. Activity tests were performed with cell extracts from the S. meliloti wild-type strain (lanes 1, 3, and 5) and PE-deficient mutant strain, KDR310 (lanes 2, 4, and 6), that had been grown in complex medium.
Complementation of S. meliloti mutants deficient in Pmt.
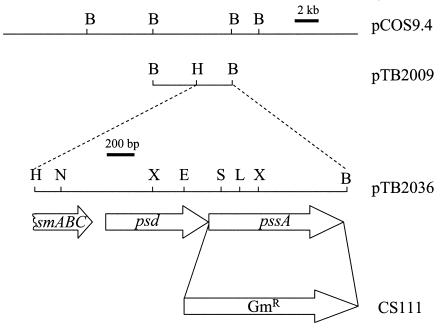
During our search for Pmt-deficient mutants of S. meliloti, we had identified nine chemically generated mutants that showed no or reduced Pmt activity when the respective cell extracts were assayed (14). A cosmid, pCOS24.1, that was able to complement mutant KDR309 (12) also complemented most other mutants (KDR296, KDR304, KDR339, KDR345, KDR365) that were devoid of Pmt activity (data not shown). However, the PE-deficient mutant KDR310 was not complemented by pCOS24.1, indicating that genes complementing KDR310 would be of another complementation group. E. coli HB101, carrying a genomic library of S. meliloti that had been constructed in the cosmid vector pLAFR3 as described previously (52), was conjugated with S. meliloti KDR310. In contrast to the PE-deficient mutant KDR310, which hardly grew on minimal medium, complemented mutants should be able to do so, and we selected for transconjugant colonies able to grow like the wild type on minimal medium. From growth-complemented mutants, the cosmids were isolated and transformed into DH5α for further analysis. Three types of overlapping cosmids were able to complement the PE-deficient phenotype of mutant KDR310. One representative, cosmid pCOS9.4, was used for further studies (Fig. 3). Subcloning of restriction fragments, comprising regions of overlapping DNA, in the broad-host-range vector pRK404 and subsequent analysis for complementation of the PE-deficient mutant KDR310 shows that a plasmid (pTB2009) containing a 6.6-kb BamHI-BamHI DNA fragment is able to restore formation of PE, MMPE, and DMPE (data not shown). A subclone of the complementing DNA containing a 2.1-kb BamHI-HindIII fragment in pRK404 (pTB2036) (Fig. 3) is also able to complement mutant KDR310 (Fig. 4, lane 4). The DNA sequence of the 2.1-kb BamHI-HindIII fragment was determined, and analysis of the sequence (2,183 bp) revealed one partial open reading frame (ORF) and two complete ORFs (Fig. 3). The complementing 2.1-kb fragment was further subcloned to analyze which of the complete ORFs was able to complement the mutant KDR310. Plasmid pTB2104, containing a 1.1-kb EcoRI-BamHI fragment including the second complete ORF, was able to complement the PE-deficient mutant (Fig. 4, lane 5), whereas plasmid pTB2086, containing a 1.4-kb HindIII-BclI DNA fragment with the first complete ORF, was not able to do so (Fig. 4, lane 6). In order to confirm these results, the second complete ORF was amplified by PCR and cloned into pMP3510 (pTB2112). Expression of this second ORF complemented PE biosynthesis in KDR310 (Fig. 4, lane 7).
FIG. 3.
Genomic region of S. meliloti 1021 complementing the PE-deficient mutant KDR310. Schematic representation of the genomic DNA insert of cosmid pCOS9.4 complementing PE deficiency. The subcloning strategy is shown with the 6.6-kb DNA region (pTB2009) and the 2.1-kb HindIII-BamHI fragment that was sequenced (pTB2036). The location and direction of one partial ORF, encoding an ABC transporter-like protein (smABC), and two complete ORFs, encoding Psd (psd) and Pss (pssA), are shown by arrows. The replacement of the pssA gene by a gentamicin resistance cassette (GmR) in mutant CS111 is indicated. Restriction sites (B, BamHI; L, BclI; E, EcoRI; H, HindIII; N, NcoI; S, SmaI; X, XhoI) used are shown.
FIG. 4.
Complementation of PE-deficient mutant KDR310. Lipids of S. meliloti strains containing different plasmids were radiolabeled with [14C]acetate during growth in complex medium and separated by one-dimensional TLC. The following strains were analyzed: S. meliloti wild type 1021 (lane 1), PE-deficient mutant KDR310 (lane 2), and KDR310 containing either pCOS9.4 (lane 3), pTB2036 (lane 4), pTB2104 (lane 5), pTB2086 (lane 6), or pTB2112 (lane 7).
Gene bank searches with the NCBI BLAST program showed that the gene product of the partial ORF (positions 1 to 387) shows homology to members of the drug peptide and lipid export family (DPL) of ATP binding cassette (ABC) transporters (9). A relative of this partial ORF, the MsbA lipid flippase of E. coli, is involved in lipid export and presumably in flipping lipids from the inner layer to the outer layer of the cytoplasmic membrane (16), raising the question whether this sinorhizobial gene might be involved in lipid transport. The first complete ORF (positions 497 to 1192), encoding a protein of 232 amino acids, was found to be preceded by a putative ribosome binding site. The predicted protein sequence contains a conserved domain typical for Psd (COG0688), but it is only distantly related to the Psd proteins from E. coli (30), B. subtilis (33), and S. cerevisiae (8, 58, 59, 60) for which functions have been shown. Psd enzymes of the latter group contain a conserved amino acid pattern (L/G)GST for E. coli and S. cerevisiae that has been described as a motif where probably the proteolytic cleavage of Psd into α- and β-fragment and the formation of the pyruvoyl group occur (62). ORFs containing the conserved domain COG0688 fall into two quite different families, and whereas functions have been shown for several representatives of the above-mentioned family, for no member of the second family, to which the putative S. meliloti Psd belongs, has a function been demonstrated. In the S. meliloti Psd, the conserved amino acid pattern of the putative cleavage site is reduced to GS.
The second complete ORF follows immediately downstream of psd, suggesting that the two ORFs might form an operon. The potential gene product of the latter ORF (positions 1205 to 2071) encodes a protein of 289 amino acids that shows the typical motif [DG(X)2AR(X)8G(X)3D(X)3D] characteristic for CDP-alcohol phosphatidyltransferases (53, 63), like phosphatidylglycerolphosphate synthases, phosphatidylinositol synthases, amino alcohol phosphotransferases, and type II Pss. The best homologues to the latter ORF for which a function has been shown are the type II Pss from Agrobacterium sp. ATCC 31749 (60% identity, 70% similarity on amino acid level; accession number AAL01116) and Helicobacter pylori (31% identity, 47% similarity on amino acid level; G64653). Therefore, this second complete ORF probably encodes a type II Pss.
The identified pssA gene codes for a protein that has Pss activity but no Pcs activity.
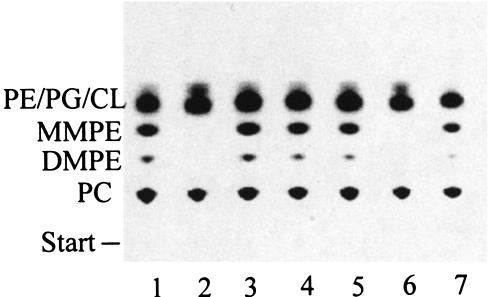
The sinorhizobial PssA protein is of the type II subgroup. Type II Pss share a conserved domain with Pcs, phosphatidylinositol synthases, and phosphatidylglycerolphosphate synthases (53). Due to this similarity, we wanted to investigate whether the sinorhizobial Pss enzyme would show Pcs activity in addition to the Pss activity and whether the Pcs enzyme would show Pss activity in addition to Pcs activity. Pss activity and Pcs activity were studied in cell-free crude extracts from E. coli BL21(DE3)/pLysS strains that had expressed either the pssA gene from S. meliloti or the pcs gene from S. meliloti or that were harboring the pET9a plasmid only. Neither Pcs activity nor Pss activity could be detected in the crude extract not expressing any sinorhizobial gene (Fig. 5), meaning that the type I E. coli Pss does not interfere with either the Pss assay or the Pcs assay (lanes 1 and 2). In the crude extract containing the Pcs of S. meliloti, only Pcs activity (lane 3) and no Pss activity (lane 4) could be detected, whereas in the crude extract containing the sinorhizobial Pss, no Pcs activity (lane 5) but only Pss activity (lane 6) was detected.
FIG. 5.
Pss and Pcs do not overlap in their substrate specificities. Lipids formed during in vitro activity tests for Pcs and Pss were separated by one-dimensional TLC. Pcs (lanes 1, 3, and 5) or Pss (lanes 2, 4, and 6) activities were assayed with cell-free crude extracts from E. coli BL21(DE3)/pLysS strains expressing either no rhizobial gene when harboring pET9a only (lanes 1 and 2), pcs of S. meliloti from pTB2559 (lanes 3 and 4), or pssA from S. meliloti from pTB2919 (lanes 5 and 6).
Replacement of the sinorhizobial pssA gene with a gentamicin resistance cassette.
When we tried to characterize the PE-deficient mutant KDR310 in more detail, we noticed that this chemical mutant reverted easily, thereby impeding a rigorous characterization. In order to be able to tackle the role(s) that PE plays in S. meliloti, we decided to substitute the whole pssA gene with a cassette conferring resistance to the antibiotic gentamicin. The genomic regions flanking the pssA gene from S. meliloti were amplified by PCR, and a gentamicin cassette was cloned between them. This whole construct was introduced into the suicide vector pK18mobsacB, and the resulting plasmid was conjugated into S. meliloti 1021, leading to the exchange of the entire pssA gene with the gentamicin cassette (Fig. 3). Mutant candidates were analyzed by in vivo labeling with [14C]acetate and subsequent separation of the radiolabeled lipids by one-dimensional TLC. Six mutants lacking PE and its methylated derivatives were identified (data not shown). Substitution of the pssA gene with the gentamicin resistance cassette was confirmed by Southern blotting (data not shown). One of the six allelic mutants (CS111) was selected for further analysis. The lipid pattern of the mutant CS111 was identical to the lipid pattern shown above for the PE-deficient chemical mutant KDR310 (Fig. 1B). In order to find out if there might be an additional activity leading to PE synthesis, we performed in vivo labeling of CS111 cells grown in complex TY medium with [32P]phosphate. When the lipid extracts were analyzed by two-dimensional TLC, no radioactivity above background levels could be detected where the PE spot was to be expected (data not shown).
Growth of the PE-deficient knockout mutant CS111.
Mutants of E. coli that are deficient in PE are conditionally lethal and require a millimolar concentration of bivalent cations like Ca2+ or Mg2+ to grow (10). We therefore expected a drastic growth phenotype for the PE-deficient S. meliloti mutant CS111 as well. Growth of CS111, wild-type S. meliloti 1021, and CS111 harboring either the complementing plasmid pTB2112 or vector pMP3510 only was compared on TY medium supplemented with different concentrations of CaCl2 (0 to 50 mM). On TY medium lacking calcium, mutant CS111 was not able to grow, whereas the wild type was able to grow for one passage but then stopped growing as well (data not shown). This result suggests that CS111 needs more calcium than the wild type to sustain growth. When additional calcium was present in the TY growth medium, a clear difference in growth between mutant CS111 and the wild type could be observed at calcium concentrations up to 7.5 mM, but this difference was getting smaller with increasing calcium concentrations (Table 4). The difference in growth is much more striking when CS111 carrying the complementing plasmid (pTB2112) is compared to CS111 harboring the empty vector (pMP3510). At 1 mM and 4.5 mM CaCl2, the generation time of the strain lacking PE was more than three times longer than the generation time of the complemented mutant CS111/pTB2112 (Table 4). With increasing calcium concentrations, the difference in generation time between CS111/pTB2112 and CS111/pMP3510 decreased, but it did remain significant. A possible explanation for this drastic difference might be that CS111/pMP3510, due to the absence of PE from its membranes, has a greater sensitivity to tetracycline than the complemented mutant CS111/pTB2112, although both strains carry a tetracycline resistance gene. Resistance to tetracycline is conferred by TetA, a transporter of the major facilitator superfamily (19). The increased sensitivity towards tetracycline of CS111/pMP3510 compared to that of CS111/pTB2112 might be due to reduced transporter activity caused by the lack of PE in the membrane, in analogy to what has been described for the major facilitator superfamily lactose transporter (LacY) from E. coli (4, 5, 6). By adding increasing amounts of calcium, which has been shown to stimulate tetracycline transport, the tetracycline hypersensitivity might be masked partially (48). Antibiotic hypersensitivity of a mutant deficient in PE has been described before for E. coli (43). Using antibiotic susceptibility disks (Oxoid), we compared the susceptibilities of CS111 and 1021 towards 18 different antibiotics on TY medium containing 0.5, 4.5, or 20 mM CaCl2. Mutant CS111 seemed to be more sensitive towards carbenicillin, neomycin, and tetracycline (data not shown), and the increased sensitivity of CS111 appeared to be more pronounced when less calcium was present in the medium. In the case of tetracycline, the explanation for the higher resistance at higher calcium concentrations again might be that calcium is stimulating tetracycline export from the cells (48). The hypersensitivity towards these antibiotics was confirmed by determining the MICs of these antibiotics for both strains when grown in TY medium containing 4.5 mM CaCl2. The PE-deficient mutant CS111 was two- to fourfold more sensitive towards these antibiotics (data not shown).
TABLE 4.
Correlation between Ca2+ concentration and generation times of different rhizobial strains when grown in complex mediuma
| Ca2+ concn (mM) | Avg generation time (min)
|
|||
|---|---|---|---|---|
| 1021 | CS111 | CS111/pTB2112 | CS111/pMP3510 | |
| 1 | 190 ± 10 | 253 ± 31 | 197 ± 15 | 680 ± 35 |
| 4.5 | 181 ± 14 | 217 ± 24 | 200 ± 25 | 624 ± 128 |
| 7.5 | 167 ± 21 | 205 ± 26 | 170 ± 20 | 373 ± 64 |
| 20 | 170 ± 8 | 180 ± 26 | 171 ± 18 | 303 ± 86 |
| 50 | 155 ± 15 | 165 ± 10 | 157 ± 6 | 227 ± 50 |
S. meliloti 1021 (wild type), PE-deficient mutant CS111, CS111/pTB2112 (complemented mutant), and CS111/pMP3510 (mutant harboring the empty vector only) were grown in complex TY medium containing between 1 and 50 mM CaCl2. Numbers in the table give the average generation time of at least three independent growth experiments ± standard deviation.
The lack of the non-bilayer-forming lipid PE leads to filamentous growth of PE-deficient mutants of E. coli (35). This growth phenotype might be a direct effect of the lack of the non-bilayer-forming lipid PE, as no other non-bilayer-forming lipid seems to be part of the S. meliloti membrane, but there is also evidence that the high negative charge density on the cell surfaces of E. coli cells lacking PE is partially responsible for these defects in cell division (37). When grown in complex medium, mutant cells should lack PE, MMPE, and DMPE, but when grown in choline-free minimal medium, they should lack PE, MMPE, DMPE, and PC, which should lead to an excess of negative charge at the membrane. Mutant cells and wild-type cells were cultivated on complex medium and obtained from the exponential growth phase before being analyzed by light microscopy for their morphologies. In addition, mutant and wild-type cells were cultivated on MOPS minimal medium with or without supplementation of 100 μM choline. Under all growth conditions mentioned, no significant difference in cell morphology could be observed when comparing the wild type, 1021, and the PE-deficient mutant CS111 (data not shown).
Lipid composition of PE-deficient mutant CS111 during growth on complex medium.
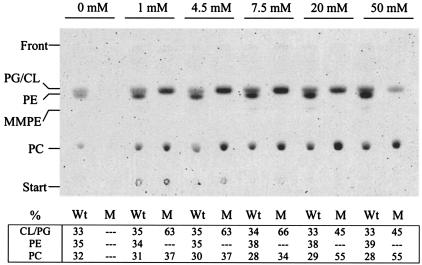
The difference in growth between the PE-deficient mutant CS111 and the wild type, 1021, at low calcium concentrations prompted us to study the membrane lipid composition of the two strains after growth in TY medium containing different concentrations of calcium (Fig. 6). Lipid composition of wild-type strain 1021 and the PE-deficient mutant CS111 was analyzed by in vivo labeling with [14C]acetate and subsequent lipid analysis by TLC. In the wild type, PG and CL comprised 33 to 36%, PE comprised 34 to 39%, and PC comprised between 28 and 32%, and notably, the wild type exhibited a similar lipid composition over the whole range of calcium concentrations used. Without added calcium in the growth medium, hardly any radioactivity was incorporated into membrane lipids of mutant CS111, which is consistent with the earlier observation that the mutant does not grow on medium lacking Ca2+. At calcium concentrations up to 7.5 mM, PG and CL comprise 63 to 66% of the membrane lipids whereas the residual 34 to 37% are PC, which is in agreement with the results obtained for lipid composition of the PE-deficient chemical mutant KDR310 (Table 2). Surprisingly, at calcium concentrations of 20 and 50 mM, PC becomes the major membrane lipid in mutant CS111 (relative amount of PC increases to 55%) and the proportion of anionic membrane lipids (PG and CL) decreases to 45% (Fig. 6). Therefore, in contrast to that of the wild type, the lipid profile of the PE-deficient mutant CS111 changes depending on the calcium concentration in the medium, and whereas at low concentrations of calcium, PE is largely replaced by the anionic phospholipids PG and CL, at concentrations of 20 mM calcium and above, the zwitterionic PC functions as a major replacement lipid for PE.
FIG. 6.
Lipid analysis of wild-type S. meliloti 1021 and Pss-deficient mutant CS111 after in vivo labeling with [14C]acetate in complex TY medium containing different concentrations of CaCl2. S. meliloti wild-type 1021 (Wt) and mutant CS111 (M) were grown in complex TY medium containing between 0 and 50 mM CaCl2. [14C]acetate-labeled lipids were extracted, separated by one-dimensional TLC, and quantified using a PhosphorImager. Relative amounts (percent) of individual lipids are given in the table at bottom.
Choline supplementation partly rescues the growth defect of PE-deficient S. meliloti mutant CS111 on minimal medium.
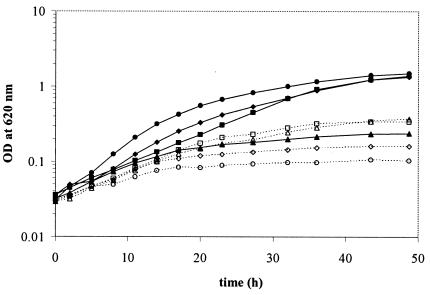
The PE-deficient sinorhizobial mutant CS111 grows surprisingly well on complex medium even at relatively low concentrations of calcium ions. Unlike E. coli, S. meliloti possesses with PC a second major zwitterionic lipid in its membranes. Since wild-type S. meliloti can synthesize PC via the Pmt pathway or via the Pcs pathway, even a mutant deficient in Pss will be able to synthesize PC via the Pcs pathway when grown in medium containing choline. We speculated that the presence of PC might make up for the lack of PE. Cultivation of S. meliloti in defined choline-free medium would completely prevent the synthesis of PC. Strains were pregrown for one passage on TY medium containing 4.5 mM CaCl2. Then, cells were washed twice with MOPS minimal medium before being cultured in the same medium in the presence of different amounts of choline. S. meliloti 1021 wild-type cells grew well in MOPS medium lacking or containing up to 10 μM choline (Fig. 7). When higher choline concentrations (100 μM or 1 mM) were present in the medium, growth of the wild type was inhibited. In contrast, growth of PE-deficient mutants was much worse on MOPS minimal medium. Choline concentrations up to 1 μM did not improve growth of the PE-deficient mutant. A partial restoration of wild-type-like growth was observed when the medium was supplemented with 10 μM choline or more. Interestingly, in minimal medium supplemented with 1 mM choline, the mutant grew even better than the wild type (Fig. 7). The addition of 0.01% (wt/vol) Casamino Acids (Difco) restored growth of the PE-deficient mutant almost to the growth rate observed on complex TY medium (data not shown). Since growth of the PE-deficient mutant CS111 was much worse than that of the wild type on MOPS minimal medium when supplemented with choline but was nearly as good as that of the wild type on complex medium and on minimal medium supplemented with small amounts of Casamino Acids, we suggest that some compound(s) other than choline and present in complex medium or in the Casamino Acids preparation is needed for optimal growth of a PE-deficient sinorhizobial mutant.
FIG. 7.
The growth phenotype of the PE-deficient mutant CS111 is partly rescued by choline supplementation to the medium. S. meliloti 1021 (solid lines, closed symbols) and the Pss-deficient mutant CS111 (dashed lines, open symbols) were grown in MOPS minimal medium without choline supplementation (circles) or in the presence of 10 μM choline (diamonds), 100 μM choline (squares), or 1 mM choline (triangles).
DISCUSSION
During a screening for mutants of S. meliloti deficient in Pmt activity, we identified a number of chemical mutants that showed reduced or no Pmt activity (14). Mutant KDR309, deficient in the sinorhizobial gene (pmtA) for Pmt, was complemented by a cosmid (pCOS24.1) containing sinorhizobial DNA (12). Most of the other Pmt-deficient chemical mutants were complemented by the same cosmid or pmtA-containing subclones of it, suggesting that the original defect of these mutants was within the sinorhizobial pmtA gene. One mutant isolated in the original screening (KDR310) was complemented by another cosmid containing sinorhizobial DNA (pCOS9.4) or by subclones containing an intact sinorhizobial pssA gene encoding for Pss. Originally we did not detect Pmt activity in KDR310, because this mutant cannot form the substrate PE needed for this reaction. When external PE is added to cell extracts of KDR310, Pmt activity can be demonstrated.
Whereas eukaryotes can form PE via the CDP-ethanolamine pathway, prokaryotes usually form PE from CDP-diacylglyceride, employing Pss and the Psd reaction. Replacement of the pssA gene with a gentamicin resistance-conferring cassette in S. meliloti eliminates the formation of PE and of methylated PE derivatives, demonstrating that the knockout of the pssA gene eliminates Pss activity and that no other pathway for PE biosynthesis exists in S. meliloti. The complementation of pssA-deficient mutants with broad-host-range plasmids containing the sinorhizobial 2.1 kb with the pssA gene (pTB2036) or only the PCR-amplified pssA gene (pTB2112) demonstrates that this single gene restores Pss activity in S. meliloti.
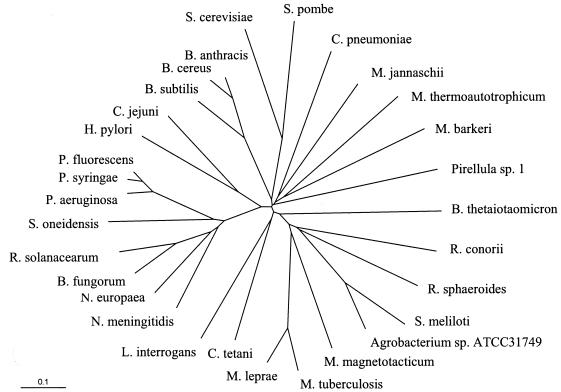
Sequence analysis of the sinorhizobial pssA gene revealed that S. meliloti Pss is a type II Pss. Type II Pss were originally thought to be specific for gram-positive bacteria (32), but recent experimental data (21, 25, 39) and the wealth of information provided by the large number of genome projects indicate that type II Pss occur in all three kingdoms of life (archaea, eubacteria, eukaryotes) (Fig. 8). They are therefore not restricted to gram-positive bacteria but can be found in such diverse organisms as Methanococcus jannaschii (archaebacteria), Leptospira interrogans (spirochetes), Pirellula sp. (Planctomycetales), Pseudomonas fluorescens (γ-proteobacteria), Neisseria meningitidis (β-proteobacteria), Helicobacter pylori (ɛ-proteobacteria), and Rhodobacter sphaeroides (α-proteobacteria). In contrast, the type I subclass of Pss seems to be restricted to certain families of the γ-proteobacteria. Homologues to the type I E. coli Pss can be found, for example, in Yersinia pestis (Enterobacteriaceae), Vibrio cholerae (Vibrionaceae), Haemophilus influenzae (Pasteurellaceae), Shewanella oneidensis (Alteromonaceae), and Pseudomonas putida (Pseudomonadaceae). In the family of the pseudomonads some strains seem to have the subclass I Pss (P. putida), others seem to have the subclass II Pss (Pseudomonas aeruginosa), and surprisingly, P. fluorescens, Pseudomonas syringae, and S. oneidensis seem to have good homologues for both subclasses of Pss.
FIG. 8.
Unrooted phylogenetic tree of subclass II Pss and Pss-like ORFs. The tree was constructed using the program ClustalW (http://www.expasy.ch/). Distances between sequences are expressed as 0.1 changes per amino acid residue. Accession numbers are as follows: Saccharomyces cerevisiae Cho1p, NP_010943;, Schizosaccharomyces pombe Pss, NP_588326; Chlamydophila pneumoniae ORF, NP_225177; Methanococcus jannaschii ORF, NP_248207; Methanobacterium thermoautotrophicum Pss, A69004; Methanosarcina barkeri ORF, ZP_00079280; Pirellula sp. ORF, NP_865741; Bacteroides thetaiotaomicron ORF, NP_811145; Rickettsia conorii ORF, NP_359963; Rhodobacter sphaeroides ORF, ZP_00008123; Sinorhizobium meliloti Pss, AAG00422; Agrobacterium sp. ATCC31749 Pss, AAL01116; Magnetospirillum magnetotacticum ORF, ZP_00056392; Mycobacterium tuberculosis Pss, P96282; Mycobacterium leprae ORF, NP_301340; Clostridium tetani ORF, NP_781048; Leptospira interrogans ORF, NP_711632; Neisseria meningitidis ORF, NP_274337; Nitrosomonas europaea ORF, NP_841370; Burkholderia fungorum ORF, ZP_00029658; Ralstonia solanacearum ORF, NP_520194; Shewanella oneidensis ORF, NP_719120; Pseudomonas aeruginosa ORF, NP_253381; Pseudomonas syringae ORF, NP_790823; Pseudomonas fluorescens ORF, ZP_00083027; Helicobacter pylori Pss, AAB66379; Campylobacter jejuni ORF, NP_282262; Bacillus subtilis Pss, NP_388109; Bacillus cereus ORF, NP_830762; Bacillus anthracis ORF, NP_843464.
For the characterization of a sinorhizobial mutant lacking Pss activity, the mutant CS111 was created in which the pssA gene was replaced with a gentamicin resistance cassette. When grown in complex medium, S. meliloti forms PG, CL, PE, MMPE, DMPE, and PC as major membrane lipids. Mutant CS111, deficient in Pss activity, lacks PE, MMPE, and DMPE. On complex medium or other media containing choline, CS111 is able to form PC via the Pcs pathway (13, 52). No additional membrane lipid is synthesized to make up for the deficiency of PE, but the relative amounts of both the anionic lipids PG and CL and the zwitterionic lipid PC increase, with the anionic lipids presenting 63 to 66% of the lipids. Interestingly, at calcium concentrations of 20 mM or higher the relative amounts of membrane lipids are different, and membranes contain more PC than anionic lipids. The conditionally lethal phenotype of E. coli mutants deficient in PE can be rescued by millimolar concentrations of certain bivalent cations, such as Ca2+, Mg2+, and Sr2+, with calcium being the most efficient ion. It has been suggested that the presence of high concentrations of these ions enables the bilayer-forming lipid CL to change into the reverse-hexagonal-phase configuration and thereby substitute for PE, which prefers the reverse-hexagonal-phase configuration (5). S. meliloti needs calcium supplementation for growth, and TY medium complemented with 4.5 mM CaCl2 is a standard growth medium for S. meliloti. Therefore, we had to use Ca2+ in our studies instead of Mg2+, which was used in the studies with E. coli. Similarly as observed in the case of Agrobacterium (25), S. meliloti seems to be much less affected by the lack of PE than E. coli, and there might be several reasons for this. First, PE forms about 70% of the membrane lipids in E. coli but only about 30% in S. meliloti. Furthermore, S. meliloti contains significant amounts of MMPE and DMPE, which are less likely to form a non-bilayer phase than PE. When PE is lost in a PssA-deficient mutant, MMPE and DMPE are lost as well; thus, the loss of PE might be less severe and could be compensated by an increase in CL. Another reason why the lack of PE is affecting S. meliloti less than E. coli might have been the presence of PC, another zwitterionic lipid, in the membranes of S. meliloti. To test this hypothesis, we studied growth of S. meliloti in minimal medium with the supplementation of choline. The supplementation with 10 μM choline or more partly complemented the growth phenotype of PE-deficient S. meliloti mutants under these conditions. Such elevated choline concentrations were impairing growth of the wild type, 1021, for an unknown reason. The observation that no drastic growth difference between the mutant and the wild type can be observed when both strains are grown in complex medium but that growth of the mutant is much worse than that of the wild type in minimal medium supplemented with choline indicates that some other factor from the complex medium is needed for optimal growth.
Interestingly, PE seems to be essential for some microorganisms, whereas it is not for others. S. cerevisiae cannot grow without it, and similarly, Ge and Taylor (21) were not able to construct an H. pylori mutant deficient in Pss, indicating that PE might be essential for this organism. In contrast, an Agrobacterium mutant deficient in Pss seemed to be unaffected in growth (25). The requirement of PE for growth of some bacteria and not in the case of some others parallels the requirement of PC for some bacteria but not for others. No specific phenotype was detected for PC-deficient mutants of R. sphaeroides and Zymomonas mobilis, whereas relatively drastic phenotypes for the PC-deficient mutants were described for S. meliloti and B. japonicum. In order to explain these observations, far more has to be learned about the roles which different membrane lipids play for membrane function, i.e., in what form they are responsible for protein activity or stability.
An interesting aspect for future studies to address is how the PE-deficient S. meliloti strain behaves during symbiosis. Mutants of B. japonicum that were deficient in Pmt and had a reduced amount of PC in their membranes were impaired in their symbiotic performance (38), and double mutants of S. meliloti that were deficient in Pcs and PmtA activity and therefore completely lacking PC were not able to form nodules on their host plant, Medicago sativa (alfalfa) (53). If the lack of PC has such a drastic effect on symbiotic performance, one might expect that the symbiotic performance of mutants deficient in the other major zwitterionic membrane lipid, PE, might be seriously affected. Such studies are currently under way.
Acknowledgments
We thank V. Röhrs for excellent technical assistance.
This research was supported by a grant (Ge 556/2-3) from the Deutsche Forschungsgemeinschaft and by grants from DGAPA/UNAM (IN200802), CONACyT-Mexico (33577-N), and the Howard Hughes Medical Institute (HHMI 55003675). C. Sohlenkamp is a Feodor Lynen Fellow of the Alexander von Humboldt Foundation, Bonn, Germany.
REFERENCES
- 1.Bardin, S., S. Dan, M. Østeras, and T. M. Finan. 1996. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 178:4540-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 3.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 4.Bogdanov, M., and W. Dowhan. 1995. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease in Escherichia coli. J. Biol. Chem. 270:732-739. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanov, M., and W. Dowhan. 2002. Functional roles of lipids in membranes, p. 1-35. In D. E. Vance and J. E. Vance (ed.), Biochemistry of lipids, lipoproteins and membranes, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 6.Bogdanov, M., J. Sun, H. R. Kaback, and W. Dowhan. 1996. A phospholipid acts as a chaperone in assembly of a membrane protein. J. Biol. Chem. 271:11615-11618. [DOI] [PubMed] [Google Scholar]
- 7.Carman, G. M., and D. S. Wieczorek. 1980. Phosphatidylglycerophosphate synthase and phosphatidylserine synthase activities in Clostridium perfringens. J. Bacteriol. 142:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clancey, C. J., S. C. Chang, and W. Dowhan. 1993. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J. Biol. Chem. 268:24580-24590. [PubMed] [Google Scholar]
- 9.Dassa, E., and P. Bougie. 2001. The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152:211-229. [DOI] [PubMed] [Google Scholar]
- 10.DeChavigny, A., P. N. Heacock, and W. Dowhan. 1991. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J. Biol. Chem. 266:5323-5332. [PubMed] [Google Scholar]
- 11.Delhaize, E., D. M. Hebb, K. D. Richards, J.-M. Lin, P. R. Ryan, and R. C. Gardner. 1999. Cloning and expression of a wheat (Tritium aestivum L.) phosphatidylserine synthase cDNA. J. Biol. Chem. 274:7082-7088. [DOI] [PubMed] [Google Scholar]
- 12.de Rudder, K. E. E., I. M. López-Lara, and O. Geiger. 2000. Inactivation of the gene for phospholipid N-methyltransferase in Sinorhizobium meliloti: phosphatidylcholine is required for normal growth. Mol. Microbiol. 37:763-772. [DOI] [PubMed] [Google Scholar]
- 13.de Rudder, K. E. E., C. Sohlenkamp, and O. Geiger. 1999. Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 274:20011-20016. [DOI] [PubMed] [Google Scholar]
- 14.de Rudder, K. E. E., J. E. Thomas-Oates, and O. Geiger. 1997. Rhizobium meliloti mutants deficient in phospholipid N-methyltransferase still contain phosphatidylcholine. J. Bacteriol. 179:6921-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ditta, G., T. Schmidhauser, E. Yacobson, P. Lu, X.-W. Liang, D. R. Finlay, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 16.Doerrler, W. T., and C. R. H. Raetz. 2002. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277:36697-36705. [DOI] [PubMed] [Google Scholar]
- 17.Dulley, J. R., and P. A. Grieve. 1975. A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal. Biochem. 64:136-141. [DOI] [PubMed] [Google Scholar]
- 18.Dutt, A., and W. Dowhan. 1977. Intracellular distribution of enzymes of phospholipid metabolism in several gram-negative bacteria. J. Bacteriol. 132:169-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckert, B., and C. F. Beck. 1989. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J. Bacteriol. 171:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge, Z., and D. E. Taylor. 1997. The Helicobacter pylori gene encoding phosphatidylserine synthase: sequence, expression, and insertional mutagenesis. J. Bacteriol. 179:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiger, O., V. Röhrs, B. Weissenmayer, T. Finan, and J. E. Thomas-Oates. 1999. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol. Microbiol. 32:63-73. [DOI] [PubMed] [Google Scholar]
- 23.Grogan, D. W., and J. E. Cronan, Jr. 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 61:429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 25.Karnezis, T., H. C. Fisher, G. M. Neumann, B. A. Stone, and V. A. Stanisisch. 2002. Cloning and characterization of the phosphatidylserine synthase gene of Agrobacterium sp. strain ATCC 31749 and effect of its inactivation on production of high-molecular mass (1→3)-β-d-Glucan (Curdlan). J. Bacteriol. 184:4114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin, E. V. 1996. A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins. Trends Biochem. Sci. 21:242-243. [PubMed] [Google Scholar]
- 27.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 28.Kusters, R., W. Dowhan, and B. de Kruijff. 1991. Negatively charged phospholipids restore prePhoE translocation across phosphatidylglycerol-depleted Escherichia coli inner membranes. J. Biol. Chem. 266:8659-8662. [PubMed] [Google Scholar]
- 29.Letts, A., L. S. Klig, M. Bae-Lee, G. M. Carman, and S. A. Henry. 1983. Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc. Natl. Acad. Sci. USA 80:7279-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Q. X., and W. Dowhan. 1988. Structural characterization of Escherichia coli phosphatidylserine decarboxylase. J. Biol. Chem. 263:11516-11522. [PubMed] [Google Scholar]
- 31.López-Lara, I. M., C. Sohlenkamp, and O. Geiger. 2003. Membrane lipids in plant-associated bacteria: their biosyntheses and possible functions. Mol. Plant-Microbe Interact. 16:567-579. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto, K. 1997. Phosphatidylserine synthase from bacteria. Biochim. Biophys. Acta 1348:214-227. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto, K., M. Okada, Y. Horikoshi, H. Matsuzaki, T. Kishi, M. Itaya, and I. Shibuya. 1998. Cloning, sequencing and disruption of the Bacillus subtilis psd gene coding for phosphatidylserine decarboxylase. J. Bacteriol. 180:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mileykovskaya, E., Q. Sun, W. Margolin, and W. Dowhan. 1998. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J. Bacteriol. 180:4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mileykovskaya, E. I., and W. Dowhan. 1993. Alterations in the electron transfer chain in mutant strains of Escherichia coli lacking phosphatidylethanolamine. J. Biol. Chem. 268:24824-24831. [PubMed] [Google Scholar]
- 37.Mileykovskaya, E., I. Fishov, X. Fu, B. D. Corbin, W. Margolin, and W. Dowhan. 2003. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo. J. Biol. Chem. 278:22193-22198. [DOI] [PubMed] [Google Scholar]
- 38.Minder, A. C., K. E. E. de Rudder, F. Narberhaus, H.-M. Fischer, H. Hennecke, and O. Geiger. 2001. Phosphatidylcholine levels in Bradyrhizobium japonicum are critical for an efficient symbiosis with the soybean host plant. Mol. Microbiol. 39:1186-1198. [PubMed] [Google Scholar]
- 39.Morii, H., and Y. Koga. 2003. CDP-2,3-di-O-geranylgeranyl-sn-glycerol:l-serine O-archaetidyltransferase (archaetidylserine synthase) in the methanogenic archaeon Methanothermobacter thermoautotrophicum. J. Bacteriol. 185:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikawa, J., Y. Tsukagoshi, T. Kodaki, and S. Yamashita. 1987. Nucleotide sequence and characterization of the yeast PSS gene encoding phosphatidylserine synthase. Eur. J. Biochem. 167:7-12. [DOI] [PubMed] [Google Scholar]
- 41.Okada, M., H. Matsuzaki, L. Shibuya, and K. Matsumoto. 1994. Cloning, sequencing, and expression in Escherichia coli of the Bacillus subtilis gene for phosphatidylserine synthase. J. Bacteriol. 176:7456-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raetz, C. R. H. 1975. Isolation of Escherichia coli mutants defective in enzymes of membrane lipid synthesis. Proc. Natl. Acad. Sci. USA 72:2274-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raetz, C. R. H., and J. Foulds. 1977. Envelope composition and antibiotic hypersensitivity of Escherichia coli mutants defective in phosphatidylserine synthase. J. Biol. Chem. 252:5911-5915. [PubMed] [Google Scholar]
- 44.Raetz, C. R. H., and E. P. Kennedy. 1972. The association of phosphatidylserine synthetase with ribosomes in extracts of Escherichia coli. J. Biol. Chem. 247:2008-2014. [PubMed] [Google Scholar]
- 45.Ruvkun, G. B., and F. M. Ausubel. 1981. A general method for site-directed mutagenesis in prokaryotes. Nature 289:85-88. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. R. Russell. 2001. Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 48.Schnappinger, D., and W. Hillen. 1996. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch. Microbiol. 165:359-369. [DOI] [PubMed] [Google Scholar]
- 49.Schweizer, H. D. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 50.Selbitschka, W., S. Niemann, and A. Pühler. 1993. Construction of gene replacement vectors for Gram− bacteria using a genetically modified sacRN gene as a positive selection marker. Appl. Microbiol. Biotechnol. 38:615-618. [Google Scholar]
- 51.Shi, W., M. Bogdanov, W. Dowhan, and D. R. Zusman. 1993. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J. Bacteriol. 175:7711-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sohlenkamp, C., K. E. E. de Rudder, V. Röhrs, I. M. López-Lara, and O. Geiger. 2000. Cloning and characterization of the gene for phosphatidylcholine synthase. J. Biol. Chem. 275:18919-18925. [DOI] [PubMed] [Google Scholar]
- 53.Sohlenkamp, C., I. M. López-Lara, and O. Geiger. 2003. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 42:115-162. [DOI] [PubMed] [Google Scholar]
- 54.Spaink, H. P., A. H. M. Wijfjes, and B. J. J. Lugtenberg. 1995. Rhizobium NodI and NodJ proteins play a role in the efficiency of secretion of lipochitin oligosaccharides. J. Bacteriol. 177:6276-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spaink, H. P. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54:257-288. [DOI] [PubMed] [Google Scholar]
- 56.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 58.Trotter, P. J., J. Pedretti, and D. R. Voelker. 1993. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268:21416-21424. [PubMed] [Google Scholar]
- 59.Trotter, P. J., J. Pedretti, R. Yates, and D. R. Voelker. 1995. Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. J. Biol. Chem. 270:6071-6080. [DOI] [PubMed] [Google Scholar]
- 60.Trotter, P. J., and D. R. Voelker. 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270:6062-6070. [DOI] [PubMed] [Google Scholar]
- 61.Verma, J. N., and H. Goldfine. 1985. Phosphatidylserine synthase from Clostridium butyricum. J. Lipid Res. 26:610-616. [PubMed] [Google Scholar]
- 62.Voelker, D. R. 1997. Phosphatidylserine decarboxylase. Biochim. Biophys. Acta 1348:236-244. [DOI] [PubMed] [Google Scholar]
- 63.Williams, J. G., and C. R. McMaster. 1998. Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of Saccharomyces cerevisiae cholinephosphotransferase. J. Biol. Chem. 273:13482-13487. [DOI] [PubMed] [Google Scholar]
- 64.Xia, W., and W. Dowhan. 1995. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:783-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yanisch-Perron, C., J. Viera, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]