Abstract
The two major capsid proteins of Lactobacillus bacteriophage A2 share their amino termini. The smaller of these (gp5A) results from translation of orf5 and proteolytic processing after residue 123. The larger form (gp5B) originates through a −1 ribosomal frameshift at the penultimate codon of orf5 mRNA, resulting in a product that is 85 amino acids longer than gp5A. Frameshifting needs two cis-acting elements: a slippery region with the sequence C CCA AAA (0 frame), and a stem-loop that begins 9 nucleotides after the end of the slippery sequence. Mutations introduced in the slippery sequence suppress the frameshift. Similarly, deletion of the second half of the stem-loop results in drastic reduction of frameshifting. Both gp5A and gp5B appear to be essential for phage viability, since lysogens harboring prophages that produce only one or the other protein become lysed upon induction with mitomycin C, though no viable phage progeny are observed.
A2 is a temperate bacteriophage that infects strains of several Lactobacillus casei-related species, including some used as probiotic components in fermented milks. It belongs to the family Siphoviridae, and its genome lies in a double-stranded DNA molecule 43,411 bp long which encodes 61 open reading frames (ORFs) and presents 3′-protruding cohesive ends (15). These may be grouped into several functional modules. Towards the center of the DNA lies the region that regulates the switch between the lytic and lysogenic cycles. Here, two adjacent, divergently oriented repressor genes, cI and cro, encode proteins that play roles superficially similar to those of the analogous proteins of bacteriophage lambda (14, 18-20). Downstream of cI is located the cassette that mediates integration of the phage DNA into the chromosome of its hosts (2), while cro is followed by the replication module of the phage (27). Downstream of this module there is a long DNA stretch that encodes a series of small polypeptides, most of which are unessential for development or lysogenization of the phage, at least under laboratory conditions (15, 20). This end of the genome is occupied by the small terminase subunit determinant, which marks the beginning of the morphogenetic region and is followed, towards the other side of the cohesive ends, by the large terminase subunit (13) and the genes that encode the structural components of the virion in the following order: capsid, head-tail connector, tail, and host recognition, all of which form a late-expressed, single operon (15). Finally, the host lysis cassette lies between the morphogenetic and integration modules.
Analysis of the virion structural proteins revealed two polypeptides of 35 and 42 kDa (as judged from sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) that were the major components of the head at a ratio of 4:1. Surprisingly, both shared their amino termini, which matched an internal sequence of orf5 in the phage genome. In line with this finding, orf5 would originate two polypeptides of different sizes, both of which would suffer the same proteolytic processing upon incorporation into the capsid. This processing would render a hypothetical 123-amino-acid polypeptide from the amino end that is postulated to be the scaffolding protein of the phage and the two polypeptides of different sizes found in the virions (15).
In this report, we confirm this hypothesis through presentation of data indicating that the small head protein, gp5A, corresponds to the translation product of orf5 after proteolytic processing, while its large counterpart, gp5B, results from a change in the frame of translation and is 85 amino acids longer than gp5A. Additionally, some requirements for the frameshift to occur are revealed. Finally, it is demonstrated that both proteins are essential for the generation of viable phages. To our knowledge, this is the first report of a −1 translational frameshift occurring in a gram-positive-specific bacteriophage, although sequence analysis of the Listeria bacteriophage PSA suggests that this situation might be more general (36). In this recent study, two +1 frameshifts in the major capsid and tail genes of PSA were reported.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and incubation conditions.
Lactobacillus casei ATCC 393 was used to propagate bacteriophage A2 on liquid MRS medium (Oxoid) supplemented with 10 mM CaCl2 and 10 mM MgSO4 (MCM) kept at 30°C without aeration. Plaque enumeration was carried out on solid MCM (1.5% agar) covered by semisolid medium (0.7% agar). Escherichia coli DH10B (Gibco-BRL) was used as the recipient for plasmid constructions. E. coli BL21(DE3)/pLysS was used in expression studies of the different versions of orf5 cloned into plasmid pET11a (31). Plasmids pLys (31) and pUC18 (35) have been described previously. E. coli was propagated in 2×TY (29) at 37°C under agitation. Ampicillin (100 μg/ml), IPTG (isopropylthiogalactopyranoside, 1 mM) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 40 μg/ml) were used when required.
Purification of A2 structural proteins and mass spectrometry of tryptic peptides.
A2 particles were purified through isopycnic centrifugation in continuous CsCl gradients and disrupted as previously described (15). The structural proteins were separated by SDS-PAGE, and the bands of interest were excised manually and then automatically processed in a ProGest protein digestion station (Genomic Solutions, Huntingdon, England) with modified porcine trypsin (sequencing degree; Promega) for 12 h at 37°C. The resulting peptides were eluted and analyzed in a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Bruker-Franzen Analytic, Bremen, Germany) equipped with the SCOUT source in the positive ion reflector mode by delayed extraction and AutoXecute acquisition software. The tryptic peptide masses were automatically transferred through the mass spectrometry BioTools program as inputs to search the NCBI database with the Mascot software (Matrix Science).
Plasmid constructions, site-directed mutagenesis, and gene overexpression.
The sequence corresponding to orf5 was amplified by PCR, placed under the P10 promoter of phage T7 in the pET11a vector, and introduced by electroporation into E. coli BL21(DE3)/pLys. Overexpression was promoted by successive addition, at 30-min intervals, of 1 mM IPTG, 500 μg of rifampin per ml, and 10 μCi of [35S]Pro-mix (methionine plus cysteine, 10 TBq/mmol; Amersham) to exponential-phase cultures growing in M5 medium (29). The cells were harvested and lysed and their proteins were separated through SDS-PAGE and detected by autoradiography (29).
Mutant orf5 variants were obtained by overlapping PCR following a published method (1). The primers employed were several quasi-identical 46-mer oligonucleotides spanning positions −30 to +9 with respect to the “slippery sequence,” which carried the desired mutations, and a 23-mer oligonucleotide complementary to the 5′ end of the 46-mer primers. The amplicons obtained were inserted into pET11a and transformed into E. coli BL21(DE3)/pLys, and the mutant genes were overexpressed as indicated above.
The β-galactosidase reporter plasmids pUOfs1 and pUOfs2 were constructed by inserting DNA segments of 93 and 158 bp, respectively (which include the 7-nucleotide-long slippery sequence) (Fig. 1) into the polylinker of pUC18. Thus, correct reading of lacZ takes place only after the frameshift occurs, following concepts developed before (24, 26, 33). β-Galactosidase activity was detected in cell extracts by using o-nitro-phenyl-β-d-galactoside (Sigma) as the substrate (29).
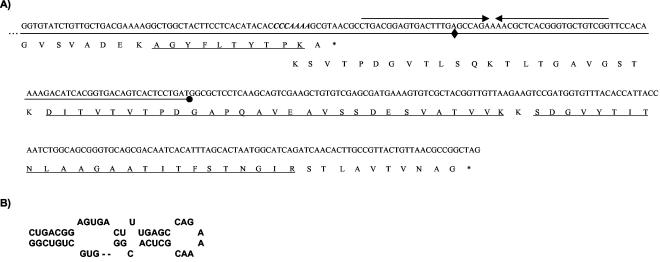
FIG. 1.
(A) DNA and deduced protein sequences of the A2 genome region surrounding the 3′ end of orf5; in the DNA, the slippery sequence is indicated in italics and bold, the loop is indicated by converging arrows, and the segments cloned in the −1 frame under the lacZ promoter of pUC18 are underlined, with their ends represented by a diamond and a dot. The underlined polypeptides correspond to those obtained by MALDI-TOF as part of gp5A and gp5B (the first of them) or as part of gp5B alone (the other two). (B) Proposed secondary structure of the mRNA frameshift-stimulatory element located downstream from the slippery sequence.
Recombinant phage generation and analysis. Mutant versions of orf5 that exclusively rendered gp5A or gp5B were amplified and inserted into pUC19E (a pUC19 derivative unable to replicate in lactobacilli, which harbors an erythromycin resistance determinant in facilitating selection in gram-positive bacteria) (23), which was subsequently introduced into an A2 lysogenic derivative of L. casei. Successive selection for erythromycin resistance and sensitivity enabled recovery of cells that had suffered one and two crossovers, respectively. Substitution of the wild type by the mutant orf5 genotype was tested by PCR amplification and sequencing of the appropriate DNA segments. Induction of A2 prophages was achieved by addition of mitomycin C (1 μg/ml) to exponential-phase cultures of the A2 lysogens. Aliquots were taken at fixed intervals, tested for viable phage generation, and subjected to SDS-PAGE followed by Western blotting with polyclonal antibodies raised against A2 virions with the BM chemiluminescence blotting substrate kit (Roche) to detect phage-specific protein bands.
RESULTS
gp5B results from a ribosomal frameshift of orf5 mRNA.
MALDI-TOF mass spectrometry of gp5A and gp5B revealed that all peptides obtained through treatment of the former with trypsin were present in gp5B, including the most carboxyl-terminal one, which finishes one amino acid before the last residue of gp5A (Fig. 1). In addition, gp5B produced two polypeptides of 2,885 and 2,514 Da, which exactly match the expected values of the tryptic peptides obtained after translation of the region located 3′ with respect to orf5 when read in the −1 frame (Fig. 1). These data indicate that gp5B is synthesized as a consequence of a −1 ribosomal frameshift that occurs at the penultimate codon of the orf5 mRNA, resulting in a protein that is 85 residues longer than gp5A. Analysis of the mRNA at the frameshift point revealed the sequence C CCA AAA (in the 0 frame) which encodes Pro-Lys. After frameshifting, the sequence would become CCC AAA A, which translates into an identical dipeptide, the substitution of the A in the wobble position of the first codon being the only change introduced.
In vitro mutagenesis of the slippery sequence and its effect on frameshifting.
The characteristics of the slippery sequence suggest that frameshifting is dependent on the compatibility of the anticodons of the tRNAs located at the ribosomal P and A sites with their complementary codons in both the 0 and −1 frames. To confirm this assumption, a series of point substitutions were introduced in the DNA sequence, and their effects on frameshifting were tested through analysis of the proportion of the resulting labeled gp5A to gp5B (Fig. 2). These experiments were performed with E. coli to take advantage of the sophisticated means for protein expression analysis available in this organism, bearing in mind that the proportion between gp5A and gp5B in E. coli is about 4:1 (Fig. 2, lane 1), which coincides with the relative concentration of both proteins in A2 virions (15). This circumstance and the similar G+C content of the genomes of A2, its host L. casei, and E. coli (46% versus 50%) might justify the extrapolation of the data obtained, although we are aware that different tRNA pools, which may occur in these two organisms, might restrict direct extrapolation of the E. coli data to L. casei.
FIG. 2.
Effect of different mutations on frameshifting. The slippery sequence is shown in bold, its point mutations are underlined, and the deletion is included in a box. In these experiments, orf5 (and its variants) was placed under the control of the P10 promoter of T7 and overexpressed in vivo in the presence of a 35S-labeled amino acid mix and rifampin. The labeled products were separated by SDS-12% PAGE and autoradiographed. The positions of the bands correspond to the sizes of gp5A and gp5B prior to processing of their 123 amino-terminal residues.
Mutagenesis of the first nucleotide of the slippery sequence from C to A or U resulted in suppression of frameshifting (Fig. 2, lanes 2 and 3), as would be expected if the tRNA at position P in the 0 frame had to recognize a compatible codon in the −1 frame (in the wild type the codon frame goes from CCA to CCC, whereas it would go from CCA to ACC or to UCC in the mutants). Similarly, no production of gp5B was observed when the second nucleotide of the slippery sequence (C) was replaced with U (Fig. 2, lane 4), confirming the involvement of the P ribosomal site in frameshifting.
Mutagenesis of the nucleotide in position 4 of the slippery sequence from A to C also resulted in frameshift elimination (Fig. 2, lane 6), indicating that the tRNA in position A of the ribosome, which recognizes the AAA codon in the 0 frame, does not withstand the change to CAA.
Finally, a 5-bp deletion was generated from nucleotides −8 to −12, considering +1 the first C of the slippery sequence. The resulting gene would naturally be located in the −1 frame after the deletion and, as expected, exclusively rendered a gp5B-like polypeptide (Fig. 2, lane 5). This confirms that the frameshift depends not only on the integrity of the slippery sequence (which remains wild type in this deletion mutant), but also on the codon-anticodon compatibility in both frames (in the mutant, the 0 frame at the slippery site is C CCC AAA, which would become CCC CAA A in the −1 frame, suppressing the compatibility in the second codon-anticodon pairing).
Influence of neighboring sequences on frameshifting.
Frameshift occurrence is usually a consequence of ribosome pausing at the slippery sequence. Among other possibilities, this can be accomplished by the presence in its 3′ vicinity of secondary structures (22). In the case of A2, a potential stem-loop was detected 9 bp after the slippery sequence (Fig. 1B). To determine its role in frameshifting, two pUC18 derivatives were constructed. The first one contained a DNA segment in its polylinker that comprised from 59 bp before the slippery sequence to the first half of the stem-loop, while the second included the entire loop (Fig. 1A). In both cases, the β-galactosidase gene was located in the −1 frame; i.e., enzymatic activity production would be expected only after frameshift occurrence. The data showed that deletion of part of the stem-loop resulted in a 79% reduction in β-galactosidase activity with respect to the clone that conserved the whole stem sequence (0.14 versus 0.66 Miller units), indicating that it plays a functional role in the process.
Effect of frameshift suppression on A2 viability.
To determine whether gp5A and/or gp5B was essential for A2 viability, bacteriophages able to produce each protein but not the other were generated. Initially, an A2 lysogen was transformed with vectors unable to replicate in L. casei, carrying orf5 with the following variations: a transversion from C to A in the first position of the slippery sequence (Fig. 2, lane 2) or the sequence carrying the 5-bp deletion described in the previous paragraph (Fig. 2, lane 5). Double recombinants were recovered in each case, and their prophages were induced with mitomycin C. Lysis of the lysogens resulted in both cases, but plaque production was very scarce from the first recombinant and completely absent in the case of the prophage that carried the deletion. Analysis of 20 progeny plaques obtained from the first mutant revealed that all of these had suffered a retromutation to the wild-type sequence, probably indicating that both mutations rendered defective prophages; i.e., gp5A and gp5B are indispensable for A2 infectivity.
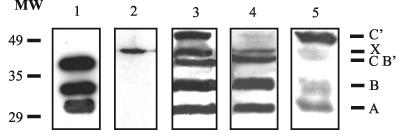
To confirm the phenotype of the mutants with respect to orf5, the corresponding lysogens were induced, and the proteins produced during the intracellular development of the phage were separated by SDS-PAGE and revealed with antibodies raised against A2 virions (Fig. 3). Lysogens harboring the wild-type phage (Fig. 3, lane 3) generated bands that corresponded to the major tail protein (labeled A in Fig. 3), the processed form of gp5A (which has lost the 123 amino acids of its amino terminus, labeled B), a third band (labeled B′ and C) that may include the unprocessed form of gp5A plus the processed gp5B polypeptide, a band which is also present in the nonlysogenic L. casei 393 (labeled X; Fig. 3, lane 2), and a fifth band that may represent the unprocessed form of gp5B (labeled C′).
FIG. 3.
Antibody-based detection (after Western blotting) of A2 structural proteins produced by lysogenic cultures of L. casei after induction with mitomycin C. Lanes: 1, CsCl-purified A2 virions; 2, nonlysogenic L. casei; 3, 4, and 5, lysogens harboring the wild-type prophage, a prophage with a point mutation in position 1 of the slippery sequence, and a prophage carrying a deletion that introduces the −1 frame (these last two correspond to the mutations labeled 2 and 5, respectively, in Fig. 2), respectively. Size standards are indicated to the left (in kilodaltons). Letters to the right identify the proteins in each band as follows: A, major tail protein; B, gp5A processed form; C and B′, unprocessed gp5A plus processed gp5B; X, unidentified protein present in nonlysogenic cultures; C′, unprocessed gp5B.
As expected, the mutant carrying the point mutation in the first nucleotide of the slippery sequence did not produce gp5B (band C′, which is made out of the unprocessed form of gp5B, is not observed in Fig. 3, lane 4). Conversely, the orf5 deletion mutant did not synthesize gp5A (bands B and B′ are absent in Fig. 3, lane 5). Curiously, no clear band representing the processed form of gp5B (band C) was observed, possibly indicating that gp5A is necessary for the processing to occur.
DISCUSSION
It was initially hypothesized that the two major polypeptides of A2 capsids originated from translation of a single gene on the basis of the following facts: these two virion proteins shared the residues located at their amino termini, and this sequence could be ascribed to an internal stretch of orf5. On the other hand, there was a better correspondence between the deduced size of the polypeptide arising from translation of orf5 (plus proteolytic processing) with the smaller form of the two virion proteins (gp5A). This would indicate that the other one (gp5B) might result from translation of a longer message, as a consequence of reading through the stop codon of this gene (gp5A would be a 277-residue polypeptide having a predicted mass of 29,742 Da; its apparent size on SDS-PAGE is 35 kDa, while that of gp5B is 42 kDa) (15). Finally, analysis of the region lying 3′ to orf5 revealed quite a long intergenic segment whose translation in the −1 frame is 38% identical to the translation product of a region that is located behind gene 10 of phage T3, which encodes its coat protein. In this case, the ribosomes undertake a −1 frameshift close to the stop codon during translation in about 10% of the transcripts, which results in a polypeptide (10B) that is 86 amino acids longer than the “canonical” one (10A) (10). A similar situation in A2 would result in production of a polypeptide 85 residues longer than gp5A, which is consistent with the difference in size of this protein and gp5B, as observed by SDS-PAGE.
MALDI-TOF mass spectrometry of both polypeptides revealed that this was indeed the case, indicating that the frameshift occurred at the very end of gp5A, in a sequence that conforms with the slippery sites found in −1 ribosomal frameshifts of eukaryotic organisms, i.e., X XXY YYZ, where X can be any nucleotide, Y indicates either A or U, and Z corresponds to A, U, or C (16). A −1 frameshift commonly involves tandem slippage of the tRNAs located at the ribosome, i.e., the presence of codons at P and A sites compatible with the respective anticodons in both frames (3, 4).
To determine the particular requirements for frameshifting in the orf5 mRNA, nucleotides situated at both the A and P sites before and after the shift of the ribosome were replaced to create a lack of complementarity between the tRNAs and the codons located in front of them in the −1 frame. Any change at this level resulted in frameshift suppression, indicating that tandem slippage occurred in the case discussed here. However, the most frequently reported slippery sequence from prokaryotic genes does not conform to the eukaryotic standard cited above, as its last nucleotide is a G. Examples include the determinant of the DNA polymerase III subunits gamma and tau of E. coli, the genes g-t of the tail assembly in phage lambda, and the FI/II-E/E′-T-U-D tail gene operon of phage P2 (9, 24, 33). All these genes are expressed in E. coli, which is devoid of a tRNA with the anticodon 3′UUC5′. Consequently, the last codon, AAG (frame 0), is recognized by the tRNA whose anticodon is 3′UUU5′, resulting in poor binding and facilitation of the ribosomal shift (3). This explains why expression of a tRNA with the anticodon 3′UUC5′ in E. coli substantially reduces dnaX frameshifting (32).
Given the high proportion of gp5B with respect to gp5A in A2 virions and the fact that its slippery sequence has an A in the Z position, it might be deduced that L. casei possesses a tRNALys with the anticodon 3′UUC5′. Other deviations from the eukaryotic rule include gene 10 of phage T7, where the third X is substituted by a Y (G GUU UUC) (12), and the cytidine deaminase gene of Bacillus subtilis, where the slippery sequence notably departs from the rule (A CGA AAG) (26). This results in a lack of complementarity between the first codons in the 0 and −1 frames, suggesting that single tRNA repairing to mRNA might occur in these cases (28, 30). The frameshift frequency varies greatly, from a modest 4% reported for the lambda g-t genes (24) to the 50% found in dnaX (22), indicating that other motifs may also be important for the frameshift to occur.
The aim of these auxiliary, cis-acting copromoting structures is to induce ribosome pausing (25), although it appears that this is not sufficient and that other, as yet undetermined factors may be needed as well for frameshifting (17). In prokaryotes, the most popular of these pause-promoting motifs is a Shine-Dalgarno-like sequence located about 10 nucleotides before the slippery sequence (9, 21, 26). An alternative or complementary system is the presence of an adjacent stem-loop 3′ to the frameshift site (12, 22). In the case of A2, no evident ribosome-binding site-like sequences were detected in the vicinity of the frameshift region, but a stem-loop is located immediately after the stop codon of gp5A. This seems to play a role in frameshift occurrence, since deletion of its second part resulted in a substantial decrease in β-galactosidase activity when it was the result of translation in the −1 frame. In the cases of T3 to T7 (11, 12), a second stem-loop was detected after the end of the gp10B coding sequence, only the first four nucleotides of the loop having been found to be essential for frameshift occurrence. There is a potential secondary structure in A2 that begins 104 nucleotides after the gp5B stop triplet. However, this does not appear to influence frameshift frequency, as a recombinant phage in which such a loop was deleted still produced both gp5A and gp5B (data not shown).
Finally, there is the question of the functionality of the dual protein system arising from the frameshift. Two situations become evident in this respect. In the case of cellular genes, it appears that the −1 product does not play an essential role. For instance, in the case of the dnaX gene of E. coli, the gamma subunit of the DNA polymerase III is not absolutely necessary for enzymatic activity, nor can it substitute for the lack of the tau subunit (6). Similarly, the cytidine deaminase of B. subtilis presents comparable specific activities irrespective of whether it is a homo- or a heterotetramer (26). However, the frameshift product is essential for viral propagation; it forms the retrotranscriptase of retroviruses (7) and an essential morphogenetic protein in phage lambda (24). Yet again, T7 may be an exception. Mutants unable to synthesize gp10B did not show any marked phenotype with respect to the wild-type phage, although it was postulated that conditions in which frameshifting would be essential for phage viability should exist, as deduced from conservation of the property in both T3 and T7 (11). A2 appears to be a typical virus in this respect. Prophages that synthesized either one of the gp5 proteins were not viable in spite of being able to promote lysis of mitomycin C-induced cultures. This finding is not surprising if one considers the large number of monomers of each type that contribute to the virion structure (calculated to be about 380 copies of gp5A and 110 copies of gp5B) (15).
In the case of retroviruses, frameshift occurrence could possibly be considered a strategy to compress as much information as possible into a very small genome. This circumstance cannot really be argued in the case of A2, since there is a long DNA stretch in its genome that is dispensable for phage development (20). An alternative possibility is that frameshifting is a means to obtain definite proportions of gp5A versus gp5B that might reflect their relative concentrations in the virions. Since these proteins share most of their sequences and gp5A is much more abundant than gp5B, it would be reasonable to speculate that several gp5A monomers would interact to form the planar faces of the icosahedral shell, whereas gp5B would occupy the edges or vertices of the facets. Thus, interaction between gp5A and gp5B could be accomplished through their identical parts (as would occur among gp5A monomers), while the extra sequence of gp5B might allow the occurrence of the curvature at the capsid edges. A similar situation has been found in phage T4 with gp23 and gp24, which, interestingly enough, share part of their sequences in spite of being encoded by separate genes (8, 34). Furthermore, in the Listeria phage PSA, where the two major head proteins are derived from a single gene through a +1 frameshift, a similar capsid architecture has been postulated (36).
Experiments to determine the positions of gp5A and gp5B in the heads of A2 are under way. In this respect, two observations merit comment. gp5B is immunodominant over gp5A (Fig. 3, lane 1; the intensities of the bands obtained after Western blotting are similar in spite of the 4:1 ratio of gp5A to gp5B in the phage particles), which suggests that the latter might occupy a protruding position in the virion, and gp5B does not appear to be processed in extracts of induced lysogens whose prophages were unable to produce gp5A (Fig. 3, lane 5), which might indicate that the proteinase involved in this processing needs preassembly of capsid precursors, as occurs with T4 prohead proteinase (T4 PPase), the analogous enzyme of phage T4 (5).
Acknowledgments
This work was funded by CICYT grants BIO2001-3621 and BMC2002-0638 from the Ministry of Science and Technology (Spain). P.G. and I.R. are holders of a fellowship and a scholarship, respectively, associated with grant BIO2001-3621.
We thank P. Barnes for English corrections. The proteomics service of the National Biotechnology Centre (CSIC) is warmly acknowledged.
REFERENCES
- 1.Adey, N. B., P. C. Stemmer, and B. K. Kay. 1996. Preparation of second generation phage libraries, p. 280-291. In B. Kay, J. Winter, and J. McCafferty (ed.), Phage display of peptides and proteins. Academic Press, San Diego, Calif.
- 2.Alvarez, M. A., M. Herrero, and J. E. Suarez. 1998. The site-specific recombination system of the Lactobacillus species bacteriophage A2 integrates in gram-positive and gram-negative bacteria. Virology 250:185-193. [DOI] [PubMed] [Google Scholar]
- 3.Baranov P. V., R. F. Gesteland, and J. F. Atkins. 2002. Recoding: translational bifurcations in gene expression. Gene 286:187-201. [DOI] [PubMed] [Google Scholar]
- 4.Baranov, P. V., O. L. Gurvich, O. Fayet, M. F. Prere, W. A. Miller, R. F. Gesteland, J. F. Atkins, and M. Giddings. 2001. RECODE: a database of frameshifting, bypassing and codon redefinition utilized for gene expression. Nucleic Acids Res. 29:264-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, L., and M. Showe. 1983. Morphogenesis of the T4 head, p. 219-245. In E. Mathews, E. Kutter, G. Mosig, and P. Berget (ed.), Bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 6.Blinkova, A., C. Hervas, P. T. Stukenberg, R. Onrust, M. E. O'Donnell, and J. R. Walker. 1993. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, tau and gamma, but only tau is essential. J. Bacteriol. 175:6018-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brierley, I. 1995. Ribosomal frameshifting of viral RNAs. J. Gen. Virol. 76:1885-1892. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S., and R. Hendrix. 1988. Control mechanisms in dsDNA bacteriophage assembly, p. 15-91. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Publishing Corp., New York, N.Y. [Google Scholar]
- 9.Christie, G. E., L. M. Temple, B. A. Bartlett, and T. S. Goodwin. 2002. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J. Bacteriol. 184:6522-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condreay, J. P., S. E. Wright, and L. J. Molineux. 1989. Nucleotide sequence and complementation studies of the gene 10 region of bacteriophage T3. J. Mol. Biol. 207:555-561. [DOI] [PubMed] [Google Scholar]
- 11.Condron, B. G., J. F. Atkins, and R. F. Gesteland. 1991. Frameshifting in gene 10 of bacteriophage T7. J. Bacteriol. 173:6998-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condron, B. G., R. F. Gesteland, and J. F. Atkins. 1991. An analysis of sequences stimulating frameshifting in the decoding of gene 10 of bacteriophage T7. Nucleic Acids Res. 19:5607-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García, P., J. C. Alonso, and J. E. Suárez. 1997. Molecular characterization of the cos region of the Lactobacillus casei bacteriophage A2. Gene product 3, gp3, specifically binds to its downstream cos region. Mol. Microbiol. 23:505-514. [DOI] [PubMed] [Google Scholar]
- 14.García, P., V. Ladero, J. C. Alonso, and J. E. Suárez. 1999. Cooperative interaction of CI protein regulates lysogeny of Lactobacillus casei by bacteriophage A2. J. Virol. 73:3920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia, P., V. Ladero, and J. E. Suarez. 2003. Analysis of the morphogenetic cluster and genome of the temperate Lactobacillus casei bacteriophage A2. Arch. Virol. 148:1051-1070. [DOI] [PubMed] [Google Scholar]
- 16.Hammell, A. B., R. C. Taylor, S. W. Peltz, and J. D. Dinman. 1999. Identification of putative programmed −1 ribosomal frameshift signals in large DNA databases. Genome Res. 9:417-427. [PMC free article] [PubMed] [Google Scholar]
- 17.Kontos, H., S. Napthine, and I. Brierley. 2001. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 21:8657-8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladero, V., P. García, J. C. Alonso, and J. E. Suárez. 1999. A2 Cro, the lysogenic cycle repressor, specifically binds to the genetic switch region of Lactobacillus casei bacteriophage A2. Virology 262:220-229. [DOI] [PubMed] [Google Scholar]
- 19.Ladero, V., P. García, J. C. Alonso, and J. E. Suárez. 2002. Interaction of the Cro repressor with the lysis/lysogeny switch of the Lactobacillus casei temperate bacteriophage A2. J. Gen. Virol. 83:2891-2895. [DOI] [PubMed] [Google Scholar]
- 20.Ladero, V., P. Garcia, V. Bascaran, M. Herrero, M. A. Alvarez, and J. E. Suárez. 1998. Identification of the repressor-encoding gene of the Lactobacillus bacteriophage A2. J. Bacteriol. 180:3474-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen, B., N. M. Wills, R. F. Gesteland, and J. F. Atkins. 1994. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 176:6842-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen, B., R. F. Gesteland, and J. F. Atkins. 1997. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50%. J. Mol. Biol. 271:47-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leenhouts, K., J. Kok, and G. Venema. 1990. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl. Environ. Microbiol. 56:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin, M. E., R. W. Hendrix, and S. R. Casjens. 1993. A programmed translational frameshift is required for the synthesis of a bacteriophage lambda tail assembly protein. J. Mol. Biol. 234:124-139. [DOI] [PubMed] [Google Scholar]
- 25.Lopinski, J. D., J. D. Dinman, and J. A. Bruenn. 2000. Kinetics of ribosomal pausing during programmed −1 translational frameshifting. Mol. Cell. Biol. 20:1095-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mejlhede, N., J. F. Atkins, and J. Neuhard. 1999. Ribosomal −1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J. Bacteriol. 181:2930-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moscoso, M., and J. E. Suarez. 2000. Characterization of the DNA replication module of bacteriophage A2 and use of its origin of replication as a defense against infection during milk fermentation by Lactobacillus casei. Virology 273:101-111. [DOI] [PubMed] [Google Scholar]
- 28.Napthine, S., M. Vidakovic, R. Girnary, O. Namy, and I. Brierley. 2003. Prokaryotic-style frameshifting in a plant translation system: conservation of an unusual single-tRNA slippage event. EMBO J. 22:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sekine, Y., N. Eisaki, and E. Ohtsubo. 1994. Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 235:1406-1420. [DOI] [PubMed] [Google Scholar]
- 31.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 32.Tsuchihashi, Z., and P. O. Brown. 1992. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 6:511-519. [DOI] [PubMed] [Google Scholar]
- 33.Weiss, R. B., D. M. Dunn, M. Shuh, J. F. Atkins, and R. F. Gesteland. 1989. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1:159-169. [PubMed] [Google Scholar]
- 34.Yanagida, M. 1977. Molecular organization of the shell of T-even bacteriophage head. II. Arrangement of subunits in the head shell of giant phages. J. Mol. Biol. 109:515-537. [DOI] [PubMed] [Google Scholar]
- 35.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 36.Zimmer, M., E. Sattelberger, R. B. Inman, R. Calendar, and M. J. Loessner. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed +1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50:303-317. [DOI] [PubMed] [Google Scholar]