Abstract
Several serious diseases are caused by biofilm-associated Staphylococcus aureus, infections in which the accessory gene regulator (agr) quorum-sensing system is thought to play an important role. We studied the contribution of agr to biofilm development, and we examined agr-dependent transcription in biofilms. Under some conditions, disruption of agr expression had no discernible influence on biofilm formation, while under others it either inhibited or enhanced biofilm formation. Under those conditions where agr expression enhanced biofilm formation, biofilms of an agr signaling mutant were particularly sensitive to rifampin but not to oxacillin. Time lapse confocal scanning laser microscopy showed that, similar to the expression of an agr-independent fluorescent reporter, biofilm expression of an agr-dependent reporter was in patches within cell clusters and oscillated with time. In some cases, loss of fluorescence appeared to coincide with detachment of cells from the biofilm. Our studies indicate that the role of agr expression in biofilm development and behavior depends on environmental conditions. We also suggest that detachment of cells expressing agr from biofilms may have important clinical implications.
Staphylococcus aureus, a leading cause of nosocomial infections worldwide, is the etiologic agent of a wide range of diseases, from relatively benign skin infections to potentially fatal systemic disorders. Many of these diseases, including endocarditis, osteomyelitis, and foreign-body related infections, appear to be caused by biofilm-associated S. aureus (12, 18, 30, and 44). Biofilms are sessile microbial communities embedded in a self-produced extracellular polymeric matrix (12, 44). There is increasing awareness that biofilms have a special clinical relevance. Biofilm-associated bacteria show an innate resistance to antibiotics (5), disinfectants (36), and clearance by host defenses (reference 43; also reviewed in reference 12). These properties likely contribute to the persistence and recalcitrance to treatment of staphylococcal biofilm infections.
Two stages of staphylococcal biofilm formation have been described (reviewed in reference 18). The first stage involves attachment of cells to a surface. This stage of biofilm formation is likely to be mediated in part by cell wall-associated adhesins, including the microbial surface components recognizing adhesive matrix molecules. The second stage of biofilm development includes cell multiplication and formation of a mature structure consisting of many cell layers. This stage is associated with the production of extracellular factors, including the polysaccharide intercellular adhesin component of the extracellular matrix.
Intercellular signaling, often referred to as quorum sensing, has been shown to be involved in biofilm development by several bacteria, including Pseudomonas aeruginosa (11), Burkholderia cepacia (22, 23), Streptococcus mutans (26, 31), and others (27, 46, 49). For example, a quorum-sensing-defective mutant of P. aeruginosa is unable to form the highly differentiated biofilm structure associated with wild-type P. aeruginosa, at least under certain conditions (11). S. aureus quorum sensing involves a system unrelated to the P. aeruginosa acyl-homoserine lactone system. The S. aureus quorum-sensing system is encoded by the accessory gene regulator (agr) locus (reviewed in references 35 and 39). The agr system contributes to virulence in model biofilm-associated infections, including endocarditis (7, 50) and osteomyelitis (3, 15), although the precise role of the agr system varies with the type of infection model used (16, 17, 54). The agr locus consists of two divergent operons driven by the P2 and P3 promoters. The P2 operon contains agrBDCA and codes for the RNAII transcript. P3 drives transcription of RNAIII, the effector molecule of the agr locus. Also, δ-hemolysin, a secreted virulence factor encoded by hld, is translated from RNAIII. AgrA and AgrC comprise a two-component regulatory system that responds to the secreted autoinducing cyclic octapeptide. The autoinducing cyclic octapeptide is processed from the agrD product by AgrB. Signaling via this system, in concert with other regulatory elements, such as SarA (6), increases transcription from both the P2 and P3 promoters, resulting in elevated intracellular concentrations of RNAIII. In batch culture, RNAIII acts to increase the expression of secreted virulence factors and decrease the expression of several surface adhesins, including protein A and the fibronectin-binding protein.
Some information about the relationship between agr expression and S. aureus biofilms is available. Pratten et al. (38) found little difference between wild-type S. aureus and an agr mutant in adherence to either uncoated or fibronectin-coated glass under flow conditions. Furthermore, expression of hld was highest near the base of the biofilm, where the highest numbers of bacteria was also observed. Shenkman et al. (42) found that RNAIII expression decreased S. aureus adherence to fibrinogen under static conditions and increased adherence to fibronectin and human endothelial cells under both static and flow conditions. Vuong et al. (51) examined the correlation between a functional agr system and the ability of S. aureus clinical isolates to adhere to polystyrene under static conditions. Only 6% of the isolates with a functional agr system formed a biofilm under these conditions compared to 78% of the agr-defective isolates. The failure of the strains with functional agr loci to form a biofilm was thought to be due in part to the surfactant properties of δ-hemolysin. Another exotoxin directly regulated by the agr system, alpha-toxin, was recently found to contribute to biofilm formation under both static and flow conditions (4). In an experimental endocarditis study, RNAIII activation was time and cell density dependent and occurred through both RNAII-dependent and -independent mechanisms (53). Taken together, these studies indicate that the precise role of agr expression in biofilm development is dependent upon the conditions under which the biofilm is grown. However, the studies were done using different strains, and it remains to be determined whether the differing results were a consequence of different conditions or different strains.
Several questions regarding the role of agr expression in biofilm behavior and function, particularly in the second stage of biofilm formation, remain to be addressed. Can quorum sensing affect the structure and development of S. aureus biofilms? Can the contribution of the agr system to biofilm development change when biofilms of the same strain are grown under different conditions? Does quorum sensing affect the antibiotic resistance of S. aureus biofilms? When and where are agr-controlled transcripts activated in biofilms, and might this have implications for the pathogenesis of biofilm-associated infections? To begin to address these questions, we have examined the effects of agr expression on biofilm development and antibiotic resistance under both static and flow conditions. We have also employed time lapse confocal scanning laser microscopy (CSLM) and green fluorescent protein (GFP) technology to examine agr-dependent expression in biofilms.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used are described in Table 1. Planktonic cultures of either S. aureus or Escherichia coli DHα were grown in Luria-Bertani (LB) broth or plated on LB agar with the appropriate antibiotics for plasmid selection or maintenance (erythromycin, 5 μg/ml; chloramphenicol, 10 μg/ml; ampicillin, 100 μg/ml) and incubated at 37°C. The medium for growth of static biofilms in microtiter dishes was Bacto Tryptic Soy Broth (TSB) (Becton Dickinson, Sparks, Md.) plus 1% glucose and 2% NaCl. We used 5% TSB as the growth medium for the spinning-disk biofilms. For most flow cell experiments, we used a medium containing 2% LB broth and 0.1% glucose. Where indicated, we used 5% TSB in flow cell experiments. When required, 5 μg of erythromycin/ml was added to the flow cell media to maintain reporter plasmids. All biofilm cultures were incubated at 37°C unless otherwise indicated.
TABLE 1.
Strains, plasmids and primers used in this study
| Strain, plasmid, or primer | Relevant characteristics | Source or reference |
|---|---|---|
| S. aureus | ||
| RN4220 | rK− mK+ | 24 |
| MN8 | tstH+; clinical isolate from nonmenstrual toxic shock syndrome case | 41 |
| AMD283 | MN8 agrD | This study |
| Plasmids | ||
| LITMUS28 | Cloning vector | New England Biolabs |
| pCR2.1-TOPO | TA cloning vector | Invitrogen |
| pKEN-GFPmut2 | gfpmut2 | 10 |
| pRSET10-B | yfp10B | R. Heim |
| pE194 | ermC | 19 |
| pCE104 | Shuttle vector containing pE194 and pUC18; Emr | 33 |
| pCE107 | pCE104 containing HindIII-SalI MN8 tstH chromosomal fragment; Emr | P. Schlievert |
| pKSV7 | pUC18-pC194 chimera; temperature sensitive; Cmr | 45 |
| pDB1 | PCR-amplified (O1 and O2) agr region from RN4220 cloned into XhoI-HindIII-digested LITMUS28 | This study |
| pDB2 | BamHI-PstI gfpmut2 from pKEN cloned into BamHI-PstI-digested LITMUS28 | This study |
| pDB3 | PCR-amplified ermC from pE194 cloned into EcoRI- and BglII-digested LITMUS28 | This study |
| pDB4 | BglIIEcoRI ermC fragment from pDB3 cloned into BglII- and EcoRI-digested pDB2 | This study |
| pDB6 | pUC18 containing XhoI (SalI)-HindIII agr fragment from pDB1 | This study |
| pDB9 | pDB6 with BglII site introduced into the RNAIII region using O35, O36 | This study |
| pDB12 | BamHI-BglII gfpmut2ermC fragment from pDB4 cloned into BglII-digested pDB9 | This study |
| pDB13 | BamHI-HindIII agr P3-gfpmut2ermC fragment from pDB12 cloned into BamHI- and HindIII-digested pCE107 | This study |
| pDB22 | agr P3-gfpmut2 reporter; PCR-amplified (O1 and O62) agr P3-gfpmut2ermC fragment from pDB13 cloned into BamHI-HindIII-digested pCE107 | This study |
| pDB39 | pUC18/pC194 shuttle vector containing BamHI-BglII yfp10B fragment from pDB44 | This study |
| pDB44 | pCR2.1-TOPO containing yfp10B PCR amplified from BamHI-digested pRSET10-B, yfp10B contains gene10 ribosome-binding site | This study |
| pDB52 | BamHI-BglII fragment (containing ermC) of pDB3 cloned into BamHI-digested pKSV7 | This study |
| pDB53 | agrCA fragment PCR amplified from MN8 (O63 and O68) and cloned into XbaI-digested pDB52 | This study |
| pDB59 | PCR-amplified (O110 and O111) agr P2-P3 region from MN8 cloned into BamHI-digested pDB39; agr P3-yfp10B; Cmr | This study |
| pDB60 | agrB-P3 fragment PCR amplified (O112 and O113) from MN8 and cloned into BamHI- and BglII-digested pDB53 | This study |
| pDB63 | PCR-amplified (O126 and O127) sar P1 region from MN8 cloned into BamHI-digested pDB39; Cmr | This study |
| pJY202 | agr P3-yfp10B reporter; pCE104 containing agr P3-yfp10BKpnI-HindIII fragment of pDB59; Emr | This study |
| pJY209 | sar P1-yfp10B reporter; pCE104 containing sar P1-yfp10BKpnI-HindIII fragment of pDB63; Emr | This study |
| Primer (5′-3′) | ||
| O1 | TATATAAGCTTGAGCTTGGGAGGGGCTCACGACC | |
| O2 | TATATCTCGAGAGCTTGCTCAAGCACCTCATAAGG | |
| O20 | ATGATCGAATTCTTATTTGTACAATTCA | |
| O35 | GCTTTTAGCATGTTTTAATATAACTAGATCTCAGAGATGTGATGGAAAATAG | |
| O36 | CTATTTTCCATCACATCTCTGAGATCTAGTTATATTAAAACATGCTAAAAGC | |
| O62 | ATAATGGATCCATTTTAACATAAAAAAATTTACAGTTAAG | |
| O63 | GCACGTCTAGAGGTATACAATGAAACGTTGTATCG | |
| O68 | ATACTCTCTAGACTATAAGAGAAAGTGTGATAG | |
| O93 | ATGGGAAGCTTCGTGGATCCTCTAGATTTAAG | |
| O94 | GTTCTTCTCCTTTACTCATATGTATATCTCC | |
| O97 | CGCTGGCAGATCTTTATTATTTGTATAGTTCATCCATGCC | |
| O110 | GGATCCACCACTCTCCTCACTGTTATTATACGA | |
| O111 | AGATCTTTTTCCATCACATCTCTGTGATCTAGT | |
| O112 | TAGATCTATCAAGGATGTGATGTTATGAAAGT | |
| O113 | AGGATCCTTTTTCATAATTTAGTCCTCCTTTGAA | |
| O126 | GGATCCGCTGATATTTTTGACTAAACCAAATG | |
| O127 | GAGATCTCCTCCCTATTTGATGCATCTTG |
Strain and plasmid construction.
The PCR primers used are listed in Table 1. Generally, we used E. coli throughout plasmid construction. The plasmids were then used to transform S. aureus RN4220, and RN4220-modified plasmids were used to transform S. aureus MN8. In all cases, S. aureus was transformed by electroporation (40).
To construct the agrD mutant, S. aureus MN8 was transformed with the temperature-sensitive plasmid pDB60, which contains an ermC erythromycin resistance cassette flanked by a 2,219-bp agrAC-containing fragment and a 2,349-bp agrB P3-containing fragment. Transformants were grown at 42°C, and a chloramphenicol-sensitive, erythromycin-resistant transformant was selected for further study. A chromosomal deletion of the agrD region was confirmed by Southern analysis, and the mutant produced only 20% of the parental levels of toxic shock syndrome toxin 1 as measured by an enzyme-linked immunosorbent assay (14, 55). Furthermore, unlike the parent, the agrD mutant containing an agr-dependent gfp expression vector showed background fluorescence in planktonic or biofilm cultures (data not shown). The sequence data for the design of primers to amplify portions of the agr locus for pDB60 were obtained from the S. aureus MRSA252 Sequencing Group at the Sanger Institute (ftp://ftp.sanger.ac.uk/pub/pathogens/sa).
The reporter plasmid pDB22 contains a fusion of the agr P3 promoter region (bp 508 to 605; GenBank accession number AF288215) with gfpmut2 (a gene coding for a bright variant of GFP). The P3 promoter fragment extends 53 bp upstream of the −35 region and is sufficient for agr-dependent transcription of RNAIII as described by Morfeldt et al. (32). The reporter plasmid pJY202 contains a fusion of nearly the entire agr P2-P3 intergenic region (bp 490 to 711; GenBank accession number AF288215) with yfp10B (a gene coding for a different bright variant of GFP). The yfp10B gene contained a T7 gene10 Shine-Delgarno sequence created as follows. The gene10 Shine-Delgarno sequence from pKEN was PCR amplified using primers O93 and O94. This PCR product was then used as a primer in a second PCR, together with primers O93 and O97 and pRSET-10B as a template, to amplify yfp10B. The fragment was cloned into a pUC18-pC194 shuttle vector to form pDB39. The reporter plasmid pJY209 contains a fusion of 257 bp of the sar P1 promoter (bp 601 to 857; GenBank accession number U46541) (8, 29) with yfp10B.
Fluorescence levels for each of these reporters were determined using flow cytometry (FACScan; Becton Dickinson). Appropriate settings for flow cytometry were determined by analyzing known mixtures of GFP-fluorescent and nonfluorescent S. aureus cells using the FL-1 channel. Ten thousand cells from stationary-phase cultures of each strain were then analyzed using identical flow cytometry settings, and the data were analyzed using CellQuest (version 3.1f) software. The average fluorescence of the strain with the agr P3 reporter plasmid pDB22 was 11 times that of the parental strain (MN8) with no reporter and ∼4 times the minimum fluorescence required for visualization by confocal microscopy. The average fluorescence of the strain with the agr P3 reporter plasmid pJY202 was 163 times that of parental cells and ∼60 times the minimum fluorescence required for visualization by confocal microscopy. The average fluorescence of the strain with the sar P1 reporter plasmid pJY209 was 40 times that of parental cells and 14 times the minimum fluorescence required for visualization by confocal microscopy. The plasmid copy number for pE194-based plasmids during growth of S. aureus at 32°C is estimated to be 10 to 25 per cell; growth at 37°C is thought to reduce the copy number (52).
Biofilm experiments.
We used three different biofilm growth systems, a static microtiter dish system, a rotating-disk reactor system, and a flow cell system. The static biofilm system was similar to that described by Christensen et al. (9). The medium described above was inoculated with overnight S. aureus cultures (1% [vol/vol]). The inoculated medium was dispensed into wells of sterile flat-bottom 96-well polystyrene tissue culture plates (Costar no. 3596; Corning Inc., Corning, N.Y.) at 0.2 ml per well. The plates were incubated for 18 h without shaking. The liquid in the wells was removed by aspiration, and the wells were washed once with 0.2 ml of sterile phosphate-buffered saline (PBS). Bacteria attached to the plastic were fixed with Bouin fixative and stained with 1% crystal violet. Excess stain was removed by washing the plates with distilled water. The crystal violet absorbance at 595 nm was measured by using a Tecan GENios microtiter dish reader (Phenix Research Products, Research Triangle Park, N.C.).
The rotating-disk reactor was similar to that described previously (20) with some modification. The reactor was inoculated with an overnight culture of S. aureus (1% [vol/vol]), and after overnight growth a flow of fresh medium was initiated (dilution rate, 0.7 h−1). After an additional 24-h incubation, polycarbonate chips with attached biofilm bacteria were removed from the spinning disk and washed three times in PBS. The remaining attached cells were dispersed into 2 ml of PBS by using a tissue homogenizer (Brinkman). Total CFU were determined by dilution and plating on LB agar.
We used a flow cell bioreactor for studies of gene expression and biofilm development (37). The flow cells were inoculated with stationary-phase cultures, and flow was initiated after a 1-h incubation at room temperature. The flow rate was laminar, with a Reynolds number of 0.17. To ensure that functional reporter plasmids were maintained during biofilm growth, we recovered bacteria by using high fluid shear to expel biofilms from flow cells with PBS and plating the material on LB agar with and without antibiotics. When S. aureus containing erythromycin-resistant reporters was removed from 6-day-old biofilms, equal numbers of colonies arose on LB agar and on LB agar with erythromycin.
The numbers of fluorescent cells in flow cell biofilms and from flow cell effluents were measured by flow cytometry. Effluent from the flow cells was collected for ∼30 min, and then the biofilms were expelled from the flow cells by the high shear force of 12 ml of fresh growth medium forced through the system by using a syringe. This expulsion of biofilm left only a thin layer of nonfluorescent cells attached to the glass surface. Both the biofilm-derived cells and cells in the biofilm effluents were dispersed by sonication (Branson Sonifier 250) at 40% power for ∼5 s. This treatment did not affect cell numbers or fluorescence in stationary-phase batch cultures of a strain carrying the agr P3-gfp reporter (pJY202). Five thousand biofilm-derived cells from each sample were analyzed for green fluorescence as described above.
Where indicated, flow cell biofilms were stained with either of two nucleic acid stains, SYTO62 (Molecular Probes, Eugene, Oreg.) or propidium iodide (Sigma Chemical Co., St. Louis, Mo.). SYTO62 penetrates and stains nucleic acids in both live and dead cells. Because SYTO62 reduces cell viability, we used it only at experimental endpoints. Propidium iodide is thought to be unable to penetrate viable cells and was thus maintained at 4 μM in the flow cell medium reservoir to allow visualization of the overall biofilm structure by time lapse CSLM. The addition of 4 μM propidium iodide to batch cultures did not affect the growth of S. aureus (pDB22), nor did it affect the detection of GFP fluorescence.
Antibiotic resistance.
Standardized planktonic antibiotic MICs were determined using the broth microdilution method as described by the NCCLS (34). We assessed the resistance of biofilm cells to antibiotics as follows. Biofilms grown in the rotating-disk reactor were washed as described above and incubated in 1 ml of LB broth with antibiotics for 5 h in 24-well tissue culture plate wells (Falcon no. 353047; Becton Dickinson Labware, Franklin Lakes, N.J.). The numbers of viable cells remaining were determined by homogenization and plating as described above.
Microscopy.
For CSLM, we used a Radiance 2100 system (Bio-Rad, Hercules, Calif.) with a Nikon Eclipse E600 microscope contained in a microscope temperature control system (Life Imaging Services, Reinach, Switzerland). The excitation wavelength used for GFP was 488 nm (argon laser), and the emission was collected at 515 ± 15 nm. To detect propidium iodide fluorescence, excitation was at 488 nm and emission wavelengths of >660 nm were collected with a 660LP filter. To detect the red fluorescent nucleic acid stain SYTO62, excitation was at 637 nm and emission wavelengths of >660 nm were collected. For the time course experiments, flow cells were incubated on the microscope stage at 37°C and z-series images were acquired at 15-min intervals. The biofilms were exposed to the scanning laser for ∼45 s at each interval. The image acquisition software was LaserSharp 2000 (Bio-Rad). Postacquisition, images were processed using Confocal Assistant or Volocity (Improvision, Lexington, Mass.) software. Nikon 20× (Plan Fluor) and 60× (Plan Apo) objective lenses were used for CSLM. For phase-contrast microscopy of flow cell biofilms, we used an Olympus BX-51 microscope with a 20× objective lens.
Fluorescence detection in batch cultures.
GFP fluorescence in batch cultures of S. aureus containing either pDB22 or pJY202 was measured in a Tecan microtiter dish reader with an excitation wavelength of 485 nm and an emission wavelength of 535 nm. Relative fluorescence units were determined by subtracting the background fluorescence (S. aureus without a GFP plasmid) from the absolute fluorescence.
RESULTS
The agr system in S. aureus biofilm development and antibiotic resistance.
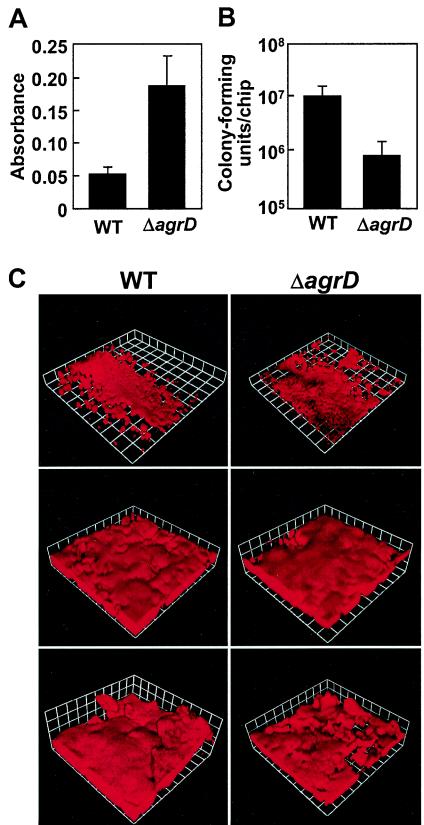
We first tested the ability of an agrD deletion mutant (AMD 283) and its parent (MN8) to form biofilms in a static biofilm system. Consistent with a previous study comparing quorum-sensing-competent and quorum-sensing-defective clinical isolates under static conditions (51), the agrD mutant formed larger biofilms than did the parent strain (Fig. 1A). We were interested in several additional questions. What is the influence of quorum sensing on the development of biofilms subjected to shear forces introduced by flow? Does quorum sensing influence antibiotic resistance in biofilms? When are quorum-induced genes expressed in the biofilm? To address these questions, we used two separate systems in which there was a continuous flow of medium. One system, a once flowthrough apparatus, allows direct microscope observation of biofilms generated under relatively low shear forces. The other system, a rotating-disk reactor, allows one to easily remove biofilms for further study but is not suited for analyzing the structure of a growing biofilm. The shear forces in a rotating-disk reactor are much higher than in a flow cell; the velocity of the medium over the biofilm surface is ∼1,500-fold higher in the rotating-disk reactor than in the flow cell.
FIG. 1.
Influence of agr on biofilm development under different growth conditions. The S. aureus parent strain, MN8 (WT), and the agrD mutant (ΔagrD) were grown in microtiter dish assays (A), spinning-disk reactors (B), and flow cells (C). (A) Microtiter dish biofilm mass as measured by crystal violet staining. The data represent the means and standard deviations of 18 replicate wells. (B) CFU recovered from rotating-disk reactor chips. The data represent the means and standard deviations of duplicate chips taken from each of four independentspinning-disk reactors (total, eight chips) for each strain. There is a significant difference between the two strains (P = 0.03; Student's t test). (C) Three-dimensional reconstructions from days 1 (top) and 5 (middle and bottom) in flow cell biofilms stained with propidium iodide. Each side of a grid square is 60 μm. Microcolony sizes of biofilms of the wild type and the agrD mutant at 48 h were not significantly different. The average surface areas covered by microcolonies in biofilms of the parent and mutant were 0.020 and 0.025 mm2, respectively (determined by measuring 14 microcolonies of each strain in four different images acquired by light microscopy from two independent flow cell experiments; P = 0.43; Student's t test).
When we used a rotating-disk reactor, the parent biofilms contained 10-fold more CFU per chip than did the agrD mutant (Fig. 1B). Microscopy showed few, if any, S. aureus cells remaining on the chips after homogenization. Furthermore, cells released from the both the parent and agrD mutant biofilms by homogenization were either individuals or pairs but not clusters.
We detected no significant differences in the structures or thicknesses of the biofilms formed in flow cells by the agrD deletion mutant and its parent over a period of 5 days (Fig. 1C). The biofilms were viewed once a day using both phase-contrast microscopy and CSLM, and representative CSLM images are shown in Fig. 1C. For both strains, biofilms covered the majority of the glass surface within 48 h, and large clusters of cells that protruded from the glass surface (microcolonies) developed and persisted for the remainder of the experiment. The images shown are from an experiment in which we used 2% LB broth plus 0.1% glucose as the growth medium. In flow cell experiments with 5% TSB, biofilms developed more rapidly, but we still could not detect any differences between the parent and agrD mutant biofilms. Taken together, the results from the three different systems indicate that the contribution of the agr system to biofilm development is dependent upon the biofilm growth conditions.
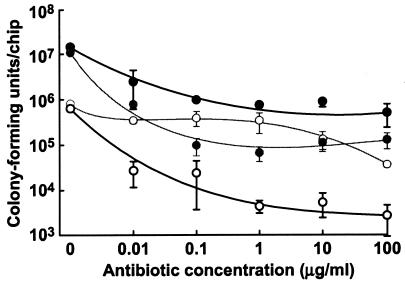
We exposed biofilms of wild-type S. aureus and the agrD mutant grown in the rotating-disk reactor to two clinically relevant antibiotics, rifampin and oxacillin. Biofilms of the parent strain, MN8, showed appreciable resistance to both antibiotics (Fig. 2). Even at the highest concentrations tested, 100 μg/ml (50 to 100 times the MIC for planktonic cells), the level of killing was <2 log units. The sensitivity of the agrD mutant to oxacillin was similar to that of the parent, but the agrD mutant was much more sensitive to rifampin than was the parent. In planktonic cultures, the MICs of rifampin and oxacillin were 0.016 and 0.16 μg/ml, respectively, for both the parent and the mutant strains. These are consistent with the published MICs of rifampin and oxacillin against sensitive S. aureus (28). Thus, at least under some conditions, agr expression is involved in biofilm resistance to certain antibiotics. This may have some clinical relevance, as rifampin is often used together with other antibiotics in cases of biofilm-associated staphylococcal infections.
FIG. 2.
Quorum sensing and biofilm resistance to antibiotics. Spinning-disk reactor biofilms of the S. aureus parent strain, MN8 (solid circles), or the agrD mutant AMD283 (open circles) were treated with rifampin (bold lines) or oxacillin (thin lines), and survival was determined by plate counts. The data are averages from two chips from each of two spinning-disk bioreactors for each strain ± the range of means.
Expression of agr P3-gfp reporters in S. aureus biofilms.
Though agr did not appear to have a significant effect on flow cell biofilm structure, we wondered whether it was expressed in this system and might still have an impact on staphylococcal biofilm biology. To visualize patterns of quorum-sensing-dependent gene expression in S. aureus flow cell biofilms, we constructed two reporter plasmids and introduced them into S. aureus MN8. These contained either the minimum P3 promoter region required for agr-dependent expression of RNAIII (32) fused to a gfp variant (pDB22) or a fragment extending farther upstream of the P3 promoter fused to a second gfp variant (pJY202). In broth culture, S. aureus MN8 containing pDB22 or pJY202 exhibited increasing fluorescence beginning in late exponential growth and continuing into stationary phase, as expected of an agr-dependent reporter. Most cells in stationary-phase cultures exhibited fluorescence. Furthermore, the agr P3-gfp plasmids were maintained in viable cells during biofilm growth in flow cells, as indicated by plating in the presence or absence of erythromycin. Thus, these constructs seemed well suited for studies of agr-dependent gene expression in biofilm cells.
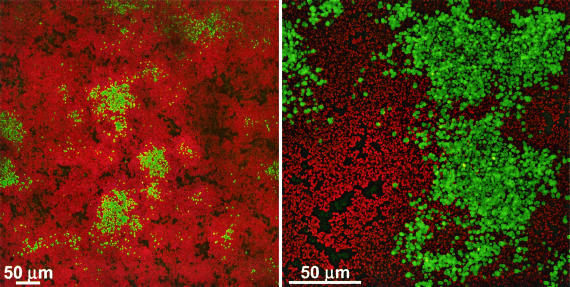
We followed expression of the agr P3-gfp reporters in flow cell biofilms by CSLM. GFP was expressed in some, but not all, biofilm cells, as exemplified by the images of a 24-h biofilm of S. aureus MN8 (pDB22) (Fig. 3). Clusters of cells could be seen on the glass surface, some of which contained patches of cells expressing the reporter. Not all cells in a cluster expressed gfp, and some clusters contained few, if any, cells expressing gfp.
FIG. 3.
Expression of the agr P3-gfp reporter in flow cell biofilms of S. aureus MN8(pDB22). The images were acquired by CSLM with either a 20× (left) or 60× (right) objective lens. The images represent a compressed z series, where multiple x-y planes from top to bottom of the biofilm are combined. Green indicates those cells expressing GFP; all other cells appear red from treatment with SYTO62.
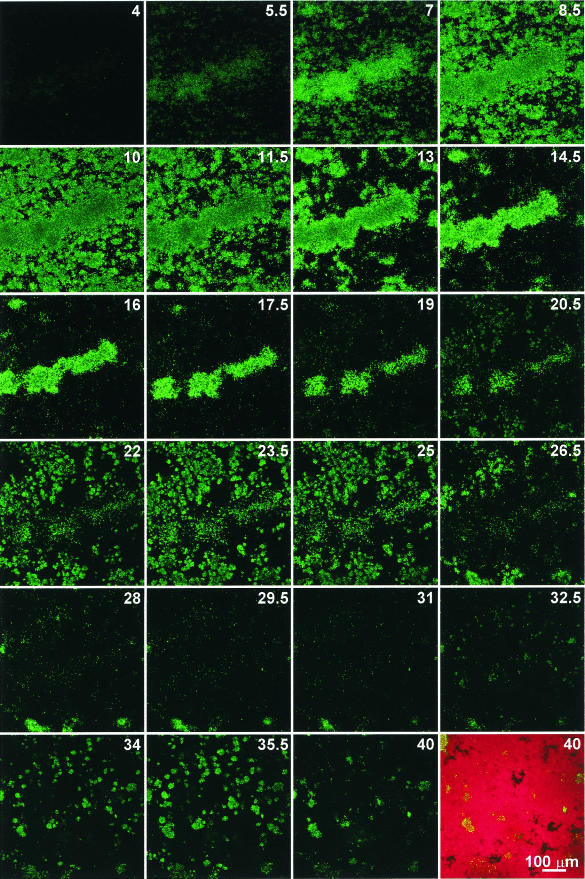
To gain insights into the complicated pattern of agr P3-gfp expression in biofilms, we followed the fluorescence of S. aureus MN8(pDB22) by using time lapse confocal microscopy. Images were acquired every 15 min over a 40-h period, and Fig. 4 shows images at 90-min intervals (the complete time lapse series can be viewed as a movie at http://www.medicine.uiowa.edu/greenberglab/Staphylococcus_aureus.htm). Inoculum cells initially exhibited fluorescence, but this fluorescence was lost by 3 h postinoculation. Fluorescent cells then began to appear 4 h after inoculation of the flow cell. There was an oscillation in gfp expression which appeared as three waves, periods in which local fluorescence increased and decreased. The first wave of fluorescence lasted ∼16 h (4 to 20 h postinoculation), with peak fluorescence at 10 h. The second wave included 20 to 31 h postinoculation, with peak fluorescence at 23 to 24 h. The third wave of fluorescence spanned the period from 32 to 40 h postinoculation, with peak fluorescence at ∼35 h. Within the span of a wave, the fluorescence peaked at different times in different areas of the region under observation. Finally, the fluorescence was progressively less intense with each wave. Despite the loss of fluorescence, most of the region under observation was covered by the biofilm (average biofilm depth, ∼30 μm), as shown by staining with SYTO62 (Fig. 4, final image). Other flow cell biofilms of S. aureus MN8(pDB22) incubated for longer periods showed only sparse fluorescence after 3 days.
FIG. 4.
Time lapse expression of the agr P3-gfp reporter in a flow cell biofilm of S. aureus MN8(pDB22). The images were acquired at 15-min intervals by using identical CSLM settings with a 20× objective lens. Sequential images taken every 90 min are shown, with the number of hours postinoculation indicated. The final image (40 h postinoculation) is shown before and after treatment with SYTO62. The images represent a compressed z series, where multiple x-y planes from top to bottom of the biofilm are combined.
During the time lapse experiment, fluorescence declined rapidly after reaching its peak within a given area of the biofilm. In some cases, brightly fluorescent cells could be observed in one image and not in the image acquired 15 min later. This loss of fluorescence was sometimes followed by the appearance of fluorescent cells in the same area of the biofilm, as exemplified by the central microcolony in Fig. 4 (7 to 17.5 h). These observations suggest that the loss of fluorescence might result from the detachment of cells from the biofilm rather than degradation of GFP, which is normally a stable protein in eubacteria (1, 47).
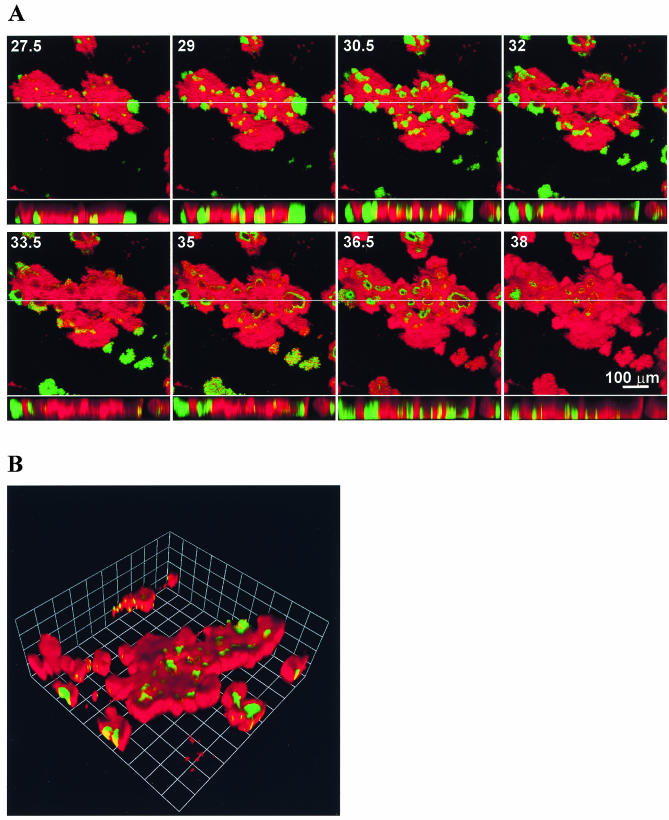
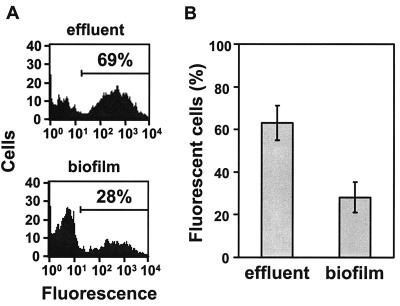
To examine the disappearance of fluorescent cells more closely, we grew a biofilm of S. aureus MN8(pJY202) in the presence of propidium iodide so that the biofilm structure and reporter expression could be imaged simultaneously (Fig. 5 and http://www.medicine.uiowa.edu/greenberglab/Staphylococcus_aureus.htm). There was a rapid loss of GFP fluorescence, usually through the entire depth of the biofilm, beginning from the centers of fluorescent clusters and progressing toward the outsides of these clusters (Fig. 5A). The loss of GFP fluorescence appeared to leave large unstained areas within the biofilm. Subsequently, fluorescence returned from the outer edges of these unstained areas toward the center. The unstained areas eventually became filled with cells expressing GFP, as well as stained biofilm matrix. This suggests that the original fluorescent cells had detached from the biofilm, leaving a void behind which was subsequently occupied as a result of new growth. We also observed that some cells along the peripheries of the microcolonies were originally green fluorescent, subsequently lost green fluorescence, and then appeared to stain with propidium iodide, suggesting that the cells were no longer viable. This disappearance and reappearance of GFP fluorescence appeared to occur simultaneously across the entire span of the nearly 500-μm-diameter microcolony in Fig. 5. A three-dimensional reconstruction of the biofilm at a single time point (Fig. 5B) shows fluorescence occurring in valleys within the biofilm, consistent with the loss of cells previously occupying those areas. This active detachment of cells expressing GFP from the biofilm should result in effluents from flow cells that contain a higher percentage of green fluorescent cells than the flow cell biofilm itself. We confirmed that this was the case using flow cytometry (Fig. 6).
FIG. 5.
Time lapse expression of the agr P3-gfp reporter in a flow cell biofilm of S. aureus MN8(pJY202). The images were acquired at 15-min intervals by using identical CSLM settings with a 20× objective lens. Green indicates those cells expressing the reporter, and the remainder of the biofilm appears red from treatment with propidium iodide. (A) Sequential images taken every 90 min are shown, with the number of hours postinoculation indicated. The larger images represent a compressed z series, where multiple x-y planes from top to bottom of the biofilm are combined. The smaller images represent x-z (side) views of the biofilm at the location indicated by the line. (B) Three-dimensional reconstruction of a z series taken ∼36.5 h after inoculation in the experiment shown in panel A. Each side of a grid square represents 60 μm.
FIG. 6.
Expression of the agr P3-gfp reporter in flow cell biofilms and effluents after 60 h. Effluents were collected for ∼30 min from flow cells. Biofilms were then expelled from the same flow cells described in Materials and Methods. All collected cells were subsequently dispersed by ultrasonication and analyzed by flow cytometry for fluorescence (FL-1 channel). (A) Flow cytometry results from one representative flow cell biofilm and its effluent. The percentages represent the cells that fell within the range of fluorescence delineated by the marker. (B) Percentages of cells from quadruplicate flow cell biofilms and their effluents that were green fluorescent ± the standard deviation. The data are significant (P < 0.001; Student t test).
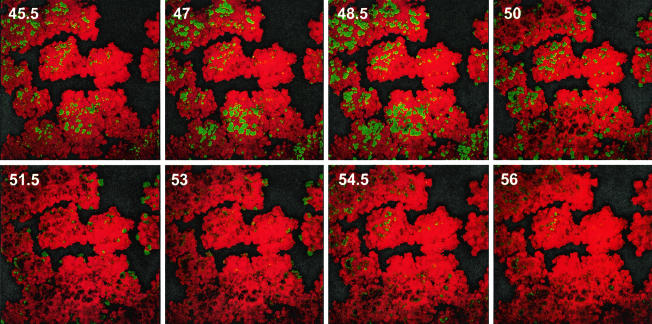
To determine whether the pattern of expression observed was unique to agr P3-gfp reporters, we also examined the expression over time of a sar P1-gfp reporter fusion. SarA has been shown to be necessary for biofilm formation (2, 48). sar P1-gfp reporters were expressed throughout growth in vitro (mid-log, late log, and stationary phases), were expressed in vivo throughout an endocarditis vegetation (8, 29), and have been used as de facto constitutive reporters to visualize staphylococcal biofilms (25). Similarly, expression of our reporter was detected throughout the growth of the organism in vitro, and its expression was not altered by mutation of agrD. Thus, it seemed suitable for use a reporter to which we could compare agr P3-gfp expression. Fluorescence from expression of the sar P1-gfp reporter was initially detected in all visible cells in the biofilm, even within very small microcolonies, suggesting that the sar P1 reporter was expressed constitutively. We observed disappearance and reappearance of fluorescence similar to that observed using the agr P3 reporter (Fig. 7 and www.medicine.uiowa.edu/greenberglab/Staphylococcus_aureus.htm). This was also demonstrated using a sar P1-gfp reporter kindly provided by A. Cheung (data not shown). This suggested that the expression patterns observed were not unique to agr P3-controlled reporters.
FIG. 7.
Time lapse expression of the sar P1-gfp reporter in a flow cell biofilm of S. aureus MN8(pJY209). The images were acquired at 15-min intervals by using identical CSLM settings with a 20× objective lens. Green indicates those cells expressing the reporter, and the remainder of the biofilm appears red from treatment with propidium iodide. Sequential images taken every 90 min are shown, with the number of hours postinoculation indicated.
DISCUSSION
We have addressed two basic questions about quorum sensing in S. aureus biofilms: what is the influence of the agr quorum-sensing system on S. aureus biofilm development and antibiotic resistance and what is the pattern of agr-dependent gene expression in an S. aureus biofilm? With regard to the first question, we used isogenic strains to show that the influence of the agr system on biofilm development depends on the conditions of the experiment. Under some conditions, agr expression appears to enhance biofilm development; under other conditions, it has no discernible affect or seems to impair biofilm development (Fig. 1). Our experiments help unify what may have been considered disparate conclusions in the literature, and they extend our understanding by showing that the different results reported elsewhere do not necessarily result from strain differences.
We show that under at least one experimental condition agr expression can influence a clinically relevant aspect of biofilm biology, antibiotic resistance (Fig. 2). Spinning-disk reactor biofilm cells of our agrD mutant were particularly sensitive to rifampin but not to oxacillin. We do not understand the basis of the enhanced biofilm sensitivity to rifampin in the quorum-sensing mutant. This is not a general effect of agr expression on rifampin resistance, because nonbiofilm cells of the mutant did not appear to be more sensitive to rifampin than the parent. The antibiotic resistance experiment was done under conditions where the agr mutant biofilm contained fewer cells than the parent biofilm. Thus, there may be a rifampin-specific biofilm inoculum effect. Regardless, it is clear that agr expression can have important and specific effects on cells in S. aureus biofilms. Because rifampin is often used in the treatment of S. aureus biofilm infections, further investigation of the influence of agr expression on biofilm sensitivity to antibiotics is warranted.
Previous reports have implicated acyl-HSL quorum sensing in the biofilm development of several species of proteobacteria under certain conditions (11, 22, 23, 27, 31, 46). For example, P. aeruginosa biofilm development into mushroom-shaped structures has been reported to be dependent on acyl-HSL signaling (11). Under conditions where biofilms do not develop mushroom-like structures, acyl-HSLs do not appear to influence biofilm structure (21). A similar situation may occur with agr-dependent quorum sensing in S. aureus. Under certain conditions, quorum sensing can play a role in biofilm development, but it may not have an obvious role in biofilm development under all conditions. As the agr system is thought to exert regulatory control over a wide range of staphylococcal genes (13), it may well be that different genes or sets of genes regulated by the agr system influence S. aureus attachment and biofilm development under some growth conditions but not others. In some cases, agr expression likely affects the attachment of inoculum cells to a surface through its regulation of cell surface-associated adhesins. This could be the case in our static and spinning-disk reactor biofilms, as stationary-phase cells were diluted into fresh medium to inoculate these systems. In other cases, agr expression may also affect biofilm maturation, perhaps through its regulation of extracellular factors, such as δ-hemolysin (51). Due to the limitations of the static and spinning-disk reactor systems, we could not evaluate agr P3 reporter expression in situ using confocal microscopy to assess its effects on biofilm maturation.
In flow cell biofilms, we did not detect an influence of agr expression on biofilm development (Fig. 1C). Thus, one obvious question was whether agr-dependent promoters were even expressed in this type of system. We found that plasmid-borne quorum-controlled agr P3-gfp reporters were expressed in these biofilms (Fig. 3 to 5), as was a sar P1-gfp reporter (Fig. 7). Though fluorescence of cells containing the sar P1 reporter was observed earlier in microcolony growth than fluorescence of cells containing agr P3 reporters, both reporter types eventually showed similar oscillating patterns of expression. In any given area, we observed cycles with peaks in fluorescence followed by rapid losses of fluorescence. Furthermore, the oscillations in areas of close proximity often appeared to occur in synchrony, either simultaneously or sequentially (Fig. 4, 5, and 7). At any given time, there appeared to be clusters of cells expressing the fluorescent reporter surrounded by nonfluorescent cells (Fig. 3, 5, and 7).
How might the complex patterns of fluorescent reporter expression in the flow cell biofilms be explained? There are at least three possible explanations for loss of GFP fluorescence. First, the GFP might be degraded, which seems unlikely considering that we have employed two versions of a GFP molecule with half-lives greater than 24 h (1, 47) with similar results. Second, cells may lyse or lose GFP through membrane disruption. We have observed a portion of the cell population, particularly in the peripheries of microcolonies, that appears to lose green fluorescence and then subsequently stains with propidium iodide (data not shown), suggesting loss of membrane integrity. These cells appear to remain part of the biofilm, together with the extracellular matrix. Third, loss of fluorescence might result from cell detachment from the biofilm. Often, cells expressing the agr P3-gfp reporter disappeared from the biofilm, leaving behind a void in propidium iodide-stained biofilms. This was followed by what appeared to be new growth of fluorescent cells in the void (Fig. 4, hours 13 to 17.5; Fig. 5A, hours 35 to 38). The fact that most of the biofilm appeared to remain intact and apparent voids were filled by new growth was consistent with our inability to detect any significant structural differences between biofilms formed by the wild type and the agrD mutant (Fig. 1C). Further evidence for fluorescent-cell detachment is our finding that the percentage of fluorescent cells in effluents from flow cells is significantly higher than the percentage of fluorescent cells within the flow cell biofilm (Fig. 6).
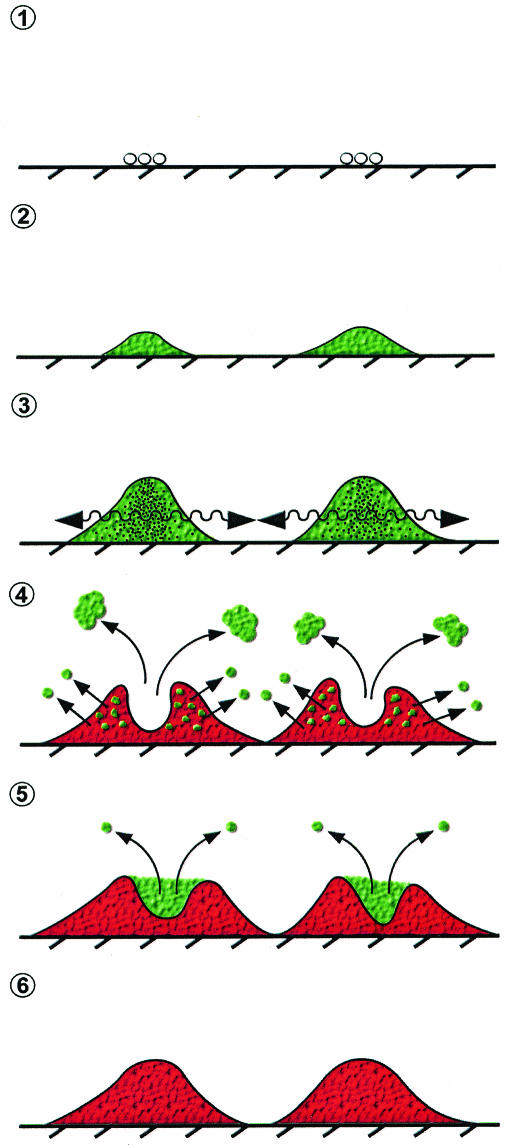
Our observations regarding oscillation of GFP fluorescence in flow cell biofilms, coupled with the fact that the oscillations in fluorescence were not unique to the expression of an agr reporter, suggest a broader model of gene expression in the biofilm that is closely related to both cell viability and detachment (Fig. 8). Only actively growing areas of the biofilm express the fluorescent reporters, and where conditions are appropriate within a sufficiently large cluster of actively growing cells, expression of quorum-controlled genes will be activated. Once cell clusters reach a sufficient size or age, most cells either detach or die. Additional growth then occurs in the voids left by detached cells. Thus, it is the cycle of cell growth, detachment, and regrowth that underlies the patterns of gene expression observed. Expression of surface-associated adhesins and agr-induced extracellular factors (51), localized shear forces, cell viability, and growth patterns may all contribute to the detachment of cells from the biofilm. Further investigation will be required to determine how these complex phenomena interact and contribute to the patterns of gene expression that we have observed.
FIG. 8.
Model of agr expression in S. aureus biofilms. After initial colonization by individual cells (step 1), microcolonies reach sufficient cell density for agr-dependent gene expression (step 2) and perhaps signaling between microcolonies (step 3, arrows). Portions of the biofilm detach through as-yet-unknown mechanisms (step 4), as either large aggregates or individual cells. Simultaneously, parts of the biofilm population become metabolically inactive and lose membrane integrity and green fluorescence. This is followed by new growth into the voids left by the detached cells (step 5), and the cycle is repeated. The microcolony finally reaches a relatively quiescent state where any growth is slow and agr expression is undetectable (step 6).
A picture that emerges from the time course studies is that in most areas of a biofilm at most times, agr, and likely agr-dependent virulence genes, is not expressed. However, cells that do express agr appear to be released from the biofilm, cells that may also be expressing agr-dependent virulence factors. Our biofilm model is reminiscent of one previously proposed for agr expression by staphylococci in abscess infections (39), where crowding within the localized infection activates the agr-dependent quorum response and synthesis of extracellular factors, enabling the staphylococci to escape the abscess and spread to new sites. If this aspect of the biofilm model, that there is dispersion of virulent staphylococci, holds true, it will have important implications for the etiology of S. aureus biofilm infections.
Acknowledgments
This research was supported by a gift from the Procter and Gamble Company, Cincinnati, Ohio, and a grant from the W. M. Keck Foundation. J.M.Y. was supported by a U.S. Public Health Training Grant (T32-AI07511).
We acknowledge Patrick Schlievert for his provision of strains and general encouragement; Kimberly Lee, Pradeep Singh, Thomas Moninger, and the Central Microscopy Research Facility for materials and assistance with confocal microscopy; and Shirley Taylor and the Creative Media Group for figure illustrations. We thank Debra Murray, Timothy Yahr, and Catherine Davis for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blevins, J. S., M. O. Elasri, S. D. Allmendinger, K. E. Beenken, R. A. Skinner, J. R. Thomas, and M. S. Smeltzer. 2003. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 71:516-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caiazza, N. C., and G. A. O'Toole. 2003. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J. Bacteriol. 185:3214-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., K. J. Eberhardt, E. Chung, M. R. Yeaman, P. M. Sullam, M. Ramos, and A. S. Bayer. 1994. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 94:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., C. C. Nast, and A. S. Bayer. 1998. Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infect. Immun. 66:5988-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 11.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 12.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed, R. C., M. L. Evenson, R. F. Reiser, and M. S. Bergdoll. 1982. Enzyme-linked immunosorbent assay for detection of staphylococcal enterotoxins in foods. Appl. Environ. Microbiol. 44:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillaspy, A., S. Hickmon, R. Skinner, J. Thomas, C. Nelson, and M. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 18.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 19.Gryczan, T. J., G. Grandi, J. Hahn, R. Grandi, and D. Dubnau. 1980. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 8:6081-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 23.Huber, B., K. Riedel, M. Kothe, M. Givskov, S. Molin, and L. Eberl. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46:411-426. [DOI] [PubMed] [Google Scholar]
- 24.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 25.Leid, J. G., M. E. Shirtliff, J. W. Costerton, and A. P. Stoodley. 2002. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect. Immun. 70:6339-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 28.Mandell, G. L., J. E. Bennett, and R. Dolin (ed.). 1995. Mandell, Douglas and Bennett's Principles and practice of infectious diseases, 4th ed. Churchill Livingstone, New York, N.Y.
- 29.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayberry-Carson, K. J., B. Tober-Meyer, J. K. Smith, D. W. Lambe, Jr., and J. W. Costerton. 1984. Bacterial adherence and glycocalyx formation in osteomyelitis experimentally induced with Staphylococcus aureus. Infect. Immun. 43:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 33.Murray, D. L., C. A. Earhart, D. T. Mitchell, D. H. Ohlendorf, R. P. Novick, and P. M. Schlievert. 1996. Localization of biologically important regions on toxic shock syndrome toxin 1. Infect. Immun. 64:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.NCCLS. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A5, 5th ed., vol. 20. NCCLS, Wayne, Pa.
- 35.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 36.Oie, S., Y. Huang, A. Kamiya, H. Konishi, and T. Nakazawa. 1996. Efficacy of disinfectants against biofilm cells of methicillin-resistant Staphylococcus aureus. Microbios 85:223-230. [PubMed] [Google Scholar]
- 37.Parsek, M. R., and E. P. Greenberg. 1999. Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Methods Enzymol. 310:43-55. [DOI] [PubMed] [Google Scholar]
- 38.Pratten, J., S. J. Foster, P. F. Chan, M. Wilson, and S. P. Nair. 2001. Staphylococcus aureus accessory regulators: expression within biofilms and effect on adhesion. Microbes Infect. 3:633-637. [DOI] [PubMed] [Google Scholar]
- 39.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone Inc., New York, N.Y.
- 40.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 41.Schlievert, P. M., and D. A. Blomster. 1983. Production of staphylococcal pyrogenic exotoxin type C: influence of physical and chemical factors. J. Infect. Dis. 147:236-242. [DOI] [PubMed] [Google Scholar]
- 42.Shenkman, B., D. Varon, I. Tamarin, R. Dardik, M. Peisachov, N. Savion, and E. Rubinstein. 2002. Role of agr (RNAIII) in Staphylococcus aureus adherence to fibrinogen, fibronectin, platelets and endothelial cells under static and flow conditions. J. Med. Microbiol. 51:747-754. [DOI] [PubMed] [Google Scholar]
- 43.Shiau, A. L., and C. L. Wu. 1998. The inhibitory effect of Staphylococcus epidermidis slime on the phagocytosis of murine peritoneal macrophages is interferon-independent. Microbiol. Immunol. 42:33-40. [DOI] [PubMed] [Google Scholar]
- 44.Shirtliff, M. E., J. T. Mader, and A. K. Camper. 2002. Molecular interactions in biofilms. Chem. Biol. 9:859-871. [DOI] [PubMed] [Google Scholar]
- 45.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 46.Steidle, A., M. Allesen-Holm, K. Riedel, G. Berg, M. Givskov, S. Molin, and L. Eberl. 2002. Identification and characterization of an N-acylhomoserine lactone-dependent quorum-sensing system in Pseudomonas putida strain IsoF. Appl. Environ. Microbiol. 68:6371-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tombolini, R., A. Unge, M. E. Davey, F. J. D. Bruijn, and J. K. Jansson. 1997. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol. Ecol. 22:17-28. [Google Scholar]
- 48.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 49.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Wamel, W., Y. Q. Xiong, A. Bayer, M. Yeaman, C. Nast, and A. Cheung. 2002. Regulation of Staphylococcus aureus type 5 capsular polysaccharides by agr and sarA in vitro and in an experimental endocarditis model. Microb. Pathog. 33:73-79. [DOI] [PubMed] [Google Scholar]
- 51.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 52.Weisblum, B., M. Y. Graham, T. Gryczan, and D. Dubnau. 1979. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J. Bacteriol. 137:635-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong, Y. Q., W. Van Wamel, C. C. Nast, M. R. Yeaman, A. L. Cheung, and A. S. Bayer. 2002. Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J. Infect. Dis. 186:668-677. [DOI] [PubMed] [Google Scholar]
- 54.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yarwood, J. M., and P. M. Schlievert. 2000. Oxygen and carbon dioxide regulation of toxic shock syndrome toxin 1 production by Staphylococcus aureus MN8. J. Clin. Microbiol. 38:1797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]