Abstract
The ula regulon, responsible for the utilization of l-ascorbate in Escherichia coli, is formed by two divergently transcribed operons, ulaG and ulaABCDEF. The regulon is negatively regulated by a repressor of the DeoR family which is encoded by the constitutive gene ulaR located downstream of ulaG. Full repression of the ula regulon requires simultaneous interaction of the repressor with both divergent promoters and seems to be dependent on repressor-mediated DNA loop formation, which is helped by the action of integration host factor. Two operator sites have been identified in each promoter. Lack of either of the two sets of operators partially relieved the repression of the other operon; thus, each promoter is dependent on the UlaR operator sites of the other promoter to enhance repression. Electrophoretic mobility shift assays with purified UlaR protein and promoter deletion analyses revealed a conserved sequence, present in each of the four operators, acting as a UlaR binding site. Glucose represses the ula regulon via at least two mechanisms, one dependent on cyclic AMP (cAMP)-cAMP receptor protein (CRP) and the other (possibly inducer exclusion) independent of it. Glucose effects mediated by other global regulators cannot be ruled out with the present information. Changes in cAMP-CRP levels affected only the expression of the ulaABCDEF operon.
l-Ascorbate is utilized by Escherichia coli K-12 under anaerobic conditions through proteins encoded by the ula regulon, previously designated yjf-sga (23, 31), located at centisome 95.3 (4). This system is formed by two divergently transcribed operons (6), the ulaG operon, proposed to encode the l-ascorbate-6-P lactonase (35), and the ulaABCDEF operon, encoding the three components of the l-ascorbate phosphotransferase transport system (UlaABC) (35) and three catabolic enzymes (UlaDEF) (31). The three gene products UlaABC (formerly known as SgaTBA) are involved in the uptake and phosphorylation of l-ascorbate (35). Intracellular l-ascorbate-6-P could be transformed by l-ascorbate-6-P lactonase into 3-keto-l-gulonate-6-P. It has been proposed that this compound is decarboxylated by UlaD to l-xylulose-5-P, which is then converted to d-xylulose-5-P by the sequential actions of UlaE (encoding 3-epimerase activity) and UlaF (encoding 4-epimerase activity) (31). Thus, the function of the gene products of the ula system is the transport of l-ascorbate and its transformation into d-xylulose-5-P (31), which is subsequently metabolized by the pentose phosphate pathway.
The ula regulon is induced by growth on l-ascorbate and is under the control of the UlaR repressor encoded by the constitutively expressed gene ulaR (6). Inactivation of this gene yields the constitutive expression of promoter fusions of the ula operons not only under anaerobic conditions but also in the presence of oxygen (6). The ula regulon also seems to be controlled by the global regulators cyclic AMP (cAMP)-cAMP receptor protein (CRP) and fumarate nitrate reductase regulator (FNR), since mutations in the corresponding genes abolished or delayed, respectively, growth on l-ascorbate (35).
The UlaR repressor belongs to the DeoR family of bacterial regulatory proteins (6). Members of this protein family are present in a variety of bacteria, like Agrobacterium tumefaciens, Bacillus subtilis, Haemophilus influenzae, Pseudomonas aeruginosa, and Lactococcus. Typically, these regulators bind to several operator sites in the promoter regions of the genes controlled. Although these proteins contain highly conserved regions, such as the second helix of the helix-turn-helix motif or a short sequence located 16 or 17 residues upstream of this motif (3), the operator arrangements of some of them are fundamentally different. In the deo system of E. coli, the repressor binds to multiple widely separated operators with a dyad symmetry sequence (21), whereas in the glp system of E. coli the repressor binds with high affinity to adjacent, tandemly arranged operators (16, 30, 33). In the case of SpoIIID of B. subtilis, the protein binds to several sites that exhibit dyad symmetry, a direct repeat, or an overlapping repeat (34). Furthermore, it has been reported that the cooperative binding of DeoR to several separate operator sites generates a loop in the DNA that is necessary for full repression. The cooperative binding presumably generates a high degree of repression, even though the intrinsic affinity of the repressor for each site is relatively low (1).
Here, we present evidence that full repression of the ula regulon requires binding of UlaR to four operator sites and the participation of integration host factor (IHF). We also demonstrate the role of CRP in the direct activation of the ulaABCDEF operon and show that ula regulon expression was not affected in an FNR mutant.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
The genotypes and sources of the bacterial strains are given in Table 1. Genetic crosses were performed by P1-mediated transduction (20).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| XL1Blue | recA1 lac endA1 gyrA96 thi hsdR17 supE44 relA1 (F′ proAB lacIq lacZΔM15 Tn10) | Stratagene |
| TE2680 | F− λ− IN(rrnD-rrnE) ΔlacX74 rplS galK2 recD::Tn10d-tet trpDC700::putA13033::(Kanr Cmrlac) | 10 |
| JMH0317 | W3110 rpoS::Tn10 | J. Membrillo-Hernandez |
| JA134 | ECL1 Lyx+ | 25 |
| JA182 | JA134 Δ(tyrR-fnr-nac-trg)17 zdd-230::Tn9 | 14 |
| JA184 | JA134 himA::cat | 14 |
| JA185 | JA134 himD3::cat | 14 |
| JA188 | JA134 rpoS::Tn10 | This work |
| BL21(DE3) | E. coli B F−ompT hsdS (rB− mB−) gal dcm λ(DE3) | Pharmacia Biotech |
| JA211 | JA134 yiaR::catΔulaR | 6 |
| JA216 | JA211 crp::cat | This work |
| Plasmids | ||
| pRS550/pRS551 | Apr Kmr; contains promoterless lacZYA | 28 |
| pET43c (+) | Nus tag bla (Carbr) lacI | Novagen |
| pET-UlaR | ulaR in pET43c (+) | This work |
| pBS-CRP | crp in pBluescript | This work |
Growth conditions and preparation of cell extracts.
Cells were grown on Luria broth (LB) or minimal medium and harvested as described previously (5). For anaerobic growth, l-ascorbate or glucose was added to a basal inorganic medium (5) at 20 mM. Casein acid hydrolysate (CAA) was routinely used at 0.5% for aerobic growth and 1% for anaerobic growth. To increase the cell yield under anaerobic conditions (31), nitrate was added to the cultures at 20 mM. When required, the following antibiotics were used at the concentrations indicated: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 50 μg/ml; carbenicillin, 50 μg/ml; and tetracycline, 12.5 μg/ml. For growth of strains carrying transcriptional fusions, tryptophan was added at 0.1 mM. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) and IPTG (isopropyl-β-d-thiogalactoside) were used at 30 and 10 μg/ml, respectively.
Cell extracts were prepared as described previously (5). The protein concentration was determined by the method of Lowry et al. (18) with bovine serum albumin as a standard.
Enzyme activities.
β-Galactosidase activity was assayed by hydrolysis of ο-nitrophenyl-β-d-galactopyranoside and expressed in Miller units (20). For this purpose, cell cultures were grown until late exponential phase unless otherwise specified. The data reported are a representative set from at least four separate experiments performed in duplicate.
DNA manipulation and site-directed mutagenesis.
Bacterial genomic DNA was obtained as described by Silhavy et al. (27). Plasmid DNA was routinely prepared by the boiling method (13). For large-scale preparation, a crude DNA sample was purified on a column (Qiagen). DNA manipulations were performed essentially as described by Sambrook and Russell (24). DNA was sequenced using an automated ABI 377 DNA sequencer and fluorescent dye termination methods. DNA fragments were amplified by PCR using E. coli chromosomal DNA as a template. When necessary, specific restriction sites were incorporated at the 5′ ends of the primers to facilitate cloning of the fragments in the appropriate vector. PCRs were performed with Pfu DNA polymerase under standard conditions. All primers used will be provided upon request.
Site-directed mutagenesis of UlaR and cAMP-CRP binding sites in probes used in the mobility shift assays was performed by PCR using primers containing the desired mutations. Site-directed mutagenesis of the cAMP-CRP binding site and insertion of 5 or 10 bp in the fragment containing the full-length ulaG and ulaA promoter region were done by crossover PCR (17).
Isolation of RNA and primer extension.
Total RNA was obtained with a Qiagen RNeasy Total RNA kit from cultures of strain JA211 grown aerobically on CAA. For determination of the 5′ ends of the ulaG and ulaA genes, primer ULA-A, complementary to an internal region of ulaG, and primer ULA-A7, complementary to an internal region of ulaA, were used. The reactions were performed with 50 μg of total RNA at 37°C for 30 min with 200 U of Moloney murine leukemia virus reverse transcriptase (Life Technologies, Inc.) and [thio-α-35S]dATP (>1,000 Ci/mmol; Amersham Pharmacia Biotech) and were followed by a 30-min chase with all four nucleotides (at 1 mM each). As a reference, double-strand sequence reactions were performed with the same primers using the dideoxy chain termination procedure of Sanger et al. (26).
Construction of lacZ fusions and deletions of the ulaG and ulaA promoters.
To create operon fusions, DNA fragments of the 5′ upstream regions of the ulaG and ulaA genes were cloned into plasmids PRS551 and pRS550 (28), respectively. The different 5′ deletion-containing fragments were obtained by PCR. The pRS plasmids carried a cryptic lac operon and genes that confer resistance to both kanamycin and ampicillin. Recombinant plasmids were selected after transformation of strain XL1-Blue as blue colonies on LB plates containing X-Gal, ampicillin, and kanamycin and were sequenced using the M13 primer to ensure that no mutation was introduced. Single-copy fusions on the E. coli chromosome were obtained by the method of Elliot (10) using strain TE2680. The transformants were selected for kanamycin resistance and screened for sensitivity to ampicillin and chloramphenicol. P1 vir lysates were made to transduce the fusions into the desired genetic background.
Expression and purification of UlaR.
UlaR was purified using the NusA gene fusion system, which yields a construct with a His6 tag at the carboxy-terminal end of the NusA protein and the recognition site for thrombin cleavage upstream of the amino-terminal end of the UlaR. For this purpose, the ulaR gene was amplified by PCR with primers ULA-R1 and ULA-R2 and cloned into the BamHI and EcoRI restriction sites of plasmid pET-43c (+), yielding plasmid pET-UlaR. Primer ULA-R1 was designed to fuse the ATG start codon of ulaR in frame with the gene encoding NusA. Overproduction of UlaR was achieved in strain BL21(DE3) carrying the recombinant plasmid pET-UlaR after induction by IPTG (1 mM) in LB-carbenicillin medium for 3 h at 37°C, and the NusA fusion protein was purified by affinity chromatography with Ni2+-nitrilotriacetic acid resin (Qiagen). For UlaR purification, the cell pellet from a 10-ml culture of strain BL21(DE3) bearing plasmid pET-UlaR was suspended in 0.5 ml of 50 mM sodium phosphate buffer, pH 8.0, containing 0.3 M NaCl and 5 mM 2-mercaptoethanol (buffer A) and was sonicated on ice. The cell lysate was centrifuged at 15,000 × g, and the supernatant was incubated with 0.5 ml of Ni2+-nitrilotriacetic acid resin for 1 h with gentle shaking. After the mixture was loaded onto a column, the resin was first washed with 50 mM sodium phosphate buffer, pH 6.0, containing 0.3 M NaCl, 5 mM 2-mercaptoethanol, and 10% glycerol (buffer B). Stepwise elution was performed with buffer B containing 100 and 250 mM imidazole to eliminate contaminant proteins. The bound protein was digested in the column with thrombin (9 U) after incubation at room temperature for 12 h. The cleaved UlaR protein was eluted with buffer A.
DNA binding studies.
For electrophoretic mobility shift assays, several PCR-amplified fragments were used as probes. These fragments were purified from acrylamide gels and labeled with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; Amersham Pharmacia Biotech).
Electrophoretic mobility shift assays were performed with crude extracts obtained as described by Nunoshiba et al. (22) or with purified UlaR protein. Acrylamide gels containing 10% glycerol were run at 4°C by using 1× Tris-borate-EDTA buffer (2). Protein samples were mixed with 32P-end-labeled DNA substrates (final concentration, ca. 2.5 nM; 10,000 to 25,000 cpm) in a 20-μl reaction volume containing 10 mM Tris-HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 10% glycerol, and 2 mM dithiothreitol. Poly(dI-dC) was used as a nonspecific competitor at 500-fold molar excess. Validation of the binding specificity was performed with unlabeled DNA. After incubation for 15 min at 30°C, a 1/6 volume of 6× gel loading buffer (2) was added, and the mixtures were loaded directly onto prerun gels.
RESULTS
Mapping the mRNA 5′ ends of ulaG and ulaABCDEF operons.
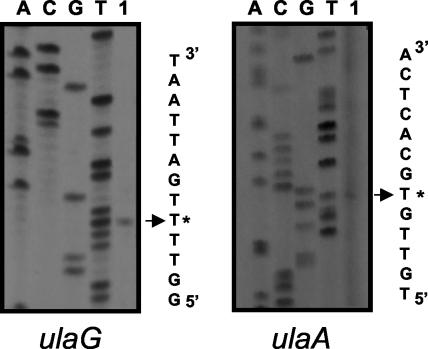
The 5′ ends of the structural genes ulaG and ulaA were determined by primer extension analysis. Total RNA was obtained from aerobic cultures on CAA of strain JA211, which constitutively expressed the ula regulon due to a mutation in the ulaR gene. For the primer extension reactions, we used primers complementary to internal regions of the ulaG and ulaA genes. The 5′ end determined for ulaG (Fig. 1) was located 43 bp upstream of the ATG codon. This putative +1 position is preceded by a promoter-like sequence including a −10 box (CAGGAT) and a −35 box (TTGCCT). This promoter region contained an AT-rich sequence between positions −40 and −60. The 5′ end determined for ulaA (Fig. 1) was located 53 bp upstream of the ATG codon, and the putative +1 was preceded by a promoter-like sequence including a −10 box (TAGAGT) and a −35 box (TTGTTG). This promoter also displayed an AT-rich sequence, in this case between positions −38 and −58. In both cases, the −10 and −35 elements were separated by 17 bp. Using the MacTargsearch program (11), we identified a potential IHF binding site extending between −103 and −150 in the coding strand of ulaG and between −102 and −149 in the coding strand of ulaA. Sequence analysis also showed four putative CRP binding sites, one centered at −110 with respect to the ulaG gene and another centered at −61 with respect to ulaA, and two additional sites overlapping the −10 or −35 promoter sequences of the ulaA gene (not shown).
FIG. 1.
Primer extension analysis of the divergently transcribed ulaG and ulaA genes. The primed-extended products using total RNA of strain JA211 growing aerobically on CAA (lanes 1) were electrophoresed with a sequencing ladder (lanes A, C, G, and T) generated by using the same template and primer. A portion of the nucleotide sequence deduced from the sequencing lanes is shown. The extended product assigned as the 5′ end of each gene is labeled with an asterisk.
Expression from the functional promoters.
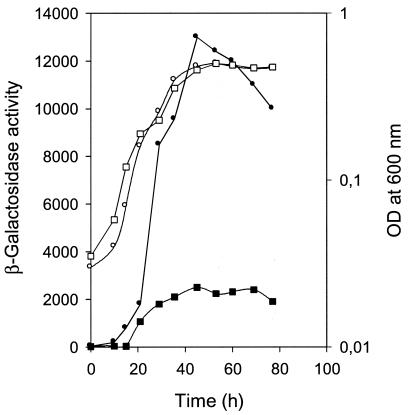
Expression of transcriptional fusions of the two operons encoding the structural genes of the ula regulon was analyzed by assaying β-galactosidase activity in strain JA134 carrying Φ(ulaG-lacZ) or Φ(ulaA-lacZ). For this purpose, we started an anaerobic culture on l-ascorbate with an aliquot of an overnight culture on CAA and monitored the growth and β-galactosidase activity. The expression of Φ(ulaA-lacZ) was around five times higher than that of Φ(ulaG-lacZ) at the end of exponential phase. In both cases, the activity increased through exponential growth, reaching a maximum during transition, and decreased slightly as the culture reached stationary phase (Fig. 2). These results imply that regulation of ula regulon expression in E. coli occurs at the transcriptional level in a growth phase-dependent manner.
FIG. 2.
Expression profiles of of Φ(ulaG-lacZ) and Φ(ulaA-lacZ) transcriptional fusions in strain JA134 growing anaerobically in l-ascorbate. At intervals, the A600 of strain JA134 bearing Φ(ulaG-lacZ) (open squares) or Φ(ulaA-lacZ) (open circles), as well as the β-galactosidase activities of the corresponding cultures (solid squares and circles, respectively), were measured. OD, optical density.
Although during growth most bacterial genes are transcribed by the RNA polymerase complex using the factor σ70, transcription induced under starvation conditions in the stationary phase depends on the factor σs encoded by the rpoS gene. Since the maximum expression of the ula regulon is seen at the transition to stationary phase, we studied the dependence of its expression on σs. We transferred, by P1 transduction, the rpoS mutation from strain JMH0317 to strain JA134, yielding strain JA188. No differences between the levels of expression of ulaG or ulaA fusions in the wild type and the rpoS-deficient strains were observed in any growth phase (data not shown). The maximum activity was also seen at the end of the exponential phase (Table 2). These results suggest that this sigma factor does not mediate the changes in ula regulon expression observed in response to the growth phase.
TABLE 2.
Expression analysis of Φ(ulaG-lacZ) and Φ(ulaA-lacZ) transcriptional fusions in different genetic backgrounds under anaerobic conditions
| Strain | Relevant phenotype | β-Galactosidase expressiona (Miller units)
|
|||
|---|---|---|---|---|---|
| Φ(ulaG-lacZ)
|
Φ(ulaA-lacZ)
|
||||
| CAA | l-Ascorbate | CAA | l-Ascorbate | ||
| JA134 | Wild type | 40 | 2,580 | 60 | 12,000 |
| JA184 | IHF− | 130 | 2,350 | 2,898 | 12,260 |
| JA185 | IHF− | 120 | 2,425 | 2,940 | 12,370 |
| JA188 | RpoS− | 55 | 2,620 | 60 | 12,300 |
| JA211 | YjfQ− | 4,850 | ND | 20,275 | ND |
| JA216 | YjfQ− CRP− | 4,720 | ND | 9,250 | ND |
| JA182 | FNR− | 45 | 2,650 | 60 | 12,375 |
ND, not determined.
Regulation of expression of the ulaG and ulaA divergent promoters by CRP, IHF, and FNR.
We tested the effect of glucose on the induction of the ula regulon by growing strain JA134 carrying Φ(ulaG-lacZ) or Φ(ulaA-lacZ) on l-ascorbate in the absence or presence of glucose. In the presence of glucose, both transcriptional fusions showed negligible β-galactosidase activity, which contrasted with the levels in the absence of glucose (Table 2; also see Fig. 7) and indicated that this sugar produced catabolite repression of the ula regulon. The addition of cAMP to a final concentration of 5 mM did not affect glucose repression (see Fig. 7), indicating that the glucose effect is due not only to the reduction of the cAMP-CRP level, as reported by Zhang et al. (35), but also to other mechanisms, such as inducer exclusion, which is mediated by the phosphoenolpyruvate-dependent sugar phosphotransferase.
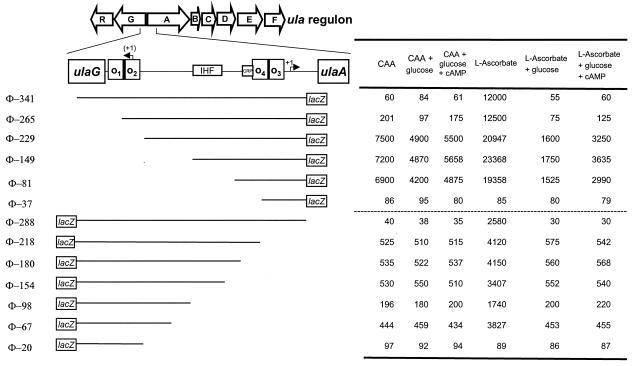
FIG. 7.
Effects of upstream deletions on ulaG and ulaA expression. The open arrows indicate the extents and directions of transcription of the genes included in the ula regulon. The regulatory elements recognized by UlaR (O1, O2, O3, and O4), IHF, and CRP between ulaG and ulaA are represented by boxes. Deleted promoter fragments are represented by lines, and the corresponding lacZ fusions are labeled on the left. The designations of the deletions are based on the number of base pairs present upstream of the 5′ end, marked by an arrow labeled +1. The 5′ end corresponding to the complementary ulaG coding strand is in parentheses. β-Galactosidase activity values under the different growth conditions are indicated in the table on the right.
In order to demonstrate the contribution of cAMP-CRP to the glucose effect on ula regulon transcription, we analyzed the expression of Φ(ulaG-lacZ) and Φ(ulaA-lacZ) in the genetic background of a crp mutant, strain JA216. This strain is a JA211 derivative and constitutively expresses ula genes, owing to a ulaR mutation. Expression of both fusions was analyzed, and only Φ(ulaA-lacZ) was reduced around twofold in the crp mutant JA216 (Table 2). This indicated that ulaG is not affected by CRP and further supports the participation of another mechanism independent of cAMP-CRP in the expression of both operons of the ula regulon.
To determine whether IHF influences the divergent promoter expression in vivo, we transferred the operon fusions to the JA134 derivative strains JA184 and JA185, which carried the himA and himD3 mutations, respectively. The β-galactosidase activities showed an increase in the basal level of Φ(ulaG-lacZ) and Φ(ulaA-lacZ) when cells were grown on CAA (Table 2). These results suggested the participation of IHF in the repression of the ula regulon.
Since Zhang et al. (35) had reported that an fnr mutant showed delayed growth on l-ascorbate under anaerobic conditions, we tested the effects of an fnr mutation on ula regulon expression. For this purpose, we transduced φ(ulaG-lacZ) and φ(ulaA-lacZ) into the fnr mutant strain JA182. β-Galactosidase analysis showed that this mutation did not affect the expression of Φ(ulaG-lacZ) and Φ(ulaA-lacZ) (Table 2), indicating that the delayed growth of this mutant on l-ascorbate is not a consequence of a direct effect of FNR on ula regulon expression.
Binding of UlaR, CRP, and IHF to the ulaG-ulaA intergenic region.
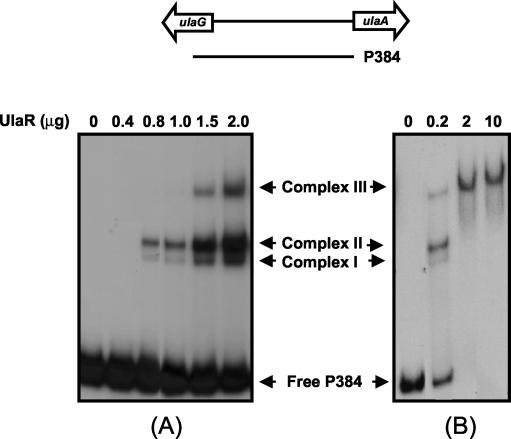
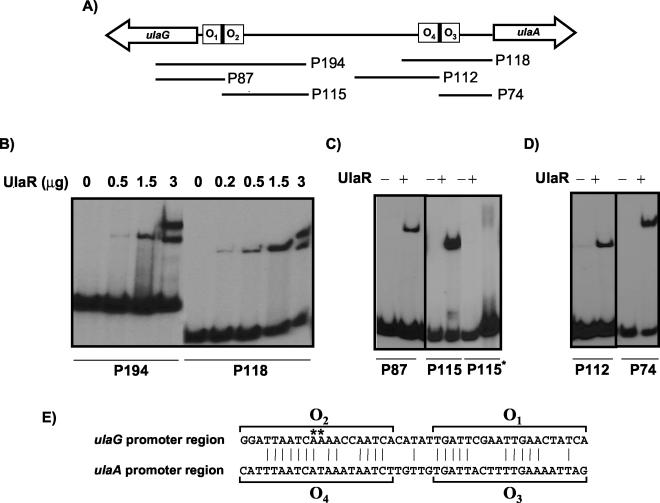
The interactions of the regulators UlaR, CRP, and IHF with the promoter regions of the ula regulon were studied by electrophoretic mobility shift assays. The repressor interactions with the complete ulaG-ulaA intergenic region were assessed using probe P384 (Fig. 3) and UlaR protein, purified as described in Materials and Methods. As the protein concentration increased, up to three different DNA-UlaR protein complexes were detected (complexes I, II, and III [Fig. 3A]), suggesting that the probe might contain multiple UlaR binding sites, probably with different affinities. When a saturating protein concentration was used, only one complex (complex III) was observed (Fig. 3B), suggesting that this complex is formed when all binding sites are occupied by UlaR. To analyze these plausible multiple DNA binding sites of UlaR, the intergenic region was split into several fragments, which were tested as probes in subsequent experiments with the purified protein. Binding assays performed with probe P194 or P118 (Fig. 4A) displayed in each case up to two complexes, depending on the concentration of protein added to the assay (Fig. 4B), suggesting the presence of two operator sites in each fragment. Sequence analysis of P194 and P118 by OMIGA revealed an extensive similarity in a region of 45 nucleotides, each of which might contain two putative operators (Fig. 4E). The proposed operators were named O1 and O2 in the ulaG promoter region and O3 and O4 in the ulaA promoter region. This finding led us to study the binding of UlaR with four probes prepared to contain only one of the operators each (Fig. 4A). As shown in Fig. 4C and D, one complex was formed between UlaR and each DNA probe, indicating that all retained UlaR binding capacity, thus giving support to the function of these operator sites. The addition of l-ascorbate to the binding mixtures did not affect the UlaR binding pattern (not shown), indicating that this compound is not the effector molecule of the UlaR repressor.
FIG. 3.
Binding of UlaR to ulaG and ulaA promoters. The line over the panels indicates the fragment corresponding to the full-length ulaG and ulaA promoter region used as a probe. The 32P-labeled DNA fragment was added at 2.5 (A) or 0.25 (B) nM to binding mixtures containing different amounts of purified UlaR protein. The migration patterns of the different DNA-protein complexes are indicated.
FIG. 4.
Binding of UlaR to different fragments of the ulaG-ulaA intergenic region. (A) Diagram of ulaG-ulaA intergenic region; the fragments used as probes after 32P labeling are indicated by lines. (B) Binding of UlaR to probes P194 and P118. (C) Binding of UlaR added (+) at 1.5 μg to probes P87, P115, and the mutated P115 (P115*). (D) Binding of UlaR added at 1.5 μg to probes P112 and P74. (E) Alignment of the nucleotide sequences (written in the direction of transcription of the ulaG and ulaA genes) of the UlaR binding sites present in fragments P194 and P118. The vertical lines indicate the conserved residues. The nucleotides forming the proposed operator sites are indicated by brackets. Nucleotide changes of the mutation present in P115* abolishing UlaR binding are shown by asterisks.
Further evidence for the binding of UlaR was obtained by site-directed mutagenesis of the O2 operator. For this purpose, P115* was constructed by PCR using primers ULA-A6 and ULA-G4, in which two A’s were changed to C (Fig. 4E). Binding experiments showed that UlaR did not bind to this mutated probe (Fig. 4C).
Sequence alignment of the four proposed operators allowed us to define a consensus motif, consisting of an inverted repeat present in each one, which might be recognized by the UlaR repressor (Fig. 5).
FIG. 5.
Alignment of the nucleotide sequences of the UlaR operator sites. The operator sequences are written in the direction of transcription of the ulaG and ulaA genes. The nucleotides indicated in the consensus sequence are present in three of the four aligned operators. The arrows indicate the inverted repeat recognized by UlaR.
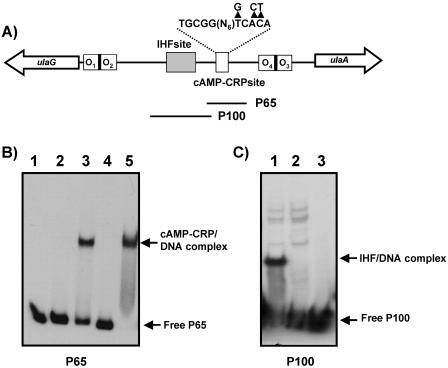
Since IHF seems to control ula regulon expression and a putative IHF binding site had been identified by computational analysis of the ulaG-ulaA intergenic region, we performed gel shift experiments with crude extracts of the wild type and the IHF mutant strain JA184 using probe P100, which contained the putative IHF site (Fig. 6A). The retarded complex observed with wild-type extracts was not present in the IHF mutant, indicating that the transcriptional factor binds to this region (Fig. 6C).
FIG. 6.
Interactions of CRP and IHF with different fragments of the ulaG-ulaA intergenic region. (A) Location of the putative IHF and CRP binding sites in the intergenic region. The sequence of the CRP site is indicated, and above it the arrowheads indicate the nucleotides changed by site-directed mutagenesis in the CRP site. (B) Binding of cAMP-CRP to probe P65, using 2 (lanes 2 and 3) or 10 (lanes 4 and 5) μg of crude extracts of strain JA211 transformed with pBS-CRP, in the absence (lanes 2 and 4) or presence (lanes 3 and 5) of 125 μM cAMP. No extract was added to lane 1. (C) Binding of IHF to probe P100 using extracts of strain JA134 (lane 1) and JA184 (lane 2) and no extract (lane 3).
Binding of cAMP-CRP to the the ulaG-ulaA intergenic region was undertaken in gel shift experiments performed with different probes and increasing amounts of cell extracts of strain JA211 overexpressing CRP from plasmid pBS-CRP in the presence of various concentrations of cAMP. The results showed that only probe P65, encompassing the putative CRP site located at −61 of ulaA, yielded a retarded complex in the presence of cAMP (Fig. 6B). The fact that no additional bands were observed with the other probes when cAMP (100 μM to 1 mM) was added to the binding mixtures indicated that no other functional CRP binding sites were present in this region. Moreover, site-directed mutagenesis of the right arm of the CRP site (Fig. 6A) located at −61 of ulaA abolished the cAMP-CRP binding capacity, confirming that this was the only functional CRP site in the region (not shown).
Deletion analysis of the ulaG-ulaA intergenic regulatory region.
To analyze the participation of the identified cis-acting motifs in the in vivo regulation of the ulaG and ulaABCDEF operons, we obtained deletion-containing fragments of the corresponding promoters. Those cloned in plasmid pRS551 were used to analyze ulaG expression, and those cloned in plasmid pRS550 were used to analyze ulaA expression. The lacZ fusions obtained were introduced into strain JA134, and β-galactosidase activity was measured after anaerobic growth in CAA in the absence or presence of l-ascorbate.
In the absence of l-ascorbate, a basal level of 60 Miller units was observed for Φ-341, corresponding to the full-length promoter of ulaA (Fig. 7). Elimination of the O1 operator site in Φ-265 increased the basal level of β-galactosidase activity only ∼3.3-fold, whereas the O1 and O2 deletion in Φ-229 displayed a higher level of uninduced activity (7,500 Miller units). The constitutive expression was maintained in Φ-149 and Φ-81 and was lost in Φ-37. In the last construct, the cAMP-CRP site located at position −61 of the ulaA promoter was also deleted. When all the ulaA transcriptional fusions were analyzed in CAA in the presence of glucose, 1.5- to 2-fold catabolite repression was seen in the constructions lacking the O1 or the O1 and O2 operator sites. Addition of cAMP partially reversed this repression. These results reinforced the idea that this promoter is regulated by the cAMP-CRP complex.
In the presence of l-ascorbate, ulaA expression was induced in Φ-341 and Φ-265, yielding β-galactosidase levels of ∼12,000 Miller units. The activity doubled in Φ-229 and decreased in Φ-81. As under noninducing conditions, deletion to position −37 almost abolished promoter function (Fig. 7). When the ulaA transcriptional fusions were analyzed in l-ascorbate plus glucose, 13- to 14-fold repression was seen in the constructions lacking the O1 and O2 operator sites, displaying β-galactosidase activities similar to those obtained when glucose was used as the only carbon source (data not shown). Although the addition of cAMP partially reversed the repression, the remaining activity was only ∼15% of the induced value. These results give further support to the previous proposal that glucose repression is also mediated by another mechanism independent of the cAMP-CRP complex.
The analysis of ulaG expression showed that under noninducing conditions, the basal level of 40 Miller units displayed by the full-length promoter (Φ-288) increased up to 525 Miller units in Φ-218, in which the O3 operator site was totally deleted and O4 was truncated. Elimination of the cAMP-CRP site in Φ-180 did not change the pattern of expression under these conditions. The low level of β-galactosidase activity observed with Φ-20 is explained by the deletion of the −35 promoter sequence. No catabolite repression by glucose was seen in any of these constructions.
In l-ascorbate cultures, the induction of ulaG was observed as long as the position −67 was retained (Fig. 7). Addition of glucose to these cultures diminished the activity in all constructions to values similar to those obtained under uninduced conditions, and no reversion was observed in the presence of cAMP. These results suggested that the glucose effect on ulaG expression is mediated by a mechanism independent of the cAMP-CRP complex.
Effect of changing DNA helical phasing between the two pairs of UlaR operators.
The results presented above suggest that full repression requires the occupancy of the four operators by UlaR and an additive effect among them that would require the proper orientation. To test whether the repression is affected by the helical phasing of the operators present in the ulaG promoter (O1 and O2) and the ulaA promoter (O3 and O4) regions, we changed their angular orientations by inserting 5 bp between the two promoters. The corresponding ulaG and ulaA transcriptional fusions were analyzed in the corresponding transfected cells grown in CAA, exhibiting basal β-galactosidase activities of 45 and 70 Miller units, respectively. Thus, efficient UlaR repression was retained after the helical phasings of both promoters were changed. Similar results were obtained when the inserted sequence was enlarged to 10 bp.
Expression of ulaR under different growth conditions.
In order to study the expression of ulaR, the fused promoter Φ(ulaR-lacZ) (6) was introduced in the genetic background of strain JA134. Analysis of β-galactosidase in anaerobic cultures on CAA or l-ascorbate in the presence or absence of glucose yielded no significant differences in the activity level (ca. 300 Miller units). This indicated a constitutive expression of this gene.
DISCUSSION
The occurrence of multiple repressor binding sites is a rather common phenomenon, as illustrated by the models of the gal operon (15, 19) or the araBAD operon (9, 12). This is also the case in the DeoR repressor family, which includes, among others, the deo operon and the sn-glycerol-3-P regulon of E. coli (8, 16, 21, 30) and the iol operon of B. subtilis (32). In these models, a single operon or two divergently transcribed operons are regulated by the cooperative binding of the specific repressor to widely separated operators located in the corresponding promoter regions (7).
In the ula regulon, four UlaR operator sites involved in the coordinated regulation of two divergently transcribed operons were identified. A characteristic of the negative control by UlaR is that the protein may bind cooperatively to every one of the four operators organized in two pairs separated by 187 bp. The pair O1-O2 is located in the ulaG promoter overlapping the transcription start site and the −10 region, and the pair O3-O4 is located in the ulaA promoter overlapping the −35 and the −10 promoter sequences. The high constitutive expression of Φ(ulaA-lacZ) only when O1 and O2 are totally deleted indicates that repression of the ulaABCDEF operon requires interaction of the repressor with these operator sites. The fact that the expression displayed further increase in the presence of l-ascorbate suggests the participation of additional UlaR operator sites in ulaA control, a role accomplished by the operators O3 and O4 located in the ulaA promoter region. In the case of the opposite promoter, deletion of the O3 and O4 operator sites also increased the β-galactosidase activity of the Φ(ulaG-lacZ) fusion, although in this case to a lower derepression ratio, as expected from a weaker promoter. The presence of l-ascorbate increased the activity to values probably corresponding to full derepression of the less efficient ulaG promoter. Thus, each promoter is dependent on the presence of the operator sites of the other promoter, suggesting that the distant operators are essential to enhance repression. When only one promoter is present, repression can also be obtained with lower efficiency.
Total repression of these operons by UlaR seems to be dependent upon repressor-mediated DNA loop formation. This is especially supported by the fact that a mutation in the himA or himD3 gene encoding the different subunits of the IHF factor partially relieved the repressor effect in the expression of ulaG or ulaA promoter fusions when cells grew in the absence of l-ascorbate. This effect was smaller in the case of ulaG, probably due to its weaker promoter activity.
As has been proposed for other operons, the DNA looping could increase the local concentration of the UlaR repressor within the promoter regions or alter the structures of the promoter regions in such a way that transcription initiation is no longer possible. In the ula regulon, changes in the helical phasing between the two sets of operators do not release the repression, as does the deletion of the pair O3-O4 in ulaG expression or the pair O1-O2 in ulaA expression. A simple explanation is that alternative loops may be formed because of the multiple binding sites. At a given interoperator distance, one specific combination of two of the four binding sites might be most favorable. When the distance is increased by half a helix turn, another combination of two binding sites may be favorable. Formation of different loops had been previously reported to be involved in DeoR regulation (8). This model of UlaR-DNA interaction proposed for the regulation of the divergent ula promoters is consistent with the different UlaR-DNA complexes observed in the gel shift experiments. It is noteworthy that, despite its apparent complexity, DNA looping is widely used in gene regulation, and although a well-established role is to increase the repression level, it has recently been reported that compared to a simple alternative regulatory mechanism, DNA looping is able to reduce the fluctuation in transcription and keep the repression level fairly constant with respect to changes in the number of repressor molecules (29).
The glucose repression of the ula regulon is mediated by at least two mechanisms, one independent of cAMP-CRP and the other mediated by this activator complex, which causes a weak, though significant, activation of the ulaA operon. Two lines of evidence support the hypothesis of this double mechanism. First, the repression exerted by glucose in Φ(ulaA-lacZ) or Φ(ulaG-lacZ) expression on the l-ascorbate cultures was not reversed by the addition of cAMP, indicating the occurrence of a mechanism other than that mediated by CRP. Second, in a strain constitutively expressing the ula regulon, a crp mutation caused a 55% reduction in ulaA operon expression, thus requiring another mechanism to account for the full reduction of either ulaA or ulaG.
In relation to the cAMP-CRP-independent mechanism, since l-ascorbate transport has been reported to be mediated by a phosphotransferase system (35), it seems likely that the phosphoryl transfer from PEP to the EIIAGlc enzyme may limit the phosphorylation of the ascorbate EIIA-specific protein (SgaA) and hence that of l-ascorbate through its specific transport protein. This impairs l-ascorbate transport and therefore the formation of the ula regulon inducer. Notice that a most accurate regulation seems to be exerted on the expression of the ulaA operon, which contains the genes involved in l-ascorbate transport necessary for the formation of a ula regulon inducer. In this context, it must be emphasized that l-ascorbate does not modify UlaR binding to the operators and that metabolism of l-xylulose-5-phosphate formed endogenously in the cell does not induce the ula regulon (6). Therefore, the other two intermediate metabolites in the pathway, l-ascorbate-6-phosphate and 3-keto-l-gulonate-6-phosphate, are the most plausible candidates to act as the effector molecule of the ula regulon. Experiments are under way to obtain these compounds, which are not commercially available, to be tested as inducers of the ula regulon.
Since addition of glucose to l-ascorbate cultures produced a repression effect on the expression of ulaA promoter deletion fusions higher than that observed on CAA, the participation of another global regulator whose expression may depend on the carbon source must be taken into account for ulaA regulation.
From our studies and those reported by other authors (16, 21, 30), we may conclude that activation by cAMP-CRP and the repressor effect caused by binding to several operator sites are general features of the genetic systems under the control of repressors belonging to the DeoR family in E. coli.
The ula regulon undergoes transcriptional regulation as a function of the growth phase, reaching a maximum at the end of exponential phase. We have demonstrated that this activation occurs in an rpoS-independent manner. At present, we do not know the biological significance of this expression pattern or the regulators involved in it. More studies need to be performed to further characterize growth phase-dependent ula regulon induction.
Acknowledgments
We gratefully acknowledge J. Membrillo-Hernandez for providing strain JMH0317.
This work was supported by grant BMC2001-3003 from the Dirección General de Investigación, Ministerio de Ciencia y Tecnología, Madrid, Spain, and by the Comissionat per Universitats i Recerca de la Generalitat de Catalunya. E.C. was the recipient of a predoctoral fellowship from the Ministerio de Educación, Cultura y Deporte (Spain).
REFERENCES
- 1.Amouyal, M., L. Mortensen, H. Buc, and K. Hammer. 1989. Single and double loop formation when deoR repressor binds to its natural operator sites. Cell 58:545-551. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 3.Beck von Bodman, S., G. T. Hayman, and S. K. Farrand. 1992. Opine catabolism and conjugal transfer of the nopaline Ti-plasmid pTiC58 are co-ordinately regulated by a single repressor. Proc. Natl. Acad. Sci. USA 89:643-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 5.Boronat, A., and J. Aguilar. 1979. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos, E., J. Aguilar, L. Baldoma, and J. Badia. 2002. The gene yjfQ encodes the repressor of the yjfR-X regulon (ula), which is involved in l-ascorbate metabolism in Escherichia coli. J. Bacteriol. 184:6065-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandanell, G. 1992. DeoR repression at a distance only weakly responds to changes in interoperator separation and DNA topology. Nucleic Acids Res. 20:5407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandanell, G., and K. Hammer. 1985. Two operator sites separated by 599 base pairs are required for deoR repression of the deo operon of Escherichia coli. EMBO J. 4:3333-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn, T. M., S. Hahn, S. Ogden, and R. F. Schleif. 1984. An operator at −280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl. Acad. Sci. USA 81:5017-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliot, T. 1992. A method for constructing single copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodrish, J. A., M. L. Schwartz, and W. R. McClure. 1990. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 18:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn, S., T. Dunn, and R. Schleif. 1984. Upstream repression and CRP stimulation of the Escherichia coli l-arabinose operon. J. Mol. Biol. 180:61-72. [DOI] [PubMed] [Google Scholar]
- 13.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 14.Ibañez, E., E. Campos, L. Baldomà, J. Aguilar, and J. Badia. 2000. Regulation of expression of the yiaKLMNOPQRS operon for carbohydrate utilization in Escherichia coli: involvement of the main transcriptional factors. J. Bacteriol. 182:4617-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irani, M. H., L. Orosz, and S. Adhya. 1983. A control element within a structural gene: the gal operon of Escherichia coli. Cell 32:783-788. [DOI] [PubMed] [Google Scholar]
- 16.Larson, T. J., J. S. Cantwell, and A. T. van Loo-Bhattacharya. 1992. Interaction at a distance between multiple operators controls the adjacent, divergently transcribed glpTQ-glpACB operons of Escherichia coli K-12. J. Biol. Chem. 267:6114-6121. [PubMed] [Google Scholar]
- 17.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin Phenol reagent. J. Biol. Chem. 193:265-273. [PubMed] [Google Scholar]
- 19.Majumdar, A., and S. Adhya. 1984. Demonstration of two operator elements in gal: in vitro repressor binding studies. Proc. Natl. Acad. Sci. USA 81:6100-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Mortensen, L., G. Dandanell, and K. Hammer. 1989. Purification and characterization of the deoR repressor of Escherichia coli. EMBO J. 8:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nunoshiba, T., E. Hidalgo, C. F. Amabile-Cuevas, and B. Demple. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 174:6054-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reizer, J., A. Reizer, and M. H. Saier. 1997. Is the ribulose monophosphate pathway widely distributed in bacteria? Microbiology 143:2519-2520. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sanchez, J. C., R. Gimenez, A. Schneider, W.-D. Fessner, L. Baldomà, J. Aguilar, and J. Badia. 1994. Activation of a cryptic gene encoding a kinase for l-xylulose opens a new pathway for the utilization of l-lyxose by Escherichia coli. J. Biol. Chem. 269:29665-29669. [PubMed] [Google Scholar]
- 26.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silhavy, T. J., M. L. Berman, and L. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 29.Vilar, J. M. G., and S. Leibler. 2003. DNA looping and physical constraints on transcription regulation. J. Mol. Biol. 331:981-989. [DOI] [PubMed] [Google Scholar]
- 30.Yang, B., and J. Larson. 1996. Action at a distance for negative control of transcription of the glpD gene encoding sn-glycerol 3-phosphate dehydrogenase of Escherichia coli K-12. J. Bacteriol. 178:7090-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yew, W.-S., and J. A. Gerlt. 2002. Utilization of l-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 184:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida, K., D. Aoyama, I. Ishio, T. Shibayama, and Y. Fujita. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 179:4591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng, G., Z. Ye, and T. J. Larson. 1996. Repressor of the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J. Bacteriol. 178:7080-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, B., R. A. Daniel, J. Errington, and L. Kroos. 1997. Bacillus subtilis SpoIIID protein binds to two sites in the spoVD promoter and represses transcription by σERNA polymerase. J. Bacteriol. 179:972-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Z., M. Aboulwafa, M. H. Smith, and M. H. Saier, Jr. 2003. The ascorbate transporter of Escherichia coli. J. Bacteriol. 185:2243-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]