Abstract
The TolC protein of Escherichia coli, through its interaction with AcrA and AcrB, is thought to form a continuous protein channel that expels inhibitors from the cell. Consequently, tolC null mutations display a hypersensitive phenotype. Here we report the isolation and characterization of tolC missense mutations that direct the synthesis of mutant TolC proteins partially disabled in their efflux role. All alterations, consisting of single amino acid substitutions, were localized within the periplasmic α-helical domain. In two mutants carrying an I106N or S350F substitution, the hypersensitivity phenotype may be in part due to aberrant TolC assembly. However, two other alterations, R367H and R390C, disrupted efflux function by affecting interactions among the helices surrounding TolC's periplasmic tunnel. Curiously, these two TolC mutants were sensitive to a large antibiotic, vancomycin, and exhibited a Dex+ phenotype. These novel phenotypes of TolCR367H and TolCR390C were likely the result of a general influx of molecules through a constitutively open tunnel aperture, which normally widens only when TolC interacts with other proteins during substrate translocation. An intragenic suppressor alteration (T140A) was isolated from antibiotic-resistant revertants of the hypersensitive TolCR367H mutant. T140A also reversed, either fully (R390C) or partially (I106N and S350F), the hypersensitivity phenotype of other TolC mutants. Our data suggest that this global suppressor phenotype of T140A is the result of impeded antibiotic influx caused by tapering of the tunnel passage rather than by correcting individual mutational defects. Two extragenic suppressors of TolCR367H, mapping in the regulatory region of acrAB, uncoupled the AcrR-mediated repression of the acrAB genes. The resulting overexpression of AcrAB reduced the hypersensitivity phenotype of all the TolC mutants. Similar results were obtained when the chromosomal acrR gene was deleted or the acrAB genes were expressed from a plasmid. Unlike the case for the intragenic suppressor T140A, the overexpression of AcrAB diminished hypersensitivity towards only erythromycin and novobiocin, which are substrates of the TolC-AcrAB efflux pump, but not towards vancomycin, which is not a substrate of this pump. This showed that the two types of suppressors produced their effects by fundamentally different means, as the intragenic suppressor decreased the general influx while extragenic suppressors increased the efflux of TolC-AcrAB pump-specific antibiotics.
Escherichia coli cells lacking the outer membrane protein (OMP) TolC display hypersensitivity to a variety of inhibitors, including bile salts, detergents, and hydrophobic antibiotics (38). This was initially thought to be due to a defect in the outer membrane permeability barrier as a result of a defective lipopolysaccharide (LPS) (32). However, it was shown that tolC and rfa (LPS core) mutations produce an additive effect on hypersensitivity, thus suggesting that tolC mutations may confer hypersensitivity independent of their effect on LPS (12). Subsequently, it was demonstrated that TolC is required for the activity of a major E. coli efflux pump that includes the AcrA and AcrB proteins (11).
Based on the pleiotropic phenotype of tolC null mutants, TolC has been implicated in other activities, including colicin import (26), α-hemolysin secretion (36), and porin regulation (23, 24). Studies of tolC null mutants also led to speculation that TolC might be involved in chromosome partitioning (14) and DNA supercoiling (8). However, no direct data have been presented to support these last two roles of TolC, and such implications presumably reflect an indirect effect of the tolC null mutation.
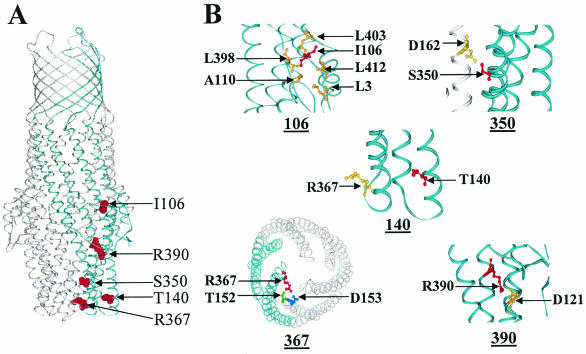
The crystal structure of TolC has been solved (18) and is strikingly distinct from those of other OMPs (Fig. 1A). It embodies two separate domains, with the outer membrane-embedded segment composed of β-strands while the region that extends deep into the periplasm consists primarily of α-helices. Both domains make up an elongated barrel structure of 140 Å, and unlike the case for porins (6), each of the three monomers makes up only one-third of the barrel. Thus, an assembled TolC trimer forms a single channel or tunnel that extends the length of the molecule with a large inner diameter of 35 Å. The crystal structure of TolC also shows tapering of the proximal (periplasmic) end due to the packing of outer and inner helices into coiled coils. It has been proposed that the opening of the proximal entrance during substrate translocation may involve uncoiling and realignment of the paired helices (18).
FIG. 1.
TolC structure. (A) TolC trimer. Positions of a single monomer and various substitutions within a monomer are shown in blue and red, respectively. (B) Close-up views of regions affected by various substitutions. Helices within the same monomer are shown in blue while those belonging to other monomers are shown in gray. Positions of the pertinent residues are shown in ball-and-stick representation; those in red represent amino acids affected by substitutions.
Like tolC, acrA mutations were initially thought to affect outer membrane permeability (29). However, it is now well established that AcrA is a component of the TolC-AcrAB efflux pump in E. coli that mediates resistance to a broad range of inhibitors that are generally hydrophobic in nature (27, 30, 43). AcrB has been shown to be a proton-substrate antiporter (42), and biochemical studies have indicated that it interacts with AcrA (16, 28, 44). Atomic structures of AcrB alone (25) and bound to inhibitors (41) have recently been solved. AcrB, like TolC, is trimeric and extends considerably into the periplasmic space, raising the intriguing possibility of direct contacts between AcrB and TolC. From genetic data and studies performed with the α-hemolysin secretion system that uses HlyBD as well as TolC (34), it has been inferred that AcrA and -B interact with TolC to make a continuous channel that transports inhibitors directly into the surrounding medium. There has never been a direct demonstration of TolC's interaction with AcrA and/or AcrB.
Only limited genetic analyses have been performed to dissect the role of individual residues in the various functions attributed to TolC. These studies include the isolation of tolC missense mutations affecting α-hemolysin secretion (35) and TolC's cell surface interactions with bacteriophage TLS and colicin E1 (13). Recently, two systematic studies conducted by V. Koronakis's group determined the mechanism of aperture opening during substrate translocation (1, 9). It is likely that during antibiotic efflux, TolC's interaction with AcrAB also influences the aperture conformation. Deletion and site-directed mutagenesis analyses have also pointed to a role for residue L412 of mature TolC in colicin import and antibiotic efflux (39, 40).
For this study, we conducted a genetic analysis and isolated tolC missense mutations affecting the antibiotic efflux phenotype. Because the isolation of hypersensitivity mutants, by definition, entails negative selection, a screening strategy was developed to enrich for missense tolC mutations. Further genetic and biochemical characterization of these mutants is expected to provide a better understanding of the role of TolC in antibiotic efflux.
MATERIALS AND METHODS
Bacterial strains, media, and biochemicals.
The E. coli K-12 strains used in this study were derived from MC4100 [F− araD139 Δ(argF-lac)U139 rpsL150 flbB5301 ptsF25 deoC1 thi-1 rbsR relA] (5). XL1-Red (endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT::Tn10) was used for the in vivo mutagenesis of plasmid DNA. Luria broth (LB) and agar (LBA) were prepared as previously described (33). ECF substrate was purchased from Amersham Pharmacia Biotech. All other biochemicals were of analytical grade.
DNA manipulations.
The tolC gene was cloned into pTrc99A (Pharmacia) essentially as described previously (35), with the exception that the forward primer used (5′-CAGGAAACAGATCATGAGGAAATTGCTCCC-3′) created a unique BspHI site (underlined) instead of the NcoI site. By the creation of the BspHI site, the second codon of the TolC signal sequence was changed from AAG (lysine) to AGG (arginine), which did not affect protein synthesis. tolC DNA was amplified from the chromosome by PCR, digested with BspHI and HindIII (created by the reverse primer), and ligated into appropriately restricted pTrc99A. In the absence of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG), the level of plasmid-borne TolC was similar to that of the chromosomally expressed TolC protein. Therefore, all experiments were carried out in the absence of IPTG. The acrAB genes were cloned into the BspHI and HindIII sites of pACYC184. For this, the acrAB region was amplified by using two primers as follows: the forward primer (5′-AGATCTCATGAACAATCCGACTTGTC-3′) created a BspHI site (underlined), while the reverse primer (5′-CTCCTTAAGCTTCGTAGGTTATGC-3′) created a HindIII site (underlined). Expression of the acrAB genes from this plasmid clone was driven from their native promoter located upstream of the acrA gene. Site-directed mutagenesis of the tolC gene was performed with a QuickChange XL mutagenesis kit (Stratagene) according to the manufacturer's protocol. In vivo mutagenesis was performed by propagating the tolC plasmid in the XL1-Red mutator strain per the manufacturer's (Stratagene) instructions.
The chromosomal tolC, acrR, acrA, and acrB genes were deleted by the method of Datsenko and Wanner (7). The primers used for these deletions are shown in Table 1. The deletion removed 20 bp upstream of the start codon while retaining the last 26 codons of the tolC gene. The deleted tolC locus was marked by genes conferring either kanamycin (Kmr) or chloramphenicol (Cmr) resistance. Two separate deletions were constructed to remove the entire coding region of acrA or acrB. For elimination of the polar effect of acrA::Kmr on acrB, the antibiotic resistance gene was removed by the pCP20-encoded recombinase (7). The resulting deletion was fully complemented by a plasmid clone carrying only the acrA gene, showing that the deletion imposed no polar effect on acrB. The acrR gene was only partially deleted because of the presence of potential promoter sequences directing the transcription of adjacent genes. This deletion removed residues 71 to 140 of the 216-residue AcrR protein. Deletions of all four genes were confirmed by PCR analysis and phenotypic tests.
TABLE 1.
Primers used to delete chromosomal genes
| Primera | Sequence (5′ to 3′) |
|---|---|
| ΔtolC-F | CGCAATAATTTTACAGTTTGATCGCGCTAAATACTGTGTAGGCTGGAGCTGCTTCG |
| ΔtolC-R | TGCGGATGTTTGCTGAACGACTGGTGCCGGGCTATCCATATCAATATCCTCCTTAG |
| ΔacrR-F | GAGATCTGGGAACTGTCAGAATCCAATATTGTGTAGGCTGGAGCTGCTTC |
| ΔacrR-R | TTCAATACAATGTTTTAACGTTTGTTCTATCATATGAATATCCTCCTTAG |
| ΔacrA-F | GACCAATTTGAAATCGGACACTCGAGGTTTACATTGTAGGCTGGCGCTGCTTCG |
| ΔacrA-R | GCAAAAATCGGGCGATCGATAAAGAAATTAGGCATATGAATATCCTCCTTAG |
| ΔacrB-F | AGTCCAAGTCTTAACTTAAACAGGAGCCGTTAAGACTGTAGGCTGGAGCTGCTTCG |
| ΔacrB-R | AGGCCGCTTACGCGGCCTTAGTGATTACACGTTGTACATATGAATATCCTCCTTAG |
F and R refer to forward and reverse primers, respectively.
Western blot analysis.
Whole-cell extracts were analyzed on mini sodium dodecyl sulfate-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Immobilon; Millipore). After transfer, the membranes were incubated for 1.5 h with a primary antibody raised against TolC, MBP, or AcrA. The membranes were washed, and a secondary antibody (goat anti-rabbit alkaline phosphatase-conjugated immunoglobulin G) was added for 1 h. The membranes were exposed to ECF substrate for 5 min and analyzed in a phosphorimager. Protein bands were quantified with the Quantity One (Bio-Rad) program.
Antibiotic, phage, and colicin sensitivity assays.
Antibiotic disk sensitivity assays were performed by placing presoaked antibiotic disks (Difco) on bacterial lawns prepared on LBA. The zones of inhibition (in millimeters) were measured after incubation of the plates for 8 h at 37°C. MICs were determined by diluting overnight bacterial cultures to 105 cells/ml in LB containing increasing concentrations of novobiocin or erythromycin. Bacterial growth after 16 h of incubation at 37°C was recorded by measuring the optical density at 600 nm. Phage sensitivity was determined by the cross-streak method. Colicin E1 sensitivity was determined by spotting twofold serial dilutions of the colicin stock solution on bacterial lawns. Killing zones were recorded after 6 to 8 h of incubation at 37°C.
RESULTS
Antibiotic efflux-defective tolC mutants.
Mutations in any one of the genes tolC, acrA, and acrB may lead to a nonfunctional efflux pump and a hypersensitive phenotype. Since the focus of this study is TolC, we therefore devised a means to enrich for tolC missense mutants. These mutant tolC alleles are expected to direct the synthesis of proteins that are defective in efflux activity due to an alteration in TolC's structure. To avoid mutations mapping in acrA and acrB, we mutagenized a plasmid carrying the tolC gene that produces TolC at a level similar to that produced from the chromosomal allele. Random tolC mutations were generated by propagating the tolC plasmid in the XL1-Red mutator strain. Plasmid DNA from five independent mutagenized pools was then transformed into a genetic background lacking the chromosomal tolC allele. Over 20,000 transformed bacterial colonies were screened by replica plating for the ability to grow on medium containing a low (5 μg/ml) or high (50 μg/ml) concentration of novobiocin, an antibiotic that is normally removed by the TolC-AcrAB pump. Initially, we focused on 126 colonies that grew on low but not high concentrations of novobiocin; colonies that failed to grow on low concentrations are likely to contain tolC null rather than missense mutations. This is because it is unlikely that a structural protein, such as TolC, could be altered so specifically by a single substitution as to completely obliterate its efflux function. Colonies that grew on medium containing a high novobiocin concentration most likely synthesized TolC with normal efflux activity.
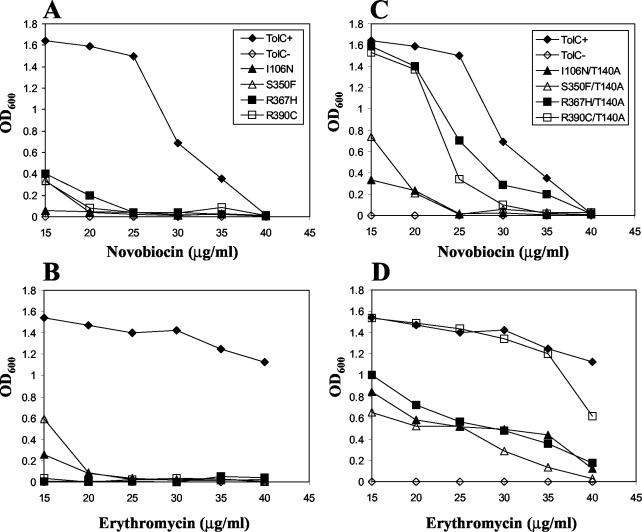
To ensure that the phenotypes were stable and expressed from plasmid DNA, we extracted the plasmids from 126 isolates and retransformed them into a tolC background. Only seven transformants expressed the desirable phenotype, i.e., they produced inhibition zones around novobiocin (30 μg) and erythromycin (15 μg) disks that were larger than those by the parental TolC+ strains but smaller than those by a tolC null strain. DNA sequence analysis of the plasmid tolC gene revealed the presence of a missense mutation in five isolates, resulting in an I106N, S350F, R367H, or R390C (two independent isolates) substitution in the mature portion of the protein. The remaining two isolates carried a frame-shift mutation close to the end of the tolC gene that resulted in the synthesis of truncated, albeit partially functional, polypeptides. These two mutants were not analyzed further. The four different missense alterations affected residues that were localized within the periplasmic α-helical domain of TolC (Fig. 1A). Mutants bearing an R367H or R390C alteration produce TolC with levels similar to those with the parental TolC+ strain (Fig. 2); the I106N and S350F substitutions resulted in somewhat reduced protein levels. It was recently shown that this reduced protein level is due to a defect in TolC's assembly (37). The MICs of novobiocin and erythromycin for a TolC+ parental strain were 35 to 40 and >40 μg/ml, respectively; for a tolC null strain, these values were between 1 and 5 μg/ml for both antibiotics. The MICs for mutants were 10 to 20 μg/ml for both antibiotics (Fig. 3A and B). The partial hypersensitivity phenotype of all TolC mutants was codominant over the wild-type TolC protein. With respect to colicin E1, mutants were either fully sensitive (R390C) or displayed a 2-, 8-, or 16-fold reduction (R367H, I106N, and S350F, respectively) in sensitivity compared to the parental TolC+ strain. On LB-blood agar plates, the hemolytic zones around individual colonies of mutants were either indistinguishable from (TolCR367H and TolCR390C) or smaller than (TolCI106N and TolCS30F) those around colonies expressing wild-type TolC.
FIG. 2.
Western blot analysis of cell extracts obtained from wild-type or mutant TolC mutants with or without the suppressor alteration. Membrane filters were treated with TolC and myelin basic protein antibodies. The positions of TolC and maltose binding protein (MBP) are shown.
FIG. 3.
Growth of TolC−, TolC+, and TolC mutant strains in LB containing different amounts of novobiocin (A and C) and erythromycin (B and D). The strains and symbols shown in panels A and C are identical to each other, as are those in panels B and D. All assays were performed in triplicate, and average values were plotted. LB-antibiotic medium was inoculated with overnight cultures (105 cell/ml), and growth (OD600) after 16 h at 37°C was recorded.
Additional substitutions at R367 and D153.
TolC's crystal structure (18) showed that when the TolC tunnel entrance is in the closed (resting) state, R367 forms a salt bridge with D153 and a hydrogen bond with T152 of the adjacent monomer subunit (Fig. 1B). The replacement of R367 with serine was shown to increase TolC-mediated conductance about 2.5-fold, whereas no effect on conductance was observed when T152 was changed to alanine (1). These results suggested that an intermonomer salt bridge between R367 and D153 is important for the control of the aperture opening by constraining of the movement of the inner coiled coils.
Despite widening the tunnel aperture, the R367S substitution was reported to have no effect on the antibiotic efflux function (1), whereas we have shown here that a substitution of R367H confers hypersensitivity (Fig. 3). It may be argued that the presence of histidine, not the absence of arginine, at position 367 is responsible for this phenotype. We therefore changed R367 to alanine and glycine by site-directed mutagenesis but observed a hypersensitivity phenotype similar to that conferred by R367H (data not shown). We also created an R367S substitution, and as expected, observed no hypersensitivity phenotype. Finally, through site-directed mutagenesis, D153 was changed to cysteine. This substitution resulted in a hypersensitivity phenotype similar to that conferred by R367H (data not shown), thus further reflecting a functional relevance for the D153-R367 salt bridge in TolC's antibiotic efflux function. These results show that (i) the absence of arginine at position 367, but not its replacement by histidine, produces the hypersensitivity phenotype; and (ii) a salt bridge between D153 and R367 is important for efflux function, even though S367 can somehow allow the protein to maintain an efflux-competent conformation despite the fact that it, like alanine and glycine, cannot form a salt bridge with D153.
Reasons for the hypersensitivity phenotype of TolC mutants.
Because the TolC protein is directly involved in the antibiotic efflux function, the observed hypersensitivity phenotype of tolC mutants is expected to be the result of a defect in this activity. However, it is also possible that aberrant assembly and insertion of mutant TolC proteins, particularly those of TolCI106N and TolCS350F, may disrupt normal outer membrane permeability function, thus allowing a larger influx of antibiotics than their removal by the TolC-AcrAB efflux pump.
In an attempt to resolve these two possibilities, we first compared the antibiotic sensitivity profile of a tolC null mutant with that of a deep rough LPS mutant in which the hypersensitivity phenotype is known to be due to a defective outer membrane permeability barrier (29). As expected, both mutants showed hypersensitivity towards novobiocin and erythromycin because these antibiotics can readily penetrate the perturbed lipid bilayer of LPS mutants and are substrates of the TolC-AcrAB efflux pump (Table 2). Interestingly, only the deep rough LPS mutant exhibited hypersensitivity to rifampin, and none of the strains showed any significant increase in sensitivity to a large hydrophilic antibiotic, vancomycin (molecular weight, 1,450). The similar sensitivity profiles of TolC+ and TolC− strains for rifampin suggested that this hydrophobic antibiotic is either not a substrate of the TolC-AcrAB efflux pump or cannot penetrate the intact outer membrane. On the other hand, the rifampin hypersensitivity of the deep rough LPS mutant showed that rifampin could readily penetrate cells with a compromised outer membrane permeability barrier. Rifampin sensitivity has also been reported for strains expressing assembly-defective OmpC and OmpF mutants (20, 22), which presumably disrupt the outer membrane. Thus, rifampin could be used to at least initially differentiate between mutants with a defective outer membrane permeability barrier and those with an efflux defect.
TABLE 2.
Zones of inhibition of bacterial strains against various inhibitorsa
| Relevant genotypeb | Zone of inhibition (mm) with indicated inhibitor
|
|||
|---|---|---|---|---|
| Novobiocin (30 μg) | Erythromycin (15 μg) | Rifampin (5 μg) | Vancomycin (75 μg) | |
| tolC+rfa+ | 7.8 | 6.7 | 8.6 | 7.4 |
| ΔtolC rfa+ | 20.8 | 14.0 | 9.0 | 6.5 |
| tolC+rfa+ ΔrcsB::Kmr | 8.5 | 6.9 | 8.5 | 7.4 |
| tolC+ Δrfa2057 ΔrcsB::Kmr | 19.8 | 12.1 | 16.3 | 8.5 |
The diameter of the paper disks was 6.5 mm. Averages from two independent experiments are shown.
The ΔrcsB::Kmr allele was used to prevent excessive capsule production due to the deletion of the LPS core genes in Δrfa2057.
Strains expressing TolCR367H and TolCR390C displayed hypersensitivity to rifampin, and surprisingly, also to vancomycin (Table 3). These profiles were distinct from that of the deep rough LPS mutant (rifampin sensitive and vancomycin insensitive) or a tolC null mutant (insensitive to both antibiotics). Therefore, while the rifampin sensitivity phenotype of a mutant expressing TolCR367H or TolCR390C suggests a breached outer membrane permeability barrier, the sensitivity to vancomycin indicates a novel mechanism for the hypersensitivity phenotype.
TABLE 3.
Antibiotic sensitivities and maltodextrin phenotypes of bacterial strains expressing various TolC mutants from plasmids
| TolC protein | Zone of inhibition (mm) with antibiotica
|
Dex phenotypec | |
|---|---|---|---|
| Rifampin (5 μg)b | Vancomycin (75 μg) | ||
| TolC+ | 10.4 | 8.0 | −/+ |
| TolC− | 10.5 | 6.5 | −/+ |
| TolCR367H | 19.8 | 18.0 | + |
| TolCR367H,T140A | 13.2 | 9.8 | −/+ |
| TolCR390C | 15.5 | 17.2 | +/− |
| TolCR390C,T140A | 10.5 | 8.6 | −/+ |
| TolCI106N | 10.8 | 7.7 | ND |
| TolCI106N,T140A | 10.0 | 8.1 | ND |
| TolCS350F | 11.4d | 20.5d | ND |
| TolCS350F,T140A | 21.8d | 17.8d | ND |
The diameter of the paper disks was 6.5 mm. Averages from three independent experiments are shown.
Note that control strains (TolC+ and TolC−) carrying plasmids show somewhat more sensitivity than those without them, as shown in Table 2.
The Dex phenotype was recorded as the ability of bacteria to grow on maltodextrin minimal medium plates after 36 h of incubation at 37°C. Plus and minus signs reflect colony sizes relative to that of a LamB+ strain, which was rated +++ under similar growth conditions. ND, not determined.
The number varied significantly (>20%) in different experiments.
Strains expressing TolCI106N showed no increase in sensitivity to either rifampin or vancomycin (Table 3). Thus, the hypersensitivity of this mutant to novobiocin and erythromycin is likely the result of an efflux defect rather than a compromised permeability barrier. As stated earlier, this efflux defect may be the result of lower TolCI106N levels in the outer membrane owing to an assembly defect (Fig. 2) (37). The antibiotic sensitivity data for TolCS350F were difficult to interpret due to large variations in the inhibition zones. This is presumably due to aberrant assembly of the mutant protein (37).
Constitutively open state of TolCR367H and TolCR390C tunnel aperture.
The vancomycin sensitivity of hemolysin-secreting TolC+ cells has been reported previously (3; also see below). This sensitivity is thought to be the result of antibiotic influx via the open state of TolC tunnels while they are engaged in hemolysin export. It is therefore conceivable that the vancomycin sensitivity phenotype of mutants expressing TolCR367H and TolCR390C is due to the influx of antibiotics through TolC tunnels that are in a constitutively open state because of mutant alterations. We further tested this notion by examining the ability of TolC mutants to grow on a medium containing maltodextrins as a sole carbon source. Maltodextrins normally enter the cell through the LamB maltoporin (29) and are excluded from other channel-forming proteins due to either a constricted pore size (6) or the presence of a plug domain (4, 10). The expression of TolCR367H and TolCR390C in a LamB− strain promoted growth on a minimal medium containing maltodextrins as a sole carbon source; hence, they had a Dex+ phenotype (Table 3). Unlike the mutant TolC proteins, the expression of wild-type TolC did not support growth on a maltodextrin minimal medium (Table 3). The vancomycin sensitivity and Dex+ phenotype of TolCR367H and TolCR390C corroborated the notion that these mutant proteins facilitate the diffusion of molecules that normally cannot pass through TolC's closed tunnels while in a resting stage.
Our argument that the constitutively open tunnel apertures of TolCR367H and TolCR390C lead to antibiotic influx should also apply to wild-type TolC when it is engaged in hemolysin translocation. We therefore tested the sensitivity of a TolC+ HlyA+ strain to vancomycin, rifampin, novobiocin, and erythromycin. As previously reported for vancomycin (3), wild-type TolC+ cells secreting hemolysin displayed increased sensitivities to rifampin, novobiocin, and to a lesser extent, erythromycin (Table 4). The similar antibiotic sensitivity profiles of the TolC+ strain, when secreting hemolysin, and mutants expressing TolCR367H or TolCR390C, without a hemolysin plasmid, suggest that TolC tunnels in the mutant strains are in a constitutively open state, thus allowing antibiotic influx.
TABLE 4.
Antibiotic sensitivities of a bacterial strain secreting hemolysina
| Antibiotic | Zone of inhibition (mm) of indicated strain
|
|
|---|---|---|
| MC4100(pACYC184) | MC4100(pHlyCABD+) | |
| Novobiocin (30 μg) | 7.6 | 12.3 |
| Erythromycin (15 μg) | 7.2 | 8.6 |
| Rifampin (5 μg) | 8.8 | 11.4 |
| Vancomycin (75 μg) | 7.5 | 18.0 |
The diameter of the paper disks was 6.5 mm. Averages from three independent experiments are shown.
Intragenic suppressors of TolCR367H.
Because the hypersensitivity phenotype resulting from R367H is thought to be the result of constitutively open tunnel apertures, second-site alterations within TolC that narrow the tunnel passage would be expected to reduce antibiotic sensitivity. With this in mind, we isolated drug-resistant revertants of a strain expressing TolCR367H. For facilitation of the isolation of intragenic suppressors, the plasmid expressing the mutant TolCR367H protein was remutagenized. Drug-resistant colonies were identified by replica plating of transformants onto novobiocin-containing medium. Plasmids isolated from colonies that grew on the antibiotic medium were reintroduced into a tolC null strain, and transformants were examined for the novobiocin resistance phenotype. Of five independently mutagenized plasmid pools, one rendered two isolates that grew on novobiocin plates upon reintroduction into the tolC null background. DNA sequence analysis revealed that in one case, a true reversion, i.e., H367R, restored the wild-type efflux activity. In the second revertant, however, a compensatory alteration mapping at position 140 of TolC resulted in a T140A substitution.
As expected, the T140A suppressor alteration significantly diminished the hypersensitivity phenotype of TolCR367H (Fig. 3). Additionally, T140A reduced the TolCR367H-mediated Dex+ phenotype and sensitivities to vancomycin and rifampin (Table 3). The specificity of suppression was studied by introducing T140A into the TolCR390C, TolCI106N, and TolCS350F backbones. Remarkably, the presence of the T140A substitution suppressed the antibiotic hypersensitivity phenotype of all mutant TolC proteins (Fig. 3C and D). As observed with TolCR367H, T140A also reduced vancomycin and rifampin sensitivities and the Dex+ phenotype of TolCR390C (Table 3). A much stronger suppression was seen for TolCR367H and TolCR390C than for TolCI106N and TolCS350F, which also confer assembly defects (37). Interestingly, the observed phenotypic suppression of TolCI106N and TolCS350F by T140A (Fig. 3) was not due to the correction of their assembly defects (Fig. 2).
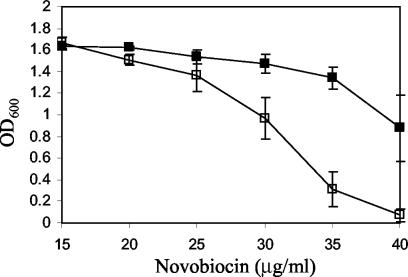
We also examined the effect of T140 on TolCR367H-mediated hypersensitivity in AcrA− AcrB+ and AcrA+ AcrB− backgrounds to see whether T140A imposed its effect independent of the AcrAB proteins. Although the absence of AcrA or AcrB exacerbated the hypersensitivity phenotype of TolCR367H, the presence of T140A only slightly reduced the antibiotic sensitivity (Table 5). Therefore, while the effect of T140A appeared to be somewhat independent of the AcrAB proteins, the bulk of the T140A-mediated reversal of antibiotic sensitivity still relies on the presence of these proteins. It is interesting that the absence of wild-type TolC in an AcrA− or AcrB− background slightly increased antibiotic sensitivities, indicating that TolC to some degree can interact with other efflux pump proteins to remove antibiotics. The T140A-mediated reversal of antibiotic resistance is, however, unlikely to be the result of a preferred interaction of the mutant TolC protein with these other efflux pump proteins, but rather the consequence of impeded antibiotic influx (Table 3). This was further supported by our observation that the presence of T140A in an otherwise wild-type TolC protein backbone increased the MIC of novobiocin (Fig. 4). Together, these observations are consistent with the notion that T140A reduces antibiotic sensitivities not by “fixing” a specific mutational defect, since no suppression specificity was observed, but rather by affecting the tunnel conformation in a general manner.
TABLE 5.
Effect of T140A in the absence of AcrA or AcrBa
| TolC protein | Zone of inhibition (mm) in presence of antibiotic
|
|||
|---|---|---|---|---|
| AcrA− AcrB+
|
AcrA+ AcrB−
|
|||
| Erythromycin (15 μg) | Novobiocin (30 μg) | Erythromycin (15 μg) | Novobiocin (30 μg) | |
| TolC+ | 16 | 18 | 16 | 19 |
| TolC− | 18 | 21 | 16 | 20.5 |
| TolCR367H | 25 | 21 | 25.5 | 20 |
| TolCR367H,T140A | 23 | 18.5 | 22 | 18.5 |
The diameter of the paper disks was 6.5 mm. Averages from three independent experiments are shown.
FIG. 4.
Growth of cultures expressing either wild-type TolC (□) or TolCT140A (▪) in LB-novobiocin. Growth conditions were identical to those described for Fig. 3. Error bars represent the standard deviations of the means of four independent experiments.
Extragenic suppressors of TolCR367H.
We also sought extragenic suppressors of TolCR367H to explore whether alterations in AcrA and AcrB can reverse the hypersensitivity phenotype. Antibiotic-resistant revertants were obtained by plating about 109 cells from four independent cultures expressing TolCR367H on LBA plates supplemented with novobiocin and erythromycin (both at 20 μg/ml); Two antibiotics were used to avoid the isolation of mutations mapping in their target genes. Revertants arose at a frequency of about 10−7. All mutations were localized in the chromosome since none moved with the plasmid. To find mutations confined to the acrAB region, we used Tn10 linked to this locus (30% by P1 transduction). In 2 mutants, of 16 examined from four independent cultures, the suppressor mutation was linked to Tn10 and hence was in the vicinity of the acrAB genes. The acrAB region from these isolates was amplified by PCR and subjected to DNA sequence analysis. In one case, we found an insertion mutation caused by the transposition of an IS2 element 83 bases upstream of the acrA start codon. This location of the IS2 disrupts the two divergent but overlapping promoters that transcribe the acrR and acrAB genes. The acrR gene is divergently transcribed from acrAB and encodes a repressor to downregulate their expression (19). The second isolate had a base pair substitution (an A-to-C transversion) 30 bases upstream of the start codon of acrR. Because neither mutation was mapped in the coding region, it is likely that they affected the expression of the acrR and/or acrAB genes.
Western blot analysis showed that strains carrying these mutations increased AcrA levels more than twofold the level expressed from the wild-type chromosomal copy (Table 6). This finding further supports the notion that acr mutations presumably uncouple the AcrR-mediated repression of acrAB. The resulting overexpression of AcrAB suppressed the hypersensitivity phenotype of TolCR367H. We verified this assertion by two additional means. First, we deleted the chromosomal copy of the acrR gene and found that this suppressed the hypersensitivity phenotype of TolCR367H (Table 6). Second, we cloned the wild-type acrAB genes into a low-copy-number vector, pACYC184, and found that the presence of this plasmid in an otherwise acrAB+ background also suppressed the hypersensitivity phenotype of TolCR367H (Table 6). As expected, both constructs produced elevated AcrA levels (Table 6). acr::IS2-, acr6-2-, ΔacrR-, and pacrAB+-mediated suppression was also observed for the other three TolC mutants (data not shown). This lack of specificity is because the suppression mechanism did not involve structural alterations among the interacting proteins, but rather elevated expression of the efflux pump proteins. It is important that unlike the T140A intragenic suppressor, these suppressors reversed hypersensitivity for only novobiocin and erythromycin, which are substrates of the TolC-AcrAB pump, but not for rifampin and vancomycin, which do not appear to be substrates of this pump.
TABLE 6.
Suppression of antibiotic sensitivity and relative AcrA levels
| Relevant strain characteristic(s) | Zone of inhibition (mm) in presence of antibiotica
|
Relative AcrA levelb | |||
|---|---|---|---|---|---|
| Novobiocin (30 μg) | Erythromycin (15 μg) | Rifampin (5 μg) | Vancomycin (75 μg) | ||
| TolCR367HacrAB+ | 10.0 | 11.4 | 18.5 | 19.0 | 1.00 |
| TolCR367Hacr::IS2 | 8.1 | 8.3 | 17.8 | 19.0 | 2.38 |
| TolCR367Hacr6-2 | 7.8 | 7.4 | 17.0 | 20.3 | 2.15 |
| TolCR367H ΔacrR::Kmr | 8.5 | 9.7 | 18.3 | 18.0 | 1.69 |
| TolCR367H(pACYC184) | 11.3c | 11.4 | 18.8 | 17.8 | 1.16 |
| TolCR367H pacrAB+ | 7.9 | 7.5 | 17.8 | 20.6 | 6.47 |
The diameter of the paper disks was 6.5 mm. Averages from three independent experiments are shown.
AcrA levels were determined from whole-cell extracts by Western blot analysis.
The presence of pACYC184 vector DNA consistently increased the sensitivity to novobiocin.
DISCUSSION
Drug-sensitive TolC mutants.
Two different classes of TolC mutants whose expression produced antibiotic sensitivity were isolated. For one class, represented by those bearing a R367H or R390C substitution, the antibiotic sensitivity was presumably caused by an effect on TolC's tunnel aperture so as to leave it in a constitutively open state. As a result of this, more antibiotics entered the cell than were expelled by cellular efflux activity. Remarkably, these TolC mutants also displayed sensitivity to a large hydrophilic antibiotic, vancomycin. This antibiotic normally cannot cross the outer membrane of a wild-type strain or even that of a deep rough LPS mutant in which the outer membrane permeability barrier is highly compromised. The drug sensitivity of the second class of TolC mutants, bearing either an I106N or S350F substitution, appeared to be the result of defective mutant TolC assembly (37).
TolC mutants resemble Dex+ porin mutants.
The data presented in this study suggest that the hypersensitivity phenotype of TolCR367H and TolCR390C was due to a larger influx of antibiotics through the constitutively open mutant tunnels rather than to a gross outer membrane permeability defect. At about 35 Å, the internal diameter of the TolC channel is much wider than that of porins (10 to 12 Å), but it does not behave as a general porin because its tunnel aperture on the periplasmic side tapers to an estimated diameter of 3.9 Å in the closed resting state (18). The opening of the TolC's tunnel aperture is proposed to require the uncoiling and realignment of tunnel entrance helices during substrate translocation (18). It is thought that in the resting stage, several critical interactions, including a salt bridge between R367 and D153 of two neighboring monomers, keep the tunnel aperture in the closed state. Given its importance in the closed state, substitutions at position 367, obliterating the salt bridge or hydrogen bonding capacity, leave the tunnel entrance in a quasi open state even in the absence of the interacting proteins of the efflux and transport systems. Thus, antibiotics gain unimpeded access through open TolC tunnels. Indeed, this access occurs even in the case of the wild-type TolC protein when it is engaged in translocating hemolysin (Table 4). The role of R390 in influencing the tunnel entrance is less clear. Although R390 of helix 8 is not located at TolC's tunnel aperture, its close vicinity to the aperture and the potential for it to form a salt bridge with D121 of helix 3 of the same monomer (Fig. 1B) could influence the tunnel entrance by affecting the movement and/or realignment of helices surrounding the aperture (Fig. 1). An R390C alteration, like those at R367, confers sensitivity to the large hydrophilic antibiotic vancomycin, suggesting that it too forces the tunnel aperture to a constitutively open state. Besides permitting the entry of large antibiotics through the open tunnel, these TolC mutants also display a Dex+ phenotype.
The permeability phenotype of TolCR367H and TolCR390C mutants due to a high level of influx is similar to that of the classical OmpC and OmpF Dex mutants, for which increased antibiotic sensitivity and the Dex+ phenotype were the result of functionally enlarged porin channels (2, 21). In these mutants, alterations either deleted residues of loop 3, which folds inward to constrict the channel passage, or affected residues that restricted loop 3 movements through electrostatic interactions. In contrast to TolCR367H and TolCR390C mutants, which showed an increased sensitivity to both hydrophilic and hydrophobic antibiotics, the Dex+ porin mutants showed an increased sensitivity primarily to hydrophilic antibiotics. This is presumably reflective of a fundamental structural difference between the two barrels; the TolC barrel is mainly engaged in the removal of hydrophobic inhibitors from the cells, while the porin barrel lets small hydrophilic molecules diffuse through it. The conversion of FhuA and FepA, which are substrate-specific and TonB-dependent OMP transporters, to general diffusion OMPs has also been reported (17, 31). These transporters do not allow general solute diffusion because of the existence of a large plug domain that blocks the inside of the barrels (4, 10). However, deletions that shorten the plug domain do allow the nonspecific diffusion of a variety of compounds, including novobiocin and rifampin (17, 31).
Assembly-defective TolC mutants and antibiotic sensitivity.
Unlike the case for TolCR367H and TolCR390C, the antibiotic sensitivity of TolCI106N and TolCS350F is primarily due to their aberrant assembly (37). I106 is part of a hydrophobic cluster, involving the mixed α/β-domain of each monomer, which packs against the helical domain of the same monomer (Fig. 1B). The replacement of I106 with a hydrophilic asparagine residue most likely destabilizes this helical packing, which not only interferes with the folding of monomers during assembly (37) but may also influence the functional activity of assembled TolC molecules. Interestingly, L412, which is also a component of the same hydrophobic cluster as I106 (Fig. 1B), has been previously noted to be important for TolC's efflux function (39, 40). Due to its location on the outer equatorial domain of TolC, the hydrophobic cluster most likely influences the overall conformation of TolC's periplasmic helical domain. The S350F substitution disrupts a hydrogen bond between the outer helices of neighboring monomers (Fig. 1B), which principally interferes with the oligomerization of TolC monomers into functional trimers (37).
T140A-mediated suppression of the hypersensitivity phenotype.
The hypersensitivity phenotypes of TolCR367H and TolCR390C were significantly diminished in the presence of the intragenic suppressor mutation T140A. Although the mutant and suppressor alterations are located at or near the tunnel aperture facing the periplasm (Fig. 1B), there is no obvious potential for direct interaction between the side chains of these residues because of the angle and long distances separating them when the aperture is in the resting stage (Fig. 1B). Moreover, since the presence of alanine at position 140 precludes any side chain interaction, it appears that the suppressor phenotype is due to the loss of a T140-mediated interaction. However, the crystal structure of TolC in its resting stage fails to reveal any T140-mediated interactions, so the role of this position in modulating the aperture conformation remains unclear. Because the T140A suppressor did not display any allele specificity and imposed its effect even on wild-type TolC, it probably acted in a general manner to diminish flux properties.
The presence of T140A reduced the hypersensitivity phenotype of TolCR367H and TolCR390C not only against the antibiotics novobiocin and erythromycin, which are substrates of the TolC-AcrAB efflux pump, but also against vancomycin, which does not appear to be removed by this pump. This general reduction in antibiotic sensitivity suggests that T140A most likely changes the conformation of helices guarding the tunnel aperture so as to reduce the influx of all antibiotics rather than to enhance a selective efflux activity of the TolC-AcrAB pump.
It is interesting that members of our laboratory previously reported the isolation of the T140A substitution among TolC mutants that secreted enzymatically inactive α-hemolysin without any apparent efflux defect (35). This pointed to a role for T140 in influencing TolC's export activity. The isolation of T140A from two independent genetic screens involving functionally distinct mutants reflects the overall importance for T140 in TolC's tunnel function. Our contention that T140A somehow narrows the tunnel entrance is consistent with the imposed phenotype in which both the export of hemolysin and the influx of antibiotics are reduced. Ironically, while T140A suppresses the R367H- and R390C-mediated hypersensitivity phenotype, R367H and R390C suppress the T140A-mediated hemolysin secretion defect (data not shown). This is entirely consistent with our view that these alterations modulate TolC's tunnel passage to either decrease (T140A) or enhance (R367H and R390C) flux properties.
The examination of position 140 in 30 different TolC homologues, which can be broadly divided into two groups based on percent identities to E. coli TolC, revealed that threonine occupies this position in all 15 sequences of the group that have identities ranging from 43 to 100%. Curiously, however, alanine is mainly found at position 140 (11 of 15) of the second group of TolC homologues, which have sequence identities in the 28 to 36% range. The natural existence of alanine raises the possibility that the suppressor alteration of T140A in our E. coli K-12 TolC mutants may emulate a structural element intrinsic to the second group of TolC homologues.
Overexpression of AcrA and AcrB suppresses the hypersensitivity phenotype of TolC mutants.
The transposition of an IS2 element and a base pair substitution in two different suppressors reduced the hypersensitivity phenotype of TolC mutants. Both mutations were located between the acrR and acrAB genes, which are divergently transcribed (19). A protein analysis showed elevated AcrA levels, which is likely due to the uncoupling of AcrR-mediated repression of acrAB transcription. The enhanced expression of AcrAB due to an insertion element-mediated insertional inactivation of the acrR gene has been reported previously (15). That the suppression was achieved as a result of the overexpression of AcrA and AcrB was independently corroborated by the construction of a multicopy acrAB+ plasmid and the deletion of the chromosomal acrR gene. Both constructs suppressed the hypersensitivity phenotype of TolC mutants. All suppressors reduced hypersensitivity against novobiocin and erythromycin, which are substrates of the TolC-AcrAB pump, but not against rifampin and vancomycin, which are not removed by this pump. This is distinct from the intragenic suppressor T140A, which could suppress the hypersensitivity phenotype against all antibiotics. This difference underscores the mechanistic distinction through which the two types of suppressors achieve their feat: the intragenic suppressor does so by reducing the influx of all antibiotics, while the extragenic and multicopy suppressors reduce antibiotic sensitivity by facilitating the efflux of TolC-AcrAB pump-specific antibiotics.
Acknowledgments
We thank Leanne Misra for critically reading the manuscript. We are grateful to Bob Kadner for valuable suggestions and to Hiroshi Nikaido for providing AcrA antibodies.
This work was supported by grants from the NIH (R01-GM066988 and R01-GM48167) to R.M. A.M.A. was supported by the BREU program.
REFERENCES
- 1.Anderson, C., E. Koronakis, E. Bokma, J. Eswaran, D. Humphreys, C. Hughes, and V. Koronakis. 2002. Transition to the open state of the TolC periplasmic tunnel entrance. Proc. Natl. Acad. Sci. USA 99:11103-11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, S. A., J. L. Occi, and B. A. Sampson. 1988. Mutations that alter the pore function of the OmpF porin of Escherichia coli K12. J. Mol. Biol. 203:961-970. [DOI] [PubMed] [Google Scholar]
- 3.Blight, M. A., A. L. Pimenta, J. C. Lazzaroni, C. Dando, L. Kotelevets, S. J. Seror, and I. B. Holland. 1994. Identification and preliminary characterization of temperature-sensitive mutations affecting HlyB, the translocator required for the secretion of haemolysin (HlyA) from Escherichia coli. Mol. Gen. Genet. 245:431-440. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 5.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 141:541-555. [DOI] [PubMed] [Google Scholar]
- 6.Cowan, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Ghosh, R. A. Paupit, J. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorman, C. J., A. S. Lynch, N. Bhriain, and C. F. Higgins. 1989. DNA supercoiling in Escherichia coli: topA mutations can be suppressed by DNA amplifications involving the tolC locus. Mol. Microbiol. 3:531-540. [DOI] [PubMed] [Google Scholar]
- 9.Eswaran, J., C. Hughes, and V. Koronakis. 2003. Locking TolC entrance helices to prevent protein translocation by the bacterial type I export apparatus. J. Mol. Biol. 327:309-315. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 11.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fralick, J. A., and L. L. Burns-Keliher. 1994. Additive effect of tolC and rfa mutations on the hydrophobic barrier of the outer membrane of Escherichia coli K-12. J. Bacteriol. 176:6404-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.German, G. J., and R. Misra. 2001. The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J. Mol. Biol. 308:579-585. [DOI] [PubMed] [Google Scholar]
- 14.Hiraga, S., H. Niki, T. Ogura, C. Ichinose, H. Mori, B. Ezaki, and A. Jaffe. 1989. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol. 171:1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawabe, T., E. Fujihira, and A. Yamaguchi. 2000. Molecular construction of a multidrug exporter system, AcrAB: molecular interaction between AcrA and AcrB, and cleavage of the N-terminal signal sequence of AcrA. J. Biochem. 128:195-200. [DOI] [PubMed] [Google Scholar]
- 17.Killmann, H., R. Benz, and V. Braun. 1996. Properties of the FhuA channel in the Escherichia coli outer membrane after deletion of FhuA portions within and outside the predicted gating loop. J. Bacteriol. 178:6913-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 19.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 20.Misra, R. 1993. A novel ompC mutation of Escherichia coli K-12 that reduces OmpC and OmpF levels in the outer membrane. Mol. Microbiol. 10:1029-1035. [DOI] [PubMed] [Google Scholar]
- 21.Misra, R., and S. A. Benson. 1988. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J. Bacteriol. 170:3611-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misra, R., M. CastilloKeller, and M. Deng. 2000. Overexpression of protease-deficient DegPS210A rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J. Bacteriol. 182:4882-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misra, R., and P. Reeves. 1987. Role of micF in the tolC-mediated regulation of OmpF, a major outer membrane protein of Escherichia coli K-12. J. Bacteriol. 169:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morona, R., and P. Reeves. 1982. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J. Bacteriol. 150:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 26.Nagel de Zwaig, R., and S. E. Luria. 1967. Genetics and physiology of colicin-tolerant mutants of Escherichia coli. J. Bacteriol. 94:1112-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-388. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:215-218. [PubMed] [Google Scholar]
- 29.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutz, J. M., J. Liu, J. A. Lyons, J. Goranson, S. K. Armstrong, M. A. McIntosh, J. B. Feix, and P. E. Klebba. 1992. Formation of a gated channel by a ligand-specific transport protein in the bacterial outer membrane. Science 258:471-475. [DOI] [PubMed] [Google Scholar]
- 32.Schnaitman, C. A., and J. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vakharia, H., G. J. German, and R. Misra. 2001. Isolation and characterization of Escherichia coli tolC mutants defective in secreting enzymatically active α-hemolysin. J. Bacteriol. 183:6908-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wandersman, C., and P. Delepelaire. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. USA 87:4746-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner, J., A. M. Augustus, and R. Misra. 2003. Assembly of TolC, a structurally unique and multifunctional outer membrane protein of Escherichia coli K-12. J. Bacteriol. 185:6540-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitney, E. N. 1971. The tolC locus of Escherichia coli K12. Genetics 67:39-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamanaka, H., H. Izawa, and K. Okamoto. 2001. Carboxy-terminal region involved in activity of Escherichia coli TolC. J. Bacteriol. 183:6961-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamanaka, H., T. Nomura, N. Morisada, S. Shioda, and K. Okamoto. 2002. Site-directed mutagenesis studies of the amino acid residues at position 412 of Escherichia coli TolC which is required for the activity. Microb. Pathog. 33:81-89. [DOI] [PubMed] [Google Scholar]
- 41.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
- 42.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 44.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]