Abstract
Cohesive interactions between Porphyromonas gingivalis and plaque-forming bacteria, such as Streptococcus oralis, are considered to play an important role in the colonization of P. gingivalis in periodontal sites. Although P. gingivalis fimbriae have been reported to mediate coaggregation with S. oralis, the S. oralis molecule involved has not been identified. We identified the coadhesin of S. oralis ATCC 9811 and purified it by affinity column chromatography. We found that the molecular mass of the purified protein was approximately 40 kDa. Dot blot and Western blot assays showed binding of the 40-kDa protein to P. gingivalis fimbriae. Further, turbidimetric assays showed that the coadhesin inhibited coaggregation between P. gingivalis and S. oralis in a dose-dependent manner. Analyses of the amino-terminal sequences of the protein and its lysyl endopeptidase-cleaved fragments revealed that the coadhesin was identical to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Next, we cloned the gene that encodes S. oralis GAPDH and found that the sequence had a high degree of homology with the sequences of GAPDHs of various bacteria, including Streptococcus gordonii and Fusobacterium nucleatum. To confirm the contribution of S. oralis GAPDH to the interaction with P. gingivalis, a recombinant GAPDH protein was generated in Escherichia coli; this protein bound to P. gingivalis fimbriae and had an inhibitory effect on coaggregation. These results suggest that S. oralis GAPDH functions as a coadhesin for P. gingivalis fimbriae. In addition, considering the high degree of homology of the GAPDHs of various bacteria, those of other plaque-forming bacteria also may contribute to the colonization of P. gingivalis.
Porphyromonas gingivalis is considered to be a prominent periodontopathogen, as it possesses a variety of virulence factors, such as proteinases, hemagglutinin, and lipopolysaccharide (28, 37, 41). In order to cause infection, it is necessary for P. gingivalis to attach to tooth surfaces, subgingival epithelium, or early colonizing gram-positive bacteria; this step constitutes the initial stage of colonization in periodontal pockets (38). In addition, the bacterium has been reported to interact with a variety of other oral gram-positive bacteria (19), including Actinomyces naeslundii (36, 43), Actinomyces viscosus (9, 12, 26, 27, 36), Streptococcus gordonii (23), Streptococcus mutans (17), Streptococcus oralis (29), and Streptococcus sanguis (39); these interactions are considered to play a vital role in the colonization of P. gingivalis in the oral cavity.
P. gingivalis possesses several cell surface components that are important for its attachment. Its fimbriae have been shown to interact with epithelial cells (15), cultured human fibroblasts (20), and saliva-coated hydroxyapatite beads (2, 32), while its vesicles have been shown to interact with collagen-coated hydroxyapatite beads (32). Further, in studies to determine which of the surface components of P. gingivalis interact with gram-positive bacteria, its fimbriae have been reported to interact with A. viscosus (12) and S. gordonii (24); its vesicles have been reported to interact with A. naeslundii (8), A. viscosus (13), and S. mutans (17); and its hemagglutinin has been reported to interact with A. viscosus (34).
On the other hand, there are only a few reports concerning the cell surface components of gram-positive bacteria that interact with P. gingivalis. Jenkinson and Demuth (16) reported that the outer membrane adhesins (SspA and SspB) of S. gordonii are related to its interaction with P. gingivalis. In addition, Yamaguchi et al. (43) found that the glycoprotein of A. naeslundii (AnAF) mediates its coaggregation with P. gingivalis. However, little is known regarding the cell components of gram-positive bacteria or the mechanism involved in their coaggregation with P. gingivalis.
In a series of studies, Nagata et al. reported that P. gingivalis 381 coaggregates with S. oralis ATCC 9811 and that this activity was inhibited by human saliva and serum and especially human fibrinogen (29, 30). The results also indicated that an arginine residue may be involved in the coaggregation (30) and that P. gingivalis 381 fimbriae mainly are responsible for the interaction with S. oralis ATCC 9811, with several domains in the carboxy terminus of fimbrillin being involved in the interaction (3). In the present study, we isolated and purified a surface component of S. oralis ATCC 9811 that was shown to bind to major fimbriae of P. gingivalis 381 and demonstrated that it is glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis 381 was obtained from stocks at the Research Laboratories of Oral Biology, Sunstar Inc., Osaka, Japan. The strain was maintained by transfer to plates containing Trypticase soy agar (BBL Microbiology Systems, Cockeysville, Md.) supplemented with 5% sheep blood, 1 mg of yeast extract (BBL)/ml, 5 μg of hemin/ml, and 1 μg of menadione/ml at 2-week intervals. The bacterial cells were cultured in prereduced Trypticase soy broth (BBL) containing 1 mg of yeast extract (BBL)/ml, 5 μg of hemin/ml, and 1 μg of menadione/ml in anaerobic system 1024 (Forma, Marietta, Ohio) in an atmosphere of 80% N2- 10% CO2- 10% H2. The cultures were incubated anaerobically at 35°C for 24 h. S. oralis ATCC 9811 was maintained as a frozen stock culture, and the bacterial cells were cultured in brain heart infusion broth (BBL) for 15 h at 35°C in air. Bacteria were harvested by centrifugation (high-speed refrigerated centrifuge SRX-201; Tomy Seiko Co. Ltd., Tokyo, Japan) at 5,000 × g for 30 min at 4°C, washed three times with 20 mM phosphate buffer containing 0.15 M NaCl (phosphate-buffered saline [PBS] [pH 6.0]), and suspended in the same buffer. Escherichia coli JM109 and BL21 (Gibco BRL, Gaithersburg, Md.), which served as host cells for the expression of recombinant GAPDH (rGAPDH), were cultured in Luria-Bertani broth (Gibco BRL) or on Luria-Bertani agar containing 50 μg of ampicillin/ml.
Identification of S. oralis components that bind to P. gingivalis fimbriae.
S. oralis molecules that bind to P. gingivalis recombinant fimbrillin (rFimA) prepared according to a previously reported method (31) were identified by the method of Lamont et al. (25). Briefly, S. oralis ATCC 9811 cells were washed in buffered KCl (5 mM KCl, 2 mM K2HPO4, 1 mM CaCl2 [pH 6.0]) and suspended in 0.1 M NaHCO3 (pH 8.1). N-Hydroxysuccinimidobiotin (Sigma Chemical Co., St. Louis, Mo.) was added to make a final concentration of 3 mg/1010 cells. After incubation for 3 h at room temperature, the cells were recovered by centrifugation (10,000 × g, 10 min) and then washed twice and resuspended in buffered KCl. The bacterial cells were mildly ultrasonicated in an ice bath for 1-min intervals with an ultrasonic disrupter (UR-200P; Tomy Seiko, Tokyo, Japan) emitting 200 W and were allowed to cool for 1 min between ultrasonic treatments. Following low-speed centrifugation (5,000 × g, 30 min), the biotinylated S. oralis extracts (150 μg) were incubated with rFimA (50 μg) for 2 h at room temperature. Rabbit anti-rFimA immunoglobulin G (IgG) (1:500; Hokkaido Systems Science Co., Ltd., Sapporo, Japan) was added, and the mixture was incubated for 30 min at room temperature. Next, the mixture was incubated with 10 μg of Affi-Gel protein A-agarose beads (Bio-Rad Laboratories, Hercules, Calif.) at 4°C for 1 h. Antibodies bound to the beads were removed by centrifugation (2,500 × g, 5 min) and washed four times in Affi-Gel protein A MAPS II binding buffer (Bio-Rad). The bound molecules were eluted with Affi-Gel protein A MAPS II elution buffer (Bio-Rad) and subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) as described below. Proteins separated by SDS-PAGE were electrotransferred by using a semidry transfer system (Trans-Blot; Bio-Rad) to a nitrocellulose membrane (Trans-Blot transfer medium; 0.2-μm-pore size; Bio-Rad). After blocking was done with Block Ace (casein solution prepared from homogenized milk; Snow Brand Co., Ltd., Sapporo, Japan) for 1 h at room temperature, the membrane was incubated with 1:2,000 avidin-horseradish peroxidase (HRP) conjugate (Bio-Rad) for 1 h at room temperature. After washing with PBS containing 0.05% Tween 20, biotinylated S. oralis components bound to rFimA were visualized by using an HRP conjugate substrate kit (Bio-Rad).
Purification of S. oralis coadhesin.
S. oralis ATCC 9811 cells were mildly ultrasonicated as described above. Following low-speed centrifugation (5,000 × g, 30 min, 4°C), the cell wall components were pelleted by centrifugation (25,000 × g, 30 min, 4°C), the pellet was dissolved with 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid (CHAPS; Wako Pure Chemical Industries, Ltd., Osaka, Japan), and a supernatant was obtained by centrifugation (25,000 × g, 30 min, 4°C). Ammonium sulfate was added to the supernatant to 30% saturation, and the mixture was stirred gently for 3 h at 4°C. The precipitated proteins were pelleted by centrifugation (25,000 × g, 30 min, 4°C). After dialysis against 50 mM Tris-HCl containing 2 M urea (pH 7.5), the samples were applied to a CNBr-activated Sepharose 4B affinity column (Amersham Pharmacia Biotech AB, Uppsala, Sweden) coupled with P. gingivalis rFimA. rFimA coupling was conducted according to the manufacturer's instructions, and the bound component was eluted with Tris-HCl containing 0.5 M NaCl.
Dot blot assay.
The binding of S. oralis components to rFimA was determined by using a dot blot assay as previously reported but with a slight modification (21). The sample (20 μg) was first adsorbed to a nitrocellulose membrane, and then the membrane was blocked with Block Ace for 1 h at room temperature and incubated with 1 mg of rFimA/ml in PBS at 4°C overnight. After three washes with PBS containing 0.05% Tween 20, the membrane was incubated with 1:1,000 rabbit anti-rFimA antibodies at 4°C overnight. After being washed, the membrane was incubated with 1:2,000 HRP-conjugated goat anti-rabbit IgG (heavy plus light chains; Zymed Laboratories Inc., San Francisco, Calif.) for 1 h at room temperature. rFimA bound to S. oralis components was visualized by using an HRP conjugate substrate kit.
Western blot assay.
The molecular mass of the purified protein was determined by SDS-PAGE, and its binding to P. gingivalis rFimA was confirmed by a Western blot assay. The purified sample was subjected to SDS-PAGE and transferred to a nitrocellulose membrane, after which the membrane was blocked with Block Ace for 1 h at room temperature and incubated with 1 mg of rFimA/ml in PBS at 4°C overnight. rFimA bound to the purified sample was detected by the same method as that described above for the dot blot assay, except with 1:400 anti-rFimA antibodies. A low-molecular-mass calibration kit for SDS electrophoresis (phosphorylase b, 97 kDa; albumin, 66 kDa; ovalbumin, 45 kDa; carbonic anhydrase, 30 kDa; trypsin inhibitor, 20.1 kDa; and α-lactalbumin, 14.4 kDa; Amersham Pharmacia Biotech, Buckinghamshire, England) was used to estimate molecular masses. Prestained SDS-PAGE standards (low range; phosphorylase b, 112 kDa; bovine serum albumin, 81 kDa; ovalbumin, 49.9 kDa; carbonic anhydrase, 36.2 kDa; soybean trypsin inhibitor, 29.9 kDa; and lysozyme, 21.3 kDa; Bio-Rad) were used for molecular mass calibration for the Western blot assay.
Coaggregation assay.
A coaggregation assay was performed by a previously reported method (29). In brief, P. gingivalis 381 and S. oralis ATCC 9811 cells were harvested and then washed with PBS (pH 6.0). Suspensions of P. gingivalis 381 and S. oralis ATCC 9811(each 0.5 ml and containing 5 × 108 cells) were mixed with the same buffer for a total volume of 2 ml in a cuvette. The progress of coaggregation was monitored at 37°C by measurement of the decrease in the A550 recorded by using a UV-visible recording spectrophotometer (UV-265FW; Shimadzu Co., Kyoto, Japan), and dA/dt was continuously calculated following the decrease in the A550. Coaggregation activity was calculated by subtracting the dA/dt of P. gingivalis 381 alone from the maximum dA/dt when both bacteria were present. For an inhibition assay, the inhibition of coaggregation was calculated by using the following equation: percent inhibition = [(coaggregation activity without inhibitor − coaggregation activity with inhibitor)/coaggregation activity without inhibitor] × 100.
Protein sequence analysis.
For amino-terminal sequence analysis, the purified protein was separated by electrophoresis and electrotransferred to a polyvinylidene difluoride membrane (Bio-Rad). In order to obtain the internal sequences, the purified protein was digested in gel with lysyl endopeptidase (Wako). The protein then was analyzed by SDS-15% PAGE and visualized by staining with 0.1% Coomassie brilliant blue R-250 (CBB) in 40% methanol-10% acetic acid. The gel was destained by treatment with 40% methanol-1% acetic acid. The stained band was cut out from the gel and placed into a microcentrifuge tube. The gel slice was washed (40% methanol for 10 min, 50 mM Tris-HCl [pH 8.5]- methanol [1:1] for 10 min, and 0.1 M Tris-HCl [pH 9.0] for 10 min) and dried by using a Speed-Vac (Savant Instruments, Holbrook, N.Y.) under vacuum conditions. The dried gel, which contained approximately 1 μg of protein, was digested with 250 ng of lysyl endopeptidase in 100 μl of 0.1 M Tris-HCl (pH 9.0)-0.05% Triton X-100 at 37°C for 16 h. After digestion, the reaction buffer was inactivated with 60% acetonitrile in 1% trifluoroacetic acid (TFA). The entire supernatant was analyzed by reverse-phase high-pressure liquid chromatography (Hitachi Co. Ltd., Tokyo, Japan) with a C18 column (Vydac 218TP5215; 2.1 by 150 mm) that was equilibrated with 0.1% TFA in high-pressure liquid chromatography-grade water and developed with a linear gradient of 0 to 60% acetonitrile in 0.1% TFA over 60 min at a flow rate of 1 ml/min at room temperature. The peaks were manually collected. The amino-terminal sequences of the purified coadhesin and its lysyl endopeptidase-cleaved fragments were determined by Edman degradation with a Procise-cLC sequencer (PE Biosystems, Branchburg, N.J.) operated according to the manufacturer's protocol.
Cloning of the gene encoding S. oralis GAPDH.
The amino-terminal sequences of the S. oralis coadhesin and its lysyl endopeptidase-cleaved fragments were aligned with those of GAPDH proteins from Streptococcus pyogenes, Streptococcus equisimilis, Streptococcus pneumoniae, and S. gordonii by using Clustal W from DDBJ (http://srs.ddbj.nig.ac.jp). DNA primers for the S. oralis GAPDH gene were determined from sequences with a high degree of homology in the amino- and carboxy-terminal regions of the above-listed GAPDHs and were as follows (both obtained from SIGMA Genosys Japan, Ishikari, Japan): forward primer, Sof1 (5′-AGTTCTGTTGAAAGG-3′); and reverse primer, Sor1 (5′-CGAAAAAGAACTCAGC-3′). The DNA fragment coding for S. oralis GAPDH was amplified by PCR (Gene Amp 2400 apparatus; PE Biosystems), and template DNA from S. oralis was purified by using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.) according to the manufacturer's instructions. PCR was performed with reaction mixtures containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 1.0 μM primer, 10 ng of template DNA, and 0.025 U of AmpliTaq Gold DNA polymerase in a final volume of 100 μl. The following cycling program was run: preheating at 94°C for 9 min and 33 cycles at 94°C for 45 s, 50°C for 1 min, and 72°C for 8 min. The gel-purified PCR product (1,143 bp) was ligated into vector pCR2.1-TOPO (TA cloning kit; Invitrogen, Carlsbad, Calif.) according to the manufacturer's protocol. The plasmid was transformed into E. coli TOP10 one-shot chemically competent cells. The presence of a DNA insert in white recombinant colonies obtained after transformation was screened by PCR amplification with primers Sof1 and Sor1 and by digestion with restriction enzyme EcoRI (New England Biolabs, Beverly, Mass.).
The nucleotide sequence of S. oralis GAPDH was amplified with vector-specific primers M13Forward (−20) (5′-GTAAAACGACGGCCAG-3′) and M13Reverse (5′-CAGGAAACAGCTATGAC-3′) and combinations of the following primers corresponding to the individual cloned sequences (all obtained from SIGMA Genosys Japan): Sof2 (5′-GTAGTTAAAGTTGGTATT-3′), Sof3 (5′-CGTTTCGACGGTACT-3′), Sor2 (5′-ACCGTCAACGTCAAGAA-3′), and Sor3 (5′-GCAGCACCAGTTGA-3′). Plasmid DNA was prepared with a plasmid minikit according to the manufacturer's instructions (Qiagen Inc., Valencia, Calif.). Each sequencing reaction was assembled by using a DYEnamic ET terminator cycle sequencing kit (Amersham Biosciences, Piscataway, N.J.) and contained 0.2 pmol of plasmid DNA, 5 pmol of primer, and 8 μl of sequencing reagent premixture in a final volume of 20 μl. The following cycling program was run: preheating at 95°C for 1 min and 25 cycles at 95°C for 20 s, 50°C for 15 s, and 60°C for 60 s. SigmaSpin postreaction purification columns (Sigma Chemical Co.) were used for removing small molecules, including unincorporated dyes or nucleotides and salts from the sequencing reactions, according to the manufacturer's instructions. The nucleotide sequence was determined by using an ABI Prism 310 genetic analyzer (PE Biosystems).
Southern hybridization.
Southern blot analysis was performed with digoxigenin (DIG) High Prime DNA labeling and detection starter kit II (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Briefly, with primers Sof2 and Sor2 (25 ng/ml), the gel-purified 909-bp gene was prepared as a probe with DIG-labeled deoxynucleoside triphosphates. S. oralis genomic DNA (4 μg) was digested overnight at 37°C in eight replications each with restriction enzymes BamHI, EcoRI, HindIII, KpnI, PstI, SacI, SalI, SmaI, SphI, and XbaI (New England Biolabs), which were selected so as not to cut into the region of the S. oralis GAPDH sequence. After each reaction product was subjected to electrophoresis, the gel was blotted by capillary transfer to a positively charged nylon membrane in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight and fixed by exposure to UV light. The membrane then was prehybridized at 42°C for 30 min with 10 ml of DIG Easy Hyb buffer and hybridized at 42°C overnight with 3.5 ml of DIG Easy Hyb buffer containing the above-described DIG-labeled probe (87.5 ng). After hybridization, the membrane was rinsed twice in wash buffer A (2× SSC containing 0.1% SDS) at room temperature and twice in wash buffer B (0.5× SSC containing 0.1% SDS) at 68°C, equilibrated with wash buffer C (100 mM maleic acid containing 150 mM NaCl and 0.3% Tween 20) for 5 min, and blocked with 1% blocking reagent (100 mM maleic acid containing 150 mM NaCl) for 1 h. For the detection of hybridizing probes, anti-DIG- alkaline phosphatase (150 mU/ml) was added to fresh blocking reagent; the sample was incubated for 30 min, washed in wash buffer C for 15 min, and equilibrated in detection buffer (0.1 M Tris containing 0.1 M NaCl [pH 9.5]). Visualization of hybridizing bands was achieved by reaction with CDP-Star—disodium 2-chloro-5-{4-methoxyspiro [1,2-dioxetane-3,2′-(5′-chloro)tricycle(3,3,1,1)decan]-4-yl}-1-phenyl phosphate.
GAPDH obtained by pH-regulated secretion from S. oralis.
GAPDH also was obtained by pH-regulated secretion from S. oralis according to a method described previously for S. gordonii (29). Frozen cells were inoculated into medium containing, per liter, 20 g of Trypticase Peptone, 5 g of yeast extract, 2 g of NaCl, 0.1 g of CaCl2, 4 g of K2HPO4, 1 g of KH2PO4, 10 g of glucose, and 0.5 g of l-arginine. The culture was controlled to maintain a constant pH of 7.5 by the addition of 2 N KOH. The culture medium was centrifuged (6,000 × g, 20 min, 4°C), and the cell-free supernatant was dialyzed against 50 mM Tris-HCl (pH 7.3). The dialyzed sample was concentrated on a 30-kDa-cutoff membrane (Amicon, Millipore Corp., Bedford, Mass.), and the concentrate was applied to a Cibacron Blue Sepharose column (3.0 by 15 cm; Amersham Pharmacia Biotech) that was equilibrated with Tris-HCl (pH 7.3). Bound GAPDH was eluted with Tris-HCl (pH 7.3) supplemented with NAD+.
Preparation of recombinant S. oralis GAPDH.
S. oralis ATCC 9811 genomic DNA was isolated as described above and used as a template for amplifying the GAPDH gene by PCR. PCR primers based on the gene sequence were selected to amplify a DNA fragment encoding all of the amino acid residues of S. oralis GAPDH. The primer sequences (with restriction sequences underlined) were as follows: forward primer, 5′-GCCCATGGTAGTTAAAGTTGGTATTAACGGT-3′; and reverse primer, 5′-GCGGATCCTTATTTAGCGATTTTTGCGAAGT-3′. The forward primer incorporated the NcoI site, whereas the reverse primer incorporated the BamHI site and a translational stop site. Plasmid pET3d (Novagen Inc., Madison, Wis.) was used as the vector for the expression of rGAPDH. The expression and purification of rGAPDH were carried out as described previously (21).
Analytical methods.
SDS-PAGE (12.5% gel) was performed by the method of Laemmli (22). Gels were stained with CBB. The protein concentrations of the samples were determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.) with bovine serum albumin as a standard.
Nucleotide sequence accession numbers.
The nucleotide sequence of S. oralis GAPDH determined in this report has been deposited in the DDBJ database under accession number AB110908. The DDBJ accession numbers for S. pneumoniae TIGR4 (40), S. gordonii FSS2 (33), S. pyogenes M1 (group A streptococcus), S. equisimilis H46A (11), Staphylococcus epidermidis ATCC 12228, Staphylococcus aureus N315, and Fusobacterium nucleatum ATCC 25586 (18) are AE007490, AF247678, AE006494, X97788, AE016745, AP003360, and AE010576, respectively.
RESULTS
Identification of S. oralis components that bind to P. gingivalis fimbriae.
For identification of the S. oralis cell components involved in the interaction with P. gingivalis fimbriae, biotinylated sonic extracts of S. oralis were incubated with P. gingivalis rFimA and then with anti-rFimA antibodies. S. oralis components immunoprecipitated with rFimA and anti-rFimA were identified by using Affi-Gel protein A-agarose beads and subjected to SDS-PAGE. A band with a molecular mass of approximately 40 kDa was detected, indicating that the S. oralis coadhesin for P. gingivalis was a 40-kDa protein (data not shown).
Purification of S. oralis coadhesin.
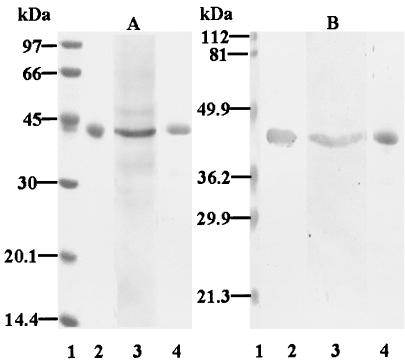
In order to purify the 40-kDa coadhesin, S. oralis ATCC 9811 cells were mildly ultrasonicated. After treatment with CHAPS and ammonium sulfate, the 40-kDa protein was purified by chromatography with a CNBr-activated Sepharose 4B affinity column coupled with P. gingivalis rFimA. The purified protein was subjected to SDS-PAGE, which revealed a single band with a molecular mass of approximately 40 kDa (Fig. 1A, lane 2).
FIG. 1.
SDS-PAGE and Western blot assay of purified S. oralis coadhesin, S. oralis GAPDH obtained by pH-regulated secretion, and S. oralis rGAPDH. Samples (5 μg) were subjected to SDS-PAGE (12.5% gels) and electrotransferred to nitrocellulose membranes. After blocking with Block Ace, the membranes were incubated with rFimA (1 mg/ml); bound rFimA was probed with rabbit anti-rFimA IgG followed by HRP-conjugated goat anti-rabbit antibodies. Bound antibodies were visualized by using an HRP conjugate substrate kit. (A) SDS-PAGE with CBB staining. (B) Western blot assay. Lanes: 1, molecular mass standard proteins; 2, purified S. oralis coadhesin; 3, S. oralis GAPDH obtained by pH-regulated secretion; 4, S. oralis rGAPDH.
Binding of S. oralis coadhesin to P. gingivalis fimbriae.
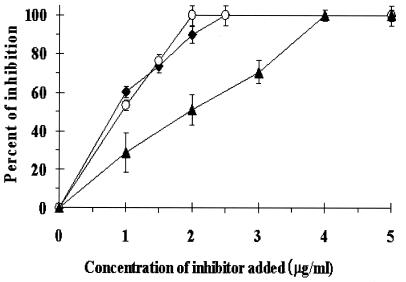
Binding of the purified S. oralis coadhesin to P. gingivalis rFimA was examined by using dot blot and Western blot assays; the coadhesin was shown to bind to P. gingivalis rFimA (Western blot assay; Fig. 1B, lane 2) (data not shown for dot blot assay). In order to examine the effect of the purified protein on coaggregation between P. gingivalis whole cells and S. oralis whole cells, an inhibition experiment was performed with a turbidimetric assay. As shown in Fig.2, the purified S. oralis coadhesin inhibited coaggregation in a dose-dependent manner and showed complete inhibition at a concentration of 2.5 μg/ml.
Binding of GAPDH obtained by pH-regulated secretion from S. oralis.
Nelson et al. (33) reported a method of purifying GAPDH from S. gordonii FSS2 by regulating pH. GAPDH obtained by the same method from S. oralis in the present study (Fig. 1A, lane 3) also bound to P. gingivalis rFimA, as shown by Western blot (Fig. 1B, lane 3) and dot blot (data not shown) assays. Further, it inhibited coaggregation between P. gingivalis 381 and S. oralis ATCC 9811 in a dose-dependent manner and showed complete inhibition at a concentration of 4.0 μg/ml (Fig. 2).
FIG. 2.
Inhibitory effects of purified S. oralis coadhesin, S. oralis GAPDH obtained by pH-regulated secretion, and S. oralis rGAPDH on coaggregation between P. gingivalis 381 and S. oralis ATCC 9811. Aliquots (0.5 ml) of suspensions of P. gingivalis 381 (5 × 108 cells) and S. oralis ATCC 9811 (5 × 108 cells) and various concentrations of inhibitors were simultaneously mixed to make a total volume of 2 ml in a cuvette. The progress of coaggregation was monitored by measurement of the decrease in the A550 at 37°C. After the decrease in the A550 was recorded, dA/dt was continuously calculated. Coaggregation activity was calculated by subtracting the dA/dt of P. gingivalis 381 alone from the maximum dA/dt when the bacteria were mixed. Symbols: ♦, purified S. oralis coadhesin; ▴, S. oralis GAPDH obtained by pH-regulated secretion; ○, S. oralis rGAPDH. Values represent means and standard deviations from three replicates.
Interaction of S. oralis rGAPDH with P. gingivalis.
Next, we generated and purified S. oralis rGAPDH. Following SDS-PAGE and CBB staining, purified rGAPDH migrated as a single component with an apparent molecular mass of 40 kDa (Fig. 1A, lane 4) and was shown to bind to P. gingivalis rFimA by Western blot (Fig. 1B, lane 4) and dot blot (data not shown) assays. rGAPDH also inhibited coaggregation between P. gingivalis 381 and S. oralis ATCC 9811 in a dose-dependent manner (Fig. 2). In addition, the inhibitory activity of rGAPDH was found to be as high as that of the purified 40-kDa S. oralis coadhesin.
Phase-contrast microscopy.
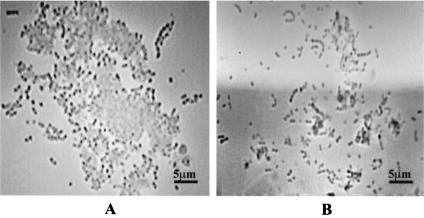
The involvement of S. oralis GAPDH in coaggregation with P. gingivalis was confirmed by phase-contrast microscopy. When P. gingivalis 381 and S. oralis ATCC 9811 were mixed at a ratio of 1:1, they demonstrated coaggregation (Fig. 3A); further, as shown in Fig. 3B, the addition of S. oralis rGAPDH (2.5 μg/ml) inhibited the coaggregation.
FIG. 3.
Phase-contrast photomicrographs of coaggregation of P. gingivalis 381 and S. oralis ATCC 9811. (A) Coaggregation of P. gingivalis 381 and S. oralis ATCC 9811 apparently has taken place. (B) The addition of S. oralis rGAPDH (2.5 μg/ml) to the mixture has inhibited coaggregation nearly completely.
Cloning and sequencing of the gene encoding the 40-kDa S. oralis coadhesin.
The amino-terminal sequence of the 40-kDa S. oralis coadhesin was determined to be VVKVGINGFGRIGXLA, and those of its lysyl endopeptidase-cleaved fragments were ALQXNFXVXEGLMTTIXAYT and VLDVDGK. A homology search for the amino acid sequence revealed that the 40-kDa coadhesin had a high degree of homology with GAPDHs of S. gordonii, S. pyogenes, S. pneumoniae, and S. equisimilis. Based on the sequences of the highly conserved regions among these streptococci, we designed the PCR primers Sof1 and Sor1, which were used to amplify the expected 1.1-kb DNA fragment; then we determined the DNA sequence (DDBJ accession number AB110908). Next, the DNA and amino acid sequences of S. oralis GAPDH were compared to those of other bacterial GAPDHs. As shown in Fig. 4, S. oralis GAPDH had a high degree of homology with GAPDHs of S. pneumoniae, S. gordonii, S. pyogenes, and S. equisimilis. In addition, its estimated amino acid sequence shared approximately 90% identity with those of S. pneumoniae, S. gordonii, S. pyogenes, and S. equisimilis and approximately 70% identity with those of S. epidermidis, S. aureus, and F. nucleatum.
FIG. 4.
Amino acid alignment of S. oralis GAPDH and other streptococcal GAPDHs. The amino acid sequence of S. oralis GAPDH was inferred from the gene sequence and compared with those of other streptococcal GAPDHs. Gap residues are indicated by dashes, residues conserved in all of the GAPDHs are indicated by asterisks, and residues conserved in three of the GAPDHs are indicated by dots. Gray boxes indicate sequences corresponding to the identified amino-terminal sequences of the purified S. oralis coadhesin and its lysyl endopeptidase-cleaved fragments.
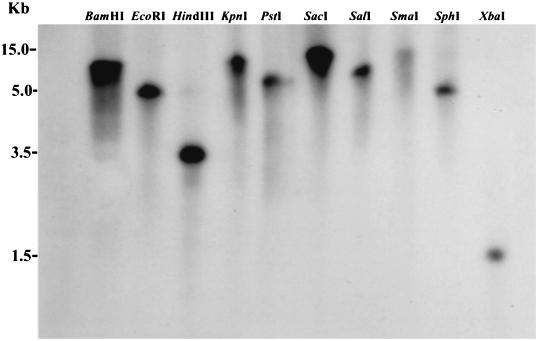
Organisms that express only a single GAPDH may possess more than a single gene (1, 14), thus, Southern hybridization was performed by using a probe consisting of only the 909-bp open reading frame amplified with Sof2 and Sor2 under high-stringency conditions at 42°C to examine whether the genome contained single or multiple copies. A Southern blot assay of the S. oralis genome revealed a single hybridizing band for 10 restriction enzyme digests (Fig. 5). These results are consistent with the existence of a single copy of the GAPDH gene. Therefore, we concluded that the GAPDH gene analyzed here was a single-copy gene of S. oralis ATCC 9811.
FIG. 5.
Copy number of the S. oralis GAPDH gene, as revealed by restriction endonuclease analysis. S. oralis DNA was digested with the indicated restriction endonucleases. Digests were subjected to electrophoresis and then blotted and probed for the GAPDH gene as described in Materials and Methods.
DISCUSSION
In a series of coaggregation studies, we have shown that P. gingivalis 381 strongly coaggregates with S. oralis ATCC 9811 (29), with the involvement of major fimbriae of P. gingivalis (3). The purpose of the present study was to identify the S. oralis coadhesin that is involved with P. gingivalis. We purified the S. oralis coadhesin for P. gingivalis fimbriae by using mild ultrasonication, CHAPS treatment, ammonium sulfate precipitation, and chromatography with an affinity column coupled with P. gingivalis rFimA; SDS-PAGE revealed that its molecular mass was approximately 40 kDa. On the basis of the amino-terminal sequences of the purified coadhesin and its lysyl endopeptidase-cleaved fragments, the S. oralis coadhesin was considered to be identical to GAPDH. Nelson et al. (33) reported that GAPDH of S. gordonii FSS2 was associated primarily with the surface during growth at pH 6.5 and then shifted to greater than 90% being secreted during growth at pH 7.5. We were able to obtain the GAPDH by pH-regulated secretion from S. oralis by using the same method.
Next, we examined the binding of the purified S. oralis coadhesin, the GAPDH obtained by pH-regulated secretion from S. oralis, and S. oralis rGAPDH to P. gingivalis rFimA. These three preparations were shown to bind to P. gingivalis rFimA by dot blot and Western blot assays and to inhibit coaggregation between P. gingivalis 381 and S. oralis ATCC 9811. These results and the high degree of DNA homology of S. oralis GAPDH with other streptococcal GAPDHs indicate that the S. oralis coadhesin is identical to GAPDH. Although GAPDH is known to be a tetrameric enzyme of the glycolytic pathway responsible for the phosphorylation of glyceraldehyde-3-phosphate to generate 1,3-bisphosphoglycerate (42), it has been reported to possess multiple binding activities. Pancholi and Fischetti (35) found that the GAPDH of group A streptococci binds to fibronectin, lysozyme, and the cytoskeletal proteins myosin and actin and noted that it may play a role in the colonization of those bacteria. Therefore, it is reasonable to speculate that S. oralis GAPDH may function as an adhesin for P. gingivalis.
As for the adhesive components of plaque-forming gram-positive bacteria involved in coaggregation with P. gingivalis, AnAF of A. naeslundii (43) and SspB of S. gordonii (5, 7, 25) have been reported to interact with the bacterium. Yamaguchi et al. (43) reported that AnAF purified from A. naeslundii KWS81 is a glycoprotein with a molecular mass in excess of 200 kDa and that a carbohydrate component mediates coaggregation with P. gingivalis. Further, Lamont et al. (25) suggested that S. gordonii SspB, with a molecular mass of 170 kDa, mediates adhesion to P. gingivalis, and it has also been shown that residues 1167 to 1193 of SspB are essential for P. gingivalis binding (5, 7). P. gingivalis has been reported to possess two distinct kinds of fimbriae, major and minor, which are different in size and antigenicity (4). Recently, it was reported that the P. gingivalis receptor for S. gordonii SspB may be a minor fimbria-protein complex with a molecular mass of 100 kDa (6); however, the S. gordonii receptor for P. gingivalis major fimbriae remains unknown. In the present study, we identified the S. oralis coadhesin that interacts with P. gingivalis major fimbriae and concluded that it is GAPDH. Chung et al. (6) postulated that the major fimbriae of P. gingivalis are required for the initial association between P. gingivalis and S. gordonii and that the binding then is stabilized by SspB-minor fimbria complex interactions. Therefore, GAPDHs of early plaque-forming bacteria, such as S. oralis, may mediate the initial adhesion of P. gingivalis in periodontal pockets.
It is well known that eubacteria and eukaryotic GAPDHs are conserved (10); this fact was confirmed by comparisons of the DNA and amino acid sequence homologies of S. oralis GAPDH with other bacterial GAPDHs. Among oral bacteria, S. oralis GAPDH had approximately 92% DNA homology with S. gordonii FSS2 GAPDH and approximately 66% homology with F. nucleatum ATCC 25586 GAPDH, indicating that GAPDHs of other oral bacteria in addition to that of S. oralis may function as adhesins for P. gingivalis. In order to elucidate the function of GAPDHs of a wide range of early plaque-forming bacteria as adhesions for P. gingivalis, further studies, including cloning of other early plaque-forming bacterial GAPDHs and identification of the binding regions within GAPDH, are necessary. However, this report is the first to show that GAPDHs of early plaque-forming bacteria may be involved in the colonization of P. gingivalis.
Acknowledgments
This work was supported by a grant-in-aid for scientific research (14370694) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Editor: V. J. DiRita
REFERENCES
- 1.Alefounder, P. R., and R. N. Perham. 1989. Identification, molecular cloning and sequence analysis of a gene cluster encoding the Class II fructose 1, 6-biphosphate aldolase, 3-phosphoglycerate kinase and a putative second glyceraldehydes 3-phosphate dehydrogenase of Escherichia coli. Mol. Microbiol. 3:723-732. [DOI] [PubMed] [Google Scholar]
- 2.Amano, A., H. T. Sojar, J.-Y. Lee, A. Sharma, M. J. Levine, and R. J. Genco. 1994. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect. Immun. 62:3372-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano, A., T. Fujiwara, H. Nagata, M. Kuboniwa, A. Sharma, H. T. Sojar, R. J. Genco, S. Hamada, and S. Shizukuishi. 1997. Prophyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J. Dent. Res. 76:852-857. [DOI] [PubMed] [Google Scholar]
- 4.Arai, M., N. Hamada, and T. Umemoto. 2000. Purification and characterization of a novel secondary fimbrial protein from Porphyromonas gingivalis strain 381. FEMS Microbiol. Lett. 193:75-81. [DOI] [PubMed] [Google Scholar]
- 5.Brooks, W., D. R. Demuth, S. Gil, and R. J. Lamont. 1997. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect. Immun. 65:3753-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, W. O., D. R. Demuth, and R. J. Lamont. 2000. Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect. Immun. 68:6758-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demuth, D. R., D. C. Irvine, J. W. Costerton, G. S. Cook, and R. J. Lamont. 2001. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 69:5736-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellen, R. P., and D. A. Grove. 1989. Bacteroides gingivalis vesicles bind to and aggregate Actinomyces viscosus. Infect. Immun. 57:1618-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellen, R. P., M. Song, and L. A. Buivids. 1992. Inhibition of Actinomyces viscosus-Porphyromonas gingivalis coadhesion by trypsin and other proteins. Oral Microbiol. Immunol. 7:198-203. [DOI] [PubMed] [Google Scholar]
- 10.Figge, R. M., M. Schubert, H. Brinkmann, and R. Cerff. 1999. Glyceraldehyde-3-phosphate dehydrogenase gene diversity in eubacteria and eukaryotes: evidence for intra- and inter-kingdom gene transfer. Mol. Biol. Evol. 16:429-440. [DOI] [PubMed] [Google Scholar]
- 11.Gase, K., A. Gase, H. Schirmer, and H. Malke. 1996. Cloning, sequencing and functional overexpression of the Streptococcus equisimilis H46A gapC gene encoding a glyceraldehyde-3-phosphate dehydrogenase that also functions as a plasmin(ogen)-binding protein. Purification and biochemical characterization of the protein. Eur. J. Biochem. 239:42-51. [DOI] [PubMed] [Google Scholar]
- 12.Goulbourne, P. A., and R. P. Ellen. 1991. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J. Bacteriol. 173:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiratsuka, K., Y. Abiko, T. Ito, H. Sasahara, and H. Takiguchi. 1992. Role of Porphyromonas gingivalis 40-kDa outer membrane protein in the aggregation of P. gingivalis vesicles and Actinomyces viscosus. Arch. Oral Biol. 37:717-724. [DOI] [PubMed] [Google Scholar]
- 14.Holland, J. P. 1983. Homologous nucleotide sequence at the 5′ termini of messenger RNAs synthesized from the yeast enolase and glyceraldehydes-3-phosphate dehydrogenase gene families. J. Biol. Chem. 258:5291-5299. [PubMed] [Google Scholar]
- 15.Isogai, H., E. Isogai, F. Yoshimura, T. Suzuki, W. Kagota, and K. Takano. 1988. Specific inhibition of adherence of an oral strain of Bacteroides gingivalis 381 to epithelial cells by monoclonal antibodies against the bacterial fimbriae. Arch. Oral Biol. 33:479-485. [DOI] [PubMed] [Google Scholar]
- 16.Jenkinson, H. F., and D. R. Demuth. 1997. Structure, function and immunogenicity of streptococcal antigen I/lI polypeptides. Mol. Microbiol. 23:183-190. [DOI] [PubMed] [Google Scholar]
- 17.Kamaguchi, A., H. Baba, M. Hoshi, and K. Inomata. 1995. Effect of Porphyromonas gingivalis ATCC 33277 vesicle on adherence of Streptococcus mutans OMZ 70 to the experimental pellicle. Microbiol. Immunol. 39:521-524. [DOI] [PubMed] [Google Scholar]
- 18.Kapatral, V., I. Anderson, N. Ivanova, G. Reznik, T. Los, A. Lykidis, A. Bhattacharyya, A. Bartman, W. Gardner, G. Grechkin, I. Zhu, O. Vasieva, L. Chu, Y. Kogan, O. Chaga, E. Goltsman, A. Bernal, N. Larsen, M. D'Souza, T. Walunas, G. Pusch, R. Haselkorn, M. Fonstein, N. Kyrpides, and R. Overbeek. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 184:2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 20.Kontani, M., H. Ono, H. Shibata, Y. Okamura, T. Tanaka, T. Fujiwara, S. Kimura, and S. Hamada. 1996. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultured human fibroblasts and matrix proteins. Infect. Immun. 64:756-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuboniwa, M., A. Amano, and S. Shizukuishi. 1998. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (Lys-gingipain). Biochem. Biophys. Res. Commun. 249:38-43. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lamont, R. J., S. G. Hersey, and B. Rosan. 1992. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol. Immunol. 7:193-197. [DOI] [PubMed] [Google Scholar]
- 24.Lamont, R. J., C. A. Bevan, S. Gil, R. E. Persson, and B. Rosan. 1993. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol. Immunol. 8:272-276. [DOI] [PubMed] [Google Scholar]
- 25.Lamont, R. J., S. Gil, D. R. Demuth, D. Malamud, and B. Rosan. 1994. Molecules of Streptococcus gordonii that bind to Porphyromonas gingivalis. Microbiology 140:867-872. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., and R. P. Ellen. 1989. Relative adherence of Bacteroides species and strains to Actinomyces viscosus on saliva-coated hydroxyapatite. J. Dent. Res. 68:1308-1312. [DOI] [PubMed] [Google Scholar]
- 27.Li, J., R. P. Ellen, C. I. Hoover, and J. R. Felton. 1991. Association of proteases of Porphyromonas (Bacteroides) gingivalis with its adhesion to Actinomyces viscosus. J. Dent. Res. 70:82-86. [DOI] [PubMed] [Google Scholar]
- 28.Mayland, D., and S. C. Holt. 1988. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol. Rev. 52:134-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagata, H., Y. Murakami, E. Inoshita, S. Shizukuishi, and A. Tsunemitsu. 1990. Inhibitory effect of human plasma and saliva on co-aggregation between Bacteroides gingivalis and Streptococcus mitis. J. Dent. Res. 69:1476-1479. [DOI] [PubMed] [Google Scholar]
- 30.Nagata, H., A. Amano, T. Hanioka, H. Tamagawa, S. Shizukuishi, and T. Miyata. 1993. Inhibition of coaggregation between Porphyromonas gingivalis and Streptococcus oralis by fibrinogen fragments. FEMS Microbiol. Lett. 114:31-36. [DOI] [PubMed] [Google Scholar]
- 31.Nagata, H., A. Sharma, H. T. Sojar, A. Amano, M. J. Levine, and R. J. Genco. 1997. Role of the carboxyl-terminal region of Porphyromonas gingivalis fimbrillin in binding to salivary proteins. Infect. Immun. 65:422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naito, Y., H. Tohda, K. Okuda, and I. Takazoe. 1993. Adherence and hydrophobicity of invasive and noninvasive strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, D., J. M. Goldstein, K. Boatright, D. W. Harty, S. L. Cook, P. J. Hickman, J. Potempa, J. Travis, and J. A. Mayo. 2001. pH-regulated secretion of a glyceraldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: purification, characterization, and cloning of the gene encoding this enzyme. J. Dent. Res. 80:371-377. [DOI] [PubMed] [Google Scholar]
- 34.Okuda, K. 1993. Attachment mechanisms and colonization, p. 139-157. In H. N. Shah, D. Mayland, and R. J. Genco (ed.), Biology of the species Porphyromonas gingivalis. CRC Press, Inc., Boca Raton, Fla.
- 35.Pancholi, V., and V. A. Fischetti. 1993. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc. Natl. Acad. Sci. USA 90:8154-8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz, S., R. P. Ellen, and D. A. Grove. 1987. Bacteroides gingivalis-Actinomyces viscosus cohesive interactions as measured by a quantitative binding assay. Infect. Immun. 55:2391-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slots, J., and R. J. Genco. 1984. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J. Dent. Res. 63:412-421. [DOI] [PubMed] [Google Scholar]
- 38.Slots, J., and R. J. Gibbons. 1978. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect. Immun. 19:254-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stinson, M. W., K. Safulko, and M. J. Levine. 1991. Adherence of Porphyromonas (Bacteroides) gingivalis to Streptococcus sanguis in vitro. Infect. Immun. 59:102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, L. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 41.van Winkelhoff, A. J., U. van der Velden, and J. de Graaff. 1987. Microbial succession in recolonizing deep periodontal pockets after a single course of supra- and subgingival debridement. J. Clin. Periodontol. 15:116-122. [DOI] [PubMed] [Google Scholar]
- 42.Winram, S. B., and R. Lottenberg. 1996. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehydes-3-phosphate dehydrogenase. Microbiology 142:2311-2320. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi, T., K. Kasamo, M. Chuman, M. Machigashira, M. Inoue, and T. Sueda. 1998. Preparation and characterization of an Actinomyces naeslundii aggregation factor that mediates coaggregation with Porphyromonas gingivalis. J. Periodontal Res. 33:460-468. [DOI] [PubMed] [Google Scholar]