Abstract
We have used representational difference analysis to identify a novel Mycobacterium avium subsp. paratuberculosis-specific ABC transporter operon (mpt), which comprises six open reading frames designated mptA to -F and is immediately preceded by two putative Fur boxes. Functional genomics revealed that the mpt operon is flanked on one end by a fep cluster encoding proteins involved in the uptake of Fe3+ and on the other end by a sid cluster encoding non-ribosome-dependent heterocyclic siderophore synthases. Together these genes form a 38-kb M. avium subsp. paratuberculosis-specific locus flanked by an insertion sequence similar to IS1110. Expression studies using Western blot analyses showed that MptC is present in the envelope fraction of M. avium subsp. paratuberculosis. The MptD protein was shown to be surface exposed, using a specific phage (fMptD) isolated from a phage-peptide library, by differential screening of Mycobacterium smegmatis transformants. The phage fMptD-derived peptide could be used in a peptide-mediated capture PCR with milk from infected dairy herds, thereby showing surface-exposed expression of the MptD protein in the host. Together, these data suggest that the 38-kb locus constitutes an M. avium subsp. paratuberculosis pathogenicity island.
Paratuberculosis, also called Johne's disease, is a severe and incurable enteritis of ruminants caused by Mycobacterium avium subsp. paratuberculosis and has a considerable economic impact on the livestock industry (21, 28). In cattle, infection most commonly occurs in newborn calves by the fecal-oral route. However, clinical symptoms, predominantly including persistent diarrhea and weight loss, are usually delayed in animals until 3 to 5 years of age (13). Paratuberculosis is prevalent in domestic animals and many species of wildlife worldwide, including primates (31). In addition, M. avium subsp. paratuberculosis has been isolated from intestinal tissue of Crohn's disease patients, and its potential role as a zoonotic pathogen is currently being investigated (12, 22).
Previous attempts to determine the molecular cause for pathogenicity of M. avium subsp. paratuberculosis have focused on the identification of antigenic proteins (16, 26) and the characterization of secreted components by using monoclonal antibodies (32). Further studies have identified an M. avium subsp. paratuberculosis-specific low-GC cassette carrying genes with putative functions related to lipopolysaccharide or extracellular polysaccharide biosynthesis (47) and have identified cross-reactive, species- and subspecies-specific epitopes (32). In addition, an M. avium subsp. paratuberculosis-specific ferric reductase which is secreted in vivo and in vitro has been described (25). Other potential virulence factors reported include a serine protease (10) and a fibronectin attachment protein (41). However, closely related homologues of these factors can be found in M. avium subsp. avium and Mycobacterium tuberculosis, and they are unlikely to be M. avium subsp. paratuberculosis specific.
In this study we have investigated the hypothesis that, as has been shown in other bacterial species, unique membrane proteins and secretory pathways are likely to be critical features involved in M. avium subsp. paratuberculosis pathogenicity (7, 17, 18, 20). We have used representational difference analysis (RDA) (30, 45, 47) to identify a 7-kb M. avium subsp. paratuberculosis-specific ABC transporter operon (mpt). Proteins from this operon were shown to contain putative membrane-spanning regions and to be expressed on the mycobacterial cell surface in vitro and in vivo. We further show that this ABC transporter operon is preceded by two putative Fur boxes and represents part of an M. avium subsp. paratuberculosis-specific 38-kb putative pathogenicity island that includes several iron uptake-related gene clusters.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and growth conditions.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1. Mycobacteria were grown on Middlebrook 7H10 agar or in Middlebrook 7H9 medium (both from Difco Laboratories, Detroit, Mich.) supplemented with oleic acid-albumin-dextrose-catalase enrichment (Difco), Tween 80 (0.05%), and kanamycin (40 μg ml−1) for Mycobacterium smegmatis transformants; for M. avium subsp. paratuberculosis culture, mycobactin (2 μg ml−1) (Synbiotics, Lyon, France) was added. Escherichia coli strains were grown in Luria-Bertani medium supplemented with appropriate antibiotics (ampicillin, 100 μg ml−1; kanamycin, 40 μg ml−1).
TABLE 1.
Characteristics of strains, phages, plasmids, and primers used in this study
| Strain, phage, plasmid, primer, peptide | Characteristics, reference, and/or sourcea |
|---|---|
| Strains | |
| M. avium subsp. paratuberculosis strain 6783 | Clinical isolate, DSM 44135 |
| 14 M. avium subsp. paratuberculosis bovine clinical isolates | Fecal and lymph node isolates from clinically diseased cattle from different herds in northern Germany |
| M. avium subsp. avium strain ATCC 25291 | DSM 44156 |
| M. smegmatis mc2155 | 42 |
| M. scrofulaceum | NCTC 10803 |
| M. bovis BCG | NCTC 5692 |
| M. bovis | NCTC 10772 |
| M. kansasii | NCTC 10268 |
| M. microti | NCTC 8710 |
| M. gordonae | NCTC 10267 |
| M. fortuitum | NCTC 10394 |
| E. coli DH5αF′ | F′/endA1 hsdR17 (rK−mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF) U169 deoR [φ80dlacΔ(lacZ)M15] (35) |
| Phage fMptD | Phage isolated from the Ph.D.-12 phage display library with the specific sequence 5′-GGG AAG AAT CAT CAT CAG CAT CAT AGG CCT CAG-3′ |
| Plasmids | |
| pUC19 | E. coli cloning vector carrying an ampicillin resistance determinant (Pharmacia) |
| pGEX5x-3 | E. coli expression vector for the construction of glutathione S-transferase fusion proteins (Pharmacia) |
| pMV361 | Integrative mycobacterial shuttle vector carrying a kanamycin resistance determinant (9) |
| pRDIII300 | Fragment RDIII300 in pUC19 (this work) |
| pRDIII301 | Fragment RDIII301 in pUC19 (this work) |
| pRDIII302 | 4-kb DpnII fragment in pGH432 obtained by hybridization with a probe derived from the 5′ end of RDIII300 (this work) |
| pRDIII303 | 4-kb DpnII fragment in pGH432 obtained by hybridization with a probe derived from the 3′ end of RDIII300 (this work) |
| pRDIII320 | Fragment RDIII300 in pMV361 (Fig. 1, inset) (this work) |
| pRDIII320Δ1 | Deletion derivative of pRDIII320 with a 3,201-bp FseI fragment deleted (Fig. 1, inset) (this work) |
| pRDIII320Δ2 | Deletion derivative of pRDIII320 with a 2,410-bp SmaI-SacI fragment deleted (Fig. 1, inset) (this work) |
| pRDIII350 | PCR fragment obtained with primers ABC3 and ABC4, cut with BamHI and EcoRI, and ligated into pGEX5x-3 |
| Primers | |
| RBam12 | 5′-GAT CCT CGG TGA-3′ (29) |
| RBam24 | 5′-AGC ACT CTC CAG CCT CTC ACC GAG-3′ (29) |
| IPIII30 | 5′-CTA TGC GCA CTG ACG CTT C-3′ (this work) |
| IPIII31 | 5′-TTC CGA AGA ATC CGA TGA G-3′ (this work) |
| ABC3 | 5′-CCG CGG ATC CGC TTA CGA CGG AGG TCA A-3′ (this work) |
| ABC4 | 5′-GCC GGA ATT CGA TGT TGA TGA GAAT CCC T-3′ (this work) |
| ISMav1 | 5′-GTA TCA GGC CGT GAT GGC GG-3′ |
| ISMav2 | 5′-CGC CAC CAG CGC TCG ATA CA-3′ |
| Peptides | |
| aMptD | Biotin-aminohexacarbonic acid-GKNHHHQHHRPQ |
| aMPr | Biotin-aminohexacarbonic acid-HSQPKQVKKASR (control peptide) |
The nucleotides in boldface indicate the restriction enzyme sites used for cloning into pGEX5x-3. DSM and NCTC strains were from the Deutsche Gesellschaft für Mikroorgansismen und Zellkulturen (Braunschweig, Germany) and the National Collection of Type Cultures (Colindale, United Kingdom), respectively.
RDA and isolation of RDA fragments.
Mycobacterial chromosomal DNA was extracted according to the method described by Bose and coworkers (6). A simplified RDA technique (45) using total genomic DNA digested with BclI was used to subtract the driver strain genome of M. avium subsp. avium ATCC 25291 from the test strain genome of M. avium subsp. paratuberculosis isolate 6783 (DSM 44135). An M. avium subsp. paratuberculosis-specific BclI fragment was identified and designated RDIII30; it was further used to identify larger fragments by Southern blot analyses of M. avium subsp. paratuberculosis DNA digested with KpnI and SacI. Hybridizing fragments were cloned and designated RDIII300 (KpnI fragment) and RDIII301 (SacI fragment). Based on the sequence information for RDIII300 and RDIII301, a third and a fourth overlapping fragment, designated RDIII302 and RDIII303, were isolated from an M. avium subsp. paratuberculosis genomic library generated by a partial DpnII digestion and ligation of fragments of 2 to 4 kb into pGH432 and pGH433 (Advanced Vectors Inc., Hopkins, Minn.). The specificity of the RDA fragment was confirmed by PCR with RDIII30-derived primers IPIII30 and IPIII31, using 14 bovine clinical M. avium subsp. paratuberculosis isolates and the reference strains of six other mycobacterial species (Table 1).
Manipulation and analysis of DNA.
Agarose gel electrophoresis, Southern blot analyses, plasmid preparation, PCR, DNA cloning, and transformation of E. coli were done by standard procedures (37). DNA-modifying enzymes were purchased from New England Biolabs (Karlsruhe, Germany). DNA sequencing was done by primer walking; primers were purchased from Invitrogen (Groningen, The Netherlands), and sequencing reactions were done by SeqLab (Göttingen, Germany). Sequencing data analyses were performed with the HUSAR 5.0 program (DKFZ, Heidelberg, Germany). Preliminary sequence data were obtained from the Institute for Genomic Research website at http://www.tigr.org. BLASTN and BLASTP engines were used through the National Center for Biotechnology Information portal (http://www.ncbi.nlm.nih.gov/BLAST). The M. avium subsp. paratuberculosis genome was searched by using Contig16, available from the University of Minnesota portal (http://www.ccgb.umn.edu/cgi-bin/common/web_blast.cgi).
Construction of recombinant mycobacterial shuttle plasmids and deletion derivatives and transformation in M. smegmatis mc2155.
The 5,367-bp SacI fragment RDIII300 (Fig. 1), containing the complete mptC, mptD, and mptE open reading frames (ORFs) and truncated mptB and mptF ORFs, was cloned into the mycobacterial shuttle vector pMV361 (Table 1) under control of the vector-based hsp60 promoter. Based on this plasmid, designated pRDIII320, deletion derivatives were generated. Deletion of a 3,201-bp FseI fragment resulted in plasmid pRDIII320Δ1 (Fig. 1), containing mptE and a partial mptF gene. Deletion of the DNA downstream of an SmaI restriction site by using a double digestion with SmaI and HpaI resulted in plasmid pRDIII320Δ2 (Fig. 1), containing an intact mptC ORF. These plasmids were used to transform M. smegmatis mc2155 by electroporation as described previously (38).
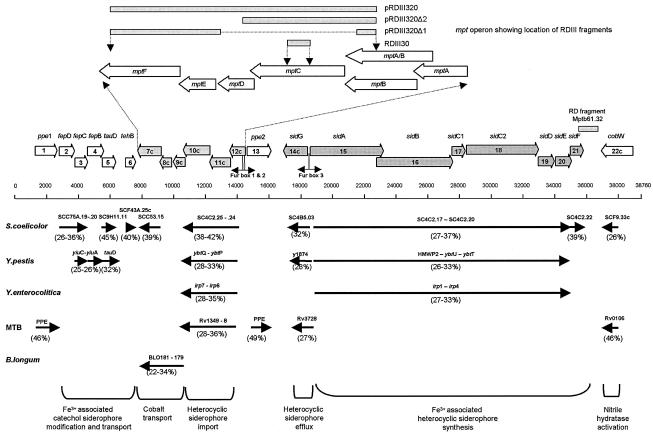
FIG. 1.
Representation of ORFs and putative Fur boxes from the M. avium subsp. paratuberculosis 38-kb locus and identity of homologues in other bacteria. The inset shows the relative locations of the mpt ORFs with respect to the deletion derivatives constructed. Numbers in parentheses represent the percent identities of homologues to putative M. avium subsp. paratuberculosis ORFs.
Protein preparations, electrophoresis, and Western blotting.
Mycobacterial whole-cell lysates were prepared from 100 mg of bacterial pellet harvested from Middlebrook liquid medium. Bacteria were resuspended in 500 μl of distilled water, and cell disruption was achieved by mechanical treatment for 190 s with zirconium beads in a mini-bead beater (Bio-Spec Products, Inc., Bartlesville, Okla.) followed by sonication. Residual cellular debris was removed by centrifugation (2,500 × g) for 10 min; total crude mycobacterial cell envelopes were obtained by ultracentrifugation (175,000 × g) for 2 h at 4°C and dissolved in 500 μl of Tris-EDTA buffer. Protein aggregates were prepared as previously described (19), and all protein preparations were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting as described earlier (19). For the detection of the MptC protein in M. avium subsp. paratuberculosis, the ECL Western blotting detection system (Amersham Pharmacia Biotech, Freiburg, Germany) was used according to the manufacturer's instructions; for other Western blots, an alkaline phosphatase-labeled goat anti-rabbit conjugate and a substrate containing nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate were used.
Preparation of antiserum.
A DNA fragment obtained by PCR with primers ABC3 and ABC4 (Table 1) was cut with BamHI and EcoRI and cloned into pGEX5x-3, resulting in plasmid pRDIII350 (Table 1). Upon induction with isopropyl-thiogalactoside (IPTG) (1 mM final concentration), inclusion bodies were formed and purified as described previously (19). Serum was raised in rabbits by an initial intracutaneous injection and two subcutaneous booster injections 3 and 6 weeks later, each with 100 μg of recombinant fusion protein in a total volume of 300 μl containing 30% adjuvant (Emulsigen-Plus; MVP Inc.).
Isolation and labeling of phages recognizing surface-exposed epitopes of an Mpt protein.
Biopanning was performed based on the Ph.D.-12 phage display library (New England Biolabs), containing random 12-mer peptides with a complexity of 1.9 × 109. Phages were panned by using whole cells or cell envelopes coupled to cyanogen bromide-activated Sepharose of M. smegmatis transformants carrying pRDIII320. After amplification in E. coli, counterselection to remove nonspecific phages was done by absorption to whole cells or cell envelopes of M. smegmatis carrying the vector pMV361 only. This selective panning with counterselection was repeated five times with high-stringency washing conditions in the last panning, as described previously (44). Both panning procedures resulted in the selection of the same phage, designated fMptD. Fluorescence labeling of phage particles was performed as previously described (44).
Enrichment of M. avium subsp. paratuberculosis from bulk milk.
Peptide aMptD (GKNHHHQHHRPQ) was synthesized based on the sequence obtained from phage fMptD with amino-terminal biotinylation and amino-hexacarbonic acid as a spacer (Fa. Affina Immuntech, Berlin, Germany). The peptide was dissolved, coupled to paramagnetic particles (Promega Inc., Madison, Wis.), and used for peptide-mediated capture PCR on bulk milk from herds previously analyzed by this method with a different peptide (44). The specificity of the amplification product was confirmed by restriction enzyme digestion (data not shown).
RESULTS
RDA-based identification and cloning of an M. avium subsp. paratuberculosis-specific ABC transporter operon.
An RDA approach comparing total genomic DNAs from M. avium subsp. avium and M. avium subsp. paratuberculosis, digested with BclI, identified a fragment of 340 bp designated RDIII30. The sequence of the RDIII30 fragment revealed one continuous ORF which encoded a putative transmembrane domain but showed no significant homology to GenBank sequences. Using the RDIII30 fragment as a probe, we identified and sequenced a further four overlapping fragments (designated RDIII300 to -303) by directed cloning of a SacI fragment and a KpnI fragment of the sizes predicted by Southern blot analyses (RDIII300 and RDIII301) and by screening a genomic library of M. avium subsp. paratuberculosis strain 6783 generated by the cloning of 2- to 4-kb fragments generated by a partial DpnII digest (RDIII302 and RDIII303). This revealed an 8,745-bp contiguous sequence (accession number AF419325) which carried six putative ORFs (designated mptA to mptF) transcriptionally orientated in tandem. A BLASTN search of this sequence showed no significant homology to sequences from any of the bacterial genomes, including that of M. avium subsp. avium, currently in GenBank. The M. avium subsp. paratuberculosis specificity of the region was further supported by PCR with RDIII30-derived primers IPIII30 and IPIII31 and amplification a fragment of 247 bp from 14 bovine M. avium subsp. paratuberculosis isolates, with no product obtained from six other mycobacterial species (data not shown). Although PCR is not a foolproof method to demonstrate sequence specificity, as a polymorphism in the primer binding region can result in the absence of an amplification product, the combined outcome of the PCR and GenBank analyses is a strong indication for the M. avium subsp. paratuberculosis specificity of the sequence.
A genomic alignment of the completed M. avium subsp. avium (TIGR-104) genome and the recently completed M. avium subsp. paratuberculosis (K-10) genome revealed that the RDIII30 sequence is part of a 38-kb M. avium subsp. paratuberculosis-specific locus (GenBank accession number AE016958, located at coordinates 4146188 through 4184947 of the M. avium subsp. paratuberculosis K-10 genome) comprising 22 ORFs orientated into three major gene clusters, provisionally designated fep, mpt, and sid (Fig. 1). The locus is bounded on one side by an insertion element with 83% homology to IS1110 (23) that is present as a single copy in M. avium subsp. paratuberculosis and in some strains of M. avium subsp. avium but not in the genome of the sequenced M. avium subsp. avium (TIGR-104) strain. The other side of the 38-kb locus contains sequence with 95% DNA homology over 23 kb to M. avium subsp. avium (TIGR-104), with the 38-kb locus starting from CGGCCCGGCGAG positioned in the M. avium genome at coordinate 5012818.
In this work the 38-kb locus was numbered for clarity as positions 1 to 38760 (the sequence with accession number AF419325 is located at position 6600 to 15344). A BLASTN search of the whole 38-kb locus sequence against GenBank confirmed the unique specificity of this region, which also includes the previously identified M. avium subsp. paratuberculosis-specific probe Mptb61.32 (43), located at positions 36208 to 36803 (Fig. 1).
Functional genomics of the mpt operon and the 38-kb locus.
The lack of homology of the carboxy-terminal region of MptA to the GenBank protein database and the close homology of the amino-terminal region of MptA and carboxy-terminal region of MptB to the products of genes rv1348 of M. tuberculosis and sc4c2.24 of Streptomyces coelicolor (Table 2) suggest that both ORFs may combine to encode a single MptA-MptB protein. The ORFs of mptA and mptB overlap by 62 bp and contain a previously identified “slippery site” (CCC CCA at positions 13599 to 13604) (43) associated with programmed translational frameshifts. A similar frameshift resulting in the expression of a single transporter protein has been previously described for a permease protein belonging to an ABC transporter operon in M. smegmatis (2). We propose, therefore, that the putative start codon for MptA-MptB is at position 14040, encoding the carboxy-terminal region of ORF mptA only (Fig. 1) and, together with mptB, encoding an ORF of 589 amino acids in length. This start codon is supported by an associated Shine-Dalgarno consensus sequence (GAAGGA) and two putative transcriptional Fur box control motifs (38-kb locus positions 14079 to 14105 and 14504 to 14531) within putative bidirectional promoters upstream of this position. Both Fur boxes have significant homology (90 and 74%) (Fig. 2) to an E. coli Fur box consensus sequence (1), and Fur box 2 also has a perfect panlindromic structure.
TABLE 2.
BLAST search results
| ORF no. | ORF name (position in Fig. 1) | Length of ORF (amino acids) | Putative function | Homologue (species) | Identity (%) | Span (amino acids) | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| 1 | ppe1 (1070-2626) | 518 | PPE signaling membrane protein | Rv3018c (M. tuberculosis) | 46 | 182 | NP_217534 |
| 2 | fepD (2684-3658) | 324 | FepD FeIII permease | SCC75A.19 (S. coelicolor) | 26 | 324 | CAB61719 |
| 3 | fepC (3654-4464) | 271 | FepC FeIII ABC transporter with ATP binding domain | YiuC (Y. enterocolitica) | 27 | 278 | AAD29087 |
| SCC75A.20 (S. coelicolor) | 36 | 224 | CAB61720 | ||||
| 4 | fepB (4461-5498) | 345 | FepB FeIII ABC transporter | YiuA (Y. pestis) | 25 | 356 | NP_670175 |
| 5 | tauD (5498-6274) | 258 | Taurine dehydrogenase | TauD (Y. pestis) | 32 | 260 | NP_671259 |
| Rv3406 (M. tuberculosis) | 33 | 270 | NP_217923.1 | ||||
| 6 | tehB (6885-7529) | 214 | TehB tellurite resistance protein, S-adenosyl-l-methionine dependent non-nucleic acid methyltransferase | SCF43A.25c (S. coelicolor) | 40 | 193 | NP_625135 |
| 7c | mptF (9162-7660) | 500 | ATP binding protein and CbiO ABC-cobalt transporter | BLO181 (B. longum) | 34 | 486 | ZP_00120392 |
| SCC53.15 (S. coelicolor) | 39 | 215 | NP_626571 | ||||
| 8c | mptE (9851-9159) | 230 | CbiQ ABC-cobalt transporter | BLO180 (B. longum) | 29 | 215 | ZP_00120392 |
| 9c | mptD (9869-10495) | 208 | Unknown | BLO179 (B. longum) | 22 | 194 | ZP_00120391 |
| 10c | mptC (12273-10492) | 593 | ATP binding protein, putative ABC transporter | Rv1349 (M. tuberculosis) | 29 | 583 | NP_215865 |
| YbtQ (Y. pestis) | 32 | 571 | NP_405475 | ||||
| Irp7 (Y. enterocolitica) | 28 | 575 | CAB46572 | ||||
| SC4C2.25 (S. coelicolor) | 38 | 585 | NP_631729 | ||||
| 11c | mptB (13628-12270) | 452 | ATP binding protein, putative ABC transporter | Rv1348 (M. tuberculosis) | 36 | 573 | NP_215864 |
| YbtP (Y. pestis) | 38 | 421 | NP_405474 | ||||
| Irp6 (Y. enterocolitica) | 34 | 508 | CAB46573 | ||||
| SC4C2.24 (S. coelicolor) | 46 | 413 | NP_631728 | ||||
| 12c | mptA (14040-13567) | 157 | ATP binding protein, putative ABC transporter | Rv1348 (M. tuberculosis) | 34 | 115 | NP_215864 |
| SC4C2.24 (S. coelicolor) | 37 | 126 | NP_631728 | ||||
| 13 | ppe2 (14702-16216) | 504 | PPE signaling membrane protein | Rv3018c (M. tuberculosis) | 49 | 160 | NP_217534 |
| 14c | sidG (18558-17026) | 510 | Metabolite efflux transporter | Y1874 (Y. pestis) | 28 | 417 | NP_669190 |
| 15 | sidA (18728-23443) | 1,571 | Non-ribosome-dependent heterocyclic siderophore synthase | Irp2 (Y. pestis) | 31 | 888 | NP_405472 |
| HMWP2 (Y. enterocolitica) | 31 | 891 | P48633 | ||||
| 16 | sidB (23232-27896) | 1,554 | Non-ribosome-dependent heterocyclic siderophore synthase | Irp2 (Y. pestis) | 27 | 980 | NP_405472 |
| HMWP2 (Y. enterocolitica) | 27 | 980 | P48633 | ||||
| SC4C2.17 (S. coelicolor) | 32 | 737 | NP_631721 | ||||
| 17 | sidC1 (27893-28738) | 281 | Non-ribosome-dependent heterocyclic siderophore synthase | SC4C2.18 (S. coelicolor) | 37 | 249 | NP_631722 |
| 18 | sidC2 (28784-33391) | 1,535 | Non-ribosome-dependent heterocyclic siderophore synthase | Irp2 (Y. pestis) | 29 | 1433 | NP_405472 |
| HMWP2 (Y. enterocolitica) | 29 | 1433 | P48633 | ||||
| SC4C2.17 (S. coelicolor) | 34 | 1535 | NP_631721 | ||||
| 19 | sidD (33388-34419) | 343 | Saccharopine dehydrogenase | SC4C2.19 (S. coelicolor) | 27 | 354 | NP_631723 |
| 20 | sidE (34495-35529) | 344 | Oxidoreductase (NAD binding) heterocyclic siderophore synthase | SC4C2.20 (S. coelicolor) | 30 | 352 | NP_631724 |
| YbtU (Y. pestis) | 32 | 242 | T17441 | ||||
| Irp3 (Y. enterocolitica) | 32 | 242 | T30343 | ||||
| 21 | sidF (35526-36278) | 250 | Thioesterase heterocyclic siderophore synthase | SC4C2.22 (S. coelicolor) | 39 | 231 | NP_631726 |
| YbtT (Y. pestis) | 33 | 230 | NP_405469 | ||||
| Irp4 (Y. enterocolitica) | 33 | 230 | T30344 | ||||
| 22c | cobW (38645-37425) | 406 | Nitrile hydratase | CobW (M. tuberculosis) | 46 | 377 | NP_214620 |
FIG. 2.
Alignment of M. avium subsp. paratuberculosis 38-kb region putative Fur boxes with an E. coli consensus sequence and Fur boxes from several other pathogens. Dots indicate identical nucleotides, and numbers in parentheses indicate the base location.
The mptA-mptB ORF is immediately followed by two overlapping ORFs, mptC and mptD. The mptC sequence contains the RDA fragment RDIII30 and codes for a predicted protein of 593 amino acids in length with a calculated molecular mass of 64 kDa. Homology searches revealed significant degrees of identity to ABC transporter proteins from S. coelicolor (38%), M. tuberculosis (29%), and Yersinia pestis (28%). The putative MptC protein contains five predicted transmembrane domains, including Walker A and B motifs (5) and a Higgins motif (24) typical of ABC transporters and in positions identical to the homologues in M. tuberculosis and Y. pestis (data not shown).
Unlike other ORFs in the 38-kb region, mptD to -F do not have functional homologues in Yersinia spp. or S. coelicolor. Even though the mptC reading frame overlaps mptD, ORFs mptD to -F are more linked by their significant protein identities (22 to 34%) and similar genomic organization to a cobalt ABC transporter cassette found in the gut commensal Bifidobacterium longum (39). MptE contains a CbiQ functional motif (E value, 2E−13) and MptF contains a CbiO functional motif (E value, 3E−39), which are associated with active transport of cobalt into the cytosol. MptD has no predicted functional motifs, but TMHMM (algorithm for the prediction of transmembrane helices in proteins, accessible with the HUSAR 5.0 program [DKFZ, Heidelberg, Germany]) predictions indicate six transmembrane domains (data not shown), suggesting that MptD is a membrane-bound component in this system.
The mpt genes are positioned between the sid and fep clusters. The sid cluster is the largest of the putative operons and includes the genes sidA to sidG and possibly a gene encoding a membrane signaling protein (ppe2). These genes are contiguous but are orientated in groups transcribed in opposing directions enclosing a Fur box located at positions 18571 to 18597 (Fig. 2) between sidG and sidA. The ORFs sidB and sidC1, sidC2 and sidD, and sidE and sidF are orientated in tandem with overlapping start and stop codons. Eleven of 12 predicted genes from these clusters have significant identities (24 to 42%) and are arranged in an organization similar to that of operons involved in the nonribosomal synthesis and membrane transport of Fe3+-associated heterocyclic siderophore peptides from Y. pestis (3), Yersinia enterocolitica (11), and S. coelicolor (4). The sid cluster is followed by a putative nitrile hydratase homologue (cobW) at the end of the 38-kb island and is flanked by an IS1110-like insertion sequence. The fep cluster is a cluster of six ORFs, with four ORFs overlapping. The first ORF encodes a homologue (46% identity) of the ppe gene family in M. tuberculosis and is a probable membrane protein involved in signaling. Adjacent to this are three overlapping ORFs, fepB to fepD (Fig. 1), with significant identity (25 to 36%) and similar organization to genes in Y. pestis and S. coelicolor that are involved in the transport of catecholate Fe3+-associated siderophore into the bacterial cytosol (40). Homologues of fepC and fepD are ABC transporters associated with the ability of catecholate siderophores to permeate through bacterial membranes. The putative FepC protein has an ATP binding motif signature (E value, 1E−38) suggesting that it is an active transport system. Homologues to FepB, such as YiuA of Y. pestis, are periplasmic substrate binding proteins associated with the transport of Fe3+ siderophores. The final two ORFs have significant functional motifs but have only minor connections with other fep systems. The first, tauD, overlapping fepB, has 32 to 37% identity to taurine deoxygenases of S. coelicolor, Y. pestis, and Pseudomonas aeruginosa. The second, tehB, contains a tellurite resistance motif (parse 6E-06) with significant homology to S-adenosyl-l-methionine-dependent non-nucleic acid methyltransferases of S. coelicolor (Table 2).
Expression of Mpt proteins and their potential role in virulence.
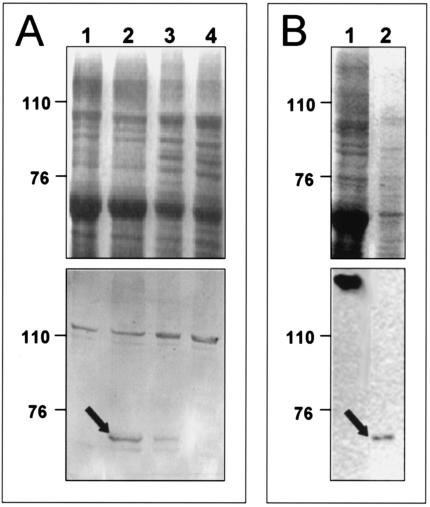
The predicted expression and localization of MptC and MptD in cell membranes were investigated by various methods. For MptC, Western blotting was performed with a serum raised in rabbits against a glutathione S-transferase-MptC fusion protein. No MptC protein was expressed by E. coli transformants (data not shown); however, a 64-kDa protein, consistent with the predicted size of MptC, could be demonstrated in envelope preparations of M. avium subsp. paratuberculosis and M. smegmatis transformed with MptC expression constructs, but not in controls (Fig. 3).
FIG. 3.
Coomassie blue-stained gel (top) and Western blot (bottom) of crude membrane preparations. (A) Lanes 1 to 4, M. smegmatis mc2155 transformants containing pMV361, pRDIII320, pRDIII320Δ2, and pRDIII320Δ1, respectively. (B) M. smegmatis (lane 1) and M. avium subsp. paratuberculosis (lane 2). The arrows indicate the position of the MptC protein detected by the serum raised against the recombinant fusion protein. The numbers on the left indicate the positions of protein markers in kilodaltons.
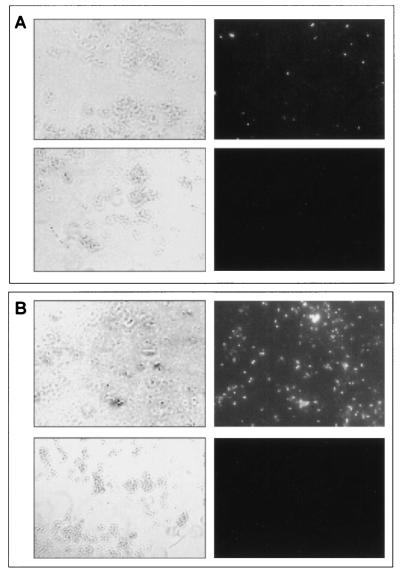
For MptD, a subtractive screening of an M13 phage peptide library was performed on envelope and whole-cell preparations of M. smegmatis transformed with the MptD expression construct pRDIII320, using M. smegmatis transformed with vector alone as a control. Both of these screens identified the same phage, designated fMptD, encoding the binding peptide designated aMptD (GKNHHHQHHRPQ). Fluorescein isothiocyanate (FITC)-labeled fMptD was used (Fig. 4A) to demonstrate specific binding of phage to the cell surface of M. smegmatis transformed with constructs expressing MptC-F (pRDIII320) but not with controls transformed with constructs expressing MptC (pRDIII320Δ2) or MptE-F (pRDIII320Δ1). In addition, FITC-labeled fMptD could detect M. avium subsp. paratuberculosis, but not M. avium subsp. avium, grown in Middlebrook 7H9 medium (Fig. 4B). These results strongly suggest that phage fMptD binds an epitope of a specific M. avium subsp. paratuberculosis protein, MptD, which is expressed and exposed on the cell surface.
FIG. 4.
Fluorescence microscopy with FITC-labeled phage fMptD as an affinity reagent. (A) M. smegmatis mc2155 transformed with pRDIII320 (top panels) and pMV361 (bottom panels) observed by light microscopy (left panels) and immunofluorescence (right panels). (B) M. avium subsp. paratuberculosis (top panels) or M. avium subsp. avium (bottom panels) observed by light microscopy (left panels) and immunofluorescence (right panels).
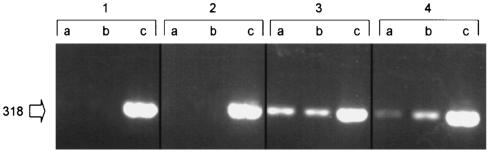
To investigate the potential role of the mpt operon in M. avium subsp. paratuberculosis infection, peptide aMptD encoded by phage fMptD was synthesized and used in a peptide-mediated capture PCR to identify M. avium subsp. paratuberculosis within bulk milk samples from infected and noninfected dairy herds (Fig. 5). This result confirms the expression of cell surface-associated MptD protein by M. avium subsp. paratuberculosis during infection of the natural host.
FIG. 5.
Peptide aMptD-mediated capture PCR from M. avium subsp. paratuberculosis-negative (panels 1 and 2) and -positive (panels 3 and 4) bulk milk samples with ISMav2-derived primers. Lanes a and b, products of PCRs of the sample; lanes c, internal positive control (sample spiked with M. avium subsp. paratuberculosis DNA). The arrow indicates the expected position of the PCR product in base pairs.
DISCUSSION
In this work we have identified three novel M. avium subsp. paratuberculosis operons (mpt, fep, and sid) present within a 38-kb locus. The absence of significant DNA homology of the entire 38-kb region to M. avium subsp. avium, the presence of a previously described short sequence (Mptb61.32) that was also identified by RDA between M. avium subsp. avium and M. avium subsp. paratuberculosis (34), the positive PCR analyses with 14 clinical M. avium subsp. paratuberculosis isolates, and the negative PCR analyses with the reference strains of six other mycobacterial species strongly suggest that this locus is M. avium subsp. paratuberculosis specific. Functional genomic analysis revealed that most ORFs from the 38-kb locus have significant identities to previously identified proteins in other pathogens. Similarities in functional motifs which are consistent with an involvement in linked molecular processes primarily concerned with uptake of iron and other essential trace elements were also found. This is supported by the presence of Fe3+-regulated transcriptional control motifs (Fur boxes) within putative promoters associated with the sid and mpt operons (11, 18), which are responsible for the translocation into the bacterial cytosol of Fe3+, bound to a peptide siderophore. The putative MptE and -F proteins have homologues from the gut commensal B. longum (39) which contain motifs associated with active transport of cobalt into the cell cytosol. Identities to homologous proteins in other species are insufficient to suggest that cobalt is the ion involved in M. avium subsp. paratuberculosis, and many other ions, such as Fe3+ and Mn2+, could be associated instead. However, the chelation of cobalt is important for intracellular survival and may be linked to cobW, located further down the 38-kb locus, as some nitrile hydratases require cobalt for activity (15). The role of the mpt operon in iron uptake is further supported, however, by the presence of two Fur boxes located immediately upstream of the mpt operon within a putative promoter region.
Homologues to proteins encoded by the sid operon are involved in the nonribosomal synthesis and membrane transport of Fe3+-associated heterocyclic siderophore peptides from Y. pestis (3), Y. enterocolitica (11), and S. coelicolor (4). In these organisms, the operons are transcriptionally controlled by binding of proteins, such as the furB gene product, onto Fur box motifs positioned within associated promoters. fur genes have also been identified in several mycobacterial species; a role of furA in virulence has been postulated, as it modulates the response to oxidative stress by regulating the expression of the catalase:peroxidase. To date, nothing is known about the function of furB, which likewise is present in several mycobacterial species (36). The sid operon described in this work contains a 19-mer Fur box homologue with four mismatches to an E. coli consensus sequence (1) within a putative bidirectional promoter sequence. This suggests that the sid operon may have biosynthetic and transcriptional control mechanisms similar to the those of the ybt operon of Yersinia spp.
Homologues of proteins encoded by the fep operon (FepC and -D) in Y. pestis and S. coelicolor are ABC transporters associated with the ability of catecholate Fe3+ siderophores to permeate through bacterial membranes into the cytosol (40). The putative FepC protein has an ATP binding motif signature, suggesting that this is part of an active transport system. Homologues to the FepB protein, such as YiuA of Y. pestis, are periplasmic binding proteins also associated with the transport of Fe3+ siderophores. The TauD homologues are taurine deoxygenases which liberate sulfur from taurine for assimilation into processes such as cysteine biosynthesis (27) and may be crucial virulence determinants in intracellular persistence when access to sulfonated compounds is limited. The sequence divergence of the predicted M. avium subsp. paratuberculosis TauD protein suggests that it may have a different specific substrate that could possibly be a catechol siderophore. The putative TehB protein has significant homology to S-adenosyl-l-methionine-dependent non-nucleic acid methyltransferases whose function could be related to the catechol-o-methyltransferase activity on catechol siderophores observed with gene Rv1703c from M. tuberculosis (14).
The final ORF in the 38-kb locus, cobW, is located at the very end of the locus between the sidA-F cluster and IS1110. Its product contains a CobW motif and significant identity with Rv0106 of M. tuberculosis and with SCF9.33c of S. coelicolor. The function of this gene is uncertain, but the presence of a CobW motif suggests that it may be an activator of nitrile hydratase, an enzyme important in production of nitric oxide and whose biosynthesis is linked to Fe3+ regulation in macrophages (15).
We have investigated the expression and location of two proteins from the mpt operon, which support our in silico analyses. We have demonstrated the expression of MptC in M. smegmatis transformants, confirmed its predicted size, and shown it to be an envelope-associated protein. The expression of MptC in cell envelopes of M. avium subsp. paratuberculosis was also shown, supporting its putative function as an ABC transporter (18). In addition, an M. avium subsp. paratuberculosis-specific phage (fMptD) was isolated and used to demonstrate the expression of MptD in M. smegmatis transformants and on the surface of M. avium subsp. paratuberculosis. This finding supports the hypothesis that this protein might form a selective pore associated with the outer membrane-like layer of the mycobacterial cell envelope. Furthermore, we developed and used a cell capture PCR technique that employs an fMptD-derived peptide with specific binding capacity for MptD to capture and identify M. avium subsp. paratuberculosis in raw milk from infected cattle. This demonstrated that the MptD protein is exposed on the surface of M. avium subsp. paratuberculosis during infection and is therefore a potential target for M. avium subsp. paratuberculosis immunization or treatment.
These results together with the predicted functions of other genes in the 38-kb region strongly suggest its association with M. avium subsp. paratuberculosis virulence and that the 38-kb locus should be considered the first pathogenicity island identified in M. avium subsp. paratuberculosis. The localization of an ABC transporter operon on a putative pathogenicity island has been previously described for Salmonella enterica serovar Typhimurium (48), and an ABC transporter also involved in iron uptake has been identified on a pathogenicity island of Streptococcus pneumoniae (7). Furthermore, the ABC transporter system of Y. pestis, YbtPQ, which also is required for iron uptake and has a significant degree of similarity to the mpt operon products of M. avium subsp. paratuberculosis, is located within a large unstable region of the Y. pestis chromosome (18).
Future studies are needed to determine the precise function of the M. avium subsp. paratuberculosis-specific fep, mpt, and sid operons. The use of specific phage-displayed peptides might be of considerable value in this field by overcoming the problems associated with the extremely slow growth of this organism and the lack of a proven targeted mutagenesis system for M. avium subsp. paratuberculosis.
The induction of protective immunity against systemic S. pneumoniae infection in mice when they are vaccinated with components of iron uptake ABC transporters (8) and the sustained reduction of challenged M. tuberculosis growth when mice are vaccinated with a phosphate ABC transporter (46) suggest that the MptD protein may be a suitable component in an M. avium subsp. paratuberculosis vaccine design. The recent development of a beige/scid mouse model able to demonstrate intestinal pathophysiologic changes upon M. avium subsp. paratuberculosis infection (33) may support such studies.
Acknowledgments
This work has been supported by the Deutsche Forschungsgemeinschaft, Bonn, Germany (Sonderforschungsbereich 280 Gastrointestinale Barriere), and by EU grant QLK 2-CT-2001-01420. Additional support has been provided by the Scottish Executive Rural Affairs Department (SERAD), Scotland, United Kingdom, and the Deutscher Akademischer Austauschdienst, Bonn, Germany. Research in the laboratory of Vivek Kapur is supported by grants from the U.S. Department of Agriculture's National Research Initiative and Agriculture Research Service, the National Institutes of Health, and the Minnesota Agriculture Experiment Station's Rapid Agricultural Response Fund. Research in the laboratory of Tim J. Bull is supported by a grant from Action Medical Research.
Editor: B. B. Finlay
REFERENCES
- 1.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barsom, E. K., and G. F. Hatfull. 1997. A putative ABC-transport operon of Mycobacterium smegmatis. Gene 185:127-132. [DOI] [PubMed] [Google Scholar]
- 3.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Bianchet, M. A., Y. H. Ko, L. M. Amzel, and P. L. Pedersen. 1997. Modeling of nucleotide binding domains of ABC transporter proteins based on a F-1-ATPase/recA topology: structural model of the nucleotide binding domains of the cystic fibrosis transmembrane conductance regulator (CFTR). J. Bioenerg. Biomembr. 29:503-524. [DOI] [PubMed] [Google Scholar]
- 6.Bose, M., A. Chander, and R. H. Das. 1993. A rapid and gentle method for the isolation of genomic DNA from mycobacteria. Nucleic Acids Res. 21:2529-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. S., S. M. Gilliland, and D. W. Holden. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40:572-585. [DOI] [PubMed] [Google Scholar]
- 8.Brown, J. S., A. D. Ogunniyi, M. C. Woodrow, D. W. Holden, and J. C. Paton. 2001. Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect. Immun. 69:6702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burlein, J. E., C. K. Stover, S. Offutt, and M. S. Hanson. 1994. Expression of foreign genes in mycobacteria, p. 239-252. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection, and control. ASM Press, Washington, D.C.
- 10.Cameron, R. M., K. Stevenson, N. F. Inglis, J. Klausen, and J. M. Sharp. 1994. Identification and characterization of a putative serine protease expressed in vivo by Mycobacterium avium subsp. paratuberculosis. Microbiology 140:1977-1982. [DOI] [PubMed] [Google Scholar]
- 11.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 12.Chiodini, R. J. 1989. Crohn's disease and the mycobacterioses: a review and comparison of two disease entities. Clin. Microbiol. Rev. 2:90-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiodini, R. J., H. J. Van Kruiningen, and R. S. Merkal. 1984. Ruminant paratuberculosis (Johne's disease): the current status and future prospects. Cornell Vet. 74:218-262. [PubMed] [Google Scholar]
- 14.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 396:190-198. [DOI] [PubMed] [Google Scholar]
- 15.Drapier, J. C., H. Hirling, J. Wietzerbin, P. Kaldy, and L. C. Kuhn. 1993. Biosynthesis of nitric oxide activates iron regulatory factor in macrophages. EMBO J. 12:3643-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Zaatari, F. A., S. A. Naser, and D. Y. Graham. 1997. Characterization of a specific Mycobacterium paratuberculosis recombinant clone expressing 35,000-molecular-weight antigen and reactivity with sera from animals with clinical and subclinical Johne's disease. J. Clin. Microbiol. 35:1794-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57:995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 19.Gerlach, G. F., C. Anderson, A. A. Potter, S. Klashinsky, and P. J. Willson. 1992. Cloning and expression of a transferrin-binding protein from Actinobacillus pleuropneumoniae. Infect. Immun. 60:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microb. Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 21.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermon-Taylor, J., T. J. Bull, J. M. Sheridan, J. Cheng, M. L. Stellakis, and N. Sumar. 2000. Causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can. J. Gastroenterol. 14:521-539. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez, P. M., N. G. Fomukong, T. Hellyer, I. N. Brown, and J. W. Dale. 1994. Characterization of IS1110, a highly mobile genetic element from Mycobacterium avium. Mol. Microbiol. 12:717-724. [DOI] [PubMed] [Google Scholar]
- 24.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 25.Homuth, M., P. Valentin-Weigand, M. Rohde, and G. F. Gerlach. 1998. Identification and characterization of a novel extracellular ferric reductase from Mycobacterium paratuberculosis. Infect. Immun. 66:710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulten, K., H. M. El Zimaity, T. J. Karttunen, A. Almashhrawi, M. R. Schwartz, D. Y. Graham, and F. A. El Zaatari. 2001. Detection of Mycobacterium avium subspecies paratuberculosis in Crohn's diseased tissues by in situ hybridization. Am. J. Gastroenterol. 96:1529-1535. [DOI] [PubMed] [Google Scholar]
- 27.Kertesz, M. A. 2000. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 28.Kreeger, J. M. 1991. Ruminant paratuberculosis—a century of progress and frustration. J. Vet. Diagn. Invest. 3:373-382. [DOI] [PubMed] [Google Scholar]
- 29.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 30.Lisitsyn, N. A. 1995. Representational difference analysis: finding the differences between genomes. Trends Genet. 11:303-307. [DOI] [PubMed] [Google Scholar]
- 31.McClure, H. M., R. J. Chiodini, D. C. Anderson, R. B. Swenson, W. R. Thayer, and J. A. Coutu. 1987. Mycobacterium paratuberculosis infection in a colony of stumptail macaques (Macaca arctoides). J. Infect. Dis. 155:1011-1019. [DOI] [PubMed] [Google Scholar]
- 32.Mutharia, L. M., W. Moreno, and M. Raymond. 1997. Analysis of culture filtrate and cell wall-associated antigens of Mycobacterium paratuberculosis with monoclonal antibodies. Infect. Immun. 65:387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mutwiri, G. K., U. Kosecka, M. Benjamin, S. Rosendal, M. Perdue, and D. G. Butler. 2001. Mycobacterium avium subspecies paratuberculosis triggers intestinal pathophysiologic changes in beige/scid mice. Comp. Med. 51:538-544. [PubMed] [Google Scholar]
- 34.Nielsen, K., and P. Ahrens. 2002. Putative in vitro expressed gene fragments unique to Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 214:199-203. [DOI] [PubMed] [Google Scholar]
- 35.Raleigh, F., A. K. Lech, and R. Brent. 1989. Selected topics from classical bacterial genetics, p. 1.4.1-1.4.14. In F. M. Ausubel et al. (ed.), Current protocols in molecular biology. Wiley Interscience, New York, N.Y. [DOI] [PubMed]
- 36.Rodriguez, G. M., and I. Smith. 2003. Mechanisms of iron regulation in mycobacteria: role in physiology and virulence. Mol. Microbiol. 47:1485-1494. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Sander, P., and E. C. Boettger. 1998. Gene replacement in Mycobacterium smegmatis using a dominant negative selectable marker, p. 207-216. In T. Parish and N. G. Stoker (ed.), Mycobacteria protocols. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 39.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schubert, S., D. Fischer, and J. Heesemann. 1999. Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J. Bacteriol. 181:6387-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Secott, T. E., T. L. Lin, and C. C. Wu. 2001. Fibronectin attachment protein homologue mediates fibronectin binding by Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 69:2075-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 43.Snyder, L., and W. Champness. 1997. Molecular genetics of bacteria, p. 55. ASM Press, Washington, D.C.
- 44.Stratmann, J., B. Strommenger, K. Stevenson, and G. F. Gerlach. 2002. Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk. J. Clin. Microbiol. 40:4244-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strommenger, B., K. Stevenson, and G. F. Gerlach. 2001. Isolation and diagnostic potential of ISMav2, a novel insertion sequence-like element from Mycobacterium avium ssp. paratuberculosis. FEMS Microbiol. Lett. 196:31-37. [DOI] [PubMed] [Google Scholar]
- 46.Tanghe, A., P. Lefevre, O. Denis, S. D'Souza, M. Braibant, E. Lozes, M. Singh, D. Montgomery, J. Content, and K. Huygen. 1999. Immunogenicity and protective efficacy of tuberculosis DNA vaccines encoding putative phosphate transport receptors. J. Immunol. 162:1113-1119. [PubMed] [Google Scholar]
- 47.Tizard, M., T. Bull, D. Millar, T. Doran, H. Martin, N. Sumar, J. Ford, and J. Hermon-Taylor. 1998. A low G+C content genetic island in Mycobacterium avium subsp. paratuberculosis and M. avium subsp. silvaticum with homologous genes in Mycobacterium tuberculosis. Microbiology 144:3413-3423. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, D., W. D. Hardt, and J. E. Galan. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]