Abstract
Escherichia coli EDL933, an O157:H7 strain, is known to colonize the streptomycin-treated CD-1 mouse intestine by growing in intestinal mucus (E. A. Wadolkowski, J. A. Burris, and A. D. O'Brien, Infect. Immun. 58:2438-2445, 1990), but what nutrients and metabolic pathways are employed during colonization has not been determined. In this study, when the wild-type EDL933 strain was fed to mice along with an EDL933 ΔppsA ΔpckA mutant, which is unable to utilize tricarboxylic acid cycle intermediates and gluconeogenic substrates for growth, both strains colonized the mouse intestine equally well. Therefore, EDL933 utilizes a glycolytic substrate(s) for both initial growth and maintenance when it is the only E. coli strain fed to the mice. However, in the presence of large numbers of MG1655, a K-12 strain, it is shown that EDL933 utilizes a glycolytic substrate(s) for initial growth in the mouse intestine but appears to utilize both glycolytic and gluconeogenic substrates in an attempt to maintain colonization. It is further shown that MG1655 predominantly utilizes glycolytic substrates for growth in the mouse intestine whether growing in the presence or absence of large numbers of EDL933. Data are presented showing that although small numbers of EDL933 grow to large numbers in the intestine in the presence of large numbers of MG1655 when both strains are fed to mice simultaneously, precolonization with MG1655 affords protection against subsequent colonization by EDL933. Moreover, in mice that are precolonized with EDL933, small numbers of MG1655 are able to grow rapidly in the intestine and EDL933 is eliminated. In situ hybridization experiments using E. coli-specific rRNA probes showed that while MG1655 is found only in mucus, EDL933 is found both in mucus and closely associated with intestinal epithelial cells. The data are discussed with respect to competition for nutrients and to the protection that some intestinal commensal E. coli strains might afford against infection by O157:H7 strains.
Escherichia coli strains of serotype O157:H7 cause outbreaks of hemorrhagic colitis and hemolytic uremic syndrome in humans (reviewed in reference 14). E. coli O157:H7 initiates infection by binding to intestinal epithelial cells and producing Shiga toxins Stx1 and/or Stx2, depending on the strain (reviewed in reference 14). Stx1 and Stx2 depurinate a critical residue in the eucaryotic 28S rRNA of 60S ribosomes, resulting in the inhibition of protein synthesis and consequent cell death (33). E. coli EDL933, an O157:H7 strain, does not normally kill streptomycin-treated mice and appears to colonize the mouse intestine by growing in intestinal mucus (38, 40), but little is known about the nutrients that are utilized for growth or the metabolic pathways involved. If these pathways were defined, it is likely that preventative measures or more effective treatments for patients infected with O157:H7 strains could be developed. With this goal in mind, we isolated an EDL933 ΔppsA ΔpckA mutant, which grows normally on glycolytic substrates but is unable to utilize tricarboxylic acid (TCA) cycle intermediates and gluconeogenic substrates for growth, and tested its ability to colonize the mouse intestine in both the presence and absence of the K-12 strain E. coli MG1655, a human commensal strain. The results of these experiments are reported in the present study.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| E. coli strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Wild type (CGSC no. 7740) | E. coli Genetic Stock Culture Collection, Yale University |
| MG1655 Strr | Spontaneous streptomycin-resistant mutant of MG1655 | This study |
| MG1655 Strr Nalr | Spontaneous nalidixic acid-resistant mutant of MG1655 Strr | This study |
| MG1655 Strr ΔppsA ΔpckA::Cm | ppsA pckA double deletion mutant of MG1655 Strr, carrying cat in the pckA deletion | This study |
| EDL933 | Wild-type O157:H7 | Alison O'Brien |
| EDL933 Strr | Spontaneous streptomycin-resistant mutant of EDL933 | This study |
| EDL933 Strr Nalr | Spontaneous nalidixic acid-resistant mutant of EDL933 Strr | This study |
| EDL933 Strr ΔppsA ΔpckA::Cm | ppsA pckA double deletion mutant of EDL933 Strr, carrying cat in the pckA deletion and retaining the pO157 virulence plasmid (see Materials and Methods) | This study |
| Plasmids | ||
| pKD3 | Template plasmid, contains chloramphenicol resistance cassette flanked by FLP recombinase target sites; bla cat | 4 |
| pKD46 | Temperature-sensitive plasmid, contains arabinose-inducible phage λ red recombinase gene for linear DNA exchange; bla | 4 |
| pCP20 | Temperature-sensitive plasmid, contains FLP recombinase gene for removal of antibiotic resistance genes; bla cat | 4 |
Bacterial growth and media.
Luria broth was made as described by Revel (32). Luria agar is Luria broth containing 12 g of Bacto agar (Difco) per liter. MacConkey agar (Difco) was prepared according to the package instructions. M9 minimal medium (22) was supplemented with either reagent-grade glucose (0.2% [wt/wt]), glycerol (0.2% [wt/wt]), gluconate (0.2% [wt/wt]), l-aspartic acid (0.4% [wt/wt]), potassium acetate (0.4% [wt/wt]), potassium fumarate (0.4% [wt/wt]), malic acid (0.4% [wt/wt]), sodium oleate (5 mM) plus 5 mg of Brij 58/ml, sodium pyruvate (0.4% [wt/wt]), or sodium succinate (0.6% [wt/wt]).
In vitro growth in mouse intestinal mucus.
Mouse intestinal mucus was isolated as previously described (3). Briefly, mice (35 days old) were fed Charles River Valley rat, mouse, and hamster formula for 5 days. The drinking water was then replaced with sterile distilled water containing streptomycin sulfate (5 g/liter). Twenty-four hours later, the mice were sacrificed by CO2 asphyxiation and their ceca were removed. The cecal contents were collected for use in growth experiments (see below), and any remaining cecal contents were washed out with sterile distilled water. Mouse cecal mucus was scraped from ceca, and 1-ml aliquots were placed into culture tubes as described previously (3). Cecal mucus was inoculated and incubated without shaking at 37°C, and samples taken at different times were plated and counted as described previously (23). Growth experiments utilizing 1-ml volumes of cecal contents were performed identically.
Mouse colonization experiments.
The method used to compare the large-intestine-colonizing abilities of E. coli strains in mice has been described previously (35, 36). Briefly, three male CD-1 mice (5 to 8 weeks old) were given drinking water containing streptomycin sulfate (5 g/liter) for 24 h to eliminate resident facultative bacteria (21). After 18 h of starvation for food and water, the mice were fed 1 ml of 20% (wt/vol) sucrose containing Luria broth-grown E. coli EDL933 strains and/or MG1655 strains, depending on the experiment. After ingestion of the bacterial suspension, both the food (Charles River Valley rat, mouse, and hamster formula) and streptomycin-water were returned to the mice, and 1 g of feces was collected after 5 and 24 h and on odd-numbered days at the indicated times. Mice were housed individually in cages without bedding and were placed in clean cages daily. Fecal samples (no older than 24 h) were homogenized in 1% Bacto tryptone, diluted in the same medium, and plated on Luria agar plates with appropriate antibiotics. Plates contained streptomycin sulfate (100 μg/ml), streptomycin sulfate (100 μg/ml) and nalidixic acid (50 μg/ml), or streptomycin sulfate (100 μg/ml) and chloramphenicol (30 μg/ml). All plates were incubated for 18 to 24 h at 37°C prior to counting. When necessary, 100 colonies from plates containing streptomycin were applied with toothpicks to plates containing appropriate antibiotics to differentiate strains. Each colonization experiment was performed at least twice, with essentially identical results. Data from typical experiments are presented.
Isolation and enumeration of MG1655 and EDL933 from mouse intestinal mucus.
E. coli EDL933 Strr and MG1655 Strr were each fed to separate sets of three mice. On day 16 postfeeding, the mice were sacrificed, and the ileum, the rest of the small intestine, the cecum, and the colon were removed from each mouse. Each section of the intestine was washed extensively with HEPES-Hanks buffer (pH 7.2), and the mucus from each section of the intestine was scraped into 5 ml of HEPES-Hanks buffer (pH 7.2) as described previously (3). Each sample was homogenized by vortexing and then plated on MacConkey agar with appropriate antibiotics. Plates were incubated for 18 to 24 h at 37°C prior to counting. The number of CFU per intestinal section was calculated from the CFU per milliliter by multiplying by the total volume (in milliliters) of each mucus sample.
Mutant construction.
Gene knockout mutants (deletions) were made as described by Datsenko and Wanner (4). Their method allows the sequential construction of double deletion mutants by use of the same chloramphenicol cassette (4). Gene knockouts were confirmed phenotypically as discussed in Results and genetically by PCR using primers specific for upstream and downstream flanking sequences. Primers used to construct the deletion mutants were designed according to the EDL933 and MG1655 genome databases. Deletions of 2,367 and 1,341 bp were made in the ppsA gene and the pckA gene, respectively. The specific PCR primers used to construct the deletions and to confirm deletions after allelic exchange are listed in Table 2.
TABLE 2.
Primers for construction of ΔppsA ΔpckA mutants
| Primer | Sequence (5′→3′) |
|---|---|
| E. coli MG1655 ΔppsA primer 1 | GCGACTAAACGCCGCCGGGGATTTATTTTATTTCTTCAGTgtgtaggctgagctgcttcga |
| E. coli EDL933 ΔppsA primer 1 | Identical to E. coli MG1655 ΔppsA primer 1, except that the first 5′ T is A |
| E. coli MG1655 and EDL933 ΔppsA primer 2 | AATGTGTTTCTCAAACCGTTCATTTATCACAAAAGGATTGTTCgcatatgaatatcctccttagb |
| E. coli MG1655 and EDL933 ΔppsA confirming primers | Forward, CCTGTGTGGTTGCAATGTCCG; reverse, TTGTTCTTCCCGTGATGCAGAC |
| E. coli MG1655 and EDL933 ΔpckA primer 1 | AGACTTTACTATTCAGGCAATACATATTGGCTAAGGAGCAGTGgtgtaggctggagctgcttcga |
| E. coli MG1655 and EDL933 ΔpckA primer 2 | GCGGGTATCTTTAATCGAGATACGTTTGCCAGTGCCGTTCCAgcatatgaatatcctccttagb |
| E. coli MG1655 and EDL933 ΔpckA confirming primers | Forward, GTTAATTATCGCATCCGGGCAG; reverse, TTACCCCGGCAGCATTGAAGTG |
Uppercase letters are ppsA, or pckA specific and lowercase letters are pKD3 specific, immediately downstream of the cat gene.
Uppercase letters are ppsA specific and lowercase letters are pKD3 specific, immediately upstream of the cat gene.
Primers flanking either the ppsA or pckA gene were used to confirm expected changes in the size of DNA fragments.
Confirmation that EDL933 Strr ΔppsA ΔpckA::Cm retained the pO157 plasmid.
The presence or absence of the EDL933 pO157 plasmid (GenBank accession number AF074613) was determined for strains MG1655, EDL933, EDL933 Cu (cured of the pO157 plasmid [38]), and the EDL933 ΔppsA ΔpckA double mutant by attempting to PCR amplify an internal 595-bp fragment of the espP gene, which is known to be in the pO157 plasmid (2). The forward primer 5′-CGGCAGAGTATCAAGAGC-3′ and the reverse primer 5′-CATTAAATGGAGTTATGCGTC-3′ were used for this amplification. The internal fragment of espP was amplified from EDL933 and the EDL933 ΔppsA ΔpckA double mutant but was not amplified from EDL933 Cu or MG1655. Therefore, the EDL933 ΔppsA ΔpckA double mutant appears to have retained the pO157 plasmid.
Preparation of histological sections for hybridization.
After the mice were sacrificed, the cecum was removed from each mouse and cleaned of fecal material as described above. Each cecum was placed in 5 ml of fixing solution containing methanol, chloroform, and acetic acid (6:3:1) for 1 h and then was transferred to 5 ml of fresh fixing solution for an additional hour. Prior to sectioning, each cecum was stored at 4°C in 70% ethanol. Mouse colonic, ileal, and small intestinal tissues were fixed in the same way. Tissues were dehydrated and embedded in paraffin prior to the preparation of 5-μm-thick cross sections. Sections were placed onto glass microscope slides. Prior to hybridization, the sections were deparaffinized by treatment with xylene (three times for 10 min each) and were dehydrated for 10 min in 96% ethanol. Before the hybridization solution was applied, the intestinal sections were circumscribed with an ImmEdge pen (Vector Laboratories, Inc., Burlingame, Calif.).
Oligonucleotide probes.
A probe specific for E. coli 23S rRNA (EC1531; 5′-CACCGTAGTGCCTCGTCATCA-3′) was used (30). The probe was labeled at the 5′ end with CY3 red fluorescent dye (cyanine dye CY3.29-OSu; Biological Detection Systems, Pittsburgh, Pa.). In addition, probe EU338 (30), which is specific for the eubacterial 23S rRNA domain, was used. This probe (5′-GCTGCCTCCCGTAGGAGT-3′) was labeled at the 5′ end with the green fluorescent compound fluorescein (Peninsula Laboratories, Inc., Belmont, Calif.).
Hybridization and microscopy.
For the visualization of bacteria in the intestinal sections, the eubacterial probe labeled with fluorescein was used in combination with the CY3-labeled E. coli-specific probe. Hybridization was carried out as described by Poulsen et al. (30), with the following modifications. The hybridization solution contained 10% formamide and 2.5 ng of each probe at pH 7.2. Washing solution I contained 10% formamide at pH 7.2. Washing solution II was at pH 7.2. After hybridization, sections were viewed by confocal microscopy.
Gas chromatography.
Short-chain volatile fatty acids were extracted from the mouse cecal mucus as described by Holdeman et al. (12). Nonvolatile acids were extracted from the mouse cecal mucus and methylated as described by Holdeman et al. (12). Short-chain volatile fatty acids and methylated nonvolatile acids were assayed in a CAPCO model 700 gas chromatograph equipped with a 1/4 in. by 6 ft column packed with 10% SP1000 on 100/120 Chromosorb W. The chromatograph was run at 135°C with helium as the carrier gas at 120 ml/min, and the acids were detected with a thermal conductivity detector at −95 mA. A Supelco volatile acid standard mix and nonvolatile acid standard mix were used as standards.
RESULTS
Growth of EDL933 in cecal mucus and in cecal contents in vitro.
Although strain EDL933 has been shown to colonize streptomycin-treated mouse large intestines and to grow in cecal mucus in vitro (38), it has not previously been tested for growth in cecal contents. To this end, E. coli EDL933 Strr was inoculated at 5 × 105 CFU/ml into both undiluted cecal mucus and cecal luminal contents at 37°C and cultures were assayed for viable counts at various times. In mucus, EDL933 Strr grew with a doubling time of about 20 min and reached a final cell density of about 109 CFU/ml. In contrast, EDL933 Strr remained relatively constant in number in cecal contents for 6 h postinoculation and was undetectable at 24 h postinoculation. Therefore, as with strain MG1655, it appears likely that EDL933 colonizes the mouse large intestine by growing in mucus.
E. coli MG1655 and EDL933 appear to utilize different nutrients in the intestine when fed to mice simultaneously.
The mouse model of large intestine colonization requires that mice be fed streptomycin sulfate in their drinking water throughout the duration of the experiment. This treatment eliminates the facultative flora and creates a niche for E. coli but leaves the obligate anaerobe population largely intact (21). When E. coli MG1655 Strr and MG1655 Strr Nalr are fed together to streptomycin-treated mice (105 CFU of each strain per mouse), the two strains colonize identically over a 15-day period, with each at a level of about 107 CFU/g of feces (data not shown). Therefore, MG1655 Strr and MG1655 Strr Nalr are considered to be isogenic, and each is regarded as the wild-type MG1655 strain. Similarly, E. coli EDL933 Strr and EDL933 Strr Nalr also colonize identically, with each at a level of about 107 CFU/g of feces (data not shown). Thus, they are also considered to be isogenic, and each is regarded as the wild-type EDL933 strain.
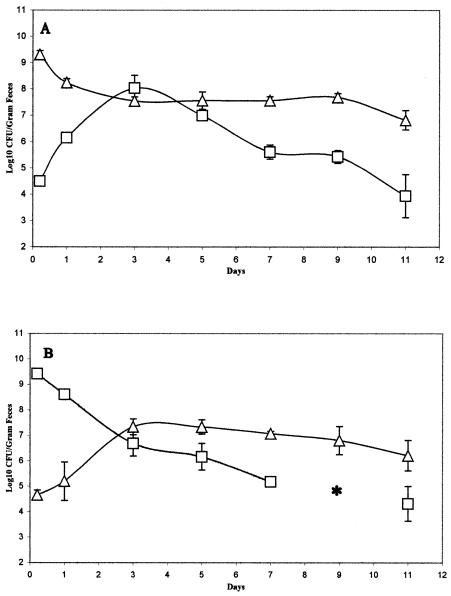
When streptomycin-treated mice are simultaneously fed 1010 CFU of an E. coli strain and 106 CFU of the same strain marked with an additional antibiotic resistance cassette for identification, the strains maintain the ratio of their original input values throughout a colonization experiment, implying that they have the same growth rate in the intestine and that the same nutrients are used with equal efficiencies (35). With this in mind, experiments were performed to determine whether EDL933 and MG1655 also have the same growth rate and nutrient use during growth in the intestine. A set of three mice was fed 1010 CFU of the wild-type MG1655 Strr strain and 105 CFU of the wild-type EDL933 Strr Nalr strain. At 5 h postfeeding, MG1655 Strr was found at an approximate level of 109 CFU/g of feces, whereas EDL933 Strr Nalr was at an approximate level of 104 CFU/g of feces, reflective of the input ratio (Fig. 1A). However, within 3 days postfeeding, EDL933 Strr Nalr had grown to a level of 108 CFU/g of feces, i.e., about the same level as MG1655 Strr, and then slowly but continuously dropped to a level about 3 orders of magnitude below that of MG1655 Strr (Fig. 1A). Since EDL933 Strr Nalr grew in the intestine from small to large numbers in the presence of large numbers of MG1655 Strr, it appeared that EDL933 was initially able to utilize one or more nutrients in the intestine better than MG1655 could, but then had difficulty maintaining itself in the presence of large numbers of MG1655. Similarly, small numbers of MG1655 Strr Nalr (105 CFU/mouse) grew to large numbers (∼107 CFU/g of feces) in the presence of large numbers of EDL933 Strr (1010 CFU/mouse) within 3 days postfeeding and remained at that level throughout the 15-day duration of the experiment (Fig. 1B). These results indicated that MG1655 was initially able to use one or more nutrients in the intestine better than EDL933 could.
FIG. 1.
Colonization of the intestine by MG1655 and EDL933 fed simultaneously to mice. Sets of three mice were fed either 1010 CFU of MG1655 Strr (▵) and 105 CFU of EDL933 Strr Nalr (□) (A) or 1010 CFU of EDL933 Strr (□) and 105 CFU of MG1655 Strr Nalr (▵) (B). At the indicated times, fecal samples were homogenized, diluted, and plated as described in Materials and Methods. When necessary, 100 colonies from MacConkey agar plates containing streptomycin were applied with toothpicks to MacConkey agar plates containing both streptomycin and nalidixic acid. The asterisk in place of the day 9 data point represents <1 colony in 100 toothpicked colonies from each mouse that were EDL933 Strr. Bars representing standard errors of the log10 means of CFU per gram of feces for each set of three mice are presented for each time point.
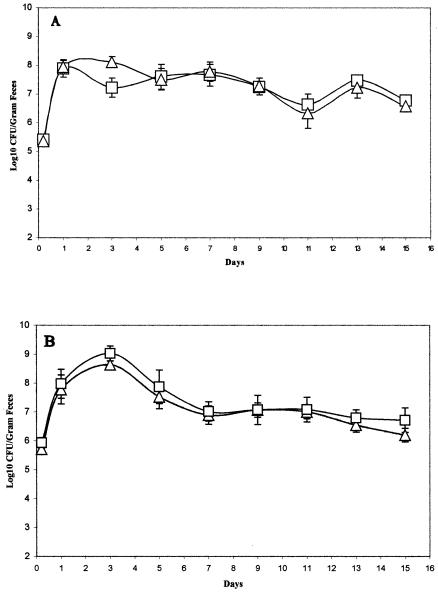
MG1655 and EDL933 ΔppsA ΔpckA double mutants colonize at the same level as their wild-type parents.
E. coli strains can utilize both glycolytic and gluconeogenic substrates for growth; however, it is not known whether they use both glycolytic and gluconeogenic substrates to colonize the mouse intestine. Phosphoenolpyruvate carboxykinase converts oxaloacetate to phosphoenolpyruvate and is encoded by the E. coli pckA gene (20). Phosphoenolpyruvate synthase is encoded by the E. coli ppsA gene and converts pyruvate to phosphoenolpyruvate (27). Since the conversion of TCA cycle intermediates to phosphoenolpyruvate is completely blocked in a ppsA pckA double mutant, it is unable to grow on gluconeogenic substrates “below” the step of phosphoenolpyruvate, but glycolytic pathways are not affected (10). MG1655 Strr ΔppsA ΔpckA::Cm and EDL933 Strr ΔppsA ΔpckA::Cm were constructed, and after it was shown that they failed to grow on gluconeogenic substrates but were still able to grow on glycolytic substrates (Table 3), each was fed to a set of three mice along with the respective wild-type parent at a level of 106 CFU/mouse. The colonizing abilities of MG1655 Strr Nalr and MG1655 Strr ΔppsA ΔpckA::Cm were virtually indistinguishable; each strain colonized at a level of about 107 CFU/g of feces (Fig. 2A). These data indicate that MG1655 predominantly utilizes glycolytic substrates for growth in the intestine when it is the only E. coli strain fed to mice. Similarly, the EDL933 Strr ΔppsA ΔpckA::Cm mutant colonized the intestine at the same level as EDL933 Strr Nalr throughout the 15 days of the experiment; each strain colonized at a level of about 107 CFU/g of feces (Fig. 2B). These data suggest that EDL933 predominantly utilizes glycolytic substrates for growth when it is the only E. coli strain fed to mice.
TABLE 3.
Growth of MG1655 and EDL933 strains on glycolytic and gluconeogenic carbon sources
| Carbon source | Growth of EDL933 or MG1655 strainsa
|
|
|---|---|---|
| Wild type | ΔppsA ΔpckA::Cm | |
| Gluconate | +++ | +++ |
| Glucose | +++ | +++ |
| Glycerol | +++ | +++ |
| Acetate | + | − |
| Aspartate | ++ | − |
| Fumarate | ++ | − |
| Malate | +++ | − |
| Pyruvate | +++ | − |
| Oleate | ++ | − |
| Succinate | ++ | − |
+++, A600 > 0.80; ++, A600 = 0.30 to 0.50; +, A600 = 0.10 to 0.20; −, A600 < 0.05. All cultures were inoculated at about 105 CFU/ml from an overnight M9 minimal medium glucose culture. Cultures were incubated at 37°C for 24 h with aeration in a shaking water bath.
FIG. 2.
Colonization of the mouse intestine by E. coli strain MG1655 and its ppsA pckA deletion mutant and EDL933 and its ppsA pckA deletion mutant. Sets of three mice were fed either 105 CFU of MG1655 Strr Nalr (□) and 105 CFU of MG1655 Strr ΔppsA ΔpckA::Cm (▵) (A) or 105 CFU of EDL933 Strr Nalr (□) and 105 CFU of EDL933 Strr ΔppsA ΔpckA::Cm (▵) (B). At the indicated times, fecal samples were homogenized, diluted, and plated as described in Materials and Methods. Bars representing standard errors of the log10 means of CFU per gram of feces for each set of three mice are presented for each time point.
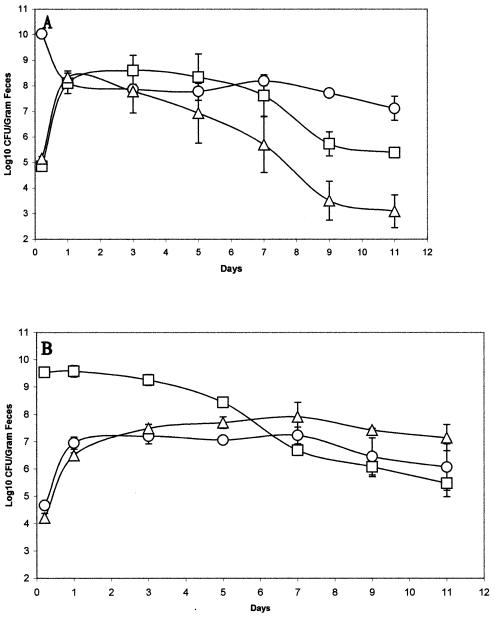
The EDL933 Strr ΔppsA ΔpckA (gluconeogenesis defective) mutant is not maintained as well as wild-type EDL933 Strr Nalr in the intestine in the presence of large numbers of MG1655 Strr.
Although strain EDL933 did not appear to utilize gluconeogenic substrates for growth when it was the only E. coli strain fed to mice (Fig. 2B), we were interested in determining whether EDL933, which had difficulty maintaining itself in the presence of large numbers of MG1655 (Fig. 1A), utilizes gluconeogenic substrates under those conditions. Therefore, mice were fed MG1655 Strr (1010 CFU/mouse) along with both EDL933 Strr Nalr (105 CFU/mouse) and EDL933 Strr ΔppsA ΔpckA::Cm (105 CFU/mouse). As shown in Fig. 3A, during day 1 postfeeding, both EDL933 Strr Nalr and EDL933 Strr ΔppsA ΔpckA::Cm initially grew equally well in the mouse intestine to levels of about 108 CFU/g of feces. However, beyond day 1 postfeeding, EDL933 Strr ΔppsA ΔpckA::Cm was not maintained as well as the wild-type EDL933 Strr strain, such that by day 7 postfeeding, it was at a level about 2 orders of magnitude below that of the wild-type strain (Fig. 3A). These data indicate that, in addition to the utilization of glycolytic substrates, EDL933 utilizes gluconeogenic substrates for growth in the presence of large numbers of MG1655 in an attempt to maintain itself in the mouse intestine. In contrast, MG1655 Strr ΔppsA ΔpckA::Cm actually had a slight advantage over the wild-type MG1655 Strr strain in the mouse intestine in the presence of large numbers of the EDL933 wild-type strain (Fig. 3B). These data indicate that MG1655 utilizes glycolytic substrates exclusively for growth in the intestine in the presence of large numbers of EDL933.
FIG. 3.
Colonization of the mouse intestine by E. coli strains MG1655 and EDL933 and their ΔppsA ΔpckA::Cm mutants. Sets of three mice were fed 1010 CFU of MG1655 Strr (○) and 105 CFU each of EDL933 Strr Nalr (□) and EDL933 Strr ΔppsA ΔpckA::Cm (▵) (A) or 1010 CFU of EDL933 Strr (□) and 105 CFU each of MG1655 Strr Nalr (○) and MG1655 Strr ΔppsA ΔpckA::Cm (▵) (B). At the indicated times, fecal samples were homogenized, diluted, and plated as described in Materials and Methods. When necessary, 100 colonies from MacConkey agar plates containing streptomycin were applied with toothpicks to MacConkey agar plates containing both streptomycin and nalidixic acid and plates containing both streptomycin and chloramphenicol to determine numbers of either EDL933 Strr or MG1655 Strr cells. Bars representing standard errors of the log10 means of CFU per gram of feces for each set of three mice are presented for each time point.
Location (vertical distribution) of EDL933 and MG1655 in the mouse intestine.
Although the ability of E. coli to grow in intestinal mucus has been correlated with its ability to colonize the mouse large intestine (18, 26, 35, 36, 39), there is little information to indicate where various E. coli strains are found in the intestine. Therefore, EDL933 Strr and MG1655 Strr were each fed to separate sets of three mice. On day 16 postfeeding, the numbers of the strains in ileal mucus, the mucus isolated from the rest of the small intestine, cecal mucus, and colonic mucus were determined. The numbers of MG1655 and EDL933 CFU were at least 10-fold higher in cecal and colonic mucus than in ileal and small intestinal mucus (Table 4). These data indicate that during the maintenance stage of colonization, both EDL933 and MG1655 grow in mucus throughout the mouse intestine, but to far larger numbers in cecal and colonic mucus.
TABLE 4.
Distribution in intestinal mucus preparations of MG1655 Strr and EDL933 Strr at 16 days postfeeding to separate mice
| Tissue from which day 16 mucus was isolated | Amt of bacteria in tissue (log10 CFU/section)c
|
|
|---|---|---|
| MG1655 Strr | EDL933 Strr | |
| Small intestinea | 4.07 ± 0.33 | 4.72 ± 0.02 |
| Ileum | 3.80 ± 0.15 | 3.62 ± 0.29 |
| Cecum | 5.59 ± 0.21 | 6.11 ± 0.65 |
| Colon | 5.67 ± 0.25 | 5.57 ± 0.54 |
| Feces (day 15)b | 7.42 ± 0.23 | 7.44 ± 0.22 |
Mucus from immediately below the stomach to the proximal ileum.
Data for feces are given as log10 CFU per gram of feces on day 15 postfeeding.
Data are means ± standard errors of the means.
Visualization of EDL933 and MG1655 in thin slices of the mouse intestine.
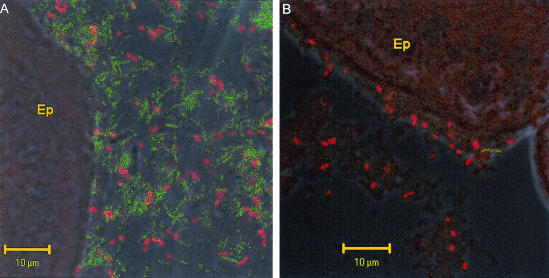
It was previously shown that when MG1655 is fed to mice, it is found in the mucus layer of the mouse cecum but is not associated with epithelial cells (see Fig. 5A in reference 23). In contrast, EDL933 has been shown to bind to intestinal epithelial cells in calves, pigs, and mice (5, 15, 37, 39). For determination of whether EDL933 is associated with mouse intestinal epithelial cells under the conditions of the present study, mice were fed EDL933 Strr (105 CFU/mouse). At 24 and 48 h postfeeding, EDL933 Strr was visualized in situ in the mucosa of the cecum, the colon, the ileum, and the middle of the small intestine by hybridization with an E. coli-specific oligonucleotide probe (see Materials and Methods). As a control, separate mice were fed MG1655 Strr (105 CFU/mouse), which was also visualized by the same method in situ in the mucosa of the cecum, the colon, the ileum, and the middle of the small intestine at 24 and 48 h postfeeding. In the cecum, EDL933 was present both in the mucus and closely associated with epithelial cells at both 24 and 48 h postfeeding (Fig. 4A and 5B). That EDL933 was closely associated with epithelial cells was especially apparent in sections in which the mucus layer had been ripped away from the epithelial cell surface (Fig. 4B). The same results were found in the colon (not shown). At least 10-fold fewer EDL933 cells were seen in the mucosa of the ileum and the middle of the small intestine than in the cecum and colon, and the majority of EDL933 cells observed were associated with epithelial cells (not shown). In contrast, MG1655 was observed in the cecum exclusively in mucus (not shown), as was observed previously (see Fig. 5 in reference 23). The same result was true of the colon, the ileum, and the middle of the small intestine, although at least 10-fold fewer MG1655 cells were observed in the ileum and the middle of the small intestine than in the cecum and colon (not shown). Since the vast majority of EDL933 cells in the intestine at 24 and 48 h postfeeding were in the cecum and colon and since EDL933 fails to grow in cecal contents (see above), it appears that the vast majority of its growth in the first 24 to 48 h occurs in cecal and colonic mucus and perhaps on epithelial cells.
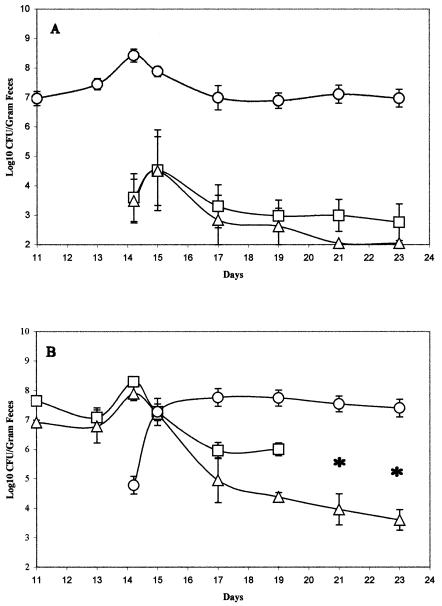
FIG. 5.
Colonization of the intestine by strains MG1655 and EDL933 when precolonized with either MG1655 or EDL933. Sets of three mice were fed either 105 CFU of MG1655 Strr on day 0 (○) and 105 CFU each of EDL933 Strr Nalr (□) and EDL933 Strr ΔppsA ΔpckA::Cm (▵) on day 14 postfeeding (A) or 105 CFU each of EDL933 Strr (□) and EDL933 Strr ΔppsA ΔpckA::Cm (▵) on day 0 and 105 CFU of MG1655 Strr Nalr (○) on day 14 postfeeding (B). At the indicated times, fecal samples were homogenized, diluted, and plated as described in Materials and Methods. When necessary, 100 colonies from MacConkey agar plates containing streptomycin were applied with toothpicks to MacConkey agar plates containing both streptomycin and nalidixic acid and plates containing both streptomycin and chloramphenicol to determine numbers of EDL933 Strr cells. Asterisks in place of data points represent days when <1 colony in 100 toothpicked colonies from each mouse were EDL933 Strr. Bars representing standard errors of the log10 means of CFU per gram of feces for each set of three mice are presented for each time point.
FIG. 4.
In situ hybridization with fluorescence-labeled oligonucleotide probes. At 24 and 48 h postfeeding, cecal mucosal sections of mice fed EDL933 Strr were hybridized with an E. coli-specific oligonucleotide probe (red) and a eubacteria-specific oligonucleotide probe (green). Cecal sections from 48 h are shown. EP, epithelial cell. E. coli cells appear red, while all other bacteria appear green. Bar, 10 μm. (A) The mucus layer is intact. (B) The mucus layer was separated from the epithelial cell during preparation.
Mouse cecal mucus does not contain high levels of TCA cycle intermediates.
For determination of whether EDL933 might use TCA cycle intermediates for growth in mouse cecal mucus in vitro, mouse cecal mucus (4 mg/ml) was analyzed by gas chromatography for fumarate, oxaloacetate, succinate, and acetate (see Materials and Methods). Fumarate, oxaloacetate, and succinate were below the limit of detection (0.2 μM) and acetate was just at the limit of detection (0.2 μM). It is therefore unlikely that the gluconeogenic substrates that EDL933 utilizes for growth in cecal mucus in vitro are TCA cycle intermediates or acetate. Furthermore, since the dry weight of mucus isolated directly from the mouse cecum is about 80 mg/ml, it would contain, at most, 4 μM concentrations of acetate, fumarate, oxaloacetate, and succinate. This translates to <1 μg of each of these substrates in cecal mucus/ml in vivo, making it highly unlikely that they are being used as gluconeogenic substrates for the growth of EDL933 in the mouse intestine.
EDL933 Strr and EDL933 Strr ΔppsA ΔpckA::Cm fail to grow in mice that are precolonized with MG1655.
Humans infected with O157:H7 strains are presumably colonized with at least one commensal E. coli strain. To mimic that situation, mice were precolonized with strain MG1655 Strr (105 CFU/mouse) and on day 14 postfeeding were fed EDL933 Strr Nalr (105 CFU/mouse) and EDL933 Strr ΔppsA ΔpckA::Cm (105 CFU/mouse). In contrast to the results of the cofeeding experiments (Fig. 3A), EDL933 Strr Nalr and EDL933 Strr ΔppsA ΔpckA::Cm failed to grow extensively in mice that were precolonized with MG1655 Strr, whereas MG1655 Strr remained at a level of about 107 CFU/g of feces (Fig. 5A). Furthermore, the wild-type EDL933 strain stabilized at a level of about 103 CFU/g of feces, whereas EDL933 Strr ΔppsA ΔpckA::Cm was completely eliminated from the intestine (Fig. 5A). Therefore, the precolonization of mice with the commensal E. coli MG1655 afforded protection against the growth of low levels of infecting wild-type EDL933 to high levels in the intestine.
Mice were also precolonized with EDL933 Strr and EDL933 Strr ΔppsA ΔpckA::Cm and were then fed MG1655 Strr Nalr (105 CFU/mouse) at day 14. Within 1 day postfeeding, MG1655 Strr Nalr grew to the level of EDL933 Strr and then colonized the mice at a level of >107 CFU/g of feces, whereas both EDL933 Strr and EDL933 Strr ΔppsA ΔpckA::Cm levels dropped continuously (Fig. 5B). Once again, the EDL933 Strr ΔppsA ΔpckA::Cm level dropped at a much faster rate than the EDL933 Strr level, further indicating that EDL933 is maintained in the presence of MG1655 by the utilization of gluconeogenic substrates (Fig. 5B). In control experiments, it was found that wild-type EDL933 not challenged with MG1655 colonizes the mouse intestine at a level of about 107 CFU/g of feces for at least 23 days (not shown), showing that it was indeed the MG1655 fed to mice at day 14 that caused the elimination of EDL933 (Fig. 5B).
DISCUSSION
The large intestine of the mouse consists of the cecum and the colon, each of which contains the mucosa and the luminal contents. The two components of the mucosa are the layer of epithelial cells on the intestinal wall and the mucus layer, which covers them. The relatively thick (up to 400 μm) mucous layer consists of mucin, a 2-MDa gel-forming glycoprotein, and a large number of smaller glycoproteins, proteins, glycolipids, lipids, and sugars (1, 6, 7, 16, 28, 29, 31, 34). Presumably, shed epithelial cells are a source of many of the smaller mucous components (29, 31). The mucus layer itself is in a dynamic state, constantly being synthesized and secreted by specialized goblet cells and degraded to a large extent by the indigenous intestinal microbes (13, 25). Degraded mucus components are shed into the intestinal lumen, forming a part of the luminal contents, and are excreted in feces (13).
The prevalent theory to explain how bacteria colonize the mammalian gut is that the approximately 500 indigenous species (24) can coexist as long as each member of the flora is able to utilize one or a few limiting nutrients better than all the others and its rate of growth during the colonization process is at least equal to the washout rate from the intestine (8, 9). The rate of growth in the intestine by a particular bacterium is presumably determined by the nature of the limiting nutrient it utilizes, and the density to which it grows is determined by the available concentration of that nutrient. E. coli strains appear to colonize the mouse intestine by growing in the mucus layer (18, 26, 35, 36, 38, 40). Consequently, if two E. coli strains compete for the same nutrient(s) in the mucus layer and neither strain is attached to epithelial cells, the one that utilizes them more efficiently will eliminate the other strain (9, 10).
Although the human and mouse large intestinal mucosa are structurally alike, the unperturbed human intestine is sensitive to as few as 50 E. coli O157:H7 cells (11), whereas the mouse intestine must be treated with streptomycin to create a niche for E. coli strains, including O157:H7. It is currently unclear whether the difference in sensitivity to O157:H7 strains is due to a difference in innate immunity, the composition of the microflora, consequences of adhesion to epithelial cells, nutrient availability, or a combination of these factors. Nevertheless, the data presented here suggest that the streptomycin-treated mouse model appears to be ideally suited to identifying differences in nutrient preferences between O157:H7 and commensal E. coli strains in the mouse intestine that might also contribute to pathogenesis in humans.
By this investigation, evidence was presented that EDL933 primarily utilizes one or more glycolytic substrates for 1 day of initial rapid growth in the mouse intestine in the presence of large numbers of MG1655 cells, suggesting either that it uses those glycolytic substrates more efficiently than MG1655 or that those substrates are unavailable to MG1655. The data also suggest that to maintain itself maximally in the intestine beyond day 1 in the presence of large numbers of MG1655, EDL933 utilizes one or more gluconeogenic substrates in addition to glycolytic substrates, i.e., EDL933 Strr ΔppsA ΔpckA::Cm colonized poorly relative to EDL933 Strr Nalr in the presence of large numbers of MG1655 (Fig. 3A). However, the utilization of gluconeogenic substrates does not allow EDL933 to remain in large numbers in the presence of MG1655; instead, it merely postpones its decline to smaller numbers (Fig. 3A). In contrast, the data presented here indicate that EDL933 utilizes glycolytic substrates exclusively for both successful initial growth as well as maintenance when it is the only E. coli strain fed to the mice, i.e., in the absence of MG1655, EDL933 Strr Nalr and EDL933 Strr Δpps ΔpckA::Cm colonized equally well (Fig. 2B). An explanation may be that in the absence of MG1655, EDL933 has sufficient glycolytic substrates to maintain the colonization state, but in the presence of large numbers of MG1655, it is unable to compete effectively for those substrates beyond the first few days and begins to utilize gluconeogenic substrates in an attempt to remain in the intestine.
At the present time, it is not known which gluconeogenic substrates EDL933 uses for growth in the mouse large intestine, but it is unlikely that acetate, fumarate, oxaloacetate, or succinate are involved since, as shown here, the concentration of each is too low in mucus to support growth. Possibilities for gluconeogenic substrates that EDL933 utilizes in mucus include the amino acids aspartate and tryptophan, which can serve as sole carbon sources for E. coli growth (19). Free amino acids are found in mouse cecal mucus (7). In addition, phosphatidylserine, present at a relatively high concentration in mouse cecal mucus preparations scraped from the intestinal wall (17), has been shown to serve as a sole source of carbon for the growth of EDL933 (17). Since EDL933 can utilize fatty acids for growth (Table 3), it is possible that the fatty acids contained in phospholipids serve as gluconeogenic substrates for EDL933 in the mouse intestine when this strain is growing in the presence of large numbers of MG1655. In addition, it was previously reported that, of the carbon sources present in intestinal mucus, E. coli K-12 utilizes gluconate and other as yet unidentified carbon sources for growth in streptomycin-treated mice (28, 36, 37). Since MG1655 is a K-12 strain, it is possible that gluconate is one of the glycolytic substrates that MG1655 utilizes better than EDL933 in the intestine.
In the present study, it was also shown that EDL933 is found both in mucus and closely associated with intestinal epithelial cells for at least 2 days (Fig. 4), whereas MG1655 is known to reside exclusively in the mucus layer (see Fig. 5 of reference 23). EDL933 may therefore be able to utilize nutrients that are available exclusively at the surfaces of the epithelial cells. One intriguing possibility is that if EDL933 damages the membranes of intestinal epithelial cells in streptomycin-treated mice, as it does in other animal models (5, 15, 37), it may be able to utilize the nutrients present in the damaged membranes for growth. This source of nutrients would presumably be unavailable to MG1655, since it is not associated with intestinal epithelial cells in vivo (23).
Another interesting finding is that small numbers of EDL933 (105 CFU/mouse) are unable to grow in the intestine to any large extent in mice that are precolonized with MG1655 (Fig. 5A), yet they are able to grow to at least the same level as MG1655 when small numbers of EDL933 (105 CFU/mouse) are fed to mice simultaneously with large numbers of MG1655 (1010 CFU/mouse) (Fig. 1A and 3A). It is unlikely that after several days in the intestine MG1655 secretes an antimicrobial that limits the growth of EDL933, since precolonized EDL933 cells are eliminated rapidly by small numbers of MG1655 fed to mice at day 14 (Fig. 5B). These findings suggest that there is competition between the two strains for nutrients. It is therefore more likely that after a few days in the intestine, MG1655 adapts to the glycolytic substrate that small numbers of EDL933 need for initial growth, and after adapting, utilizes this substrate at least as well as EDL933 or that the substrate is depleted by EDL933 during its rapid growth phase. In either case, these data raise the possibility that in humans colonized with commensal strains such as MG1655, exposure to EDL933 would not result in disease, whereas other humans could be colonized with commensal strains that do not compete well with EDL933 for glycolytic substrates and disease would ensue. For determination of whether this hypothesis has merit, the experiments presented here will have to be repeated with a number of commensal E. coli strains that have been isolated from humans. Future studies will be designed to address this issue as well as to determine the glycolytic and gluconeogenic substrates that EDL933 utilizes in the mouse intestine in the presence of large numbers of MG1655 and the glycolytic substrates that MG1655 utilizes in both the presence and absence of EDL933.
Acknowledgments
We thank April Anderson for a critical reading of the manuscript.
This work was supported by a grant from the NIH (RO1-AI 48945) to T.C. and P.S.C. W.E.N. was supported by grant H-401 from the University of Rhode Island Agricultural Experiment Station to P.S.C.
Editor: B. B. Finlay
REFERENCES
- 1.Allan, A. 1981. Structure and function of gastrointestinal mucus, p. 637-639. In L. R. Johnson (ed.), Physiology of the gastrointestinal tract. Raven Press, New York, N.Y.
- 2.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, P. S., and D. C. Laux. 1995. Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods Enzymol. 253:309-314. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean-Nystrom, E. A., B. T. Bosworth, W. C. Cray, Jr., and H. W. Moon. 1997. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect. Immun. 65:1842-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forstner, G. G. 1970. [1-14C]glucosamine incorporation by subcellular fractions of small intestine mucosa. J. Biol. Chem. 245:3584-3592. [PubMed] [Google Scholar]
- 7.Franklin, D. P., D. C. Laux, T. J. Williams, M. C. Falk, and P. S. Cohen. 1990. Growth of Salmonella typhimurium SL5319 and Escherichia coli F-18 in mouse cecal mucus: role of peptides and iron. FEMS Microbiol. Ecol. 74:229-240. [Google Scholar]
- 8.Freter, R., H. Brickner, M. Botney, D. Cleven, and A. Aranki. 1983. Mechanisms that control bacterial populations in continuous-flow culture models or mouse large intestinal flora. Infect. Immun. 39:676-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freter, R., H. Brickner, J. Fekete, M. M. Vickerman, and K. E. Carey. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 39:686-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldie, A. H., and B. D. Sanwal. 1980. Genetic and physiological characterization of Escherichia coli mutants deficient in phosphoenolpyruvate carboxykinase activity. J. Bacteriol. 141:1115-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin, P. M. 1998. Epidemiology of Shiga toxin-producing Escherichia coli infections in humans in the United States, p. 15-22. In J. P. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 12.Holdeman, L. V., E. P. Cato, and W. E. C. Moore (ed.). 1977. Chromatographic procedures for analysis of acid and alcohol products, p. 134-140. In Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 13.Hoskins, L. 1984. Mucin degradation by enteric bacteria: ecological aspects and implications for bacterial attachment to gut mucosa, p. 51-65. In E. C. Boedecker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 14.Kaper, J. B., and A. D. O'Brien. 1998. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 15.Karpman, D., H. Connell, M. Svensson, F. Scheutz, P. Alm, and C. Svenborg. 1997. The role of lipopolysaccharide and Shiga-like toxin in a mouse model of Escherichia coli O157:H7 infection. J. Infect. Dis. 175:611-620. [DOI] [PubMed] [Google Scholar]
- 16.Kim, Y. S., A. Morita, S. Miura, and B. Siddiqul. 1984. Structure of glycoconjugates of intestinal mucosal membranes. Role of bacterial adherence, p. 99-109. In E. C. Boedecker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 17.Krivan, H. C., D. P. Franklin, W. T. Wang, D. C. Laux, and P. S. Cohen. 1992. Phosphatidylserine found in intestinal mucus serves as a sole source of carbon and nitrogen for salmonellae and Escherichia coli. Infect. Immun. 60:3943-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licht, T. R., T. Tolker-Nielsen, K. Holmstrøm, K. A. Krogfelt, and S. Molin. 1999. Inhibition of Escherichia coli precursor 16S rRNA processing by mouse intestinal contents. Environ. Microbiol. 1:23-32. [DOI] [PubMed] [Google Scholar]
- 19.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikov, M. Riley, M. Schaechter, and H. E. Umbarger, (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 20.Medina, V., R. Pontarollo, D. Glaeske, H. Tabel, and H. Goldie. 1990. Sequence of the pckA gene of Escherichia coli K-12: relevance to genetic and allosteric regulation and homology of E. coli phosphoenolpyruvate carboxykinase with the enzymes from Trypanosoma brucei and Saccharomyces cerevisiae. J. Bacteriol. 172:7151-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, C. P., and M. Bohnhoff. 1963. Changes in the mouse's enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin-treatment. J. Infect. Dis. 113:59-66. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Møller, A. K., M. P. Leatham, T. Conway, P. J. M. Nuijten, L. A. M. de Haan, K. A. Krogfelt, and P. S. Cohen. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, W. E. C., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neutra, M. R. 1984. The mechanism of intestinal mucous secretion, p. 33-41. In E. C. Boedecker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 26.Newman, J. V., R. Kolter, D. C. Laux, and P. S. Cohen. 1994. The role of leuX in Escherichia coli colonization of the streptomycin-treated mouse large intestine. Microb. Pathog. 17:301-311. [DOI] [PubMed] [Google Scholar]
- 27.Niersbach, M., F. Kreuzaler, R. H. Geerse, P. W. Postma, and H. J. Hirsch. 1992. Cloning and nucleotide sequence of the Escherichia coli K-12 ppsA gene, encoding PEP synthase. Mol. Gen. Genet. 231:332-336. [DOI] [PubMed] [Google Scholar]
- 28.Peekhaus, N., and T. Conway. 1998. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 180:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potten, C. S., and T. D. Allen. 1977. Ultrastructure of cell loss in intestinal mucosa. J. Ultrastruct. Res. 60:272-277. [DOI] [PubMed] [Google Scholar]
- 30.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine from rRNA in situ hybridization. Infect. Immun. 62:5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quastler, H., and F. G. Sherman. 1959. Cell population in the intestinal epithelium of the mouse. Exp. Cell. Res. 17:420-438. [DOI] [PubMed] [Google Scholar]
- 32.Revel, H. R. 1966. Restriction and nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology 31:688-701. [DOI] [PubMed] [Google Scholar]
- 33.Saxena, S. K., A. D. O'Brien, and E. J. Ackerman. 1989. Shiga toxin, Shiga-like toxin II variant, and ricin are all single-site RNA N-glycosidases of 28S RNA when microinjected into Xenopus oocytes. J. Biol. Chem. 264:596-601. [PubMed] [Google Scholar]
- 34.Slomiany, A., S. Yano, B. I. Slomiany, and G. B. J. Glass. 1978. Lipid composition of the gastric mucus barrier in the rat. J. Biol. Chem. 253:3785-3791. [PubMed] [Google Scholar]
- 35.Sweeney, N. J., P. Klemm, B. A. McCormick, E. Moller-Nielsen, M. Utley, M. A. Schembri, D. C. Laux, and P. S. Cohen. 1996. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect. Immun. 64:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sweeney, N. J., D. C. Laux, and P. S. Cohen. 1996. Escherichia coli F-18 and K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 64:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzipori, S., F. Gunzer, M. S. Donnenberg, L. de Montigny, J. B. Kaper, and A. Donohue-Rolfe. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadolkowski, E. A., J. A. Burris, and A. D. O'Brien. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 58:2438-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wadolkowski, E. A., D. C. Laux, and P. S. Cohen. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect. Immun. 56:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wadolkowski, E. A., L. M. Sung, J. A. Burris, J. E. Samuel, and A. D. O'Brien. 1990. Acute renal tubular necrosis and death of mice orally infected with Escherichia coli strains that produce Shiga-like toxin type II. Infect. Immun. 58:3959-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]