Abstract
Streptococcus mutans is a biofilm-forming bacterium that is adapted to tolerate rapid and dramatic fluctuations in nutrient availability, carbohydrate source, and pH in its natural environment, the human oral cavity. Dissecting the pathways used to form stable biofilms and to tolerate environmental stress is central to understanding the virulence of this organism. Here, we investigated the role of the S. mutans relA gene, which codes for a guanosine tetraphosphate and guanosine pentaphosphate [(p)ppGpp] synthetase/hydrolase, in biofilm formation and acid tolerance. Two mutants in which relA was insertionally inactivated or replaced by an antibiotic resistance determinant were constructed. Under normal growth and stress conditions, the mutants grew slower than the wild-type strain, although the final yields were similar. The mutants, which were still able to accumulate (p)ppGpp after the induction of a stringent response, showed significant reductions in biofilm formation on microtiter plates or hydroxylapatite disks. There was no difference in the sensitivities to acid killing of the parent and relA strains grown in planktonic cultures. However, when cells were grown in biofilms, the mutants became more acid resistant and could lower the pH through glycolysis faster and to a greater extent than the wild-type strain. Differences in acid resistance were not correlated with increases in F-ATPase activity, although bacterial sugar:phosphotransferase activity was elevated in the mutants. Expression of the luxS gene was increased as much as fivefold in the relA mutants, suggesting a link between AI-2 quorum sensing and the stringent response.
During periods of nutrient starvation, bacteria undergo a “stringent response” that is characterized by the inhibition of rRNA synthesis, the induction or repression of metabolic pathways according to physiological needs, and the induction of stationary-phase survival genes (12). Under nutrient-limiting conditions, the synthesis of guanosine tetraphosphate and guanosine pentaphosphate [(p)ppGpp], which function as chemical messengers that allow bacteria to adapt their metabolism from a growth mode to a survival mode, is triggered by the binding of uncharged tRNAs to ribosomes. In Escherichia coli, relA encodes the (p)ppGpp synthetase and spoT is a bifunctional enzyme with both (p)ppGpp synthetase and (p)ppGpp hydrolase activities (12). In gram-positive bacteria, RelA is a bifunctional enzyme with (p)ppGpp synthetic and degradative activities (29, 31, 51).
Recently, a few reports have demonstrated that the relA gene product is involved in a variety of fundamental physiological processes, including biofilm formation in Listeria monocytogenes and E. coli (4, 44), competence development in Bacillus subtilis (21), and quorum-sensing in Pseudomonas aeruginosa (47). It was shown that in Lactococcus lactis, increased levels of (p)ppGpp lead to enhanced tolerance of multiple stresses (38), and in L. monocytogenes, a relA mutant strain that failed to accumulate (p)ppGpp displayed an osmosensitive phenotype (34). Therefore, it appears that there is a connection between intracellular levels of (p)ppGpp and stress tolerance.
Streptococcus mutans, the principal etiological agent of human dental caries, is an “obligate” biofilm bacterium that is well adapted to tolerate rapid environmental fluctuations in nutrient availability, carbohydrate source, and pH (7, 9). In oral biofilms, organisms must deal with rapid and often substantial changes in nutrient availability, pH, oxygen tension, and redox environment and other stressors. Of these variables, pH and carbohydrate have been shown to have the most profound impact on supragingival plaque ecology and the development of dental caries. Carbohydrate concentrations can go from roughly 10 μM during fasting periods to well in excess of 10 mM during intake of heavily sweetened foodstuffs. Concurrent with the influx of dietary carbohydrates is a dramatic fall in pH to values as low as 4 and lower. This so-called feast-or-famine lifestyle (11) demands that organisms such as S. mutans, which take advantage of periods of high carbohydrate concentrations and low pHs to emerge in cariogenic plaque, thrive when environmental conditions are less favorable for competing species in the biofilms. Thus, stress tolerance and regulation of metabolism are key virulence attributes of this organism. In the past few years, a number of studies have sought to identify genes that are required by S. mutans to form stable biofilms and tolerate acid stress (3, 18, 20, 23, 24, 27, 32, 53). Some of these gene products appear to play a dual role in stress tolerance and biofilm formation, further demonstrating a connection between these important virulence factors (23, 26). Here, RelA-deficient strains were constructed and used to demonstrate a link between the stringent response and pathways required for biofilm formation and acid tolerance.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli DH10B was grown in Luria broth at 37°C. S. mutans strains were grown in brain heart infusion (BHI) broth in a 5% CO2 atmosphere at 37°C. If appropriate, ampicillin (100 μg ml−1), chloramphenicol (30 μg ml−1), or kanamycin (50 μg ml−1 for E. coli or 500 μg ml−1 for S. mutans) was added to the medium. To evaluate the capacity of the mutant strains to grow under different stress conditions, we grew strains overnight in BHI broth, diluted 50-fold into fresh BHI broth, and incubated at 37°C (control), at a high temperature (44°C), at a low pH (pH 5.0, 37°C), or at 37°C in the presence of 0.0025% H2O2. For amino acid starvation studies, cells were grown in the chemically defined medium FMC (45) to an optical density at 600 nm (OD600) of about 0.35. Cells were then collected by centrifugation, and aliquots were resuspended in experimental (FMC minus supplemented amino acids) or control medium (complete FMC) for different periods of time. The ability of cells to form stable biofilms was assessed by growing the cells in biofilm medium (BM) (28) broth in 96-well polystyrene microtiter plates (Costar 3595; Corning, Inc., Corning, N.Y.) for 24 or 48 h. For scanning electron microscopy (SEM), biofilms were grown in BM on the surface of hydroxylapatite (HA) disks that were deposited in the wells of a 24-well polystyrene tissue culture plate (Falcon 3047; Becton Dickinson, Lincoln Park, N.J.). Biofilms used for acid killing experiments, RNA extractions, or reporter gene assays were grown in the wells of 24-well polystyrene plates.
DNA methods.
Chromosomal DNA was prepared from S. mutans UA159 and its derivatives as previously described (10). Plasmid DNA was isolated from E. coli by using QIAGEN (Chatsworth, Calif.) columns, and restriction and DNA-modifying enzymes were obtained from Invitrogen (Gaithersburg, Md.) or New England Biolabs (Beverly, Mass.). PCRs were carried out with 100 ng of chromosomal DNA by using Taq DNA polymerase, and PCR products were purified with the QIAquick kit (QIAGEN). DNA was introduced into S. mutans by natural transformation (36) and into E. coli by the calcium chloride method (39). Southern blot analyses were carried out under high-stringency conditions as detailed by Sambrook et al. (39).
Construction of strains.
To insertionally inactivate the relA gene, a DNA fragment containing the gene of interest was amplified by PCR and directly cloned onto pGEM5 (Promega, Madison, Wis.) as a SphI-SpeI fragment. A kanamycin resistance (Kanr) gene that is flanked by strong transcription terminators (ΩKan) (35) was cloned into an NcoI site located 0.5 kbp from the relA start codon. A second relA mutant was constructed by replacing an 1,125-bp EcoRV fragment containing the 5′ end of relA with the ΩKan element, eliminating the possibility that any portion of the RelA could be produced. Plasmids containing the Kanr gene marker were then isolated and used to transform S. mutans. Transformants were selected on BHI agar containing the appropriate antibiotic, and Southern blot analysis was used to confirm that the desired gene inactivation had occurred by double-crossover recombination.
To express the relA gene in trans, the full-length relA gene was amplified by PCR with primers containing BamHI (5′ primer) and XbaI (3′ primer) restriction sites and cloned onto pMSP3535 (8, 15) that had been digested with BamHI and XbaI. The resulting plasmid, pJL69, contains the intact relA gene under the control of the lactococcal nisin promoter. Expression of relA was induced with 10 μg of nisin ml−1, and the production of RelA protein was confirmed by Western blotting. To construct a reporter gene fusion using the relA promoter region, a 0.5-kbp fragment containing the putative promoter region was cloned directly in front of a promoterless chloramphenicol acetyltransferase (CAT) gene (cat) in pCW24 (13) to yield plasmid pJL55. The reporter gene fusion was then excised from plasmid pJL55, subcloned onto the S. mutans integration vector pBGK (48), and integrated into the chromosome of the wild-type strain in a single copy by double-crossover recombination at the gtfA locus. The integration of the reporter gene fusion into the S. mutans chromosome was confirmed by Southern blot analysis.
SEM.
Biofilms grown on HA disks for 24 h were rinsed by gently dipping the disks in PBS buffer (50 mM sodium phosphate buffer [pH 7.0], 0.85% NaCl) and fixed with Trump fixative solution overnight. Fixed samples were then dehydrated through a graded series of ethanol concentrations, mounted, and sputter coated with gold-palladium. Samples were analyzed by SEM (Hitachi S-4000; Hitachi Instruments Inc., San Jose, Calif.) at several magnifications (×500 to ×13,000) at the University of Florida EM core laboratory (Biotechnology Program, University of Florida, Gainesville, Fla.).
(p)ppGpp measurements.
Strains were grown in FMC medium to an OD600 of 0.25. Two 1.3-ml aliquots were transferred to Eppendorf tubes, and control cultures were labeled with 100 μCi of carrier-free 32Pi (Amersham Biosciences, Piscataway, N.J.) ml−1 for 1.5 h, at which point the cultures had reached an OD600 of 0.45. To trigger a stringent response, the test cultures were grown to an OD600 of 0.45, at which point serine hydroxamate (1 mg ml−1) was added, inducing growth arrest, and the samples were labeled for 1.5 h with 100 μCi of carrier-free 32Pi ml−1. Extracts of the cells were obtained by concentrating the samples 20 times before adding 1 volume of 13 M formic acid, followed by three freeze-thaw cycles. Extracts were centrifuged at 13,000 rpm for 2 min in a microcentrifuge (model 5415C; Eppendorf, Westbury, N.Y.), and 30 μl of supernatant fluid was spotted on a polyethyleneimine-cellulose plate (Sigma, St. Louis, Mo.) for separation by thin-layer chromatography in 1.5 M KH2PO4 (pH 3.4) of the phosphorylated guanosine nucleotides. Spots were identified by comigration with similar extracts from E. coli cultures that had undergone a stringent response or with [32P]GTP as a standard (Amersham).
RNA methods.
Total RNA was extracted from S. mutans strains grown in batch cultures that had been subjected to amino acid starvation or from biofilm-grown and planktonic cells that were cultured in tissue culture plates by using a previously described protocol (13). RNAs were transferred to nylon membranes (GeneScreen Plus; NEN Life Science Products, Boston, Mass.) by a slot blot apparatus (LTI) as described by Sambrook et al. (39). For Northern blot analysis, total RNA was separated on a 0.9% formaldehyde gel as described elsewhere (2). The integrity of the RNA was assessed, and confirmation that equal amounts of RNA were analyzed in Northern blots was done by agarose gel electrophoresis and ethidium bromide staining. RNAs were UV cross-linked to nylon membranes, and the membranes were probed with internal fragments of the desired genes that had been labeled with psoralen-biotin by using the Brightstar labeling kit (Ambion, Inc., Austin, Tex.). All hybridizations and washes were carried out under high-stringency conditions. Signals obtained on autoradiographs were quantified with an IS1000 digital imaging system (Alpha Innotech Corp., San Leandro, Calif.).
Acid killing experiments.
For acid killing of planktonic cells, strains were grown in BHI broth to the mid-exponential phase (OD600 ≅ 0.5) or the stationary phase, washed once with 0.1 M glycine buffer (pH 7.0), and resuspended in one-half of the original volume of 0.1 M glycine buffer (pH 2.8) for up to 60 min. For acid killing of biofilm-grown cells, cultures were grown in a 24-well polystyrene (flat-bottom) tissue culture plate containing BM. Cells were incubated at 37°C in a 5% CO2 atmosphere for 24 or 48 h and then subjected to acid killing. Briefly, cells in the planktonic phase were removed by discarding the medium and blotting the plate on absorbent paper. Biofilms were then incubated in 0.1 M glycine (pH 2.8) for up to 90 min. For each time point, biofilms from two independent wells were resuspended in the glycine solution by repeated pipetting, transferred to a 1.5-ml Eppendorf tube, and dispersed by vortexing at high speed for 30 s. Dispersed cells were serially diluted, plated on BHI agar plates, and incubated for 2 days before colonies were counted. Cell viability at each time point was expressed as the percentage of viable cells (CFU ml−1) at time zero.
Physiological assays.
The ability of S. mutans to lower the pH through glycolysis was monitored with a pH meter (5). Briefly, biofilms grown in 24-well polystyrene plates were removed by repeated pipetting, washed once with ice-cold water, and concentrated 10-fold in 50 mM KCl-1 mM MgCl2. The suspension was then titrated with 0.1 M KOH to a pH of 7.2, and pH drops were initiated by adding glucose to a final concentration of 55.6 mM. For F-ATPase assays, biofilm-grown cells were permeabilized with toluene and incubated with 5 mM ATP in ATPase buffer as previously described (5). Samples were removed at various intervals and assayed for inorganic phosphate released from ATP with the Fiske-Subbarow reagent (Sigma). ATPase activity was expressed as nanomoles of PO4 min−1 mg of protein−1. To measure glucose transport by the phosphoenolpyruvate:sugar phosphotransferase system (PTS), cells obtained from biofilms were washed and permeabilized with acetone:toluene (9:1). Permeabilized cells were assayed by the method of LeBlanc et al. (22). PTS activity was expressed as nanomoles of NADH oxidized in a phosphoenolpyruvate-dependent manner min−1 mg of protein−1.
CAT assay.
Cell-free lysates were prepared as previously described (13). CAT activity was measured by a spectrophotometric method (40) using the colorimetric substrate 5,5′-dithio-bis-nitrobenzoic acid (DTNB; Boehringer Mannheim, Indianapolis, Ind.). One unit of CAT activity was defined as the amount of enzyme needed to acetylate 1 nmol of chloramphenicol min−1.
Protein electrophoresis and Western blotting.
Whole-cell lysates for protein analysis were obtained by homogenization in the presence of glass beads with a Bead Beater (Biospec, Bartlesville, Okla.) as previously described (13). Protein lysates (10 μg per lane) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto Immobilon-P membranes (Millipore, Bedford, Mass.), and subjected to Western blot analysis by standard techniques (2). Membranes were incubated with Streptococcus dysgalactiae subsp. equisimilis anti-RelA (anti-RelSeq) polyclonal antisera, a kind gift from H. Malke, Institute for Molecular Biology, Jena University, Jena, Germany, at a dilution of 1:500. The protein concentration of samples was determined by the bicinchoninic acid assay (Sigma).
RESULTS
Inactivation of relA leads to relaxed phenotype and reduction in growth rate.
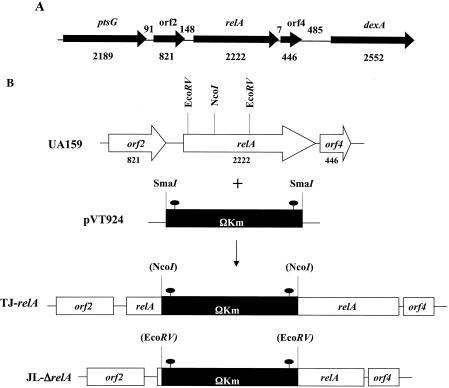
The gene encoding the putative RelA protein was identified in the S. mutans UA159 genome (1). The S. mutans relA gene (Smu2044; nucleotides 170983 to 171554) encodes a 740-amino-acid protein with a predicted molecular mass of 84 kDa that shared high levels of identity to the RelSeq protein (82% identity, 90% similarity). Located 7 bp 3′ of the relA stop codon was an open reading frame (ORF) predicted to encode a conserved d-tyrosyl tRNA deacylase, while 148 bp upstream of the relA gene, an ORF coding for a conserved hypothetical protein (Fig. 1A) was revealed by BLAST searches.
FIG. 1.
Schematic diagram of the relA locus and construction of two relA mutant strains. (A) ptsG is predicted to code for a putative PTS system EII enzyme, while dexA codes for a dextranase precursor. ORF2 codes for a hypothetical protein, and ORF4 is predicted to code for a conserved d-tyrosyl tRNA deacylase. Arrows indicate the direction of transcription. The numbers below the schematic indicate the sizes of the ORFs in base pairs, and the numbers between ORFs indicate the sizes of the intergenic regions in base pairs. (B) The relA gene was mutated either by insertional inactivation (TJ-relA) or by allelic replacement of the gene encoding the N-terminal portion of RelA by a cassette containing the ΩKan element (JL-ΔrelA). The filled ovals indicate the transcriptional and translational terminators of the ΩKan element.
In order to investigate the role of RelA in biofilm formation and acid tolerance, two relA mutant strains were generated (Fig. 1B). In the first mutant (TJ-relA), a polar kanamycin cassette (ΩKan element) was used to insertionally inactivate the gene between codons 167 and 168, so theoretically this strain could express a truncated RelA containing almost one-half of the N-terminal portion of the (p)ppGpp synthetase domain and about two-thirds of the N-terminal portion of the hydrolase domain, which overlaps the synthetase domain. The C-terminal portion of RelA is devoted to the regulation of the activity of the protein. Of note, the putative 167-amino-acid truncated RelA could not be detected by Western blot analysis using an anti-RelSeq antibody. Nevertheless, to be certain that a strain completely devoid of RelA was examined, a second mutant was included in our analyses. That strain, JL-ΔrelA, was generated by allelic replacement of the 5′ portion of relA with the ΩKan element, which was integrated 39 bp 3′ of the relA start codon. The correct integration of ΩKan, which is flanked by transciption terminators, was confirmed by Southern blotting, and a lack of production of RelA was confirmed by Western blotting with anti-RelSeq antibody (data not shown).
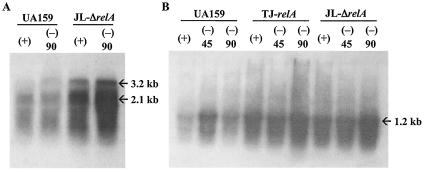
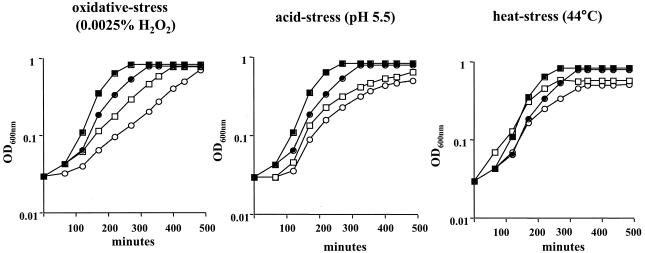
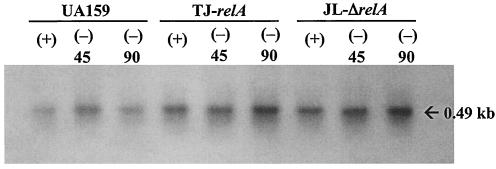
To confirm that the relA knockout strains had a relaxed phenotype, two S. mutans genes coding for elongation factor G and ribosomal protein L1, which have been shown to be down-regulated during the stringent response in B. subtilis (6), were used to probe RNAs isolated from the wild-type and mutant strains that had been subjected to amino acid starvation. Transcription of both genes was enhanced at least fourfold in the relA mutant strains (Fig. 2). When grown in liquid media (BHI broth, BM, or FMC), both relA mutants grew more slowly than the wild-type strain yet were able to reach similar final OD600s. Interestingly, the difference in the growth rates of the relA mutants was more pronounced in FMC medium than in the nutritionally rich medium BHI broth (data not shown). The ability of TJ-relA to grow under stress conditions was also assessed. Under all conditions tested (heat, acid, and oxidative stress), the mutant strain grew more slowly than the wild-type strain (Fig. 3).
FIG. 2.
Northern blot analysis of S. mutans UA159 and its RelA-deficient strains (TJ-relA and JL-ΔrelA). Cells were grown in complete FMC (+) or amino acid-depleted FMC (−) for 45 and 90 min. RNA (10 μg per lane) was isolated, separated in a 0.9% formaldehyde gel, transferred to a nylon membrane, and hybridized to probes specific for elongation factor G (A) and ribosomal L1 (sll) (B) genes.
FIG. 3.
Growth curves of S. mutans wild type (squares) and TJ-relA (circles) grown in BHI broth at 37°C (solid symbols) or under oxidative, heat, or acid stress conditions (open symbols). Typical results representative of at least three experiments are shown.
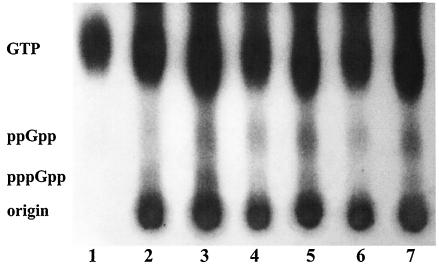
Detection of (p)ppGpp in TJ-relA and JL-ΔrelA strains.
To determine whether disruption of the relA gene resulted in alterations in the levels of (p)ppGpp pools, the accumulation of (p)ppGpp was assessed (Fig. 4). Both wild-type and mutant strains accumulated (p)ppGpp after the induction of a stringent response with serine hydroxamate (an ∼60% increase). Most importantly, this result indicates that strains lacking RelA were still able to synthesize and accumulate (p)ppGpp.
FIG. 4.
Accumulation of (p)ppGpp in S. mutans UA159 (lanes 2 and 3), TJ-relA (lanes 4 and 5), and JL-ΔrelA (lanes 6 and 7). Cells were grown in FMC media and labeled with 32Pi (100 μCi ml−1) before (lanes 2, 4, and 6) and after (lanes 3, 5, and 7) being treated with serine hydroxamate (1 mg ml−1) to induce the stringent response. Lane 1 shows the [32P]GTP standard.
Effects of relA inactivation on biofilm formation.
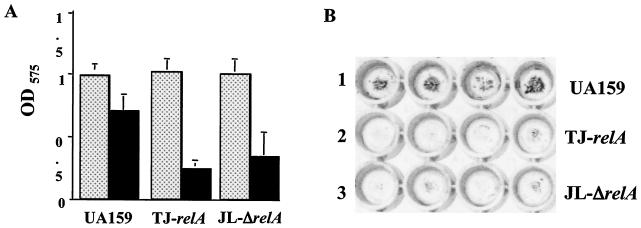
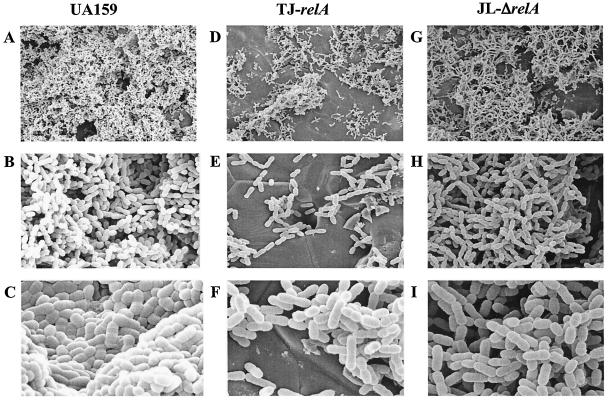
The capacity of TJ-relA and JL-ΔrelA to form stable biofilms was compared with that of the wild-type strain. As stated above, although the relA mutants displayed slower growth in BM, both strains grew equally well over a 24- or 48-h period compared to the parental strain. The data obtained after 48 h showed that the relA mutants, when grown in the presence of glucose, had a reduced capacity to form biofilms on the surfaces of microtiter plates, indicating that relA is required for efficient biofilm formation (Fig. 5). Comparable results were obtained with 24-h biofilms (data not shown). To further investigate the biofilm phenotypes of these strains, SEM analysis of biofilms grown on the surfaces of HA disks was performed. After 24 h, the wild-type strain was able to form a nearly uniform, thick layer of cells. In contrast, biofilms of TJ-relA and JL-ΔrelA contained fewer cells, with microcolonies being spread around the disks and large parts of the disks being uncolonized (Fig. 6).
FIG. 5.
Biofilm formation by S. mutans UA159 (wild type), TJ-relA, and JL-ΔrelA. Cultures were grown in a 96-well microtiter plate containing BM at 37°C in 5% CO2 for 48 h. Growth (dotted bars) and biofilm formation (solid bars) were measured at absorbances of 600 and 575 nm, respectively. (A) The graph shows the averages and standard deviations for three independent experiments. (B) Crystal violet-stained biofilms of strains UA159 (lane 1), TJ-relA (lane 2), and JL-ΔrelA (lane 3) on polystyrene plates. Typical results for three independent experiments are presented.
FIG. 6.
Scanning electron micrographs comparing biofilm formation of S. mutans UA159 (wild type), TJ-relA, and JL-ΔrelA on the surfaces of HA disks. Images shown were obtained at ×1,500 (A, D, and G), ×6,000 (B, E, and H), and ×13,000 (C, F, and I) magnification.
Acid tolerance by relA mutant strains.
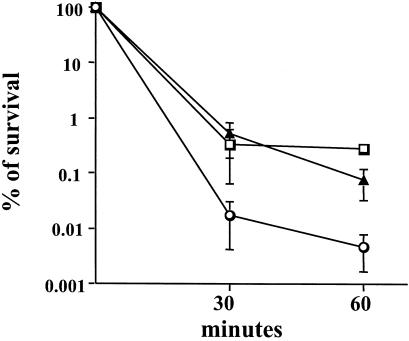
To determine whether the RelA-deficient strains had altered resistance to low pH, acid killing experiments were performed with planktonic or biofilm-grown cells, which were submitted to acidification at pH 2.8. Acid killing of mid-exponential-phase cultures revealed no significant differences in acid resistance between the wild-type and the mutant strains (data not shown). Similar results were obtained with stationary-phase cells (overnight growth), although exponentially growing cells were much more sensitive to acid challenge than stationary-phase cells were (data not shown). Interestingly, when the strains were cultured as biofilms, both RelA-deficient strains became more resistant to acid killing than the wild-type strain (Fig. 7). To assess whether the enhanced acid resistance of biofilm-grown cells was related to the biofilm architecture or diffusion limitation, acid killing was also carried out on biofilms that were dispersed by repeated pipetting. The data obtained were comparable to what was found for intact biofilms, indicating that biofilm-grown cells were inherently more resistant to acid killing (data not shown). To evaluate whether the acid-resistant phenotype of the relA mutant biofilms was a result of a general acquisition of stress tolerance, biofilm-grown cells of the TJ-relA and UA159 strains were subjected to killing by 0.2% H2O2. No significant differences in resistance to peroxide killing were found between these two strains (data not shown).
FIG. 7.
Survival of S. mutans wild-type (squares), TJ-relA (triangles), and JL-ΔrelA (circles) strains after acid challenge at pH 2.8. Cells from 48-h biofilms grown on the surfaces of polystyrene plates were collected and subjected to acid killing in 0.1 M glycine (pH 2.8). Cell viability at each time point is expressed as the percentage of viable cells (CFU ml of culture−1) at time zero.
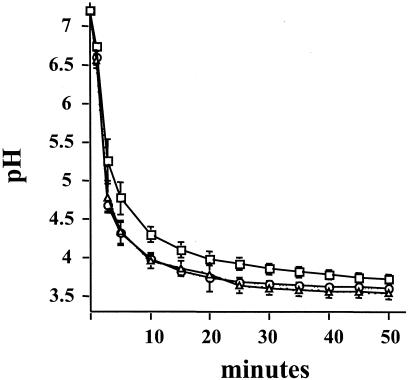
In agreement with the acid-resistant phenotype, biofilm cells from both the TJ-relA and the JL-ΔrelA strains were able to lower the pH through glycolysis faster than, and to values lower than, those achieved by the wild-type strain (Fig. 8 and Table 1). To assess whether the basis for the enhanced acid tolerance and glycolytic capacity was an increase in the proton-translocating F-ATPase or the capacity to transport sugar through the PTS, the activity of these enzyme systems was measured. Biofilm-grown cells from the parent and mutant strains showed comparable levels of F-ATPase activity. In contrast, glucose PTS activity was approximately 70% higher in the relA mutant strains grown in biofilms (Table 1) and 40% higher in planktonic cultures (data not shown) than in the parent cultured under the same conditions.
FIG. 8.
pH drop profile of S. mutans wild-type (squares), TJ-relA (triangles), and JL-ΔrelA (circles) strains grown in biofilms. Biofilms grown for 24 h on the surfaces of polystyrene plates were resuspended in ice-cold water and collected by centrifugation. Cells were resuspended in a 50 mM KCl-1 mM MgCl2 solution, and pH drops were initiated by the addition of glucose. pH drops were continuously monitored for 50 min. Datum points were collected every 10 s, and data for every 2 min and 30 s are presented. The graph shows the averages and standard deviations for three independent experiments.
TABLE 1.
Acid tolerance responses of S. mutans UA159, TJ-relA, and JL-ΔrelA biofilms
| Strain | pH after 50 min of glycolysisa | ATPase activity (nmol of PO4 min−1 mg of protein−1)a | Glucose PTS activity (nmol min−1 mg of protein−1)b |
|---|---|---|---|
| UA159 | 3.727 ± 0.0639 | 2.391 ± 0.301 | 130.7 ± 45.33 |
| TJ-relA | 3.545 ± 0.0755 | 2.231 ± 0.468 | 233.65 ± 56.7 |
| JL-ΔrelA | 3.608 ± 0.0548 | 2.177 ± 0.455 | 218.1 ± 74.92 |
Mean ± standard deviation for three independent experiments.
Mean ± standard deviation for five independent experiments.
Complementation of relA mutation restores wild-type phenotype.
Sequence analysis revealed that an ORF starting 7 bp from the relA stop codon encodes a putative d-tyrosyl tRNA deacylase. Considering the small degree of separation between relA and this putative gene, it seems likely that the two genes were cotranscribed and that mutation of relA with ΩKan would create a polar effect on the downstream gene. The tight linkage of the genes may be significant, since it has been demonstrated that in E. coli, d-tyrosyl deacylase provides protection against the toxic effects of d-aminoacyl tRNA molecules that accumulate during amino acid starvation (17). To confirm that the lack of RelA was responsible for the biofilm-defective and acid-tolerant phenotypes of strains TJ-relA and JL-ΔrelA, the intact relA gene was provided in trans on plasmid pJL69 after induction with subinhibitory concentrations of nisin. The expression of RelA from pJL69 in both relA background strains was confirmed by Western blotting (data not shown). The introduction of relA onto pJL69 in strains TJ-relA and JL-ΔrelA restored the levels of (p)ppGpp accumulated during exponential growth and the capacity to form biofilms, and the acid tolerance capacity of the complemented strains was indistinguishable from that of S. mutans UA159 (data not shown). Controls included the wild-type strain and relA strains carrying the vector plasmid that were treated identically to the experimental cultures. Of note, we also determined that concentrations of nisin up to 16 μg ml−1, which is one-half of the MIC, have no effect on the stress tolerance of S. mutans.
Expression of relA is constitutive.
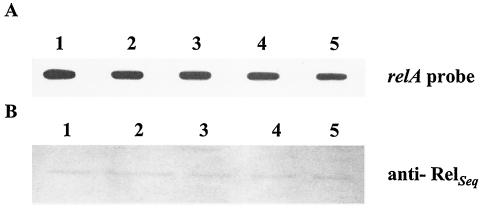
relA expression was assessed by slot blot and promoter fusions in the wild-type strain. The detection of individual transcripts by Northern blot analysis proved unsatisfactory, most likely as a result of low-level expression of relA coupled with rapid degradation of the relA mRNA (data not shown). Therefore, quantitative detection of relA mRNA was accomplished by slot blot analysis. RNA was isolated from batch-grown cultures that were subjected to amino acid starvation and from biofilm or planktonic cells grown in tissue culture plates. Under all circumstances, there were no detectable differences in the levels of relA mRNAs, suggesting that relA expression is not regulated at the transcriptional level (Fig. 9A). In agreement with these findings, a transcriptional fusion to a promoterless cat gene demonstrated that transcription from the relA promoter was not affected by amino acid starvation (data not shown). Finally, Western blot analysis with an anti-RelSeq antibody confirmed the results obtained by the RNA analysis and indicated that RelA levels correlate well with transcription levels of the relA gene (Fig. 9B).
FIG. 9.
Expression of relA mRNA and RelA from S. mutans UA159 in response to amino acid starvation. (A) Slot blot of total RNA; (B) Western blot analysis of RelA levels with an anti-RelSeq polyclonal antibody diluted 1:500. RNAs (10 μg per lane) or total protein (10 μg per lane) was isolated from samples grown in complete FMC (1), in FMC minus supplemented amino acids for 45 min (2), and in FMC minus supplemented amino acids 90 min (3) and from biofilm (4) or planktonic (5) cells grown in BM.
Transcriptional profile of genes involved in stress tolerance and biofilm formation in relA mutant strains.
S. mutans strains lacking ClpP or LuxS or producing lower levels of DnaK were all acid sensitive and showed impaired biofilm formation (23, 24, 32; J. A. C. Lemos and R. A. Burne, unpublished data; Z. T. Wen and R. A. Burne, submitted for publication). In an attempt to elucidate the basis for the enhanced acid tolerance and biofilm-defective phenotypes of the RelA-deficient strains, the transcription of clpP, luxS, and dnaK was examined in the wild-type and mutant strains by Northern blotting. The absolute amounts of clpP and dnaK mRNA did not appear to be affected in the mutant strains and thus could not readily be connected to the biofilm-defective or acid-resistant phenotypes (data not shown). However, the expression of luxS was elevated in the mutant strains approximately threefold compared to that in UA159 and was increased up to fivefold after cells were subjected to amino acid starvation (Fig. 10).
FIG. 10.
Northern blot hybridization with luxS. Cells were grown in complete FMC (+) or amino acid-depleted FMC (−) for 45 and 90 min. RNA (10 μg per lane) was isolated, separated in a 0.9% formaldehyde gel, transferred to a nylon membrane, and hybridized to a luxS-specific probe.
DISCUSSION
Four factors are known to be critical in the development of dental caries by oral bacteria: the abilities of the bacteria to adhere to the tooth surface, to form biofilms, to metabolize carbohydrates, and to tolerate fluctuations in the environment, especially with regard to pH and nutrient limitation (9). Some recent reports have linked the stringent factor RelA with environmental stress tolerance in and biofilm formation by bacteria (4, 34, 38, 44). The data presented here show that the RelA-deficient strains of S. mutans had an impaired capacity to form biofilms and exhibited enhanced resistance to acid killing that was specific to cells grown in biofilms.
A (p)ppGpp-null phenotype has been obtained in E. coli with the isolation of a relA spoT double mutant (52). In gram-positive bacteria, insertional inactivations of relA genes have generated truncated RelA proteins that retain residual levels of (p)ppGpp (30, 34, 38, 42, 44), apparently by virtue of the production of a truncated RelA protein that retains synthetase activity. In our TJ-relA strain, the relA gene was inactivated by insertion of an antibiotic resistance marker at codon 168. Although we could not detect immunoreactive, truncated RelA protein by Western blotting, we cannot exclude the possibility that the cells produced a low level of a 167-amino-acid protein that retained some (p)ppGpp synthetase activity. The creation of the ΔrelA strain, lacking all RelA protein, demonstrated that it was possible to isolate a relA null mutant of S. mutans.
To the best of our knowledge, relA null mutants have not been isolated in gram-positive bacteria; unsuccessful attempts to isolate a viable S. dysgalactiae subsp. equisimilis relA (relSeq) deletion mutant have been reported (30). In the Streptococcus pyogenes genome (16), Steiner and Malke (42) identified two candidate proteins (Spy0873 and Spy1125) containing putative GTP pyrophosphokinase domains that shared limited sequence homology with RelA. In searching the S. mutans genome, we have found three ORFs with putative GTP pyrophosphokinase domains (Smu926, Smu1046c, and Smu1206c) that share similarity with Spy0873 and Spy1125, albeit the similarities are limited. Thus, it is possible that there are alternative sources of (p)ppGpp production in S. mutans besides RelA. In support of this idea, both the TJ-relA and the JL-ΔrelA strains were still able to synthesize and accumulate (p)ppGpp in a pattern similar to that of the wild type strain. It has been demonstrated that the S. dysgalactiae subsp. equisimilis relA gene (also known as relSeq) encodes a strong (p)ppGpp 3′-pyrophosphohydrolase and a weaker (p)ppGpp synthethase (29). Since the levels of (p)ppGpp depend on its rates of synthesis and degradation, we speculate that there are other sources of (p)ppGpp production and that RelA may constitute the only (p)ppGpp hydrolase enzyme in S. mutans. Most importantly, though, our results clearly indicate that insertional inactivation or deletion of relA does not yield a (p)ppGpp-null phenotype.
It has been suggested that transcription of relA in L. monocytogenes is induced upon adhesion to a model surface (44). Using reporter gene fusions as well as Northern and Western blot analyses, we did not detect the induction of relA during amino acid starvation or in biofilm-grown cells, demonstrating that increased synthesis of RelA protein is not a primary control point for (p)ppGpp production and that allosteric regulation of RelA may be sufficient to control the levels of (p)ppGpp in S. mutans. In fact, Wendrich et al. (50) demonstrated that RelA hops between blocked ribosomes, allowing low concentrations of RelA (1 for 200 ribosomes) to synthesize (p)ppGpp at levels that are correlated with the number of “starved ribosomes.” Therefore, it may not be necessary for the cells to increase RelA production in response to stress or starvation to mount an effective stringent response. Whether the other (p)ppGpp synthases are differentially expressed in response to environmental stress is not known.
In the present study, the capacity to form biofilms was clearly impaired in both relA mutant strains. During the preparation of this paper, Yoshida and Kuramitsu (53) explored the effects of the inactivation of relA on biofilm formation by S. mutans GS-5. In contrast to the data presented here, biofilm formation by their relA mutant (strain AYRA1) was equivalent to that of the parental strain in a microtiter plate assay (53). However, there are critical differences between the mutants and growth conditions used in these two studies. First, the GS-5-derived mutant was cultured in a chemically defined medium containing 0.5% glucose, whereas our UA159-derived mutant was cultured in BM containing 0.3% glucose. Perhaps more importantly, AYRA1 was constructed by single-crossover insertion of a suicide plasmid in a way that could allow the production of a 233-amino-acid version of RelA. Truncated versions of RelA that were even smaller have been shown to have residual (p)ppGppsynthetase and hydrolase activities (30, 34, 42). Consequently, in addition to differences in growth conditions, the retention of RelA-dependent (p)ppGpp production or degradation in AYRA1 may account for behavior unlike that of the relA mutants described in this study.
It is intriguing that biofilms of the relA mutants became more resistant to acid killing but planktonic cells did not display the same phenotype. It has been demonstrated that bacterial biofilms are partially protected from external agents by multiple mechanisms, including diffusion limitation (14). Zhu et al. (54) reported that after acid shock, the majority of live S. mutans cells were found in deep layers of the biofilms, and Li et al. (25) showed that cells from thicker and denser biofilms had higher resistances to killing by low pH than planktonic cells. However, it appears that in S. mutans, the physical barrier of biofilms may play only a limited role in acid diffusion. Indeed, the relA mutants in dispersed biofilms were as resistant to acid killing as the same strains in intact biofilms and more acid resistant than the parent strain, suggesting that changes in cellular physiology, and not biofilm structure, were responsible for the increased acid resistance. Along these lines, Li et al. (25) have suggested that the increased acid resistance of S. mutans biofilms results from physiological changes mediated, at least in part, by extracellular signals rather than diffusion limitation.
Recently, it was demonstrated that a quorum-sensing system essential for genetic competence in S. mutans (comABCDE genes) is also involved in biofilm formation (27). The inactivation of relA does not seem to affect DNA genetic transformation, as evidenced by our ability to readily transform the mutants with a plasmid DNA. Thus, the impaired biofilm-forming capacity of the relA mutants could not be associated with a reduction in genetic competence and therefore is not likely to result from altered expression of the com genes. More recently, it was demonstrated that the inactivation of the S. mutans luxS gene, which is involved in quorum sensing in a wide range of bacteria, leads to impaired biofilm formation (32; Wen and Burne, submitted for publication), as well as an acid-sensitive phenotype (Wen and Burne, submitted for publication). In this report, it was shown that the expression of luxS was induced by amino acid starvation and that the levels of luxS were three- to fivefold higher in the relA mutants. It is possible that alterations in the levels of LuxS could affect pathways that are required for biofilm formation and acid tolerance, perhaps providing some insight into the molecular basis for the phenotypes of the relA mutants. Lending strength to this hypothesis, Wen and Burne (submitted for publication) demonstrated that the expression of acid stress-related genes such as ffh (a signal recognition particle subunit), as well as that of the genes encoding the DNA repair enzymes RecA, SmnA, and Nth, was down-regulated in an luxS mutant. Further studies will be essential for understanding the relationships between RelA and LuxS.
The acid resistance of S. mutans has been directly related to increased glycolytic capacities and increased activity of the proton-translocating F-ATPase (5, 19). The F-ATPase appears to be essential for pH homeostasis in S. mutans and is often considered to be the major determinant of acid tolerance in this organism (37). Cells isolated from biofilms of the RelA-deficient strain showed no increase in F-ATPase activity. However, glucose PTS activity was higher in the relA mutant strains, suggesting that PTS genes may be under RelA stringent control. In oral streptococci, the PTS is the primary sugar transport system, particularly at low sugar concentrations (46), but the PTS also plays important roles in the regulation of gene expression (33, 43). Thus, the effects of elevated PTS activity on acid tolerance in the relA mutants in biofilms could be explained in at least two ways. First, increases in the PTS activity may have imparted to the mutants an enhanced capacity to effectively scavenge residual sugars in the biofilms, allowing energy generation and enhancing survival. Whether this could occur at pH 2.8 in glycine buffer is not known. Perhaps a more palatable explanation is that pleiotropic effects on gene expression caused by changes in expression of the PTS components altered the acid resistance properties in the RelA-deficient background. It is also noteworthy that the inactivation of the ccpA gene in S. mutans, which encodes a global regulator of catabolite repression in gram-positive bacteria, has been shown to affect biofilm formation in B. subtilis and S. mutans (41, 49). Alterations in the DNA binding activity of CcpA induced by PTS-dependent changes in carbohydrate flux could also provide a partial explanation of the linkage between altered carbohydrate metabolism in the RelA strain and diminished biofilm formation.
In summary, we showed that RelA plays an important role in biofilm formation and acid tolerance and thus intimately regulates the virulence of S. mutans. Analysis of genes under the control of RelA by means of transcriptomics or proteomics may lead to the discovery of essential determinants of acid tolerance and biofilm formation for S. mutans.
Acknowledgments
This work was supported by grant RO1 DE13239 from the NIDCR.
We thank H. Malke for providing the anti-RelSeq polyclonal antibody, F. Bennett for assistance with the SEM analysis, and J. Abranches and Z. T. Wen for critical reading of the manuscript.
Editor: V. J. DiRita
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, E. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Baev, D., R. England, and H. K. Kuramitsu. 1999. Stress-induced membrane association of the Streptococcus mutans GTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect. Immun. 67:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balzer, G. J., and R. J. McLean. 2002. The stringent response genes relA and spoT are important for Escherichia coli biofilms under slow-growth conditions. Can. J. Microbiol. 48:675-680. [DOI] [PubMed] [Google Scholar]
- 5.Belli, W. A., and R. E. Marquis. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57:1134-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhardt, J., J. Weibezahn, C. Scharf, and M. Hecker. 2003. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res. 13:224-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowden, G. H., and I. R. Hamilton. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 9:54-85. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 9.Burne, R. A. 1998. Oral streptococci… products of their environment. J. Dent. Res. 77:445-452. [DOI] [PubMed] [Google Scholar]
- 10.Burne, R. A., K. Schilling, W. H. Bowen, and R. E. Yasbin. 1987. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J. Bacteriol. 169:4507-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsson, J. 1983. Regulation of sugar metabolism in relation to feast-and-famine existence of plaque, p. 205-211. In B. Guggenheim (ed.), Cariology today. Karger, Basel, Switzerland.
- 12.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. C. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and A. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 13.Chen, Y.-Y., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 15.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferri-Fioni, M. L., E. Schmitt, J. Soutourina, P. Plateau, Y. Mechulam, and S. Blanquet. 2001. Structure of crystalline d-Tyr-tRNA(Tyr) deacylase. A representative of a new class of tRNA-dependent hydrolases. J. Biol. Chem. 276:47285-47290. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez, J. A., P. J. Crowley, D. G. Cvitkovitch, L. J. Brady, I. R. Hamilton, J. D. Hillman, and A. S. Bleiweis. 1999. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology 145:357-366. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton, I. R., and N. D. Buckley. 1991. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol. Immunol. 6:65-71. [DOI] [PubMed] [Google Scholar]
- 20.Hanna, M. N., R. J. Ferguson, Y. H. Li, and D. G. Cvitkovitch. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 183:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 184:3923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeBlanc, D. J., V. L. Crow, L. N. Lee, and C. F. Garon. 1979. Influence of the lactose plasmid on the metabolism of galactose by Streptococcus lactis. J. Bacteriol. 137:878-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemos, J. A. C., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemos, J. A. C., Y. Y. M. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y. H., M. N. Hanna, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mechold, U., M. Cashel, K. Steiner, D. Gentry, and H. Malke. 1996. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J. Bacteriol. 178:1401-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mechold, U., and H. Malke. 1997. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J. Bacteriol. 179:2658-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mechold, U., H. Murphy, L. Brown, and M. Cashel. 2002. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of RelSeq, the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184:2878-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 34.Okada, Y., S. Makino, T. Tobe, N. Okada, and S. Yamazaki. 2002. Cloning of rel from Listeria monocytogenes as an osmotolerance involvement gene. Appl. Environ. Microbiol. 68:1541-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry, D., and H. K. Kuramitsu. 1981. Genetic transformation of Streptococcus mutans. Infect. Immun. 32:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quivey, R. G., Jr., W. L. Kuhnert, and K. Hahn. 2000. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42:239-274. [DOI] [PubMed] [Google Scholar]
- 38.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517-528. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 41.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 38:1004-1016. [DOI] [PubMed] [Google Scholar]
- 43.Stulke, J., and W. Hillen. 1998. Coupling physiology and gene regulation in bacteria: the phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften 85:583-592. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, C. M., M. Beresford, H. A. Epton, D. C. Sigee, G. Shama, P. W. Andrew, and I. S. Roberts. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J. Bacteriol. 184:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terleckyj, B., N. P. Willett, and G. D. Shockman. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vadeboncoeur, C., and M. Pelletier. 1997. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 19:187-207. [DOI] [PubMed] [Google Scholar]
- 47.van Delden, C., R. Comte, and A. M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen, Z. T., and R. A. Burne. 2001. Construction of a new integration vector for use in Streptococcus mutans. Plasmid 45:31-36. [DOI] [PubMed] [Google Scholar]
- 49.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wendrich, T. M., G. Blaha, D. N. Wilson, M. A. Marahiel, and K. H. Nierhaus. 2002. Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10:779-788. [DOI] [PubMed] [Google Scholar]
- 51.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26:65-79. [DOI] [PubMed] [Google Scholar]
- 52.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 53.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu, M., S. Takenaka, M. Sato, and E. Hoshino. 2001. Influence of starvation and biofilm formation on acid resistance of Streptococcus mutans. Oral Microbiol. Immunol. 16:24-27. [DOI] [PubMed] [Google Scholar]