Abstract
Salmonella enterica serovar Typhimurium, similar to various facultative intracellular pathogens, has been shown to respond to the hostile conditions inside macrophages of the host organism by inducing stress proteins, such as DnaK. DnaK forms a chaperone machinery with the cochaperones DnaJ and GrpE. To elucidate the role of the DnaK chaperone machinery in the pathogenesis of S. enterica serovar Typhimurium, we first constructed an insertional mutation in the dnaK-dnaJ operon of pathogenic strain χ3306. The DnaK/DnaJ-depleted mutant was temperature sensitive for growth, that is, nonviable above 39°C. We then isolated a spontaneously occurring revertant of the dnaK-dnaJ-disrupted mutant at 39°C and used it for infection of mice. The mutant lost the ability to cause a lethal systemic disease in mice. The impaired ability for virulence was restored when a functional copy of the dnaK-dnaJ operon was provided, suggesting that the DnaK/DnaJ chaperone machinery is required by Salmonella for the systemic infection of mice. This result also indicates that with respect to the DnaK/DnaJ chaperone machinery, the cellular requirements for growth at a high temperature are not identical to the cellular requirements for the pathogenesis of Salmonella. Macrophage survival assays revealed that the DnaK/DnaJ-depleted mutant could not survive or proliferate at all within macrophages. Of further interest are the findings that the mutant could neither invade cultured epithelial cells nor secrete any of the invasion proteins encoded within Salmonella pathogenicity island 1. This is the first time that the DnaK/DnaJ chaperone machinery has been shown to be involved in bacterial invasion of epithelial cells.
Salmonella enterica serovar Typhimurium causes gastroenteritis in humans and a lethal systemic disease in mice that serves as a model for human typhoid fever (for a review, see reference 28). Following oral infection, Salmonella colonizes the intestinal tract, penetrates the intestinal epithelium, and migrates to the mesenteric lymph nodes, where it is phagocytosed by professional phagocytic cells, such as macrophages, replicates intracellularly, and subsequently is disseminated to the spleen and liver (6, 9, 27, 32).
Specific virulence factors encoded within Salmonella pathogenicity islands (SPIs) are required for the development of salmonellosis. SPI1 and SPI2 are major SPIs that encode structurally similar but functionally distinct type III secretion systems that translocate effector proteins directly into host cells to contribute to pathogenesis (25). SPI1 functions are involved in the initial stage of salmonellosis, that is, the invasion of nonphagocytic cells and the penetration of gastrointestinal epithelium. SPI1-encoded proteins induce membrane ruffling of epithelial cells, thereby promoting bacterial uptake into enterocytes (7, 12). SPI2-encoded factors function at a later stage of infection with Salmonella; that is, SPI2-encoded proteins are associated with survival within macrophages and subsequent systemic infection to reach and proliferate within the lymphatics and bloodstream (5, 24, 41). However, much less is known about the mechanisms by which SPI2-encoded proteins alter host responses to Salmonella.
Intracellular pathogens, including S. enterica serovar Typhimurium, which maintain long-term resistance within host phagocytes, elicit a variety of genetic programs to help them adapt to the hostile environmental conditions encountered within phagosomes. Prominent among these programs is the heat shock response (3, 35, 50). The heat shock response is well known to be induced by damaged proteins that accumulate after the exposure of cells to a variety of stress conditions, including a sudden elevation in temperature. It can be speculated that intracellular pathogens are subject to a considerable amount of protein misfolding and denaturation, resulting in the induction of the heat shock response within phagosomes. The heat shock proteins have been functionally divided into two groups, molecular chaperones and proteases. Evidence supporting the essential role of the heat shock proteins in Salmonella pathogenesis was initially demonstrated by insertional mutation of the protease gene, htrA, which resulted in attenuation of the bacteria in BALB/c mice (26). The htrA gene encodes a periplasmic serine protease that is necessary for the degradation of abnormally folded proteins transported into the periplasmic space (44).
Various investigators recently reported that the ATP-dependent proteases ClpXP (52) and Lon (45), which are also known to be heat shock proteins, are essentially involved in systemic infection with S. enterica serovar Typhimurium in BALB/c mice. ClpXP and Lon are required for the survival and growth of S. enterica serovar Typhimurium within macrophages and cope with the accumulation of damaged proteins within phagosomes. Furthermore, it was shown that the Lon protease controls Salmonella invasion through the negative regulation of SPI1 gene expression (46).
DnaK forms a chaperone machinery with the cochaperones DnaJ and GrpE and is involved in many cellular processes, such as DNA replication of the bacterial chromosome, RNA synthesis, protein transport, cell division, and autoregulation of the heat shock response (13, 22). Furthermore, this chaperone machinery nonspecifically interacts with many unfolded and misfolded proteins and therefore assists in proper folding, helps in refolding, prevents aggregation, and mediates the degradation of misfolded proteins (19, 40). In Escherichia coli, the DnaK/DnaJ chaperone machinery and cellular ATP-dependent proteases, such as Lon, ClpXP, and HslVU, constitute the cellular network for the de novo folding and quality control of proteins (19, 47).
To elucidate the role of the DnaK/DnaJ chaperone machinery in the pathogenesis of S. enterica serovar Typhimurium, we constructed a dnaK insertional mutation in pathogenic strain χ3306, assessed its virulence in animal systems, and assayed the characteristics associated with virulence in vitro. Consequently, we found that the DnaK/DnaJ chaperone machinery is required by S. enterica serovar Typhimurium for invasion of epithelial cells and survival within macrophages and is essential for causing a systemic infection in the host.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Bacterial cells were routinely grown in L broth (1% Bacto Tryptone [Difco, Detroit, Mich.], 0.5% Bacto Yeast Extract [Difco], 0.5% sodium chloride [pH 7.4]) with shaking or on L agar. The media were supplemented with chloramphenicol (25 μg ml−1), ampicillin (25 μg ml−1), and/or nalidixic acid (25 μg ml−1), when necessary.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Reference or source |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| χ3306 | Nx-resistant derivative of SR-11 | 17 |
| CS2021 | dnaK::Cm in χ3306 | This study |
| CS2069 | Spontaneous Ts+ revertant of CS2021 | This study |
| CS2501 | CS2021 harboring pTKY608 | This study |
| CS2635 | χ3306 harboring pTKY608 | This study |
| CS2773 | CS2069 harboring pTKY608 | This study |
| E. coli | ||
| DH5α | F−recA endA gyrA thi hsdR supE relA Δ(lacZYA-argF) deoR Δφ80 lac(ΔlacZ)M15 | Our collection |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2Tc::MuKm pir | 51 |
| Plasmids | ||
| pT7blue-2 | Cloning vector | Our collection |
| pMW119 | Low-copy-number plasmid | Our collection |
| pCVD442 | pir-dependent suicide vector carrying sacB | 8 |
| pTKY505 | pT7blue-2 with a 948-bp fragment containing part of dnaK | This study |
| pTKY508 | pTKY505 carrying dnaK::Cm | This study |
| pTKY509 | pCVD442 with an EcoRI-SalI fragment containing dnaK::Cm | This study |
| pTKY608 | pMW119 carrying the dnaK-dnaJ operon of χ3306 | This study |
Nx, nalidixic acid; Tc, tetracycline; Km, kanamycin; Cm, chloramphenicol.
DNA techniques.
DNA purification, ligation, restriction analysis, PCR amplification, DNA sequencing, and agarose gel electrophoresis were carried out as previously described (52).
Insertional inactivation of dnaK and dnaJ in S. enterica serovar Typhimurium χ3306.
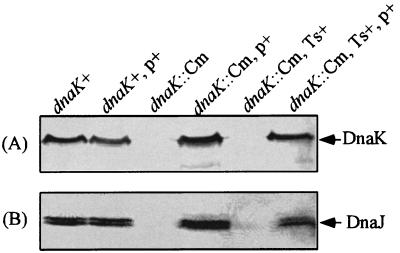
To block the expression of the dnaK and dnaJ genes, which are in the same operon, a polar mutation was introduced into the upstream gene, dnaK, as follows. On the basis of the previously determined sequence of the dnaK gene of S. enterica serovar Typhimurium strain LT2 (GenBank accession number U58360), we designed PCR primers SK (5′-TTATGGATGGAACGCAGGCACG-3′) and AK (5′-TATCAGACACGGACAGGCCAG-3′) to amplify the region from nucleotides 524 to 1471 of dnaK. The internal region of the dnaK gene of strain χ3306 was amplified with these primers and subsequently cloned into pT7blue-2. The resultant plasmid, pTKY505, was cleaved at the Aro51HI site in the cloned fragment, filled, and ligated to a chloramphenical resistance (Cm) cassette, which was generated from BamHI-digested pNK2882 and filled. The resultant plasmid, pTKY508, was digested with SalI and SmaI, generating a dnaK::Cm fragment that was ligated to the SalI and SmaI sites of pCVD442, a transferable suicide vector. The resultant mutator plasmid, pTKY509, was introduced into strain SM10λpir, which allows replication of the suicide vector by transformation. The chromosomal dnaK gene was replaced by the dnaK::Cm construct by conjugative crossover as previously described (51). A double-crossover event resulting in a dnaK::Cm mutant was initially assessed by its sensitivity to ampicillin, a resistance marker for which is found on the suicide vector. Allelic exchange was checked by direct sequencing of the dnaK::Cm region in the resultant strain, CS2021, by PCR amplification with primers SK and AK, and by immunoblotting with anti-E. coli DnaK serum (Fig. 1).
FIG. 1.
Immunoblot analysis of whole-cell proteins prepared from strains χ3306 (dnaK+), CS2635 (dnaK+/pTKY608), CS2021 (dnaK::Cm), CS2501 (dnaK::Cm/pTKY608), CS2069 (dnaK::Cm, Ts+ suppressor), and CS2773 (dnaK::Cm/pTKY608, Ts+ suppressor) with an antiserum against either DnaK protein (A) or DnaJ protein (B). Plasmid pTKY608 contains the complete dnaK-dnaJ operon of S. enterica serovar Typhimurium χ3306. p+, pTKY608; Ts+, Ts+ suppressor.
Cloning of the dnaK-dnaJ operon of S. enterica serovar Typhimurium χ3306 into a low-copy-number plasmid.
The dnaK-dnaJ region, including its promoter, of strain χ3306 was amplified by PCR with a sense primer (5′-GGCCGACGGAATTCGTTAACAC-3′) and an antisense primer (5′-GAGGATAGCATGCGTTTAACAG-3′) and then cloned into a low-copy-number plasmid, pMW119.
Immunoblot analysis.
Equivalent numbers of bacterial cells were suspended in sample buffer (31), boiled for 5 min, and subjected to sodium dodecyl sulfate- 10% polyacrylamide gel electrophoresis. Separated proteins on the gels were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). Proteins were reacted with rabbit anti-E. coli DnaK (1:25,000) antibody or anti-E. coli DnaJ antibody (1:25,000), followed by alkaline phosphatase-conjugated anti-rabbit immunoglobulin G as the secondary antibody. Enzymatic reactions were carried out in the presence of 300 μg of nitroblue tetrazolium (Dojin, Kumamoto, Japan) ml−1 and 150 μg of bromochloroindolylphosphate (Amresco, Solon, Ohio) ml−1.
Two-dimensional gel electrophoresis of secreted proteins.
For preparation of proteins secreted into the medium, the bacterial culture was incubated at 30°C overnight and then centrifuged to remove cells. The filtered supernatant was mixed with prechilled trichloroacetic acid (TCA) (final concentration, 10%), chilled on ice for 15 min, and centrifuged at 10,000 × g for 20 min. The pellet was washed once with 5% cold TCA and then with acetone. The acetone washing was repeated twice to completely remove TCA from the precipitate. The pellet was solubilized in sample buffer containing 8 M urea, 0.5% Nonidet P-40, 10 mM dithiothreitol, and 0.2% Bio-Lyte 3/10 (Bio-Rad, Hercules, Calif.). Isoelectric focusing in the first dimension was performed with a Protean IEF cell (Bio-Rad). Samples were focused in polyacrylamide gels within a pH range of 3 to 10 according to the manufacturer's instructions and resolved in the second dimension on sodium dodecyl sulfate-10% polyacrylamide slab gels. Total proteins were stained with Coomassie brilliant blue.
Identification of proteins on two-dimensional gels.
Protein spots of interest on the two-dimensional gels were excised, destained, and digested in situ with endopeptidase Lys-C. After digestion overnight at 37°C, samples were centrifuged and further purified by using Zip-TipC18 pipette tips (Millipore). An aliquot of a sample was taken for analysis by matrix-assisted laser desorption ionization mass spectrometry.
Assay for mouse virulence.
Bacterial cells grown in L broth at 37°C to late exponential growth phase were centrifuged at 8,000 × g for 10 min at room temperature and suspended in phosphate-buffered saline (PBS) (pH 7.0) containing 0.01% gelatin. The actual number of bacteria present was determined by counting of viable cells. Seven-week-old female BALB/c mice (Charles River Japan, Yokohama, Japan) were orally or subcutaneously inoculated. At various times after inoculation, the spleens, Peyer's patches, or popliteal lymph nodes (PLNs) were aseptically removed and then homogenized in PBS-gelatin. The numbers of viable bacteria in the organs of the infected mice were determined by plating serial 10-fold dilutions of the homogenates on L agar plates. Colonies were routinely counted 24 h later.
Assay for survival and growth within macrophages.
RAW264.7 and J774.1 macrophages were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Grand Island, N.Y.) containing 10% fetal calf serum (FCS) and 4 mM l-glutamine at 37°C. A total of 4 × 10 5 cells in each well of 24-well plates were challenged with S. enterica serovar Typhimurium strains at a multiplicity of infection of 10. The plates were centrifuged for 5 min at 500 × g to enhance and synchronize infection. The cells were incubated for 30 min at 30 or 37°C to permit phagocytosis, and the free bacteria were removed by three washes with Hank's balanced salt solution (HBSS) (Sigma, St. Louis, Mo.). DMEM containing 10% FCS and 100 μg of gentamicin ml−1 was added, and the cells were incubated for 1.5 h at 30 or 37°C. The cells were washed with prewarmed HBSS three times and then incubated with DMEM containing 10% FCS and 10 μg of gentamicin ml−1 at 30 or 37°C. The wells were sampled at various times after inoculation by aspirating the medium, performing three washes with HBSS, and lysing the contents of each well with PBS containing 0.2% Triton X-100. Triplicate samples were plated individually after appropriate dilution.
Assay for invasion of epithelial cells.
Intestine-407 tissue culture cells were maintained in DMEM supplemented with 10% FCS. Cells (2 × 105) were used to seed 24-well tissue culture plates to obtain about 90% confluent monolayers on the following day. For the preparation of bacterial cultures, a single colony was inoculated into L broth and grown to an optical density at 600 nm of 0.5 with shaking at 30°C. Bacterial cells were washed with HBSS and used to inoculate monolayers previously washed with HBSS at a multiplicity of infection of 10. The monolayers were incubated for 2 h at 30°C, washed thoroughly with HBSS, and lysed with 0.2% Triton X-100 in PBS to determine the total numbers of bacteria associated with the tissue culture cells. Alternatively, to assess the numbers of intracellular bacteria, the infected tissue culture cells were further incubated for 3 h in DMEM containing 100 μg of gentamicin ml−1 to eliminate extracellular bacteria before lysis with the Triton X-100 solution. Bacterial numbers were determined by plating the lysates on L agar plates after appropriate dilution.
RESULTS
Construction of a DnaK/DnaJ-depleted mutant of S. enterica serovar Typhimurium.
DNA sequence analysis of the dnaK region of S. enterica serovar Typhimurium strain LT2 (GenBank accession number U58360) has revealed that dnaK is linked to dnaJ, similar to the dnaK-dnaJ operon in E. coli (2). Therefore, a genetically defined dnaK-dnaJ mutant was constructed as described in Materials and Methods. The disruption of the dnaK gene in the resultant mutant strain, CS2021, was confirmed by immunoblotting analysis in which antiserum specific for the E. coli DnaK protein failed to recognize a DnaK protein in cell lysates from the mutant strain (Fig. 1A, third lane) but did recognize a band corresponding to approximately 70 kDa in cell lysates from the wild-type strain (first lane) and the dnaK::Cm mutant strain carrying pTKY608, in which the dnaK-dnaJ operon is regulated by its own promoter (fourth lane). To examine whether the insertion of the Cm cassette affects the expression of dnaJ, lysates from wild-type and dnaK::Cm mutant strains were subjected to immunoblotting analysis with antiserum specific for the E. coli DnaJ protein (Fig. 1B). The absence of a band corresponding to DnaJ suggests that the insertion of the Cm cassette in dnaK results in a polar effect on the expression of dnaJ (Fig. 1B, third lane). The introduction of plasmid pTKY608 carrying the dnaK-dnaJ operon into the dnaK::Cm mutant strain results in the detection of DnaJ (Fig. 1B, fourth lane). Since it is well known that DnaK requires an interaction with DnaJ to function as a molecular chaperone (13, 16, 34, 48), we decided to use mutant strain CS2021 carrying the dnaK::Cm allele to study the role of the DnaK/DnaJ chaperone machinery in S. enterica serovar Typhimurium.
Isolation of a suppressor of the temperature-sensitive phenotype of the dnaK::Cm mutant.
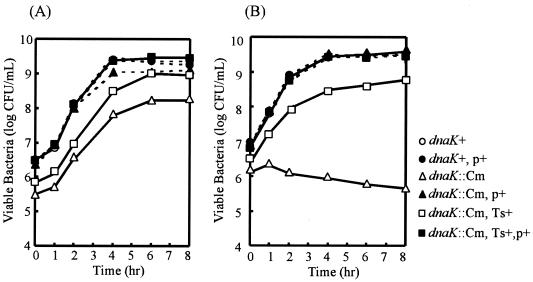
dnaK::Cm mutant strain CS2021 was initially characterized for its growth and viability at different temperatures. The mutant forms small colonies on L agar plates at 30 and 37°C but is nonviable above 39°C (data not shown). The growth curves for the dnaK::Cm mutant strain and the isogenic wild-type strain at 30 and 39°C are shown in Fig. 2. There is no difference in the rates of growth of the wild-type strain at the different temperatures examined. On the contrary, the growth of the dnaK::Cm mutant strain is slower and reaches lower stationary-phase cell densities at even the permissive temperature of 30°C. Its growth clearly is inhibited at 39°C. The growth defect of strain CS2021 was completely complemented when the functional dnaK and dnaJ genes in the low-copy-number vector pTKY608 were provided.
FIG. 2.
Growth curves for strains χ3306 (dnaK+), CS2635 (dnaK+/pTKY608), CS2021 (dnaK::Cm), CS2501 (dnaK::Cm/pTKY608), CS2069 (dnaK::Cm, Ts+ suppressor), and CS2773 (dnaK::Cm/pTKY608, Ts+ suppressor). Bacterial cultures grown in L broth overnight at 30°C were diluted 1:500 into fresh L broth and incubated at 30°C (A) or 39°C (B). Bacterial cells were diluted in L broth at various times and then plated on agar plates. Viable counts were determined after incubation of the plates for 24 h at 30°C. p+, pTKY608; Ts+, Ts+ suppressor.
Since the defect in the growth of the dnaK::Cm mutant strain at 39°C would not permit us to evaluate its virulence by infection of mice, we decided to isolate spontaneously occurring mutants of the strain at 39°C. This strategy was based on the hypothesis that the cellular requirements for DnaK/DnaJ at a high temperature might not be identical to the cellular requirements for DnaK/DnaJ in the pathogenesis of S. enterica serovar Typhimurium. Four isolates capable of forming colonies on L agar plates at 39°C were characterized for growth at different temperatures. These isolates had similar growth curves at different temperatures, and strain CS2069 was chosen as a representative (Fig. 2). In the present study, none of the strains that suppressed the temperature-sensitive phenotype (Ts+ suppressor strains) had a growth rate that was similar to that of wild-type cells even at 30°C. Therefore, we decided to use suppressor strain CS2069 for further analysis by infection of mice.
The DnaK/DnaJ chaperone machinery is essential for the virulence of S. enterica serovar Typhimurium in BALB/c mice.
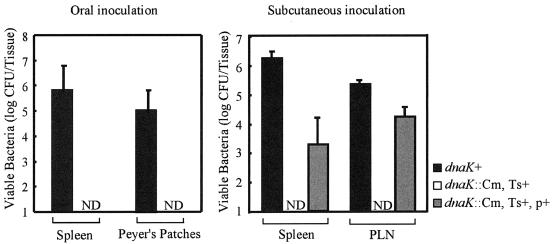
One assessment of the virulence of S. enterica serovar Typhimurium is the ability of bacteria to establish a lethal systemic infection in mice. To test whether the DnaK/DnaJ chaperone machinery is required for systemic infection of mice with S. enterica serovar Typhimurium, we determined the ability of the dnaK::Cm mutant to colonize the organs of BALB/c mice. Mice were inoculated orally with 5 × 108 CFU of strain χ3306 (dnaK+) or CS2069 (dnaK::Cm, Ts+ suppressor). The numbers of bacteria in Peyer's patches and spleens were assessed on day 5 after inoculation (Fig. 3, left panel). Mice infected with χ3306 had more than 105 bacteria in both the spleens and the Peyer's patches on day 5 after infection. All five mice infected with χ3306 died at day 6. In contrast, mice infected with mutant strain CS2069 had less than 10 bacteria in both the spleens and the Peyer's patches on day 5 after infection. These results suggest that mutant strain CS2069 was highly attenuated in mice.
FIG. 3.
Numbers of viable bacteria in the tissues of BALB/c mice after infection with S. enterica serovar Typhimurium strains χ3306 (dnaK+), CS2069 (dnaK::Cm, Ts+ suppressor), and CS2773 (dnaK::Cm/pTKY608, Ts+ suppressor). On day 5 after oral inoculation with 5 × 108 organisms or day 3 after subcutaneous inoculation with 2 × 105 organisms, the numbers of bacteria recovered from the spleens, Peyer's patches, and PLNs were determined. The error bars indicate the standard deviations of the means of bacterial counts recovered from five mice. ND, <10 CFU of viable bacteria per tissue. p+, pTKY608; Ts+, Ts+ suppressor.
BALB/c mice also were inoculated subcutaneously with 2 × 105 cells of strain χ3306 or CS2069, and the viable numbers of bacteria in the spleens and PLNs were determined on day 3 after challenge (Fig. 3, right panel). Wild-type strain χ3306 colonized the spleens and PLNs in large numbers and resulted in death 4 days after infection. Again, the DnaK/DnaJ-depleted strain, CS2069, exhibited an impaired ability to cause systemic infection of mice with S. enterica serovar Typhimurium.
To confirm that the impaired ability of strain CS2069 to cause a systemic infection in mice is due to the depletion of the DnaK/DnaJ chaperone machinery, a functional dnaK-dnaJ operon was provided in trans by the introduction of a low-copy-number plasmid carrying the operon of χ3306 and tested for complementation of the dnaK::Cm mutation by assessing the ability to colonize the spleens and PLNs of BALB/c mice infected subcutaneously. The viable numbers of bacteria recovered from the organs on day 3 after inoculation are shown in Fig. 3, right panel. The impaired ability of CS2069 (dnaK::Cm, Ts+ suppressor) to cause a systemic infection was restored when we provided a functional copy of the dnaK-dnaJ operon in strain CS2773 (dnaK::Cm/pTKY608, Ts+ suppressor). The fact that the virulence of strain CS2069, which carries the Ts+ suppressor mutation, could be restored by providing a functional copy of the dnaK-dnaJ operon supports our hypothesis that the cellular requirements for the DnaK/DnaJ chaperone machinery to grow at a high temperature may not be identical to the cellular requirements for the machinery in the pathogenesis of S. enterica serovar Typhimurium. On the basis of these results, we concluded that the DnaK/DnaJ chaperone machinery is essential for the systemic infection of mice by Salmonella.
Depletion of the DnaK/DnaJ chaperone machinery impairs the ability of S. enterica serovar Typhimurium to survive within macrophages.
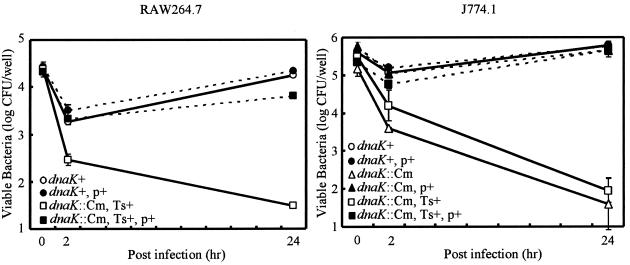
Depletion of the DnaK/DnaJ chaperone machinery greatly reduces the ability of S. enterica serovar Typhimurium to cause systemic disease in BALB/c mice by both the natural and the subcutaneous routes of infection. One of the most likely factors contributing to the reduced ability of the Salmonella mutant to cause systemic infection is the reduced capacity to survive the bactericidal action of professional killing cells, such as macrophages, in mice. To examine whether the dnaK::Cm mutant can survive the killing mechanism and proliferate within macrophages, strains CS2021 (dnaK::Cm) and CS2069 (dnaK::Cm, Ts+ suppressor) were examined for their ability to survive and proliferate within cultured macrophages. Murine macrophage-like J774.1 cells were challenged with either χ3306, CS2021 (dnaK::Cm), or CS2069 (dnaK::Cm, Ts+ suppressor) for 24 h at 30°C, and the number of viable bacteria within macrophages was determined. As shown in Fig. 4, the number of viable χ3306 bacteria was decreased during the first 2 h of incubation, indicating that the initial interaction with macrophages was the most bactericidal. After the initial decrease, the wild-type parent strain grew and increased in number almost 10-fold in J774.1 cells for 24 h after phagocytosis. In contrast, strains CS2021 (dnaK::Cm) and CS2069 (dnaK::Cm, Ts+ suppressor) were more rapidly killed than the wild-type parent strain during the first 2 h of incubation in J774.1 cells. Neither mutant strain grew during the 24 h following phagocytosis, suggesting that the DnaK/DnaJ-depleted mutant is extremely sensitive to the killing mechanism of macrophages. The loss of the ability of strains CS2021 and CS2069 to survive and grow within macrophages was fully complemented when a functional dnaK-dnaJ operon on low-copy-number plasmid pTKY608 was provided.
FIG. 4.
Fate of the S. enterica serovar Typhimurium virulent strain and the dnaK-dnaJ-disrupted mutant within macrophages after phagocytosis. RAW264.7 or J774.1 cells were challenged with strains χ3306 (dnaK+), CS2635 (dnaK+/pTKY608), CS2021 (dnaK::Cm), CS2501 (dnaK::Cm/pTKY608), CS2069 (dnaK::Cm, Ts+ suppressor), and CS2773 (dnaK::Cm/pTKY608, Ts+ suppressor). The ability of bacteria to survive within RAW264.7 or J774.1 cells was examined at 37 and 30°C, respectively. The data are the means of triplicate determinations, and the error bars indicate the standard deviations of the means. p+, pTKY608; Ts+, Ts+ suppressor.
The effect of DnaK/DnaJ depletion on the intracellular survival of S. enterica serovar Typhimurium also was examined with a different macrophage cell line, RAW264.7. Since RAW264.7 cells grow less well at 30°C, assessment of the ability of the DnaK/DnaJ-depleted mutant to survive and grow in RAW264.7 cells was performed with Ts+ suppressor mutant CS2069 at 37°C. The results shown in Fig. 4 indicate that mutant strain CS2069 could not survive or grow within macrophages, whereas the wild-type parent strain grew after an initial decrease, similar to its pattern of growth within J774.1 cells. Complementation with a functional dnaK-dnaJ operon on low-copy-number plasmid pTKY608 resulted in the restoration of wild-type-like survival and proliferation within RAW264.7 cells. The results of the assays of survival within cultured macrophages revealed that the DnaK/DnaJ chaperone machinery of S. enterica serovar Typhimurium is critically important for surviving the killing mechanism and growing within macrophages, resulting in a systemic infection. The results also suggest that the cellular requirements for the DnaK/DnaJ chaperone machinery at a high temperature may not be identical to the cellular requirements for the machinery in escaping the bactericidal mechanisms of macrophages.
Depletion of the DnaK/DnaJ chaperone machinery impairs the ability of S. enterica serovar Typhimurium to invade epithelial cells.
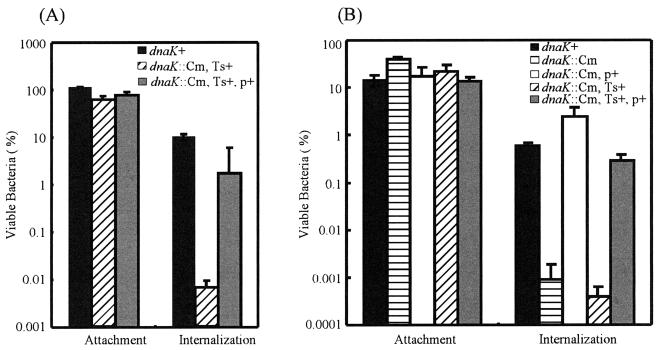
We next examined the ability of the DnaK/DnaJ-depleted mutant to invade epithelial cells. Cultured intestine-407 cells were used to assess the ability of mutant strains CS2021 (dnaK::Cm) and CS2069 (dnaK::Cm, Ts+ suppressor) to invade at 30 and 37°C. As shown in Fig. 5, both mutant strains adhered to monolayers of intestine-407 cells at levels equivalent to those of parent strain χ3306. However, the depletion of the DnaK/DnaJ chaperone machinery greatly reduced the ability of S. enterica serovar Typhimurium to enter cultured epithelial cells. Both CS2021 and CS2069 mutant strains were approximately 1,000-fold less invasive than the wild-type parent strain. The loss of the invasiveness of the mutants was fully complemented when a functional dnaK-dnaJ operon on low-copy-number plasmid pTKY608 was provided. These results suggest that the DnaK/DnaJ chaperone machinery does not function at the step of the adhesion of S. enterica serovar Typhimurium to epithelial cells but is critically important for the subsequent invasion of host cells.
FIG. 5.
Effect of the dnaK disruption on invasion by S. enterica serovar Typhimurium. The ability of strains χ3306 (dnaK+), CS2021 (dnaK::Cm), CS2501 (dnaK::Cm/pTKY608), CS2069 (dnaK::Cm, Ts+ suppressor), and CS2773 (dnaK::Cm/pTKY608, Ts+ suppressor) to adhere to and enter cultured intestine-407 cells at 37°C (A) or 30°C (B) was examined as described in Materials and Methods. The data are the means and standard deviations for each strain tested in triplicate. p+, pTKY608; Ts+, Ts+ suppressor.
Proteome analysis of proteins secreted from the DnaK/DnaJ-depleted mutant.
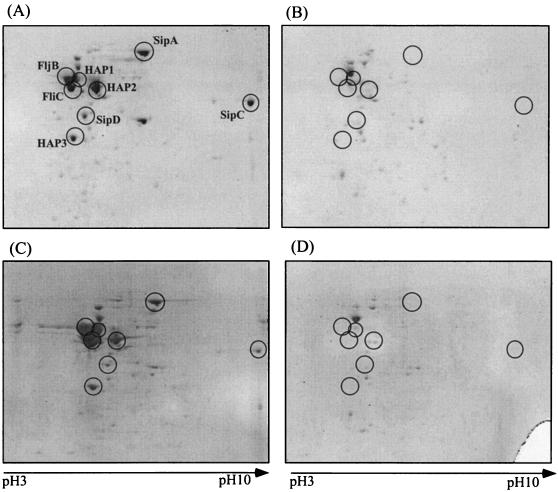
The invasion of nonphagocytic cells by S. enterica serovar Typhimurium has been shown to be mediated by invasion proteins SipA, SipB, SipC, and SipD, which are secreted by the SPI1-encoded type III secretion system and translocated into the mammalian cell plasma membrane or cytosol. To examine the effect of the depletion of DnaK/DnaJ on these invasion proteins, we performed proteome analysis, comparing the proteins secreted from mutant strain CS2021 grown at 30°C and suppressor strain CS2069 grown at 37°C with those from parent strain χ3306. As shown in Fig. 6, the predominant proteins secreted into the culture medium of S. enterica serovar Typhimurium χ3306 grown at 30 and 37°C are the SPI1-encoded proteins SipA, SipC, and SipD and the flagellum-related proteins FliC, FljB, HAP1, HAP2, and HAP3. The flagellar proteins are known to be exported by the type III secretion system specific for flagellar biogenesis (39). Interestingly, none of the proteins secreted by the two systems was detected in the culture media of mutant strain CS2021 (dnaK::Cm) and suppressor strain CS2069 (dnaK::Cm, Ts+ suppressor), suggesting that the DnaK/DnaJ chaperone machinery is essential for the expression, stabilization, and/or secretion of both SPI1-encoded invasion proteins and flagellar proteins in S. enterica serovar Typhimurium.
FIG. 6.
Two-dimensional gel electrophoresis patterns of proteins secreted into the medium by S. enterica serovar Typhimurium strains χ3306 (wild type) grown at 30°C (A) or 37°C (C), CS2021 (dnaK::Cm) grown at 30°C (B), and CS2069 (dnaK::Cm, Ts+ suppressor) grown at 37°C (D). Protein spots enclosed in circles were analyzed by mass spectrometry as described in Materials and Methods and are interpreted in the text.
DISCUSSION
It was hypothesized that the DnaK/DnaJ chaperone machinery plays an important role in allowing pathogenic bacteria to adapt to the hostile environment of the host macrophage phagosome, where bacteria are threatened by oxidative or nonoxidative bactericidal mechanisms. This hypothesis was based on evidence such as the elevated expression of DnaK along with other heat shock proteins in facultative intracellular pathogens, such as S. enterica serovar Typhimurium (3), Yersinia enterocolitica (50), Legionella pneumophila (30), and Brucella abortus (35), growing within macrophage phagosomes after phagocytosis. Later, a study with an insertional mutation in the Brucella suis homologue of the dnaK gene led to the conclusion that DnaK contributes to the intracellular multiplication of B. suis, since the dnaK mutant could survive but failed to multiply within U937-derived phagocytes (29). Although that study demonstrated the participation of the DnaK/DnaJ chaperone machinery in the interaction of bacterial cells with macrophages, the potential roles of the machinery in pathogenesis could not be examined because of the temperature sensitivity for growth of the mutant.
In the present study, isolation of the dnaK::Cm Ts+ suppressor mutant allowed us to examine the possible role of the DnaK/DnaJ chaperone machinery in the pathogenesis of S. enterica serovar Typhimurium in the mouse model. During the course of infection in mice, serovar Typhimurium colonizes many different organs, including the Peyer's patches of the small intestine, mesenteric lymph nodes, spleen, and liver, and causes a severe systemic infection, which can be fatal. The virulence assay after infection of BALB/c mice by oral and subcutaneous routes demonstrated that the DnaK/DnaJ-depleted mutant apparently lost the ability to colonize and cause systemic disease in mice (Fig. 3). It was recently reported that the individual disruption of genes for the ClpXP and Lon proteases, which are heat shock proteins, results in attenuation and a chronic persistent infection without causing an overwhelming systemic infection in mice (45, 52). In contrast, the DnaK/DnaJ-depleted mutant was completely cleared from the mice by day 6 and day 4 after oral and subcutaneous infections, respectively. Since the impaired ability of strain CS2069 (dnaK::Cm, Ts+ suppressor) to cause a systemic illness in mice was restored when a functional copy of the dnaK-dnaJ operon was provided, it can be concluded that the DnaK/DnaJ chaperone machinery is critically important for the pathogenesis of S. enterica serovar Typhimurium.
To gain a better understanding of why the DnaK/DnaJ-depleted mutant lost the ability to colonize and cause systemic disease in mice, a variety of virulence properties were examined. Since the colonization and systemic growth of Salmonella in mouse spleen and liver are associated with the ability to survive and replicate within murine macrophages (9), we examined the fate of the DnaK/DnaJ-depleted mutant after phagocytosis by cultured macrophage cell lines J774.1 and RAW264.7. The results showed that mutant strains CS2021 (dnaK::Cm) and CS2069 (dnaK::Cm, Ts+ suppressor) apparently lost the ability to survive and replicate within both lines of macrophages (Fig. 4), suggesting that the DnaK/DnaJ chaperone machinery of S. enterica serovar Typhimurium is essential for withstanding the killing mechanisms of macrophages and proliferating intracellularly.
To kill bacteria by phagocytosis, bacteria are first engulfed by endocytosis into phagosomes, which then fuse with lysosomes to form phagolysosomes. Most studies have shown that the majority of Salmonella species inhibit phagosome-lysosome fusion and replicate in phagosomes (4, 20, 42, 49). Macrophages have developed an arsenal of oxygen-dependent and -independent mechanisms to effect killing; these include the production of toxic oxygen derivatives, such as hydrogen peroxide, superoxide anions, and hydroxyl radicals, via the respiratory burst (21). In addition, the internalized organisms also are exposed to vacuolar acidification (15). We examined the sensitivity of the DnaK/DnaJ-depleted mutant to hydrogen peroxide, which mimics the oxidative killing mechanism. There was no significant difference in sensitivity to hydrogen peroxide between the mutant and the wild-type parent (results not shown). We next examined the sensitivity of the DnaK/DnaJ-depleted mutant to an acidic environment. The mutant was killed to a significantly higher extent below pH 3.5; e.g., the survival fractions of wild-type and mutant cells were 50 and 0.2%, respectively, for 30 min at pH 3.25 (data not shown). At pHs 4.0 to 7.6, there was no difference in viability between the wild type and the DnaK/DnaJ-depleted mutant. A previous study with macrophages after phagocytosis of killed Salmonella cells documented that acidification of phagosomes to a pH of <5.0 occurred within minutes after phagocytosis and that fusion with lysosomes resulted in further acidification to a pH of <4.5 (1). In contrast, macrophage phagosomes containing live Salmonella cells maintained the compartment at pHs 5.5 to 6.0 for 1 to 2 h and required 4 to 5 h to reach a pH of <5.0. Therefore, the increased sensitivity of the DnaK/DnaJ-depleted mutant observed only under acidic conditions (<pH 3.5) would not explain the impaired ability of the mutant to survive within macrophages, as shown in Fig. 5.
After uptake by macrophages, S. enterica serovar Typhimurium replicates in a membrane-bound compartment, referred to as the Salmonella-containing vacuole (SCV) (39), which is segregated from the late endocytic pathway (20, 37, 42, 49). The interactions between Salmonella and macrophages are extremely complex, and replication within the SCV is a multifactorial process involving numerous bacterial genes. These include the spv operon located on the virulence plasmid (18, 33), the PhoP/PhoQ regulon (14, 38), and the SPI2 genes (23, 49). Recently, it was demonstrated that the regulator controlled by the PhoP/PhoQ two-component system makes a major contribution to trafficking of the SCV in macrophages (14). To understand how the DnaK/DnaJ chaperone machinery is involved in the intracellular proliferation of S. enterica serovar Typhimurium, future studies must focus on the possibility that it may function in SCV biogenesis in macrophages.
Of further interest is the finding that the DnaK/DnaJ-depleted mutant could not invade cultured epithelial cells (intestine-407) (Fig. 5). It is known that the invasion of epithelial cells by S. enterica serovar Typhimurium is mediated by the SPI1-encoded invasion proteins (7, 12). It was previously demonstrated that the SPI1-encoded proteins and flagellum-related proteins, which are also secreted by a type III secretion system, are the predominant proteins secreted into culture media (46). Proteome analysis of the secreted proteins from DnaK/DnaJ-depleted mutant cells revealed that none of the SPI1-encoded proteins was secreted into the culture medium (Fig. 6). Among the SPI1-encoded proteins are SipA to SipD, which are translocated into the mammalian cell cytosol and trigger host signal transduction pathways, resulting in profuse actin cytoskeletal rearrangements and membrane ruffles at the point of bacterial cell-host cell contact that ultimately lead to bacterial uptake (10, 11). Therefore, the inability of the DnaK/DnaJ-depleted mutant to invade intestine-407 cells could have been due to the loss of Sip proteins, which are essential for bacterial entry.
The synthesis of the SPI1-encoded type III secretion apparatus and most of its secreted effectors is controlled by a complex regulatory system. Many studies have shown that the SPI1-encoded transcriptional regulators HilD, HilC, HilA, and InvF act in an ordered fashion to coordinately activate the expression of SPI1 (for a review, see reference 36). HilD and HilC derepress hilA transcription. HilA activates invF as well as other SPI1 genes and therefore plays a key role in coordinating the expression of the SPI1-encoded type III secretion system. Since none of the SPI1-encoded proteins was secreted from the dnaK::Cm mutant, it is likely that the DnaK/DnaJ chaperone machinery is involved in the expression of SPI1 through interactions with regulators such as hilD, hilC, hilA, and/or invF. Alternatively, the type III secretion apparatus may not function in DnaK/DnaJ-depleted cells. Studies of the mechanism by which this chaperone machinery regulates the SPI1-encoded type III secretion system are now in progress.
Proteome analysis also revealed that none of the flagellar proteins was secreted from DnaK/DnaJ-depleted cells (Fig. 6). In agreement with this finding, we observed that the mutant was nonmotile. Nonflagellar mutants derived from S. enterica serovar Typhimurium χ3306 by the inactivation of fliC, encoding the flagellar filament, remained virulent in BALB/c mice (unpublished data), suggesting that the flagellar defect caused by DnaK/DnaJ depletion is not responsible for the loss of virulence of the mutant. For E. coli, DnaK and DnaJ were previously reported to be required for flagellum synthesis by affecting the transcription of the flhD and flhC genes, which are at the apex of the flagellar regulon (43). In contrast, the dnaK::Cm mutation did not affect the transcription of flhD and flhC in S. enterica serovar Typhimurium (unpublished data). The regulatory mechanism of flagellum synthesis by the DnaK/DnaJ chaperone machinery in Salmonella will be published elsewhere.
Acknowledgments
We thank A. Tokumitsu, C. Yoshikawa, M. Suzuki, C. Kodama, and M. Matsui for technical assistance. We also thank Debbie Ang for critical reading of the manuscript.
This research was supported by grants-in-aid for scientific research (13470058) and research on priority areas (15019017) from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government.
Editor: B. B. Finlay
REFERENCES
- 1.Alpuche-Aranda, C. M., J. A. Swanson, W. P. Loomis, and S. I. Miller. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. USA 89:10079-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, J. C. A., and E. A. Craig. 1984. Major heat shock gene of Drosophila and the E. coli heat inducible dnaK gene are homologous. Proc. Natl. Acad. Sci. USA 81:848-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchimeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248:730-732. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier, N. A., and F. Heffron. 1991. Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect. Immun. 59:2232-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 6.Clark, M., M. A. Jepson, N. L. Simmons, and B. H. Hirst. 1994. Preferential interaction of Salmonella typhimurium with mouse Peyer's patch M cells. Res. Microbiol. 145:543-552. [DOI] [PubMed] [Google Scholar]
- 7.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnenberg, M. S., and J. B. Kapler. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4312-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finlay, B. B., and P. Cossart. 1997. Exploitation of mammalian cell functions by bacterial pathogens. Science 276:718-725. [DOI] [PubMed] [Google Scholar]
- 11.Galán, J. E. 1999. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr. Opin. Microbiol. 2:46-50. [DOI] [PubMed] [Google Scholar]
- 12.Galán, J. E., and D. Zhou. 2000. Striking a balance: modulationof the actin cytoskeleton by Salmonella. Proc. Natl. Acad. Sci. USA 97:8754-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamer, J., G. Multhaup, T. Tomoyasu, J. S. McCarty, S. Rudiger, H. J. Schonfeld, C. Schirra, H. Bujard, and B. Bukau. 1996. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J. 153:607-617. [PMC free article] [PubMed] [Google Scholar]
- 14.Garvis, S. G., C. R. Beuzon, and D. W. Holden. 2001. A role for the PhoP/Q regulon in inhibition of fusion between lysosomes and Salmonella-containing vacuoles in macrophages. Cell. Microbiol. 3:731-744. [DOI] [PubMed] [Google Scholar]
- 15.Geisow, M. J., P. D'Arcy-Hart, and M. R. Young. 1981. Temporal changes of lysosome and phagosome pH during phagolysosome formation in macrophages: studies by fluorescent spectroscopy. J. Cell Biol. 89:645-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgopoulos, C., D. Ang, K. Liberek, and M. Zylicz. 1990. Properties of the Escherichia coli heat shock proteins and their role in bacteriophage lambda growth, p. 191-221. In R. I. Morimoto, A. Tissiéres, and C. Georgopoulos (ed.), Stress proteins in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Gulig, P. A., and R. Curtiss III. 1987. Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 55:2891-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulig, P. A., and T. J. Doyle. 1993. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect. Immun. 61:504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-580. [DOI] [PubMed] [Google Scholar]
- 20.Hashim, S., K. Mukherjee, M. Raje, S. K. Basu, and A. Mukhopadhyay. 2000. Live Salmonella modulate expression of Rab proteins to persist in a specialized compartment and escape transport to lysosomes. J. Biol. Chem. 275:16281-16288. [DOI] [PubMed] [Google Scholar]
- 21.Hassett, D. J., and M. S. Cohen. 1989. Bacterial adaptation to oxidative stress: implication for phagocytosis and interaction with phagocytic cells. FASEB J. 3:2574-2582. [DOI] [PubMed] [Google Scholar]
- 22.Hendrick, J. P., and F. U. Hartl. 1993. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 62:349-384. [DOI] [PubMed] [Google Scholar]
- 23.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 24.Hensel, M., Shea, R. S. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 25.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, K., I. Charles, G. Dougan, D. Pickard, O'Gaora, P. Costa, G. Ali, I. T. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 27.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, B. D., and S. Falkow. 1996. Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 14:533-561. [DOI] [PubMed] [Google Scholar]
- 29.Köhler, S., J. Teyssier, A. Cloekkaert, B. Rouot, and J. P. Liautard. 1996. Participation of the molecular chaperone DnaK in intracellular growth of Brucella suis within U937-derived phagocytes. Mol. Microbiol. 20:701-712. [DOI] [PubMed] [Google Scholar]
- 30.Kwaik, Y. A., B. I. Eisenstein, and N. C. Engeleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Leung, K. Y., and B. B. Finlay. 1991. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 88:11470-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Libby, S. J., M. Lesnick, P. Hasegawa, E. Weidenhammer, and D. G. Guiney. 2000. The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell. Microbiol. 2:49-58. [DOI] [PubMed] [Google Scholar]
- 34.Liberek, K., and C. Georgopoulos. 1993. Autoregulation of the Escherichia coli heat shock response by the DnaK and DnaJ heat shock proteins. Proc. Natl. Acad. Sci. USA 90:11019-11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, J., and T. A. Ficht. 1995. Protein synthesis in Brucella abortus induced during macrophage infection. Infect. Immun. 63:1409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lostroh, C. P., and C. A. Lee. 2001. The Salmonellla pathgenicity island-1 type III secretion system. Microb. Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 37.Meresse, S., O. Steele-Mortimer, B. B. Finlay, and J. P. Gorvel. 1999. The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J. 16:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morimoto, R., A. Tissieres, and C. Georgopoulos. 1994. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 93:7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rathman, M., L. P. Barker, and S. Falkow. 1997. The unique trafficking pattern of Salmonella typhimurium-containing phagosomes in murine macrophages is independent of the mechanism of bacterial entry. Infect. Immun. 65:1475-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, W., Y. Zhou, J. Wild, J. Adler, and C. A. Gross. 1992. DnaK, DnaJ, and GrpE are required for flagellum sysnthesis in Escherichia coli. J. Bacteriol. 174:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takaya, A., M. Suzuki, M. Matsui, T. Tomoyasu, H. Sashinami, A. Nakane, and T. Yamamoto. 2003. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar Typhimurium infection of mice. Infect. Immun. 71:690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomoyasu, T., A. Mogk, H. Langen, P. Goloubinoff, and B. Bukau. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397-413. [DOI] [PubMed] [Google Scholar]
- 48.Tomoyasu, T., T. Ogura, T. Tatsuta, and B. Bukau. 1998. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 30:567-581. [DOI] [PubMed] [Google Scholar]
- 49.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto, T., T. Hanawa, and S. Ogata. 1994. Induction of Yersinia enterocolitica stress proteins by phagocytosis with macrophage. Microbiol. Immunol. 38:295-300. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto, T., T. Hanawa, S. Ogata, and S. Kamiya. 1996. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect. Immun. 64:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto, T., H. Sashinami, A. Takaya, T. Tomoyasu, H. Matsui, Y. Kikuchi, T. Hanawa, S. Kamiya, and A. Nakane. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in a persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 69:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]