Abstract
The gingipains have been implicated in the pathogenicity of Porphyromonas gingivalis, a major etiologic agent of chronic periodontitis. Mature gingipains often present as a membrane-bound glycosylated proteinase-adhesin complex comprising multiple adhesin domains (HA1 to -4) and a catalytic domain. Using recombinant adhesin domains, we were able to show that patients with chronic periodontitis produce significantly more immunoglobulin G reactive with gingipain domains than a corresponding group with healthy periodontium. Titers were predominantly directed toward the carbohydrate epitopes shared between the gingipains and the lipopolysaccharide of P. gingivalis with little recognition of the peptide backbone of the catalytic domains. Distribution of titers to peptide epitopes of the adhesin domains was as follows: HA4 ≈ HA1 > HA3 ≫ HA2. No correlation was observed between markers of disease severity and titers to individual adhesins within the disease group. Posttreatment titers showed no change or a decrease in titers for the majority of patients except for titers to the HA2 domain which showed marked increases in a few responding patients. Since the HA2 domain is important in hemoglobin binding and acquisition of essential porphyrin, boosting titers of antibodies to this domain may have the potential to control the growth of this organism.
Destructive periodontitis is considered to represent an immunopathological response to a complex microflora that develops adjacent to the supporting tissues of an affected dentition. A hallmark of the protracted inflammation in a progressing lesion is the failure of the epithelial attachment to the tooth and the migration of epithelial components down the tooth root, creating a cleft or pocket favorable for an abundant anaerobic microbial flora. The inflammatory reaction in the underlying connective tissue leads to degradation of the structural matrix, including the supporting bone, culminating in mobilization and eventual exfoliation of the tooth. Progressive periodontal disease of humans and animals has been linked to a group of gram-negative anaerobic organisms including Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythensis (Bacteroides forsythus), Fusobacterium nucleatum, Actinobacillus (Haemophilus) actinomycetemcomitans, and treponeme species (2). P. gingivalis has been implicated as an important periodontal pathogen due to its high incidence and relative levels in human disease (16, 44) and its virulence in monoinfected animals (13, 17). The virulence of P. gingivalis has been attributed to several components produced by the microorganism, including a cysteine proteinase complex known as the gingipain proteinases (11, 22).
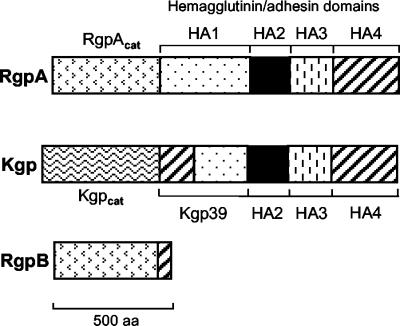
The gingipains are a group of Arg- and Lys-specific proteinases and are the products of three separate genes in P. gingivalis: rgpA, rgpB, and kgp (6). Fully processed gingipains exist in multiple forms (Fig. 1) and, in a strain-dependent fashion, are partitioned either as soluble forms in the culture supernatant or as membrane-associated forms on the cell surface or on extracellular vesicles released from the organism (35). Lower-molecular-weight forms (such as RgpB and processed RgpA in certain strains) are proteolytically released into the growth media. The higher-molecular-weight forms (RgpA and Kgp) are membrane associated and comprise a catalytic domain and three to four hemagglutinin/adhesin domains (HA1 to HA4) in strong noncovalent associations (9, 35). Membrane-associated forms of the gingipains have been reported to be glycosylated with carbohydrate moieties that are cross-reactive with those in P. gingivalis lipopolysaccharide (LPS) (7, 46).
FIG. 1.
Domains of the gingipain complex. Highly homologous regions (similarly hatched regions) occur between the mature proteins of RgpA, RgpB, and Kgp in P. gingivalis strain HG66 (31, 34). Rgpcat and Kgpcat denote the catalytic domains of the arginine-specific and lysine-specific gingipains, respectively. The hemagglutinin/adhesin domains are denoted as HA1 to HA4. Kgp39 is a fusion protein between the N-terminal region of HA4 and the C-terminal region of HA1.
Apart from providing P. gingivalis with a general proteolytic tool for degrading proteinaceous nutrients for growth, gingipains also function as important components in binding to the host tissues (4, 26), in evasion of the host immune response (33), in encouraging inflammation and vascular permeability for the exudation of nutrients and erythrocytes (18), and in the acquisition of iron and porphyrin essential for growth of the organism (25, 30).
Due to the important role of the gingipains in the virulence and nutrient acquisition by the organism, the host immune responses to the gingipain domains may play a critical role in the incidence and severity of chronic periodontitis (21, 28, 43). We provide here a comprehensive analysis of the antibody responses to individual gingipain domains in subjects with chronic periodontitis pre- and posttreatment, which may lead to the identification of specific targets for protective immunization.
MATERIALS AND METHODS
Patient and controls.
Blood samples were obtained with informed consent from 22 adult chronic periodontitis patients (8 females and 14 males; age range, 35 to 68 years; mean, 48 ± 9 years). Fourteen patients had generalized periodontitis and eight patients had localized periodontitis, with no history of systemic diseases known to affect the periodontium or of antimicrobial use within the last 6 months and no record of periodontal therapy, including subgingival scaling, root planing, or relevant surgery in the previous 3 years. All patients showed a diagnosis of mild to severe periodontitis with individual maximal attachment loss (AL; 5 to 11 mm), individual maximal probing depths of 4 to 11 mm, and resorption of alveolar bone as assessed by radiographic means. For the majority of patients, a further blood sample was obtained at the completion of the nonsurgical treatment protocol (2 to 8 weeks from the commencement of therapy). A control group of 12 adults with healthy periodontium was recruited from staff of the Institute of Dental Research (seven males and five females; age range, 27 to 64 years; mean, 45 ± 12 years), with no systemic disease, clinical signs of significant AL, or radiographic evidence of bone loss. None of the control subjects had probing depths of >3 mm.
Serum IgG isolation.
Serum was separated from venous blood and stored frozen in aliquots at −70°C until use. Once thawed, sodium azide was added to a final concentration of 10 mM, and the samples were stored at 4°C. The immunoglobulin G (IgG) fraction was purified from sera by protein G affinity chromatography. Protein G columns (Pharmacia, Uppsala, Sweden) were equilibrated with 20 mM sodium phosphate (pH 7.0) and then loaded with a 1/10 dilution of patient sera in the same buffer. Loaded columns were washed until 280-nm UV readings returned to baseline. IgG was eluted with 0.1 M glycine-HCl (pH 2.7). IgG fractions were adjusted to neutral pH with a 1/20 volume addition of 2 M Tris buffer (pH 9.0). IgG concentrations were determined with Coomassie Plus protein assay reagent (Pierce Biotechnology) with bovine serum albumin as the standard.
MAbs.
Monoclonal antibodies (MAbs) 5A1, 2B2, and 3B3 were prepared in mice against gingipains as described previously (9). In denatured samples, MAb 5A1 recognizes the HA2, HA1, and Kgp39 gingipain domains (Fig. 1) (12). 2B2 recognizes the HA1, HA3, and Kgp39 domains and 3B3 recognizes the LPS component from P. gingivalis. Possible cross-reactivities of MAb 5A1, 2B2, and 3B3 with extracts of other plaque bacteria suspected to have a role in periodontal disease and within the same genus (namely, F. nucleatum, Prevotella intermedia, Prevotella melanogenica, and Porphyromonas endodontalis) were not detected by enzyme-linked immunosorbent assay (ELISA) or Western blot (data not shown).
Gingipain complex purification.
Cultures of P. gingivalis ATCC 33277 were grown in modified CDC broth (10 g of tryptone, 10 g of tryptone soy broth, 10 g of yeast extract [Oxoid, Ltd.], and 5 g of NaCl/liter, supplemented with 0.4 g of l-cysteine, 5 mg of hemin, 2 mg of menadione, and 20 ml of horse serum/liter) at 37°C in an anaerobic atmosphere of 5% H2, 5% CO2, and 90% N2. Culture purity was regularly checked by Gram stain, PCR with r16S P. gingivalis-specific primers, and replating onto sheep blood-CDC agar plates. Gingipains were isolated by the method of Ciborowski et al. (5) with modifications. Briefly, cells were harvested at late-exponential-growth phase by centrifugation and washed three times with phosphate-buffered saline (PBS). All steps were carried out at 4°C unless stated otherwise. Surface proteins were extracted by gentle stirring for 3 h in Tris-CaCl2 buffer (50 mM Tris, 2 mM CaCl2 [pH 8.0]) containing 0.75% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}. After particulate removal by ultracentrifugation (140,000 × g, 90 min), the supernatant was passed through an ion-exchange Resource-Q column (Pharmacia) preequilibrated with Tris-CaCl2 buffer plus 1% CHAPS at a flow rate of 1 ml/min. After a wash step, bound protein was eluted with 1 M NaCl in Tris-CaCl2-CHAPS buffer. The eluate was then dialyzed back into Tris-CaCl2 buffer before the final affinity purification step through an arginine-Sepharose column (Pharmacia). After adsorption onto the matrix, the column was washed with 0.5 M NaCl in Tris-CaCl2 buffer. The Kgp-rich fraction was eluted with 0.75 M l-lysine in Tris-CaCl2, and RgpA/Kgp complex was eluted with 1 M l-arginine in Tris-CaCl2 buffer.
LPS purification.
The phenol-water extraction method used was similar to that described by Westphal et al. (47). Briefly, 10 g (wet weight) of P. gingivalis was resuspended in 100 ml of distilled H2O (dH2O) at 65 to 68°C. An equal volume of isothermal 90% phenol was slowly added with vigorous stirring. After 15 min of continuous stirring, the mixture was cooled on ice for 10 min and then centrifuged to separate the phases. The upper aqueous phase was carefully removed, and the phenol phase was reextracted as described above. Aqueous phases were pooled and dialyzed extensively against dH2O at 4°C and then lyophilized. The pellet was resuspended in 30 ml of digest buffer (20 mM Tris, 2 mM MgCl2, 20 mM NaCl, 0.1 mM dithiothreitol [pH 8.0]). DNase I and RNase A were added (to 100 and 25 μg/ml, respectively), and the solution was incubated at 37°C for 2 h with gentle stirring. Proteinase K and CaCl2 were subsequently added to the mixture (to 100 μg/ml and 1 mM, respectively), and the digest incubated for a further 3 h at 37°C. Contaminating enzymes and peptides were removed by refluxing the mixture through a series of neutral water-saturated phenol wash steps. Dialysis against dH2O was then used to remove low-molecular-weight moieties from the aqueous phase before the samples were lyophilized and weighed. For Escherichia coli LPS preparations, a final ultracentrifugation (105,000 × g, 3 h) and dH2O wash step were used to pellet the LPS prior to lyophilization. Purity was checked with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), staining with Coomassie blue for protein contamination, and silver staining for the LPS (14). Western blots with MAbs 5A1 and 2B2 were used to confirm the removal of proteinaceous gingipain contaminants from the P. gingivalis LPS preparation.
Deglycosylation of the gingipains.
The procedure was essentially similar to that described by Sojar and Bahl (45). Briefly, 800 μg of salt-free lyophilized gingipain was incubated with 500 μl of 10% (vol/vol) toluene in anhydrous trifluoromethanesulfonic acid (TFMS) at 0°C for 2 h under nitrogen in a glass vial with occasional mixing. Unreacted TFMS was neutralized by the slow addition of 1 ml of 60% (vol/vol) pyridine aqueous solution at −20°C. Pyridine salts were removed by dialysis. SDS-PAGE, along with Western blots with MAbs 2B2, 5A1, and 3B3, was used to confirm the integrity of the peptide backbone and the loss of the carbohydrate epitopes (data not shown).
Expression and purification of recombinant adhesin domains.
The HA2 domain was cloned, expressed, purified, and characterized as previously described (12). The DNA sequence, which translates to the adhesin domains of the RgpA complex, namely, the polyadhesin domain (PAD), was PCR amplified from genomic DNA. Appropriate restriction sites (NdeI and SapI) were incorporated into the primers (Table 1) to facilitate PCR product insertion into the Impact T7 expression vectors (New England Biolabs). This PAD clone was then used as the PCR template for the subsequent subcloning of PAD subdomains (HA1, HA3, and HA4; see Fig. 2) into Impact T7 vectors. Clone integrity for all constructs was confirmed via dideoxy sequencing with reference to rgp1 (GenBank accession no. U15282) (31). Use of the Impact T7 vectors resulted in all clones being fused via an intein linker to a C-terminal chitin-binding motif, allowing the preferential purification of full-length products on a chitin column and subsequent removal of the domain via intein self-cleavage in the presence of reducing agents, resulting in no-tag recombinant products. Vectors were transformed into the E. coli expression hosts ER2566 or JM109(DE3). Cultures were grown at 16°C in order to minimize proteolytic degradation of the recombinant products. T7 RNA polymerase-mediated overexpression was induced at an optical density at 600 nm (OD600) of 1.0 by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 0.5 mM. At 20 h postinduction, cells were harvested by centrifugation, and the cell pellets were stored at −70°C. Lysis was achieved via grinding (mortar and pestle) frozen pellets at 4°C in the presence of alumina (type 5-A). The pastes of cells were resuspended in Tris-Triton X-100 buffer (buffer A; 50 mM Tris, 250 mM NaCl, 0.1% Triton X-100 [pH 8.0]). Cleared lysates (centrifugation and 0.8-μm-pore-size filtration) were loaded onto a 5-ml preequilibrated chitin chromatography column. The column was washed with buffer B (buffer A plus 1.12 to 2 M NaCl [construct dependent]) until the absorbance at 280 nm stabilized. Triton X-100 was then flushed out with buffer C (50 mM Tris, 100 mM NaCl [pH 8.0]). Intein-mediated release of adhesin domains was induced by the addition of dithiothreitol to 30 mM, followed by incubation overnight. The purity of the released products was checked with SDS-PAGE and confirmed by Western blots with MAbs 5A1 and 2B2 and N-terminal protein sequencing.
TABLE 1.
Primer sets used for cloning of adhesin domains
| Clone | Primer sequence (5′ to 3′)
|
|
|---|---|---|
| Forwarda | Reverseb | |
| rPAD | gaaCATATGCGCAGCGGTCAGGCC | tccaGCTCTTCAGCACTTTACAGCGAGTTTCTC |
| rHA1 | cgaacttCATATGAGCGGTCAGGCCGAG | ↓GCGCTTGCCGTTGGCCTTGATCTC |
| rHA2 | See Paramaesvaran et al. (30) | |
| rHA3 | cgtacgtaCATATGCCTCAAAGTGTATGG | ↓ACGTACATCGTTTGCAGGTTCGATCG |
| rHA4 | cgtgatCATATGGCCAACGAAGCCAAGG | tccaGCTCTTCAGCACTTTACAGCGAGTTTCTC |
NdeI restriction sites are underlined, and irrelevant bases for ease of restriction digest are in lowercase.
SapI restriction sites are underlined and blunt ends for ligation into SapI and SmaI sites on the vector are indicated by down arrows respectively. Irrelevant bases for ease of restriction digest are in lowercase.
FIG. 2.
Diagram of the regions of the cloned protein with respect to the rgpA gene. Small arrows denote the position of primers for the particular cloned domain.
Protein-Sepharose preparation.
Protein samples to be coupled to N-hydroxsuccinimide-activated Sepharose beads (Pharmacia) were dialyzed into or added to coupling buffer (50 mM sodium borate, 0.5 M NaCl [pH 8.0]). All steps were carried out at 4°C as per manufacturer's instructions. Briefly, ligands in coupling buffer were added to beads and left to couple overnight. Excess unreacted groups on the matrix were blocked with 1 M ethanolamine-HCl buffer (pH 8.0) for 3 h. The beads were washed with Tris-buffered saline-Tween20 (TBST; 50 mM Tris, 0.5 M NaCl, 0.1% Tween 20 [pH 7.5]) prior to use in depletion assays.
Western blotting and immunoprobing.
Known amounts of protein antigen or whole bacterial cell culture were standardized (OD600 of 0.5), separated (SDS-PAGE), and then electroblotted onto 0.2-μm-pore-size nitrocellulose membranes (Bio-Rad Corp.). Membranes were blocked with 2% bovine serum albumin in PBS plus 10 mM NaN3 overnight at room temperature. A multichannel blotting apparatus (Milliblot-MP; Millipore Corp.) was used to probe the membranes with 1:40 dilution of sera or 50 ng of MAb/ml in TBST for 3 h at room temperature. When probing against a recombinant product was done, sera were first depleted of anti-E. coli antibodies by a 2-h incubation with pan-E. coli protein extract attached to NHS-Sepharose beads. Likewise, anti-PAD depleted sera were prepared by prior incubation with PAD-Sepharose beads and anti-LPS depleted sera used in the Western blots were prepared by prior incubation with 10 μg of P. gingivalis LPS/ml. Alkaline phosphatase (AP)-conjugated anti-IgG or anti-mouse (Dako Corp.) antibodies were used at a 1:10,000 dilution in TBST for 2 h at room temperature before final color development with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium as substrates (in 20 mM Tris-0.2 mM MgCl2 [pH 9.5] for 5 to 10 min at room temperature).
N-terminal sequencing.
Protein bands were blotted onto polyvinylidene difluoride membranes and then sequenced by automated Edman degradation by using an Applied Biosystems 494 Procise protein sequencing system (Australian Proteome Analysis Facility, Sydney, Australia). Performance of the system was assessed routinely against 10 pmol of β-lactoglobulin standards.
General ELISA.
ELISAs were performed in polystyrene microtiter wells with a working volume of 100 μl/well. The overnight washing or blocking cycles performed between each step were done with a Bio-Rad Model 1250 Immunowasher and were performed in TBST at room temperature. Proteins were coated overnight at room temperature in 50 mM sodium bicarbonate-137 mM NaCl (pH 9.0) with 10 mM NaN3. Quadruplicate 1:3 dilutions for five dilution points of human sera in TBST were added to the blocked plates, followed by incubation for 1.5 h at 37°C. The starting concentration for each serum and antigen was determined in a preliminary ELISA to ensure at least three dilution points were located in the linear range of the assay (32). Prior to use in ELISA assays against recombinant products, human sera were depleted of anti-E. coli antibodies as described above. None of the sera showed reactivity against recombinant host E. coli LPS. A 1:5,000 dilution of AP-conjugated goat anti-human IgG (γ-chain specific) in TBST was incubated with well samples for 1.5 h at 37°C. Similarly, murine MAbs 5A1 or 2B2 were detected by using a 1:5,000 dilution of AP-conjugated rabbit anti-mouse IgG (Dako Corp.). The AP activity was assayed as the amount of hydrolysis of 5 mM p-nitrophenol phosphate achieved in 60 min at 37°C (in 20 mM Tris, 1 mM MgCl2, 10 mM NaN3 [pH 9.5]), whereby the OD405 nm was monitored in a Bio-Rad Benchmark microplate reader (linear OD range, 0 to 3.5). A standardization matrix was used to determine the optimal concentration of coating antigen required to achieve an approximate OD405 of 2.0 after 1 h of incubation at 37°C with the same dilution of MAbs. The standardized concentration of protein was then used in subsequent ELISA assays with the patient sera. Antibody titers in serum were defined as the dilution required to give an OD405 of 0.5, at which point the titration curves enter the linear range.
Materials.
All chemicals were purchased from Sigma-Aldrich Ltd. (Sydney, New South Wales, Australia) unless otherwise specified.
Statistics.
Comparisons between antibody titers in serum to each domain of RgpA were analyzed by using the nonparametric, two-tailed, matched-pair, t test with a Bonferroni posttest correction. For comparison of titers between healthy and disease groups, the nonparametric two-tailed Wilcoxon Mann-Whitney test was used, and correlations between titers and disease indices were analyzed by nonparametric Spearman two-tailed analysis with 95% confidence intervals as calculated with Prism 3.03 (Graphpad Software, Inc., San Diego, Calif.). The correlation coefficients were expressed as rSp, and the P significance values were determined by using Spearman's correlation. For correlation analyses, levels of periodontal disease severity were assigned the following diagnosis scores (Dx score): 0, no periodontitis present with no gingival AL; 1, mild periodontitis (0.5 mm ≤ AL ≤ 3.0 mm); 3, moderate periodontitis (3.5 mm ≤ AL ≤ 5.5 mm); and 5, advanced periodontitis (AL ≥ 6 mm). If the lesions were detected at more than two sites per quadrant of the mouth, the lesions were classified as generalized periodontitis and given a Dx score of 2, 4, or 6 for mild, moderate, or advanced periodontitis, respectively (Table 2).
TABLE 2.
Derivation of Dx scoresa
| Level of AL in periodontitis patientsb | Dx score |
|---|---|
| Gingivitis | 0 |
| Mild | |
| L | 1 |
| G | 2 |
| Moderate | |
| L | 3 |
| G | 4 |
| Advanced | |
| L | 5 |
| G | 6 |
The severity of periodontal disease was graded according to the degree of loss of gingival attachment around the teeth and the localization of these lesions in the mouth.
Mild AL was defined as 0.5 mm ≤ AL ≤ 3 mm; moderate AL was defined as 3.5 mm ≤ AL ≤ 5.5 mm; and advanced AL was defined as AL ≥ 6 mm. L, localized lesions, detected at a frequency of ≤2 sites per quadrant; G, generalized lesions, detected at a frequency of >3 sites per quadrant.
RESULTS
Profile of antibody recognition of gingipain components.
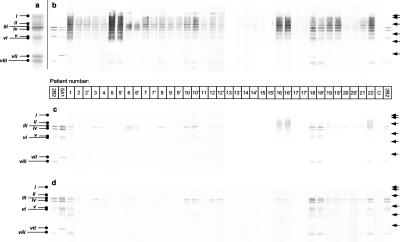
Purified gingipain from P. gingivalis strain ATCC 33277 is a high-molecular-weight complex comprising RgpA and Kgp. Component parts of the complex dissociated in SDS-PAGE denatured under reducing conditions were N terminally sequenced and are identified as shown in Fig. 3a. Western blots of sera from patients with periodontal disease against the native gingipain complex indicated antibody recognition of multiple domains (Fig. 3b). In the periodontally healthy group, two individuals showed only trace recognition of the gingipains. The extensive smearing of the strongly reactive sera resembled a reaction to a lipid-carbohydrate component within the blot. Reducing SDS-PAGE did not strip the proteins of the MAb 3B3 (an anti-LPS MAb) antigenic determinant, suggesting that a lipid or glycosidic linkage existed between the LPS/carbohydrate epitope and the proteins (data not shown). Sera preabsorbed with P. gingivalis LPS showed significantly less reactivity with native gingipain preparations (Fig. 3c), and a similar profile was seen with chemically deglycosylated gingipains (Fig. 3d). Further, preabsorption with both P. gingivalis LPS and the recombinant PAD (rPAD) removed essentially all of the reactivity against the native gingipain complex (data not shown), indicating that the catalytic domains of the protein complex are weak immunogens and that naturally occurring anti-gingipain antibodies are predominantly directed against the HA domains, including the polysaccharide components.
FIG. 3.
SDS-PAGE of the gingipain complex and the corresponding patient sera reactivity in Western blots. (a) N-terminal sequencing by automated Edman degradation identified the composition of the bands in the denaturing (boiled and reduced) SDS-PAGE preparation of gingipain as RgpA (i), Kgpcat (ii), RgpAcat plus HA1 (iii), Kgp39 (iv), truncated HA1 + HA4 (v), and truncated RgpAcat (vi), HA2 (vii), and HA3 (viii). (b to d) Western blots of whole sera from disease patients against the gingipain complex (b), sera predepleted of anti-P. gingivalis LPS antibodies (c), and sera against the deglycosylated gingipain complex (d). 2B2 and 5A1 denote MAbs recognizing HA1, HA3, and Kgp39 and HA1, HA2, and Kgp39, respectively. Disease patients are numbered in the bar below panel b; number-prime values denote posttreatment serum from the same numbered patient. The “C” denotes serum from one representative control subject. Right arrows represent molecular weight markers: 118, 82, 50.4, 33.4, 26.7, and 19.6 kDa, respectively.
Western blotting of patient sera against purified recombinant adhesins showed a semiquantitative recognition profile for each adhesin domain (Fig. 4). Both rHA1 and rHA4 were recognized by a large number of disease patients, whereas rHA3 was poorly detected. There was no recognition of rHA2 by Western blotting (data not shown).
FIG. 4.
Patient antibodies against recombinant adhesin products as shown by Western blotting. Purified recombinant proteins were separated by denaturing SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were probed with 1:40 dilutions of individual patients (numbered below the blots) sera as described in Materials and Methods. There was no recognition of rHA2.
Quantitative analysis of antibody responses to gingipain components.
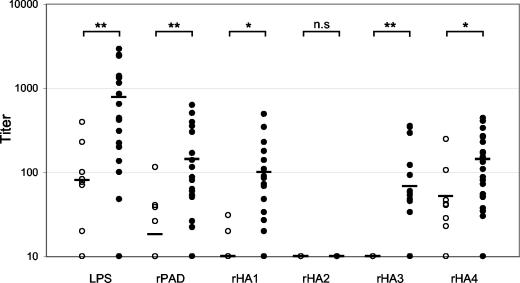
Using recombinant adhesin domains and purified P. gingivalis LPS, the hierarchy of titers by ELISA to the component domains of gingipain complex was determined to be as follows: LPS > rHA4 ≈ rHA1 > rHA3 ≫ rHA2 (Fig. 5). ELISA titers from the disease group showed significantly higher IgG reactivity against most gingipain components than for periodontally healthy controls (Fig. 5). Mean titers to P. gingivalis LPS were significantly higher in the disease group than titers to the adhesin domains of RgpA (Table 3). In nondiseased subjects, titers to P. gingivalis LPS were also detected in 7 of 12 of the group, albeit at a lower mean level. No subject in our study cohort was reactive to purified expression strain E. coli LPS as determined by Western blotting (data not shown), ruling out the possible contribution of titer to E. coli LPS should there be LPS contamination of the purified recombinant adhesins.
FIG. 5.
IgG titers to domains of RgpA. Open symbols denote the periodontally healthy subjects, and solid symbols denote the disease patients. Bar in each column represents the mean group titer to each domain. The unpaired, nonparametric Mann-Whitney test was used to analyze differences in mean titer between healthy and disease groups for each domain (n.s, not significant; ✽, P < 0.05; ✽✽, P < 0.01).
TABLE 3.
Comparison of mean titers to each domain of RgpA in the disease group
| Clone | Comparison of mean titera of clone:
|
||||
|---|---|---|---|---|---|
| LPS | rPAD | rHA1 | rHA2 | rHA3 | |
| rPAD | ** | ||||
| rHA1 | ** | NS | |||
| rHA2 | *** | ** | *** | ||
| rHA3 | ** | * | NS | * | |
| rHA4 | * | NS | NS | *** | * |
Analyzed with the nonparametric, paired Wilcoxon test using 95% confidence intervals with Bonferroni's posttest correction. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Relationship of antibody titers to disease indices.
There were significant positive correlations between disease indices and titers to LPS and the recombinant products: rPAD, rHA1, rHA3, and rHA4 (Table 4) when both healthy and disease group titers were included in the correlation matrix. Titers to rHA2 could not be measured in either group under similar assay conditions. However, after prolonged development (24 h) of the ELISA, four patients and one healthy subject did show very low titers to rHA2 (data not shown).
TABLE 4.
Correlation between total antibody titers (including control and disease groups) to individual domains of RgpA and disease indices, including Dx score, mean probe depth of the periodontal pocket, and mean gingival AL around the teeth
| Disease index | Titer correlation (rSp value)a
|
|||||
|---|---|---|---|---|---|---|
| LPS | rPAD | rHA1 | rHA2b | rHA3 | rHA4 | |
| Dx score | 0.546* | 0.454† | 0.455† | Low | 0.524† | 0.432** |
| Probing depth | 0.450† | 0.466† | 0.454† | Low | 0.516† | 0.443† |
| AL | 0.485† | 0.525† | 0.509† | Low | 0.550* | 0.454† |
The correlation coefficient (rSp) was calculated by using Spearman's nonparametric, two-tailed analysis with 95% confidence intervals. Significance (determined by using Spearman's correlation) is indicated as follows: *, P < 0.05; †, P < 0.01; and **, P < 0.001.
Titers to rHA2 were not detected under the same ELISA conditions as for the other domains.
Exclusion of the healthy control group from the correlation matrix resulted in the loss of all significant correlations between titers and disease severity (data not shown).
Effect of therapy.
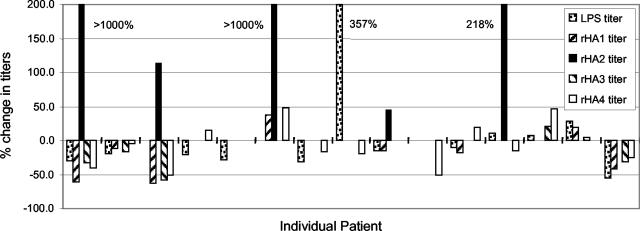
Clinically, most patients responded well to treatment with reduction of inflammation and probing depth in the majority of cases. The periodontal therapy received by the participants was subgingival root planing, which usually results in a minor bacteremia, thereby inoculating the patient with the contents of the periodontal pocket. Subgingival root planing has been associated with the establishment of periodontal health for the primary reason that etiological agents are removed from the tissue environment, but it is also possible that therapy effectively inoculates the host, leading to enhanced protection. This was evaluated by analysis of the effect of therapy on titer of antibodies to the gingipain domains. There were no consistent trends for changes in antibody titer, with the exception of four patients in which low initial titers to HA2 were markedly elevated posttherapy (Fig. 6).
FIG. 6.
Percent change in pre- and posttreatment antibody titers to each gingipain component in individual patients. Patients with undetectable pretreatment titers to rHA2 were assigned a titer of 1 to enable the calculation of percentage increase posttreatment.
DISCUSSION
Since the high-molecular-weight forms of RgpA and Kgp localize primarily to the outer membrane in association with LPS (35, 46), copurification of these moieties was possible despite the use of extensive detergent washes by ion-exchange chromatography and affinity purification chromatography. Other studies of the gingipain complex from different strains of P. gingivalis have indicated an intimate association between LPS and gingipain (46) and glycosylation of the protein that resulted in cross-reactivity between MAbs to LPS and to the carbohydrate moieties on these enzymes (7). It is therefore important to differentiate the anti-LPS/carbohydrate reactivity from that of the peptide-directed anti-gingipain antibodies when titers to the complex are being investigated. To do this, we used individual recombinant adhesin domains to determine the relative titers since it has been shown that recombinant proteins expressed in E. coli host do not undergo posttranslational glycosylation (19).
Antibodies to P. gingivalis antigens are predominantly of the IgG class (23, 29). Our finding of minimal humoral response to the peptide portion of the catalytic domains was supported by other studies (15, 43). Anti-gingipain antibody responses in patients with periodontitis were reported to be directed primarily toward the adhesin domains (15), and deglycosylation of the RgpAcat domain resulted in the abolition of antibody reactivity by sera from a group of periodontitis patients (43). Immune recognition of the carbohydrate moieties surrounding the catalytic domain has been suggested to provide immune evasion of the peptide backbone, enabling the preservation of its function (43).
The only previous study comparing IgG titers to the purified gingipain complex with disease severity did not delineate the cross-reactivity of antibodies against the carbohydrate epitopes in LPS and in the glycosylated gingipain protein complex (28). It was reported in that study that there were elevated titers of IgG2 and IgG4 subclasses against the whole gingipain complex, but no significant difference existed between the control and disease groups. The IgG2 subclass is known to be directed primarily toward polysaccharide antigens (39) and to LPS (37); hence, the titers of IgG2 reported by O'Brien-Simpson et al. (28) were presumably directed toward carbohydrate epitopes present in the gingipain preparations. The data obtained here demonstrated a significantly higher level of anti-LPS antibodies than for the anti-gingipain peptide, and there were significant differences in mean titers between healthy and disease groups. Preabsorption with P. gingivalis LPS and deglycosylation of the gingipain complex resulted in a marked decrease in antibody reactivity toward the gingipain by Western blot. Since P. gingivalis O antigens in LPS from different strains have been reported to be immunologically related (27, 41), the titers to LPS reported here were presumably representative of the LPS titer to the whole species.
Further dissection of the gingipain antibody response by O'Brien-Simpson et al. (28) indicated a significant positive correlation between the IgG2 subclass titer and a marker of disease severity whereas the IgG4 subclass titer showed a significant negative correlation. The correlation analyses were carried out with the exclusion of low-responding disease patients. Although we found significant positive correlation between titers to gingipains and the presence of disease (Table 4), no significant correlations or trends were observed between markers of disease severity and the titers of anti-adhesin or anti-LPS antibodies within our total disease group. These differences may be explained by differences in the criteria for inclusion of subjects in the correlation analysis. It would be expected that the presence of a healthy cohort with low antibody titers against P. gingivalis in an analysis will skew the correlation toward the origin, resulting in a highly significant correlation. With a sample size of 22 patients, the present study demonstrated 80% power of detecting a statistically significant correlation between severity and titer of rSp ≥ 0.5 using Spearman's correlation with a 95% confidence interval.
By using a series of truncation recombinant expression clones spanning most of the HA1 domain and a set of overlapping synthetic oligopeptides of the same region, Kelly et al. (21) reported four regions within HA1 that were immunogenic. Similarly, two minor and two major immunogenic regions were also detected by synthetic peptides located within the N-terminal half of HA4 (28). In the present study, both rHA1 and rHA4 were readily detected by patient antibodies. It is noteworthy, however, that the recognition of rHA4 (Fig. 4) was greater than for the corresponding domain in the native gingipain preparations (Fig. 3). The differences seen in these semiquantitative Western blots may be due to the relatively small amount of antigen present in the native gingipain sample (Fig. 3a). HA4 has been reported to be strongly associated with LPS or to be heavily glycosylated (46), and possible epitope masking of native HA4 by this association cannot be ruled out. Recombinant proteins expressed in E. coli hosts are not known to be glycosylated (19); hence, epitope masking is not an issue with these recombinant products. As a control for possible E. coli LPS contamination of the purified recombinant products, in particular rHA4, purified expression host E. coli LPS was found to be not recognized by any patient or healthy subjects by Western blot (data not shown). Titers by ELISA to recombinant HA1 and HA4 domains were found to be significantly elevated in patients with periodontitis compared to controls. Titers to rHA3 were intermediate in value, whereas titers to rHA2 were low or undetectable.
The HA2 domain has been reported to play a critical role in hemoglobin binding (25, 38) and heme capture (30), which are essential for the growth of P. gingivalis. Although other genes, including tlr (42), hemR (20), hmuR (40), and those of the ihtABCDE locus (8), have also been implicated in the transport of captured heme, HA2 seems to be the major high-affinity receptor (30). It was reported to be the major participant in hemoglobin binding by the organism via a deletion mutant study (38) and via cell surface labeling (30). It was, therefore, surprising to find that HA2 was poorly recognized compared to other domains of the RgpA-Kgp complex. It was also of note that therapy led in some cases to a pronounced increase in titer to this domain, a finding compatible with protective immunity induced in the rat model of periodontitis (10).
Many studies have reported that antibody titers to P. gingivalis remained unchanged or decreased after periodontal therapy (1, 3) or, in one instance, there was an initial increase in titer, followed by a net reduction of 50% by the end of 1 year posttreatment (24). Titers to LPS have been reported to decrease posttherapy (36). At the completion of nonsurgical periodontal treatment, we found that gingipain-specific titers generally remained unchanged or decreased, a finding consistent with previous anti-whole-cell antibody observations. The exception was titers to HA2. Whether the trends remained the same if the posttreatment sera were collected at a fixed time point after the commencement of treatment is unclear.
A prime objective of the present study was to investigate antibody responses to gingipain domains in patients with periodontitis. Our data suggest that anti-LPS, anti-HA1, and anti-HA4 serum IgG antibodies are an early and consistent response in subjects who are susceptible to periodontal disease, but the titers do not relate to disease severity and, therefore, may not be protective. Anti-HA3 antibody responses were lower compared to other domains, whereas anticatalytic and anti-HA2 antibodies ranged from very low to undetectable levels in untreated periodontitis patients. Although the catalytic domain may have a glycosylated shield for immune evasion, elevation of titers to the HA2 domain in some patients posttreatment suggests a potential target for antibody-mediated neutralization of the high-affinity, HA2-mediated heme acquisition pathway essential for the growth of this organism.
Acknowledgments
This study was supported by the National Health and Research Council of Australia and the Australian Dental Research Fund for postgraduate scholarship and grants to K.-A. Nguyen.
The support of the staff from the Periodontics Department at the United Dental Hospital is greatly appreciated. We thank Derek Harty and Catherine Rathsam for help with techniques and critical comments.
Editor: V. J. DiRita
REFERENCES
- 1.Aukhil, I., D. E. Lopatin, S. A. Syed, E. C. Morrison, and C. J. Kowalski. 1988. The effects of periodontal therapy on serum antibody (IgG) levels to plaque microorganisms. J. Clin. Periodontol. 15:544-550. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw, D. J., and P. D. Marsh. 1999. Use of continuous flow techniques in modeling dental plaque biofilms. Methods Enzymol. 310:279-296. [DOI] [PubMed] [Google Scholar]
- 3.Chen, H. A., B. D. Johnson, T. J. Sims, R. P. Darveau, B. J. Moncla, C. W. Whitney, D. Engel, and R. C. Page. 1991. Humoral immune responses to Porphyromonas gingivalis before and following therapy in rapidly progressive periodontitis patients. J. Periodontol. 62:781-791. [DOI] [PubMed] [Google Scholar]
- 4.Chen, T., K. Nakayama, L. Belliveau, and M. J. Duncan. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect. Immun. 69:3048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciborowski, P., M. Nishikata, R. D. Allen, and M. S. Lantz. 1994. Purification and characterization of two forms of a high-molecular-weight cysteine proteinase (porphypain) from Porphyromonas gingivalis. J. Bacteriol. 176:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, E. C. Reynolds, and J. Aduse-Opoku. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodontal Res. 34:464-472. [DOI] [PubMed] [Google Scholar]
- 7.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramonov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dashper, S. G., A. Hendtlass, N. Slakeski, C. Jackson, K. J. Cross, L. Brownfield, R. Hamilton, I. Barr, and E. C. Reynolds. 2000. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J. Bacteriol. 182:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeCarlo, A. A., and G. J. Harber. 1997. Hemagglutinin activity and heterogeneity of related Porphyromonas gingivalis proteinases. Oral Microbiol. Immunol. 12:47-56. [DOI] [PubMed] [Google Scholar]
- 10.DeCarlo, A. A., Y. Huang, C. A. Collyer, D. B. Langley, and J. Katz. 2003. Feasibility of an HA2 domain-based periodontitis vaccine. Infect. Immun. 71:562-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeCarlo, A. A., Jr., L. J. Windsor, M. K. Bodden, G. J. Harber, B. Birkedal-Hansen, and H. Birkedal-Hansen. 1997. Activation and novel processing of matrix metalloproteinases by a thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J. Dent. Res. 76:1260-1270. [DOI] [PubMed] [Google Scholar]
- 12.DeCarlo, A. A., M. Paramaesvaran, P. L. Yun, C. Collyer, and N. Hunter. 1999. Porphyrin-mediated binding to hemoglobin by the HA2 domain of cysteine proteinases (gingipains) and hemagglutinins from the periodontal pathogen Porphyromonas gingivalis. J. Bacteriol. 181:3784-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, R. T., B. Klausen, N. S. Ramamurthy, L. M. Golub, C. Sfintescu, and R. J. Genco. 1992. Periodontopathic potential of two strains of Porphyromonas gingivalis in gnotobiotic rats. Arch. Oral Biol. 37:813-819. [DOI] [PubMed] [Google Scholar]
- 14.Fomsgaard, A., M. A. Freudenberg, and C. Galanos. 1990. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J. Clin. Microbiol. 28:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genco, C. A., J. Potempa, J. Mikolajczyk-Pawlinska, and J. Travis. 1999. Role of gingipains R in the pathogenesis of Porphyromonas gingivalis-mediated periodontal disease. Clin. Infect. Dis. 28:456-465. [DOI] [PubMed] [Google Scholar]
- 16.Griffen, A. L., M. R. Becker, S. R. Lyons, M. L. Moeschberger, and E. J. Leys. 1998. Prevalence of Porphyromonas gingivalis and periodontal health status. J. Clin. Microbiol. 36:3239-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt, S. C., J. Ebersole, J. Felton, M. Brunsvold, and K. S. Kornman. 1988. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science 239:55-57. [DOI] [PubMed] [Google Scholar]
- 18.Imamura, T. 2003. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74:111-118. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins, N., R. B. Parekh, and D. C. James. 1996. Getting the glycosylation right: implications for the biotechnology industry. Nat. Biotechnol. 14:975-981. [DOI] [PubMed] [Google Scholar]
- 20.Karunakaran, T., T. Madden, and H. Kuramitsu. 1997. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J. Bacteriol. 179:1898-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, C. G., V. Booth, H. Kendal, J. M. Slaney, M. A. Curtis, and T. Lehner. 1997. The relationship between colonization and haemagglutination inhibiting and B-cell epitopes of Porphyromonas gingivalis. Clin. Exp. Immunol. 110:285-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mooney, J., E. Adonogianaki, M. P. Riggio, K. Takahashi, A. Haerian, and D. F. Kinane. 1995. Initial serum antibody titer to Porphyromonas gingivalis influences development of antibody avidity and success of therapy for chronic periodontitis. Infect. Immun. 63:3411-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouton, C., M. Desclauriers, H. Allard, and M. Bouchard. 1987. Serum antibodies to Bacteroides gingivalis in periodontitis: a longitudinal study. J. Periodontal Res. 22:426-430. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama, K., D. B. Ratnayake, T. Tsukuba, T. Kadowaki, K. Yamamoto, and S. Fujimura. 1998. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol. Microbiol. 27:51-61. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama, K., F. Yoshimura, T. Kadowaki, and K. Yamamoto. 1996. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J. Bacteriol. 178:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni Eidhin, D., and C. Mouton. 1994. The lipopolysaccharide of Porphyromonas gingivalis is not antigenically cross-reactive with that of other species. J. Dent. Res. 73:661-670. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien-Simpson, N. M., C. L. Black, P. S. Bhogal, S. M. Cleal, N. Slakeski, T. J. Higgins, and E. C. Reynolds. 2000. Serum immunoglobulin G (IgG) and IgG subclass responses to the RgpA-Kgp proteinase-adhesin complex of Porphyromonas gingivalis in adult periodontitis. Infect. Immun. 68:2704-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawa, T., Y. Kusumoto, S. Hamada, J. R. McGhee, and H. Kiyono. 1990. Bacteroides gingivalis-specific serum IgG and IgA subclass antibodies in periodontal diseases. Clin. Exp. Immunol. 82:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paramaesvaran, M., K. A. Nguyen, E. Caldon, J. A. McDonald, S. Najdi, G. Gonzaga, D. B. Langley, A. DeCarlo, M. J. Crossley, N. Hunter, and C. A. Collyer. 2003. Porphyrin-mediated cell surface heme capture from hemoglobin by Porphyromonas gingivalis. J. Bacteriol. 185:2528-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavloff, N., J. Potempa, R. N. Pike, V. Prochazka, M. C. Kiefer, J. Travis, and P. J. Barr. 1995. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J. Biol. Chem. 270:1007-1010. [DOI] [PubMed] [Google Scholar]
- 32.Peterman, J. H., and J. E. Butler. 1989. Application of theoretical considerations to the analysis of ELISA data. BioTechniques 7:608-615. [PubMed] [Google Scholar]
- 33.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000 24:153-192. [DOI] [PubMed] [Google Scholar]
- 34.Potempa, J., J. Mikolajczyk-Pawlinska, D. Brassell, D. Nelson, I. B. Thogersen, J. J. Enghild, and J. Travis. 1998. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J. Biol. Chem. 273:21648-21657. [DOI] [PubMed] [Google Scholar]
- 35.Potempa, J., R. Pike, and J. Travis. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schenck, K., K. Helgeland, and T. Tollefsen. 1987. Antibodies against lipopolysaccharide from Bacteroides gingivalis before and after periodontal treatment. Scand. J. Dent. Res. 95:112-118. [DOI] [PubMed] [Google Scholar]
- 37.Schenck, K., and T. E. Michaelsen. 1987. IgG subclass distribution of serum antibodies against lipopolysaccharide from Bacteroides gingivalis in periodontal health and disease. Acta Pathol. Microbiol. Immunol. Scand. [C] 95:41-46. [DOI] [PubMed] [Google Scholar]
- 38.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis: construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 39.Siber, G. R., P. H. Schur, A. C. Aisenberg, S. A. Weitzman, and G. Schiffman. 1980. Correlation between serum IgG2 concentrations and the antibody response to bacterial polysaccharide antigens. N. Engl. J. Med. 303:178-182. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, W., T. Olczak, and C. A. Genco. 2000. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 182:5737-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sims, T. J., R. E. Schifferle, R. W. Ali, N. Skaug, and R. C. Page. 2001. Immunoglobulin G response of periodontitis patients to Porphyromonas gingivalis capsular carbohydrate and lipopolysaccharide antigens. Oral Microbiol. Immunol. 16:193-201. [DOI] [PubMed] [Google Scholar]
- 42.Slakeski, N., S. G. Dashper, P. Cook, C. Poon, C. Moore, and E. C. Reynolds. 2000. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol. Immunol. 15:388-392. [DOI] [PubMed] [Google Scholar]
- 43.Slaney, J. M., M. Rangarajan, J. Aduse-Opoku, S. Fawell, I. Darby, D. Kinane, and M. A. Curtis. 2002. Recognition of the carbohydrate modifications to the RgpA protease of Porphyromonas gingivalis by periodontal patient serum IgG. J. Periodontal Res. 37:215-222. [DOI] [PubMed] [Google Scholar]
- 44.Slots, J., and M. A. Listgarten. 1988. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J. Clin. Periodontol. 15:85-93. [DOI] [PubMed] [Google Scholar]
- 45.Sojar, H. T., and O. P. Bahl. 1987. A chemical method for the deglycosylation of proteins. Arch. Biochem. Biophys. 259:52-57. [DOI] [PubMed] [Google Scholar]
- 46.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5:83-91. [Google Scholar]