Abstract
Background
We have previously shown that lipophilic components (LPC) of the brown seaweed Ascophyllum nodosum (ANE) improved freezing tolerance in Arabidopsis thaliana. However, the mechanism(s) of this induced freezing stress tolerance is largely unknown. Here, we investigated LPC induced changes in the transcriptome and metabolome of A. thaliana undergoing freezing stress.
Results
Gene expression studies revealed that the accumulation of proline was mediated by an increase in the expression of the proline synthesis genes P5CS1 and P5CS2 and a marginal reduction in the expression of the proline dehydrogenase (ProDH) gene. Moreover, LPC application significantly increased the concentration of total soluble sugars in the cytosol in response to freezing stress. Arabidopsis sfr4 mutant plants, defective in the accumulation of free sugars, treated with LPC, exhibited freezing sensitivity similar to that of untreated controls. The 1H NMR metabolite profile of LPC-treated Arabidopsis plants exposed to freezing stress revealed a spectrum dominated by chemical shifts (δ) representing soluble sugars, sugar alcohols, organic acids and lipophilic components like fatty acids, as compared to control plants. Additionally, 2D NMR spectra suggested an increase in the degree of unsaturation of fatty acids in LPC treated plants under freezing stress. These results were supported by global transcriptome analysis. Transcriptome analysis revealed that LPC treatment altered the expression of 1113 genes (5%) in comparison with untreated plants. A total of 463 genes (2%) were up regulated while 650 genes (3%) were down regulated.
Conclusion
Taken together, the results of the experiments presented in this paper provide evidence to support LPC mediated freezing tolerance enhancement through a combination of the priming of plants for the increased accumulation of osmoprotectants and alteration of cellular fatty acid composition.
Keywords: Arabidopsis thaliana, Ascophyllum nodosum, Freezing tolerance, Chemical priming, Soluble sugars, Metabolite profiling, Microarray analysis
Background
Environmental factors such as low temperature, drought, and salinity are the major abiotic stress factors that adversely affect the growth and development of plants limiting crop productivity [1]. Freezing stress affects plant growth directly by the inhibition of metabolic reactions and indirectly through osmotic, oxidative and other secondary stresses.
A number of chemicals have been tested for inducing freezing tolerance. Whereas exogenous application of cryoprotectants such as sorbitol and polyethylene glycol were found to be either phyto-toxic or only marginally effective in imparting frost tolerance [2], chemicals such as choline chloride and ethanolamine have been shown to be effective in improving frost tolerance, in wheat and tomato seedlings, respectively [3,4]. More recently, some acrylic compounds were found to protect crops from freezing damage by forming an inert layer on the plant surface [2]. However, the environmental safety of these chemicals and their impact on various ecosystems is unclear. Therefore, the identification of naturally occurring plant-based products to impart freezing stress tolerance to crop plants would be ideal.
Extracts of Ascophyllum nodosum, have been shown to stimulate shoot growth and branching [5], increase root growth and lateral root development [6], improve nutrient uptake [7], enhance resistance to diseases [8], and mitigate the effects of environmental stresses such as drought, salinity and frost [9,10]. Burchett et al. [11] reported that application of a commercial formulation of A. nodosum extract enhanced winter hardiness and increased frost resistance in winter barley. Another study in grapes using an extract of the Tasmanian Giant Bull kelp Durvillea potatorum (Labill.)] also improved plant freezing tolerance [12].
The role of proline in low temperature tolerance in plants has been reported [13-15] and a number of plant species, that are inherently tolerant to freezing temperatures (such as barley, rye, winter wheat, grape, potato and Arabidopsis), accumulate high levels of proline when exposed to low temperatures [16,17]. Proline plays multiple roles in frost tolerance; such as a mediator of osmotic adjustment [18], stabilizer of proteins and membranes [19], inducer of osmotic stress-related genes [20], scavenger of reactive oxygen species (ROS) [21], source of reduction equivalents during stress recovery [22], and readily available source of nitrogen and carbon during post-recovery growth [23].
In an earlier study, we demonstrated that A. nodosum extracts (ANE) and its lipophilic component (LPC) significantly enhanced freezing tolerance in Arabidopsis thaliana[24]. Electrolyte leakage measurements revealed that the LT50 value of LPC treated plants was lowered by 3°C while cell viability staining demonstrated a 30-40% reduction in area of damaged tissue in extract treated plants as compared to water controls. Additionally, ANE treatment caused a two-fold increase in the transcription of the cold response genes, cor15a, rd29a and transcription factor cbf3.
Several expression profile studies in Arabidopsis have established that large scale changes occur in gene expression during cold acclimation and subsequent freezing tolerance [25-27]. Freezing temperatures activate a number of cold-responsive genes which encode a diverse array of proteins such as dehydrins, lipid transfer proteins, molecular chaperones, anti-freeze proteins, enzymes involved in respiration and metabolism of carbohydrates, phenylpropanoids and antioxidants, late-embryogenesis-abundant proteins and others, each with a presumed function in tolerance to the dehydration caused by freezing [28-32].
Microarrays provide an advantage of allowing parallel quantification of the gene expression at the whole genome level. Here, we report changes in the metabolome and global transcript profile in plants, after treatment with the LPC and subsequent exposure to freezing temperatures and post-freezing recovery.
Results
Proline estimation
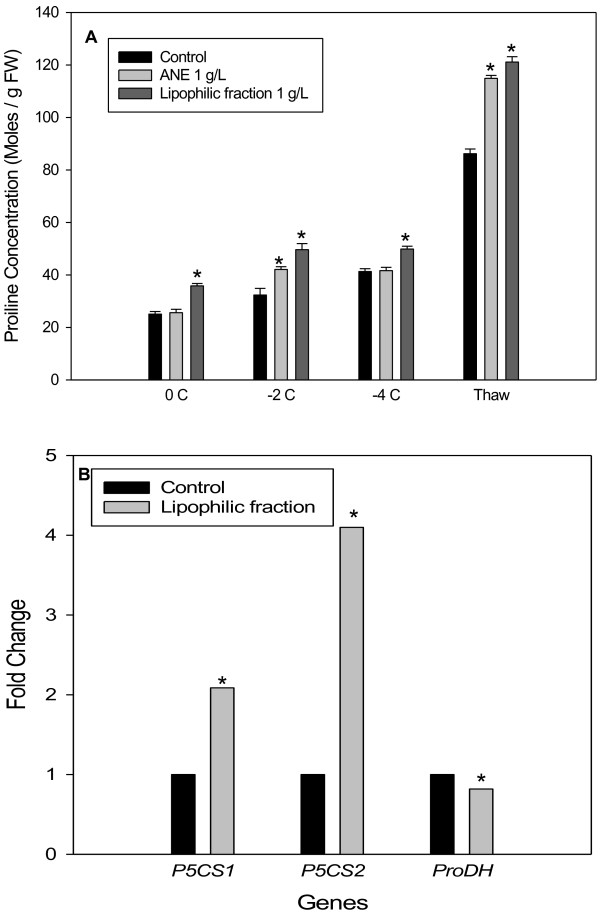
Plants treated with ANE or LPC accumulated higher amounts of proline during freezing stress, as compared to water controls (Figure 1A). Freezing stress was initiated by spraying plants with ice-cold water in an incubator set at 0°C and progressively cooled at the rate of 1°C per 24h until desired temperature was reached. The proline content of control plants after exposure to a freezing stress of 0°C for 24 h was 25.06 micro moles g-1 fresh weight versus 35.82 micro moles g-1 fresh weight in LPC treated plants. At this temperature, plants treated with ANE did not show a significant change in proline content, as compared to control plants. However, when plants were exposed to −2°C for 24 h, ANE- and LPC-treated plants accumulated higher amounts of proline (42.11 and 49.62 micro moles g-1 fresh weight, respectively) as compared to the untreated plants (32.32 moles g-1 fresh weight). A further lowering of temperature to −4°C did not significantly affect proline accumulation in any of the treatments. During thawing, Arabidopsis plants accumulated large amounts of proline amounting to 86.20, 114.89 and 121.10 micro moles g-1 fresh weight in control, ANE- and LPC-treated plants, respectively. Overall, LPC treated plants accumulated significantly higher amounts of proline at all temperatures (Figure 1A).
Figure 1.
Proline accumulation and expression of proline metabolism genes in wild-type (WT) Arabidopsis: (A) effect of ANE and LPC on the accumulation of proline in the leaves of wild type Arabidopsis (Col-0) in response to freezing stress (Data are the means ± SE) and (B) Real time-PCR analysis of expression of genes encoding Δ1-pyrroline-5-carboxylate synthetase (P5CS) and prolinedehydogenase (ProDH). Bar with * is significantly different (P ≤ 0.05) from that of the control treatment.
Transcriptional analysis of proline biosynthesis and degradation genes
Analysis of transcript abundance for genes involved in proline biosynthesis (P5CS1 and P5CS2) and degradation (ProDH) in −2°C treatments revealed that ANE and LPC treatment affected the expression of these genes (Figure 1B). There was a two-fold increase in P5CS1 and a four-fold increase in P5CS2 transcripts as compared to plants that did not receive LPC treatment. A marginal decrease in the expression of ProDH was also observed (Figure 1B).
Proline mutant studies
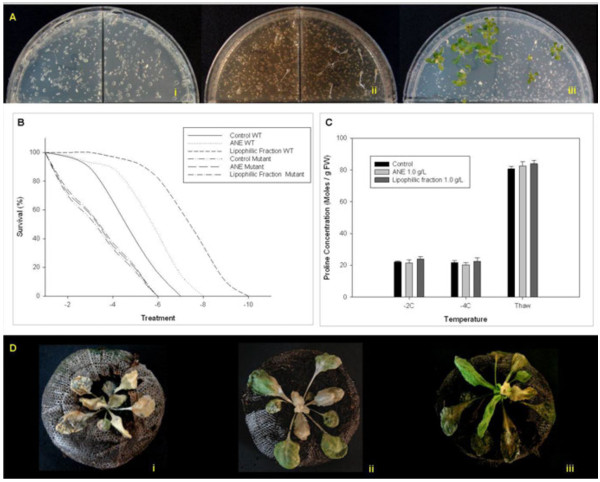
The role of proline in ANE-mediated freezing tolerance in Arabidopsis, was confirmed by using p5cs-1 mutants, deficient in proline accumulation during stress. Observations of p5cs-1 mutants revealed that application of ANE and LPC did not alter the sensitive phenotype of p5cs-1 mutants (Figure 2). When the temperature was lowered to −7.5°C, the wild-type, water control plants showed 100% mortality, while the wild-type plants treated with LPC showed considerably less damage and were able to recover from stress-induced damage (Figure 2).
Figure 2.
Ascophyllum nodosum extracts did not rescue the freezing sensitive phenotype of p5cs1 mutants: (A) Petri Plate Freezing Tolerance Assay with wild-type Arabidopsis (left partition of Petri plate) and p5cs1 mutants (right partition of Petri plate) treated with (i) Control (ii) Ascophyllum nodosum extract (ANE) and (iii) lipophilic component of ANE (LPC), (B) survival rate at different freezing temperatures of wild type Arabidopsis and p5cs1 mutant plants treated with ANE or LPC in Petri Plates, (C) proline accumulation of p5cs1 mutants treated with ANE or LPC in response to freezing in peat pellets, and (D) Phenotypic responses of p5cs1 mutant plants treated with (i) water control (ii) ANE (1.0 g L-1) and (iii) LPC (1.0 g L-1) to a temperature of −2°C for 24 h in Peat pellet freezing assay. Bars are the means ± SE.
Soluble sugar estimation and sugar mutant studies
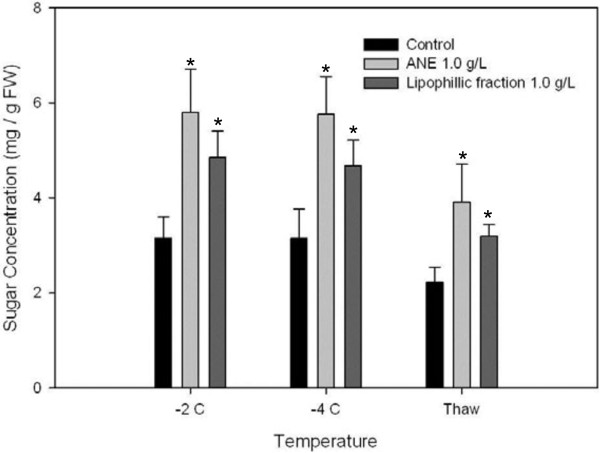
Wild-type plants treated with ANE or LPC accumulated higher amounts of total soluble sugars in response to freezing stress, as compared to untreated controls (Figure 3). At -2°C, ANE-treated plants showed an almost two-fold increase in soluble sugars (5.79 mg g-1 fresh weight) while plants treated with LPC showed a 1.5-fold increase (4.89 mg g-1 fresh weight) as compared to untreated controls (3.15 mg g-1 fresh weight). Levels of soluble sugars remained unchanged in the treated and control plants at −4°C. Soluble sugar concentrations were significantly higher in treated plants (3.91 and 3.20 mg g-1 fresh weight of ANE- and LPC-treated plants, respectively), as compared to untreated controls (2.22 mg g-1 fresh weight) (Figure 3).
Figure 3.
Accumulation of soluble sugars in wild-type Arabidopsis plants treated with ANE or LPC under different freezing and thawing regimes. Data are the means ± SE, Bar with * is significantly different (P ≤ 0.05) from that of the control treatment.
Confirmation of the role of soluble sugars in ANE-mediated freezing tolerance using Arabidopsis mutant sfr4
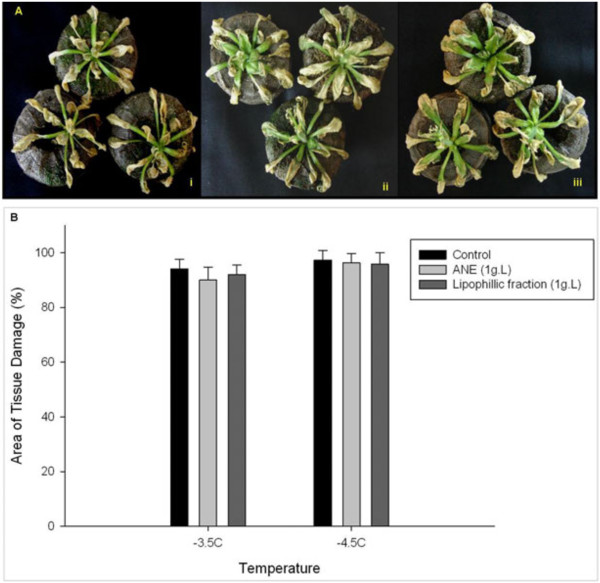
The extent of tissue damage in sfr4 mutants revealed that ANE or LPC failed to impart freezing tolerance to plants which are defective in sugar accumulation (Figure 4). Mutant plants treated with ANE or LPC showed similar levels of damage to untreated controls when exposed to freezing temperatures (Figure 4A). At −3.5°C, 92% of the leaf area of control plants was damaged, compared to 90% and 87% in ANE- and LPC-treated plants, respectively. The percentage area of tissue damage was 96% and 95%, respectively, for ANE and LPC treated plants at a temperature of −4.5°C, while it was 97% for control plants (Figure 4B). These results confirm that sugar accumulation is essential in the development of ANE-mediated freezing tolerance in Arabidopsis.
Figure 4.
Sugar accumulation is required for ANE-mediated freezing tolerance in Arabidopsis: (A) sensitive phenotype of sfr4 mutant plants treated with (i) water control (ii) ANE (1.0 g L-1) and (iii) LPC (1.0 g L-1) to a temperature of −2.5°C for 24 h in Peat Pellet Freezing Tolerance Assay, and (B) extent of freezing-induced tissue damage in controls and treated sfr4 mutant plants as revealed by trypan blue staining; comparison of the area of tissue damage in the trypan blue stained leaves using the image processing and analysis software Image-j®. Data are means ± SE.
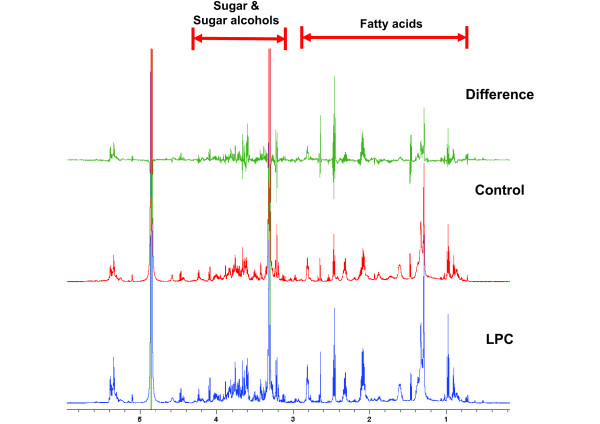
1H NMR metabolite profiles of ANE extracts
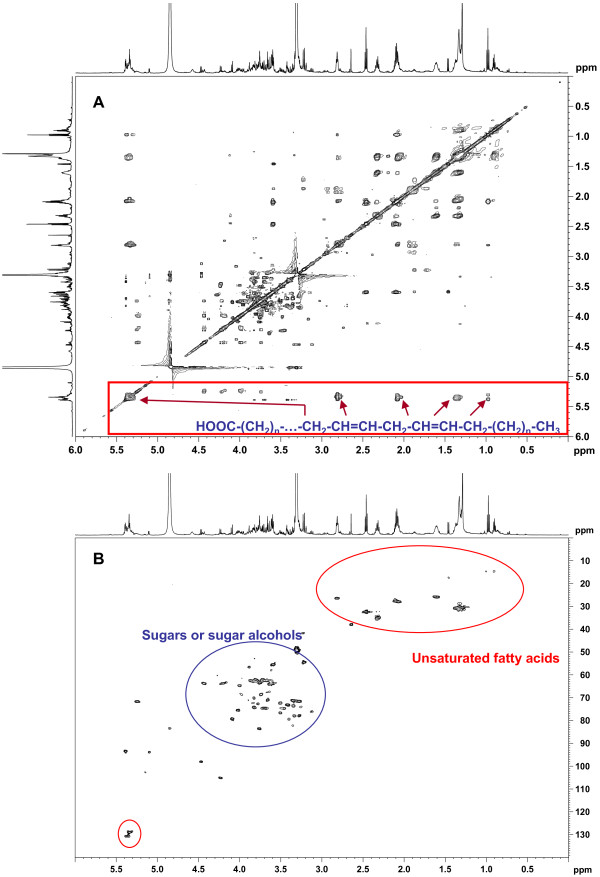
Application of LPC elicited significant changes in the metabolome of Arabidopsis in response to freezing stress. At ambient temperature, the application of ANE or LPC had minimal effect on the metabolic profile of Arabidopsis. However, when exposed to −2°C for 24 h, significant increases in peaks with 1H NMR chemical shifts (δ) at 0.8 to 1.6, 2.1 to 2.8, 3.2 to 3.9, and 5.2 to 5.5 ppm were observed in the plants treated with LPC, as compared to control (Figure 5). During the thawing period, the changes in peak intensities were observed at resonances similar to freezing treatment (δ 0.8 to 1.6, 2.1 to 2.8, 3.2 to 3.9, and 5.2 to 5.5 ppm), but the peak intensity increases were much lower than during freezing treatment. In order to better understand the metabolite changes resulting from LPC treatment, 2D NMR experiments such as COSY, TOCSY, HSQC, and HMBC were run on samples from LPC, treated plants. The proton and carbon connectivity information revealed by these 1H-1H or 1H-13C correlation spectra indicated that the major metabolite changes are due to unsaturation of fatty acids (peaks at δ 0.8 to 1.6, 2.1 to 2.8, and 5.2 to 5.5 ppm) and sugar or sugar alcohols (peaks at δ 3.2 to 3.9, and 5.2 to 5.5 ppm) (Figure 6).
Figure 5.
1H-NMR metabolite profile spectrum of wild-type Arabidopsis plants treated with LPC during freezing temperature of −2°C for 24 h.
Figure 6.
2D NMR spectra of metabolites extracted from LPC-treated plants, upon exposure to freezing stress of −2.5°C for 24 h: (A) 2D NMR TOCSY spectrum showing characteristic proton correlations in unsaturated fatty acids and (B) 2D NMR HSQC spectrum showing characteristic proton and carbon signals of unsaturated fatty acids and sugars or sugar alcohols.
Global transcriptional changes elicited by ANE
The transcript level changes during the post freezing recovery period were analyzed in ANE-treated Arabidopsis plants thawed at 4°C for 24h after a freezing treatment of −2°C for 24h (Additional file 1: Figure S1). LPC treatment affected the expression of about 5% (1113 genes) of the Arabidopsis genome (P≤0.05; change, ≥1.5 fold) when, exposed to −2°C for 24h (Additional file 1: Figure S1A). Using the gene expression data from control plants as the calibrator data set, we determined that about 2% (463 genes) of the differentially expressed genes were up-regulated and 3% (650 genes) were down-regulated during freezing stress. During post freezing recovery period, examination of the expression ratios of genes indicated that a relatively small portion of the genome was differentially expressed in plants treated with LPC as compared to controls. We found that 398 genes (1.65%) were differentially expressed more than 1.5 fold (P≤0.05) with 131 genes (0.54%) up-regulated and 267 genes (1.11%) genes down-regulated during post freezing recovery period (Additional file 1: Figure S1C).
Functional classification
The genes that were differentially expressed during freezing or post-freezing recovery were classified into different functional categories, using the web-based tool Classification Super-Viewer, BLAST searches, and literature reports. The gene lists were classified according to the absolute gene number and normalized frequency. Genes associated with abiotic or biotic stimuli, transcription factor activity, response to stress, etc. ranked highest among the up-regulated genes (Additional file 1: Figure S2A), while those responsible for extra-cellular, cell wall, receptor binding, etc. ranked highest among down-regulated genes during freezing stress (Additional file 1: Figure S2C). During the post-freezing recovery period, functional classification of the differentially expressed genes showed an opposite trend to that of freezing, with extra-cellular, DNA-RNA metabolism, cell wall, etc. ranking highest in the up-regulated ones (Additional 1: Figure S3A) while response to abiotic and biotic stimuli, response to stress, cell wall, etc. in down-regulated ones (Additional file 1: Figure S3C).
When the genes were classified according to the actual number of genes which were differentially expressed, ‘unknown category’ (other molecular functions, other biological processes, other metabolic processes, other cellular processes, etc.) ranked at the top (Additional file 1: Figure S2B, S2D, S3B and S3D), indicating there are many genes altered by treatment with LPC during freezing stress and post-freezing recovery period. A list of selected differentially expressed genes (either up-regulated or down-regulated) and their functional categories are given in Tables 1 and 2.
Table 1.
A list of selected up-regulated genes and their functional categories
| No | Locus | FC | Gene ontology class | Gene annotation detail |
|---|---|---|---|---|
|
Response to stress and/or stress stimuli | ||||
| 1 |
At1g55920 |
2 |
response to cold |
AtSerat2;1_SAT1_SAT5__AtSerat2;1 (SERINE ACETYLTRANSFERASE 1) |
| 2 |
At3g22840 |
1.94 |
response to cold |
ELIP1_ELIP__ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding |
| 3 |
At1g10760 |
1.6 |
cold acclimation |
SEX1_GWD_SOP__SEX1 (STARCH EXCESS 1) |
| 4 |
At1g09780 |
1.5 |
response to cold |
2,3-biphosphoglycerate-independent phosphoglyceratemutase, putative/phosphoglyceromutase, putative |
| 5 |
At4g26080 |
1.73 |
response to heat |
ABI1__ABI1 (ABA INSENSITIVE 1); calcium ion binding/protein serine/threonine phosphatase |
| 6 |
At4g26850 |
1.61 |
response to heat |
VTC2__VTC2 (VITAMIN C DEFECTIVE 2) |
| 7 |
At1g73330 |
3.09 |
response to water deprivation |
ATDR4__ATDR4 (Arabidopsis thaliana drought-repressed 4) |
| 8 |
At3g11410 |
2.61 |
response to water deprivation |
AHG3_ATPP2CA__AHG3/ATPP2CA (ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA); protein binding/protein serine/threonine phosphatase |
| 9 |
At5g62470 |
2.25 |
response to salt stress |
AtMYB96_mybcov1_MYB96__MYB96 (myb domain protein 96); DNA binding/transcription factor |
| 10 |
At3g61890 |
2.21 |
response to salt stress |
ATHB-12_ATHB12__ATHB-12 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 12); transcription factor |
| 11 |
At2g46680 |
1.93 |
response to water deprivation |
ATHB-7_ATHB7__ATHB-7 (ARABIDOPSIS THALIANA HOMEOBOX 7) |
| 12 |
At5g57050 |
1.87 |
response to osmotic stress |
ABI2__ABI2 (ABA INSENSITIVE 2) |
| 13 |
At5g59780 |
1.84 |
response to salt stress |
AtMYB59_MYB59__MYB59 (myb domain protein 59); DNA binding/transcription factor |
| 14 |
At5g37500 |
1.82 |
response to water deprivation |
GORK__GORK (GATED OUTWARDLY-RECTIFYING K+ CHANNEL); cyclic nucleotide binding/inward rectifier potassium channel/outward rectifier potassium channel |
| 15 |
At4g34990 |
1.80 |
response to salt stress |
AtMYB32__AtMYB32 (myb domain protein 32); DNA binding/transcription factor |
| 16 |
At1g29395 |
1.76 |
response to water deprivation |
COR414-TM1__COR414-TM1 (cold regulated 414 thylakoid membrane 1) |
| 17 |
At1g02930 |
1.70 |
response to water deprivation |
ATGSTF6_ATGSTF3_ERD11_GST1_GSTF6__ATGSTF6 (EARLY RESPONSIVE TO DEHYDRATION 11); glutathione transferase |
| 18 |
At3g19580 |
1.68 |
hyperosmotic salinity response |
AZF2__AZF2 (ARABIDOPSIS ZINC-FINGER PROTEIN 2) |
| 19 |
At3g08720 |
1.67 |
response to salt stress |
ATS6K2_S6K2_ATPK19_ATPK2__ATPK19 (ARABIDOPSIS THALIANA PROTEIN KINASE 19); kinase |
| 20 |
At2g30870 |
1.65 |
response to water deprivation |
ATGSTF10_ATGSTF4_ERD13__ATGSTF10 (EARLY DEHYDRATION-INDUCED 13); glutathione transferase |
| 21 |
At4g30660 |
1.64 |
hyperosmotic salinity response |
hydrophobic protein, putative/low temperature and salt responsive protein, putative |
| 22 |
At3g55730 |
1.61 |
response to salt stress |
AtMYB109_MYB109__MYB109 (myb domain protein 109); DNA binding/transcription factor |
| 23 |
At3g03190 |
1.57 |
response to oxidative stress |
ATGSTF11_ATGSTF6__ATGSTF11 (GLUTATHIONE S-TRANSFERASE F11); glutathione transferase |
| 24 |
At5g01600 |
1.74 |
response to hydrogen peroxide |
ATFER1__ATFER1 (FERRETIN 1); ferric iron binding |
| 25 |
At2g16060 |
1.87 |
response to hypoxia |
AHB1_ARATH GLB1_ATGLB1_GLB1_NSHB1__AHB1 (ARABIDOPSIS HEMOGLOBIN 1) |
| 26 |
At1g05200 |
1.56 |
response to light stimulus |
ATGLR3.4_GLR3.4_GLUR3__ATGLR3.4 (Arabidopsis thaliana glutamate receptor 3.4) |
| 27 |
At5g64330 |
1.59 |
response to light stimulus |
NPH3_JK218_RPT3__NPH3 (NON-PHOTOTROPIC HYPOCOTYL 3) |
| 28 |
At1g21590 |
1.63 |
response to stress |
protein kinase family protein |
| 29 |
At2g35980 |
1.56 |
response to stress |
YLS9_NHL10__YLS9 (YELLOW-LEAF-SPECIFIC GENE 9) |
| 30 |
At2g38470 |
2.0 |
defense response to fungus |
WRKY33__WRKY33 (WRKY DNA-binding protein 33); transcription factor |
| 31 |
At1g12820 |
1.87 |
response to molecule of bacterial origin |
AFB3__AFB3 (AUXIN SIGNALING F-BOX 3); auxin binding/ubiquitin-protein ligase |
| 32 |
At2g19190 |
1.80 |
defense response to bacterium |
FRK1__FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE 1); kinase |
| 33 |
At5g52450 |
1.73 |
response to nematode |
MATE efflux protein-related |
| 34 |
At4g12480 |
1.65 |
defense response to fungus |
pEARLI 1__pEARLI 1; lipid binding |
| 35 |
At1g24020 |
1.63 |
response to biotic stimulus |
MLP423__MLP423 (MLP-LIKE PROTEIN 423) |
| 36 |
At2g24570 |
1.63 |
defense response to bacterium |
WRKY17__WRKY17 (WRKY DNA-binding protein 17); transcription factor |
| 37 |
At5g64120 |
1.58 |
defense response to fungus |
peroxidase, putative |
| 38 |
At5g58600 |
1.55 |
defense response to fungus |
PMR5__PMR5 (POWDERY MILDEW RESISTANT 5) |
| 39 |
At3g11660 |
1.54 |
defense response to virus |
NHL1__NHL1 (NDR1/HIN1-like 1) |
| 40 |
At5g50200 |
2.53 |
response to wounding |
ATNRT3.1_NRT3.1_WR3__WR3 (WOUND-RESPONSIVE 3); nitrate transmembrane transporter |
| 41 |
At5g13930 |
1.51 |
response to wounding |
CHS_TT4__ATCHS/CHS/TT4 (CHALCONE SYNTHASE); naringenin-chalcone synthase |
|
Sugar Accumulation | ||||
| 1 |
At1g56600 |
3.19 |
carbohydrate biosynthetic process |
ATGOLS2 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 2); transferase, transferring glycosyl groups/transferase, transferring hexosyl groups |
| 2 |
At5g18470 |
2.63 |
sugar binding |
curculin-like (mannose-binding) lectin family protein |
| 3 |
At1g02460 |
2.46 |
carbohydrate metabolic process |
glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein |
| 4 |
At3g46970 |
1.85 |
carbohydrate metabolic process |
ATPHS2/PHS2 (ALPHA-GLUCAN PHOSPHORYLASE 2); phosphorylase/transferase, transferring glycosyl groups |
| 5 |
At4g19810 |
1.84 |
carbohydrate metabolic process |
glycosyl hydrolase family 18 protein |
| 6 |
At1g48100 |
1.84 |
carbohydrate metabolic process |
glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein |
| 7 |
At5g41670 |
1.79 |
pentose-phosphate shunt |
6-phosphogluconate dehydrogenase family protein |
| 8 |
At5g66460 |
1.75 |
carbohydrate metabolic process |
(1–4)-beta-mannanendohydrolase, putative |
| 9 |
At3g18080 |
1.72 |
carbohydrate metabolic process |
glycosyl hydrolase family 1 protein |
| 10 |
At2g44450 |
1.68 |
carbohydrate metabolic process |
glycosyl hydrolase family 1 protein |
| 11 |
At4g27830 |
1.62 |
carbohydrate metabolic process |
glycosyl hydrolase family 1 protein |
| 12 |
At4g09020 |
1.61 |
starch catabolic process; alpha-amylase activity |
ATISA3_ISA3__ATISA3/ISA3 (ISOAMYLASE 3); alpha-amylase |
| 13 |
At3g61490 |
1.61 |
carbohydrate metabolic process |
glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein |
| 14 |
At1g10760 |
1.58 |
starch catabolic process |
SEX1_GWD_SOP__SEX1 (STARCH EXCESS 1) |
| 15 |
At4g15210 |
1.54 |
starch catabolic process |
BMY1_ATBETA-AMY_AT-BETA-AMY_RAM1__ATBETA-AMY (BETA-AMYLASE); beta-amylase |
| 16 |
At2g40840 |
1.5 |
starch catabolic process |
DPE2__DPE2 (DISPROPORTION ATING ENZYME 2); 4-alpha-glucanotransferase/heteroglycan binding |
| 17 |
At1g03310 |
1.5 |
starch metabolic process |
ATISA2/BE2/DBE1/ISA2 (DEBRANCHING ENZYME 1); alpha-amylase/isoamylase |
|
Lipid metabolism | ||||
| 1 |
At4g39670 |
4.88 |
glycolipid binding; molecular function unknown |
glycolipid binding/glycolipid transporter |
| 2 |
At2g37870 |
2.25 |
lipid binding; lipid transport |
protease inhibitor/seed storage/lipid transfer protein (LTP) family protein |
| 3 |
At2g24560 |
2.04 |
lipid metabolic process |
carboxylesterase |
| 4 |
At3g22120 |
2.01 |
lipid transport; lipid binding |
CWLP (CELL WALL-PLASMA MEMBRANE LINKER PROTEIN); lipid binding |
| 5 |
At5g55450 |
1.98 |
lipid transport; lipid binding |
protease inhibitor/seed storage/lipid transfer protein (LTP) family protein |
| 6 |
At4g10955 |
1.75 |
lipid metabolic process |
lipase class 3 family protein |
| 7 |
At4g12500 |
1.72 |
lipid binding |
protease inhibitor/seed storage/lipid transfer protein (LTP) family protein |
| 8 |
At5g64080 |
1.71 |
lipid binding |
protease inhibitor/seed storage/lipid transfer protein (LTP) family protein |
| 9 |
At2g38530 |
1.61 |
phospholipid transfer to membrane |
LTP2 (LIPID TRANSFER PROTEIN 2); lipid binding |
|
ABA Responsive | ||||
| 1 |
At1g63840 |
3.27 |
Response to ABA stimulus; endome- mbrane system |
Zinc finger (C3HC4-type RING finger) family protein |
| 2 |
At5g01540 |
3.22 |
response to abscisic acid stimulus |
lectin protein kinase, putative |
| 3 |
At3g61890 |
2.21 |
response to abscisic acid stimulus |
ATHB-12 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 12); transcription factor |
| 4 |
At1g69270 |
2.04 |
abscisic acid mediated signaling |
RPK1 (RECEPTOR-LIKE PROTEIN KINASE 1); kinase |
|
Others | ||||
| 1 |
At1g21120 |
3.96 |
O-methyltransferase activity |
O-methyltransferase, putative |
| 2 |
At1g78410 |
3.79 |
Response to oxidative stress; Molecular function unknown |
VQ motif-containing protein |
| 3 |
At2g21210 |
3.31 |
Molecular function unknown |
auxin-responsive protein, putative |
| 4 |
At5g06760 |
3.18 |
Molecular function unknown |
late embryogenesis abundant group 1 domain-containing protein/LEA group 1 domain-containing protein |
| 5 |
At1g73330 |
3.09 |
response to water deprivation |
ATDR4__ATDR4 (Arabidopsis thaliana drought-repressed 4) |
| 6 |
At1g24140 |
2.67 |
anchored to membrane |
matrixin family protein |
| 7 |
At3g11410 |
2.61 |
response to water deprivation |
AHG3_ATPP2CA__AHG3/ATPP2CA (ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA); protein binding/protein serine/threonine phosphatase |
| 8 |
At2g15390 |
2.05 |
cell wall biogenesis; Fucosyltransferase activity |
FUT4 (fucosyltransferase 4); fucosyltransferase |
| 9 |
At1g55920 |
1.97 |
response to cold |
AtSerat2;1_SAT1_SAT5__AtSerat2;1 (SERINE ACETYLTRANSFERASE 1) |
| 10 |
At3g22840 |
1.94 |
response to cold |
ELIP1_ELIP__ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding |
| 11 |
At3g55130 |
1.84 |
ATPase activity, coupled to transmembrane movement of substances |
ATWBC19 (WHITE-BROWN COMPLEX HOMOLOG 19); ATPase, coupled to transmembrane movement of substances |
| 12 | At4g26850 | 1.61 | L-ascorbic acid biosynthetic process | VTC2 (VITAMIN C DEFECTIVE 2) |
Table 2.
A list of selected down-regulated genes and their functional categories
|
No |
Locus |
FC |
Gene ontology class |
Gene annotation detail |
|---|---|---|---|---|
| Carbohydrate adjustments | ||||
| 1 |
At5g49360 |
3.07 |
hydrolase activity, hydrolyzing O-glycosyl compounds |
BXL1__BXL1 (BETA-XYLOSIDASE 1); hydrolase, hydrolyzing O-glycosyl compounds |
| 2 |
At2g19800 |
2.93 |
inositol oxygenase activity |
MIOX2__MIOX2 (MYO-INOSITOL OXYGENASE 2) |
| 3 |
At4g30280 |
2.82 |
glucan metabolic process |
ATXTH18_XTH18__ATXTH18/XTH18 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18); hydrolase, acting on glycosyl bonds |
| 4 |
At5g52050 |
2.49 |
drug transporter activity |
MATE efflux protein-related |
| 5 |
At1g02640 |
2.49 |
hydrolase activity, hydrolyzing O-glycosyl compounds |
BXL2__BXL2 (BETA-XYLOSIDASE 2); hydrolase, hydrolyzing O-glycosyl compounds |
| 6 |
At1g23480 |
2.45 |
transferase activity, transferring glycosyl groups |
ATCSLA03_ATCSLA3_CSLA03__ATCSLA03 (Cellulose synthase-like A3); transferase, transferring glycosyl groups |
| 7 |
At3g27540 |
2.42 |
transferase activity, transferring glycosyl groups |
glycosyltransferase family 17 protein |
| 8 |
At3g06770 |
2.36 |
polygalacturonase activity |
glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein |
| 9 |
At1g10550 |
2.31 |
glucan metabolic process |
XET_XTH33__XTH33 (xyloglucan:xyloglucosyltransferase 33); hydrolase, acting on glycosyl bonds |
| 10 |
At1g10400 |
2.29 |
transferase activity, transferring glycosyl groups |
UDP-glycosyltransferase/transferase, transferring glycosyl groups |
| 11 |
At4g25810 |
2.24 |
glucan metabolic process |
XTR6_XTH23__XTR6 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 6); hydrolase, acting on glycosyl bonds |
| 12 |
At1g23870 |
2.17 |
trehalose-phosphatase activity |
ATTPS9_TPS9__ATTPS9 (Arabidopsis thaliana trehalose-phosphatase/synthase 9); transferase, transferring glycosyl groups/trehalose-phosphatase |
| 13 |
At1g12240 |
2.12 |
sucrose catabolic process, using beta-fructofuranosidase |
ATBETAFRUCT4_VAC-INV__ATBETAFRUCT4/VAC-INV (VACUOLAR INVERTASE); beta-fructofuranosidase/hydrolase, hydrolyzing O-glycosyl compounds |
| 14 |
At5g56870 |
2.09 |
beta-galactosidase activity |
BGAL4__BGAL4 (beta-galactosidase 4); beta-galactosidase |
| 15 |
At2g47930 |
2.06 |
anchored to membrane |
AGP26__AGP26/ATAGP26 (ARABINOGALACTAN PROTEINS 26) |
| 16 |
At4g30290 |
2.05 |
glucan metabolic process |
ATXTH19__ATXTH19 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19); hydrolase, acting on glycosyl bonds |
| 17 |
At5g27350 |
2.04 |
sugar porter activity |
SFP1__SFP1; carbohydrate transmembrane transporter/sugar:hydrogen ion symporter |
| 18 |
At2g32540 |
2.02 |
polysaccharide biosynthetic process |
ATCSLB04_ATCSLB4_CSLB04__ATCSLB04 (Cellulose synthase-like B4); transferase/transferase, transferring glycosyl groups |
| 19 |
At3g62720 |
1.86 |
polysaccharide biosynthetic process |
ATXT1__ATXT1; transferase/transferase, transferring glycosyl groups |
| 20 |
At4g37800 |
1.77 |
glucan metabolic process |
xyloglucan:xyloglucosyltransferase, putative/xyloglucanendotransglycosylase, putative/endo-xyloglucantransferase, putative |
| 21 |
At1g11260 |
1.65 |
sugar porter activity |
STP1__STP1 (SUGAR TRANSPORTER 1); carbohydrate transmembrane transporter/sugar:hydrogen ion symporter |
| 22 |
At1g75220 |
1.5 |
sugar porter activity |
integral membrane protein, putative |
|
Lipid metabolism | ||||
| 1 |
At4g22753 |
2.84 |
catalytic activity |
SMO1-3_ATSMO1_SMO1__SMO1-3 (STEROL 4-ALPHA METHYL OXIDASE); catalytic |
| 2 |
At5g52050 |
2.49 |
drug transporter activity |
MATE efflux protein-related |
| 3 |
At1g17420 |
2.37 |
lipoxygenase activity |
LOX3__LOX3 (Lipoxygenase 3); iron ion binding/lipoxygenase/metal ion binding/oxidoreductase, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen |
| 4 |
At3g47560 |
2.27 |
catalytic activity |
esterase/lipase/thioesterase family protein |
| 5 |
At5g24150 |
2.22 |
sterol biosynthetic process |
SQP1__SQP1 (Squalenemonooxygenase 1) |
| 6 |
At3g23470 |
2.1 |
lipid biosynthetic process |
cyclopropane-fatty-acyl-phospholipid synthase |
| 7 |
At1g28600 |
1.94 |
lipid metabolic process |
lipase, putative |
| 8 |
At5g18630 |
1.93 |
lipid metabolic process |
lipase class 3 family protein |
| 9 |
At5g24210 |
1.92 |
lipid metabolic process |
lipase class 3 family protein |
| 10 |
At1g02660 |
1.79 |
lipid metabolic process |
lipase class 3 family protein |
| 11 |
At3g23510 |
1.77 |
lipid biosynthetic process |
cyclopropane fatty acid synthase, putative/CPA-FA synthase, putative |
| 12 |
At4g26790 |
1.73 |
lipid metabolic process |
GDSL-motif lipase/hydrolase family protein |
| 13 |
At2g42690 |
1.68 |
lipid metabolic process |
lipase, putative |
| 14 |
At1g28670 |
1.67 |
lipid metabolic process |
ARAB-1__ARAB-1 (Arabidopsis lipase); carboxylesterase |
| 15 |
At4g13050 |
1.55 |
fatty acid biosynthetic process |
acyl-(acyl carrier protein) thioesterase, putative/acyl-ACP thioesterase, putative/oleoyl-(acyl-carrier protein) hydrolase, putative/S-acyl fatty acid synthase thioesterase, putative |
| 16 |
At1g64400 |
1.5 |
fatty acid biosynthetic process |
long-chain-fatty-acid--CoA ligase, putative/long-chain acyl-CoA synthetase, putative |
|
Others | ||||
| 1 |
At1g12610 |
6.03 |
gibberellin biosynthetic process |
DDF1__DDF1 (DWARF AND DELAYED FLOWERING 1); DNA binding/transcription factor |
| 2 |
At2g37180 |
2.36 |
membrane |
RD28_PIP2;3_PIP2C__RD28 (plasma membrane intrinsic protein 2;3); water channel |
| 3 |
At3g30775 |
2.22 |
proline catabolic process |
PRODH_ERD5_ATPDH_ATPOX_AT-POX_PRO1__ERD5 (EARLY RESPONSIVE TO DEHYDRATION 5); proline dehydrogenase |
| 4 |
At4g19420 |
2.03 |
carboxylic ester hydrolase activity |
pectinacetylesterase family protein |
| 5 |
At4g22200 |
1.95 |
regulation of membrane potential |
AKT2/3_AKT2_AKT3__AKT2 (Arabidopsis K+ transporter 2); cyclic nucleotide binding/inward rectifier potassium channel |
| 6 |
At5g21100 |
1.92 |
endomembrane system |
L-ascorbate oxidase, putative |
| 7 | At1g19670 | 1.59 | chlorophyllase activity | ATCLH1_ATHCOR1_CORI1__ATCLH1 (CORONATINE-INDUCED PROTEIN 1) |
Pathway analysis
LPC-mediated differential gene expression in Arabdopsis
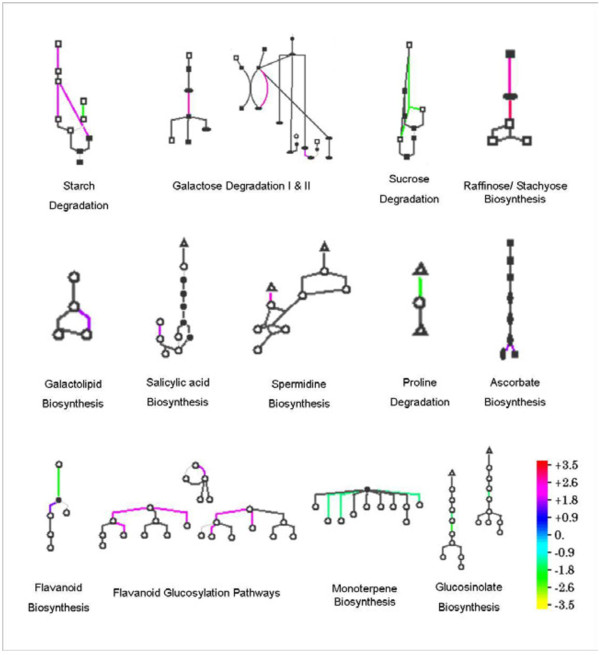
Standardized AraCyc-defined metabolic pathways were used to study the metabolic changes in LPC-treated Arabidopsis plants during freezing and post-freezing recovery. To examine the similarities and differences in the concerned metabolic pathways during the freezing stress and post-freezing recovery periods, entire biosynthetic pathways of Arabidopsis were analyzed. During freezing, out of the total 1113 genes which were differentially expressed (either up-regulated or down-regulated), 334 genes were identified by the Pathway Tools Omics Viewer, and of these 179 genes were assigned specific pathway. On the other hand, 138 genes out of total 398 genes were identified and 66 genes were assigned pathway during the post freezing recovery period. We found that specific pathways were affected significantly by LPC treatment, with little overlap during freezing and post-freezing recovery (Figure 7). The major known pathways found to be affected were soluble sugar accumulation, lipid metabolism, hormonal balance, flavanoid/terpene biosynthesis, proline degradation, and energy homeostasis (Figure 8).
Figure 7.
Diagram of representative metabolic pathways regulated by the LPC treatment during freezing stress. Each pathway is shown as a glyph consisting of nodes and lines, which represent the metabolites and reactions, respectively. Expression-level change of each reaction is shown in a color relative to the expression level, as indicated in the color scale bar. Triangle, amino acids; square, carbohydrates; diamond, proteins; open circle, others; closed circle, phosphorylated.
Figure 8.
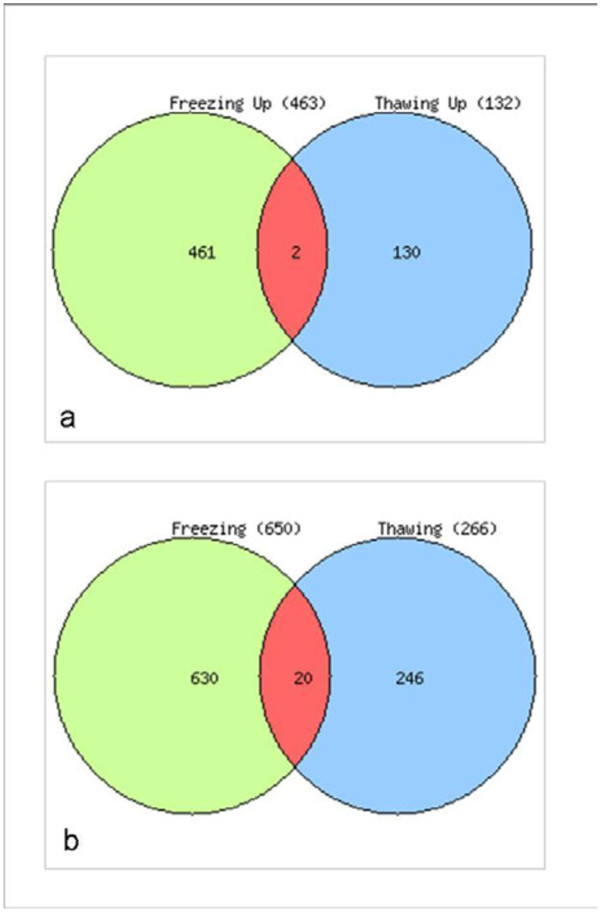
Venn diagram showing the number of significantly (a) up or (b) down-regulated and the shared genes during freezing stress and post-freezing recovery period.
Major changes observed were the multi-step promotion of the starch degradation pathway (SEX1, SEX4, AT1G03310, AT4G09020, AT4G15210, AT3G46970), inactivation of sucrose degradation (AT1G12240.2.12), induction of galactose degradation pathways I & II (MUR4, AT2G34850), stimulation of raffinose/stachyose biosynthesis (UGE2, UGE5, GolS2, GolS3), and down-regulation of trehalose synthesis (TPS8, TPS9), and gluconeogenesis (AT1G09780) (Figure 8).
A major pathway involved in galactolipid biosynthesis and lipid trafficking in thylakoid membranes (DGD1) was specifically affected (Figure 8). Other changes in lipid metabolism included down-regulation of sterol/cholesterol biosynthesis (AT5G24150), and down regulation of long chain fatty acid synthesis (AT1G01120). Additionally, the salicylic acid biosynthesis pathway was found to be stimulated in ANE-treated plants upon exposure to freezing stress. AT1G18870, which encodes a protein with isochorismate synthase activity, was found to be activated during freezing stress (Figure 8). Mutant studies of this gene's function suggest that its function is redundant with that of ICS1 (AT1G7410). Moreover, a UDP-glycosyltransferase (AT2G43820) which is induced by salicylic acid during biotic stress, was also found to be activated during this period. On the other hand, the isochorismate synthase gene, ICS1 (AT1G7410) was found to be specifically down-regulated during post-freezing recovery.
During freezing, the hormonal balance pathway responsible for conjugation of cytokinins involving N-glucosylation was highly induced while the same pathway was specifically down-regulated during the post-freezing recovery period. UDP-glycosyltransferases like UGT73B2, UGT76C2, UGT76C1, AT2G43820, and At1G24100 were highly affected by the treatment. Down-regulation of the gibberellin inactivation pathway was also observed, with GA2-oxidase (GA2ox1), a dioxygenase involved in GA inactivation being specifically down-regulated.
A polyamine biosynthetic gene (AT5G15950) involved in spermine/spermidine biosynthesis was up-regulated in LPC treated plants. Pathway analysis of proline dehydrogenase (prodh), the gene involved in degradation of proline, showed that application of LPC down-regulated the gene thereby maintaining a higher level of proline throughout the freezing and post-freezing recovery periods. The ascorbate biosynthetic gene (VTC2), involved in the response to jasmonic acid stimulus (Figure 8), and L-aspartate oxidase (At5g14760), involved in the early steps of NAD biosynthesis, were both up-regulated. Thus, the free radical scavenging pathway was also found to be activated. Furthermore, genes responsible for flavanoid biosynthesis (AT5G13930, 4CL1) and glucosylation (UGT73B2) were up-regulated, while monoterpene (TPSO3) and glucosynalate (At1G24100) biosynthetic genes were down-regulated by freezing stress in plants treated with LPC (Figure 8).
Specificity of LPC treatment at the transcriptional level
A rather limited gene overlap was found between the ANE treatment and the control, as shown in the Venn diagram (Figure 7). When the gene lists were mined for common candidates during freezing and thawing, there were only two and 20 genes in common in the up- and down-regulated classes, respectively. A list of common genes which showed a similar pattern of expression (either up-regulated or down-regulated) during freezing and post-freeze thawing are given in Additional file 2: Table S2.
Heat map
To get a broad overview of the expression pattern of individual genes during freezing and post-freezing recovery, a heat map was generated using the web-based tool, Heat Mapper Plus. A representative sample of the heat map of the global changes in the transcript levels of genes during freezing and the post-freezing recovery period is given in Additional file 1: Figure S4.
Verification of microarray data by quantitative RT–PCR
Several transcripts of selected up-regulated genes based on microarray analysis were quantified independently by RT–PCR (Additional file 2: Table S3). They include a glycolipid transportor (At4g39670), VQ motif-containing protein (At1g78410), ABA responsive zinc finger family protein (At1g63840), galactinol synthase (At1g56600), a drought-repressed protein (At1g73330), the cell wall protein precursor (At2g20870), an expansin protein (At2g40610), a gibberellin-responsive protein (At1g74670), the lipid transfer protein (LTP) (At4g22490), and an invertase/pectin methylesterase inhibitor (At5g20740). The RT–PCR results of all 10 genes showed similar differential expression patterns to those obtained by microarray analysis (Data not shown).
Discussion
The results presented in this paper show that lipophilic components of A. nodosum extracts (LPC) induced specific biochemical changes leading to enhanced tolerance to freezing stress in Arabidopsis thaliana. LPC-treatment of the roots resulted in enhanced freezing tolerance in the aerial parts of the plant, which implicates LPC in inducing systemic metabolic responses.
Proline is one of several compounds with roles as compatible solutes that accumulate in response to freezing and drought stress, [33] and osmotic stress [16]. Our results on the quantification of proline accumulation in wild-type and LPC treated Arabidopsis plants showed that application of ANE or LPC, resulted in a 50% increase in free proline in response to freezing, as compared to control plants.
In higher plants, proline is synthesized via the glutamate pathway or the ornithine pathway [18,22,33]. The glutamate pathway is predominant during abiotic stress and under nitrogen limiting conditions [33,34], while the ornithine pathway takes effect under nitrogen abundance [33]. Plants accumulate proline in response to freezing-induced osmotic stress by simultaneous activation of proline biosynthesis (via the P5CS1 pathway) and/or down regulation of proline degradation (via the ProDH pathway) [35]. The reciprocal regulation of the Δ1-pyrroline-5-carboxylate synthetase (P5CS), a rate-limiting enzyme in proline biosynthesis, and proline dehydrogenase (ProDH), responsible for proline degradation, is a key mechanism in the control of cytosolic proline concentration [34].
The results of our experiments with p5cs1 mutant plants confirmed the role of proline in ANE-mediated freezing tolerance, in that the p5cs1 mutants, treated with ANE, LPC and the water control, all showed similar freezing damage. These observations were in agreement with the results of the gene expression studies. Results of the gene expression analysis suggested that increased proline concentration in the LPC-treated plants was a result of a coordinated increase in P5CS1 and P5CS2 transcripts and, in part, due to decreased in ProDH.
Several studies have suggested that cold-induced sugar accumulation enhances the degree of plant freezing tolerance. In Arabidopsis, a large increase in the degree of freezing tolerance is positively correlated with soluble sugar content [36,37]. Similarly, increased sucrose levels in transgenic Arabidopsis plants, over-expressing a gene for sucrose phosphate synthase, equated with freezing tolerance [38]. Conversely, sensitive to freezing 4 (sfr4) mutants exhibited an impaired freezing tolerance response due to a reduced accumulation of sugar levels at low temperature relative to the wild type [39,40]. In LPC-treated plants, increased accumulation of soluble carbohydrates was brought about by multiple mechanisms such as inducing polysaccharide degradation (i.e. starch, galactose etc.), promoting the biosynthesis of soluble carbohydrates (i.e. glucose, fructose, Raffinose/stachyose), and inactivating the sucrose degradation pathways.
Recently, Uemura et al. [40] showed that the sensitivity of the Arabidopsis mutant sfr4 to freezing was due to its low sugar content, as manifested by loss of osmotic responsiveness. Additionally, it has been demonstrated that exogenous sucrose, at low concentrations, serves as a substrate for low temperature-induced metabolic alterations, while at higher concentrations it has a direct cryoprotective effect on cellular membranes [41]. Although an increased concentration of cytosolic soluble sugar has been observed in response to freezing-induced osmotic stress, in many plant species [36], it is unclear whether the accumulated sugars act as osmolytes, or serve as a source of energy and carbon, that fuels the metabolic changes leading to enhanced freezing tolerance [42].
A positive correlation between sugar accumulation and freezing tolerance has been widely documented in many plant species, including Arabidopsis [36,43]. In the present study, application of ANE or LPC resulted in a significantly enhanced accumulation of total soluble sugars in response to freezing, as compared to untreated controls. The results were further strengthened by analyzing the sensitivity of sfr4 mutant plants to freezing.
1H NMR analysis of the Arabidopsis metabolome, during freezing, revealed that the application of LPC altered biochemical pathways resulting in the accumulation of specific metabolites, leading to enhanced freezing tolerance. The 1H NMR spectra of plants treated with LPC was dominated by peaks with chemical shifts (δ) around 0.8 to 4.0 ppm. These chemical shifts primarily represented 2 major groups of compounds; 0.8 – 2.8 ppm represents lipophilic components like fatty acids and sterols, while resonance at 3.0 to 4.0 ppm represented carbohydrates, sugars, sugar alcohols and organic acids [44]. Our results are in agreement with previous reports. These findings showed that multiple primary metabolites could act collectively, as compatible solutes, ameliorating the osmotic stress caused by freezing.
1H NMR peaks with chemical shifts (δ) around 0.8 to 2.8 ppm, represented the lipophilic components such as fatty acids and sterols, were another major group of metabolites which showed specific changes during freezing stress in LPC-treated plants. Fatty acids are major components of cellular membranes, suberin, and cutin waxes that provide structural barriers to the environment. At low temperature, plant membranes undergo a transition from a liquid crystalline state to a gel-like phase with reduced fluidity contributing to ion leakage and deactivation of membrane proteins [44]. Fatty acids contribute to inducible stress resistance through the remodeling of membrane fluidity [45]. The ability to adjust membrane fluidity by modulating levels of unsaturated fatty acids is a feature of freezing-tolerant plants. Fatty acid unsaturation is thought to reduce the propensity of cellular membranes to undergo freezing-induced, non-bilayer phase formation, thus enhancing membrane integrity and cellular function during freezing [44].
Metabolite profiling revealed an increase in unsaturated fatty acids in plants treated with LPC when exposed to freezing. The spectral peaks (or peak groups) at δ = 5.4-5.2 ppm (protons on double bond carbons), 2.8, 2.3, 2.1, 1.6, 1.4-1.2 ppm (protons on alkyl chain), and 1.0-0.8 ppm (terminal methyl group of alkyl chain) are indicative of unsaturation of fatty acid. We have previously reported that application of ANE or LPC resulted in less tissue damage and electrolyte leakage, as compared to controls, thereby improving the LT50 values [24]. The metabolite profiles suggested that that LPC-mediated freezing tolerance in Arabidopsis may be the result of a combination of metabolic adjustments and increased fatty acid content in Arabidopsis.
Chemical components in the extracts of A. nodosum caused a rapid biochemical response leading to an increased accumulation of osmoprotectants (proline and soluble sugars) and unsaturated fatty acids. The results of the present study suggested that application of these extracts elicited responses reminiscent of a priming effect. Priming is a phenomenon whereby previous exposure to biotic or abiotic stress stimuli makes a plant more resistant to future incidents [46]. Chemical priming is a novel strategy, wherein application of certain chemicals mimic moderate stress stimuli, through physiological and/or hormonal reactions and brings about a priming reaction in the target plants. Exogenous application of chemicals inducing a priming effect, in abiotic stress tolerance has been previously reported [47]. Application of a non-protein amino acid, β-aminobutyric acid (BABA), was shown to induce a priming response in Arabidopsis towards salt stress tolerance (through induction of ABA-dependant elements such as RAB18and RD29A) and salicylic acid-dependent disease tolerance (through PR-1, PR-5 response) [47]. The results presented in this paper suggest the possibility of a chemical priming effect in Arabidopsis, as a reaction to the application of ANE or LPC, resulting in enhanced freezing tolerance.
Further, we used a whole-genome approach to determine the ANE-mediated freezing tolerance in Arabidopsis plants. Application of LPC significantly altered gene expression for 1% of the Arabidopsis genes. In this study, we focused on gene expression related to three aspects: (1) response to stress and/or stress stimulus; (2) compatible osmolyte accumulation; (3) changes in membrane lipid profile in response to stress.
Around 40 annotated genes were found to be up-regulated in our focus category with four genes (At1g10760, At1g55920, At1g09780, At3g22840) found to be directly involved in low temperature stress tolerance in Arabidopsis [48-51]. Another major group of induced genes was those expressed in response to osmotic stress (like water deprivation, salt stress etc.). The ability to survive periods of desiccation is an important adaptation to freezing temperatures and many of the ‘water-deprivation controlled’ genes are also required for maximum survival during freezing stress [52]. Many of the genes expressed during cold acclimation are also inducible by drought stress, and are likely to play a role in protection against cellular dehydration, which occurs during both freezing and drought conditions [53]. This finding is consistent with previous observations, that those plants which are able to efficiently manage osmotic stress, are more tolerant to freezing temperatures [54].
Genes activated in response to biotic stress and/or stimuli were also activated owing to the fact that both biotic and abiotic stress stimuli share a common signaling cascade to bring about physiological responses [55]. For example, salicylic acid (SA) has long been known as a signal molecule in the induction of defense mechanisms in plants [56-58]. Exogenous application of SA has been shown to improve freezing tolerance in wheat by regulating the ice nucleation activity of apoplastic proteins [59,60]. Ding et al. [61] have shown that 0.01 mM methyl salicylate and methyl jasmonate treatment improved the cold tolerance of tomato fruits. Moreover, SA was shown to accumulate during low temperatures in chilling-resistant Arabidopsis plants [62].
Similarly, several studies have suggested that cold-induced sugar accumulation enhances the degree of plant freezing tolerance. In Arabidopsis, a large increase in the degree of freezing tolerance is positively correlated with soluble sugar content [36,37]. Increased sucrose levels in transgenic Arabidopsis plants over-expressing a gene for sucrose phosphate synthase equated with freezing tolerance [38]. Conversely, sensitive to freezing 4 (sfr4) mutants exhibited an impaired freezing tolerance response due to a reduced accumulation of sugar levels at low temperature, relative to the wild type [39,40]. In LPC-treated plants, increased accumulation of soluble carbohydrates was brought about by multiple mechanisms such as induction of polysaccharide degradation (i.e. starch, galactose etc.), promoting biosynthesis of soluble carbohydrates (i.e. glucose, fructose, raffinose/stachyose) and inactivating the sucrose degradation pathways.
Starch is the main carbohydrate store in plants. Regulation of starch metabolism, in particular in response to environmental cues, is of primary importance for carbon and energy flow in plants [63]. Along with photosynthesis, starch degradation also plays a significant role in cold-induced sugar accumulation and enhanced freezing tolerance in Arabidopsis [43,48,64]. Starch-related glucan/water dikinases encoded by the Arabidopsis ‘Starch Excess’ 1 and 4 genes (SEX1, SEX4), regulate starch degradation in plastids by phosphorylating starch, thereby ensuring better accessibility by starch-degrading enzymes during cold induced starch degradation. SEX1 plays an essential role in the cold-induced starch degradation, sugar accumulation, and freezing tolerance enhancement during an early phase of cold acclimation [48]. SEX4/DSP4 phosphatase activity has been shown to be regulated by the redox state [65] and unfavourable environmental stress conditions alter the redox balance within the cells. It is believed that SEX4/DSP4 phosphatase activity may be regulated in response to environmental stress and might be associated to MsK4/AtK-1, a plastid-localized protein kinase associated with starch granules, which is an important regulator that adjusts carbohydrate metabolism during environmental stress [63]. SEX 1 and SEX 4 were found to be activated in LPC-treated plants during freezing stress, as compared to control plants.
Raffinose family oligosaccharides (RFOs) such as raffinose and stachyose are accumulated during the process of cold acclimation, when plants acquire increased frost tolerance [66-68]. Arabidopsis plants with higher rates of raffinose biosynthesis demonstrated increased accumulation of raffinose and galactinol upon cold acclimation and exhibited higher freezing tolerance [67]. Molecular mechanisms by which RFOs influence cellular freezing tolerance are not clear, but it has been shown previously that raffinose can stabilize isolated chloroplast thylakoid membranes during a freeze–thaw cycle [69]. Galactinol synthase (GolS) catalyses the first committed step in the biosynthesis of RFOs [70] and, therefore its expression, provides an experimental tool to assess the level of RFOs during freezing stress, to analyze the function of RFOs as osmoprotectants [67]. LPC application stimulated the raffinose/stachyose biosynthetic genes GolS2 and GolS3 during freezing.
LPC treatment specifically induced a major pathway involved in galactolipid biosynthesis and lipid trafficking in thylakoid membranes controlled by the gene DGD1. Other changes include down-regulation of sterol/cholesterol biosynthesis, regulation of Acyl-CoA thioesterases and down regulation of long chain fatty acid synthesis (Figure 8).
Membranes are the major injury sites during freezing stress [71,72], and membrane lipids undergo substantial changes when plants are exposed to freezing temperatures [73]. It has been well established that membrane polar lipid composition is one of the important factors controlling the structure and efficiency of thylakoid membranes via specific lipid–protein interactions and/or the dynamic properties of the lipid bilayer [74,75]. The galactolipids constitute the bulk (close to 80%) of the thylakoid lipid matrix and, within green plant parts, 70 to 80% of the lipids are associated with photosynthetic membranes. During freezing, dramatic alterations take place in plastid membranes, decreasing the monogalactosyldiacylglycerol (MGDG) content and increasing digalactosyldiacylglycerol (DGDG) content [76,77]. The ratio of DGDG to MGDG is critical for correct protein folding, insertion and intracellular protein trafficking in the chloroplast [78] during temperature stress [79]. DGD1 (digalactosyldiacylglycerol synthase 1) is the enzyme involved in the conversion of MGDG to DGDG in photosynthetic membranes of the chloroplast [80]. The dgd1 mutant of Arabidopsis was impaired in galactolipid assembly, as suggested by a 90% reduction in digalactosyl lipid content [80]. DGD1 (digalactosyldiacylglycerol synthase 1) was found to be highly expressed in LPC-treated plants during freezing stress. Taken together, it may be concluded that activation of digalactosyldiacylglycerol synthase 1 (DGD1) resulted in efficient conversion of MGDG to DGDG, thereby maintaining improved stability of membranes and reduced ion leakage in LPC treated plants.
An early response to low temperature stimulus, by tolerant plant species like winter barley oats, is decreased in membrane fluidity [71]. A major adaptation among the winter tolerant species is the ability to maintain the fluidity of membranes by reducing the ratio of free sterol to total phospholipids. This is achieved by a decrease in the free sterols and an increase in the proportion of phospholipids [81]. Sterol/cholesterol biosynthesis was found to be highly down regulated in LPC-treated plants during freezing stress. A further modification observed in the lipid metabolism of LPC treated plants was the regulation of Acyl-CoA thioesterase.
Conclusions
We found that lipophilic components of ANE (LPC) modulated specific metabolic pathways in A. thaliana resulting in enhanced tolerance to freezing stress. Response to stress and/or stress stimuli, accumulation of compatible osmoprotectants, and regulation of the membrane lipid profile, were the major adaptations observed in LPC-treated plants during freezing stress. There was little overlap in the genes involved in tolerance to freezing stress versus post freezing recovery, indicating the specificity of the mode of action of LPC during freezing stress. A number of genes in the ‘unknown category’ (other molecular functions, other biological processes, other metabolic processes, other cellular processes, etc.) were differentially expressed, indicating that a number of potential genes had been altered by LPC treatment during freezing stress and post-f reezing recovery period. Further studies are needed to dissect the specific genes identified in this study-involved in specific pathways of stress signaling/response, osmolyte accumulation, and lipid metabolism and their roles in A. nodosum extract-mediated, enhanced freezing tolerance in A. thaliana.
Methods
Plant material and Ascophyllum nodosum extract
Seeds of Arabidopsis thaliana (col-0) were purchased from Lehle Seeds (Round Rock, TX). Murashige and Skoog [82] basal salt mixture (Cat No: M5524), sucrose and agar, were purchased from Sigma Aldrich (Oakville, ON).
Preparation of A. nodosum extract (ANE) and its lipophilic fraction (LPC)
A commercial formulation of powdered, alkaline extract of A. nodosum was provided by Acadian Seaplants Limited, Dartmouth, Nova Scotia, Canada. An aqueous solution of the commercial formulation of A. nodosum extract (hereafter referred to as ANE) was prepared by dissolving 1 g of the extract powder in 20 ml of sterile distilled water by constant stirring for 15 min. The solution was then sterilized using a 0.22 μm filter (Corning Inc. NY, USA) and stored in sterile glass centrifuge tubes at 4°C until further use.
The lipophilic fraction (hereafter referred to as LPC) was prepared from a methanol extract of ANE by sequential fractionation with hexane, chloroform and ethyl acetate as described previously by Rayirath et al. [83]. The ethyl acetate sub-fraction, rich in lipophilic components such as fatty acids and sterols, was dried under a stream of nitrogen and stored in sterile glass centrifuge tubes at −80°C until further use. This fraction, re-suspended in a minimal quantity of methanol (100 μl/g equivalent) and diluted with sterile distilled water, was used as LPC for further experiments.
Phenotypic studies and sample collection
Petri dish freezing tolerance assay
The in vitro Petri dish freezing tolerance assay, used to investigate the plant responses in ANE-induced freezing tolerance, was conducted as described by Xin and Browse (1998). Briefly, sterilized Arabidopsis seeds were planted on solidified Murashige and Skoog basal salt medium (Murashige and Skoog 1962), containing 1% sucrose and supplemented with different concentrations of ANE or LPC. Required concentrations of filter-sterilized ANE (0.5 gL-1) or LPC (1.0 gL-1 equivalent of ANE) were added to molten (50°C) media, and plated in partitioned petri dishes (Fisher Scientific, Ottawa, ON). Two sets of controls, one set of plates with distilled water used to dissolve ANE, and a second set with equal amount of methanol used to dissolve LPC (100 μl L-1 medium) were maintained. Seeds were evenly distributed in the Petri dishes by placing individual seeds with a 100 μL micro pipette at the rate of 10–15 seeds per partition. The petri dishes were incubated at 22/18°C day/night temperatures, 16:8 photoperiod and light intensity 100 μmol photons m-1 s-1 for 10 days.
Ten days after germination, Petri dishes were transferred to a temperature-controlled incubator, set to −2 ± 0.1°C. To achieve uniform freezing, the Petri dishes were equilibrated at −2°C for 24 h before further lowering the temperature. After equilibration, the temperature of the chamber was progressively lowered at the rate of 1°C per day until the desired sub-zero temperatures were attained. The temperature was monitored by wireless temperature sensor modules (Traceable® Remote sensor module, Fisher Scientific, Ottawa, ON, Canada) placed inside the Petri plates. At each temperature, five Petri plates per treatment were withdrawn from the chamber, thawed at 4°C for 12 h, in the dark, and returned to the original growth conditions (16:8 photoperiod, 22/18°C day/night temperatures). Survival of plants (by degree of chlorosis and shoot damage) was recorded visually, 48 h after returning to the original growth conditions.
In vivo freezing assay
The Arabidopsis ecotype Col-0 plants were grown in peat pellets (Jiffy-7®, Jiffy Products Ltd, NB, Canada) in a greenhouse set at 22/18°C day/night temperatures and 16:8 photoperiod with an irradiance of 100 μmol photons m-1 s-1. Three-week-old plants were used in the experiments. Extract treatments were administered 48 h prior to freezing treatment by irrigating with 1.0 g L-1 of ANE or LPC (1.0 g L-1 equivalent made up with sterile distilled water) at the rate of 20 ml per plant. The control plants were grown under identical conditions as the plants used in the extract treatment, except they received an equal volume of methanol (100 μl/g equivalent), made up with distilled water, but without the LPC. Forty eight hours after treatment, plants were transferred to a low temperature incubator set to 0°C. Freezing was initiated by spraying ice cold water, and the temperature of the chamber was progressively lowered at the rate of 1°C every 24 h until the desired sub-zero temperature was attained. Temperature was monitored through wireless temperature sensor modules placed at different points inside the freezing cabinet. At each treatment temperature, fifteen randomly selected plants from each treatment were withdrawn from the chamber, thawed at 4°C for 12 h in the dark, and then returned to the original growth conditions of the green house. Two days later, survival of plants was recorded visually according to the degree of chlorosis and leaf damage. Leaf samples, for biochemical assays, collected from plants undergoing the peat pellet freezing tolerance, were flash frozen in liquid nitrogen, ground to fine powder and stored at −80°C until further analysis.
Proline estimation
To study the role of proline in ANE-mediated freezing tolerance, the proline content was measured in acidic extracts and quantified spectrophotometrically using an acid-ninhydrin reagent with proline as the standard [84]. The proline concentrations of treated plants were compared with untreated controls using Tukey’s HSD test with P≤0.05 and n≥15. Three independent experiments were performed with similar results.
Proline mutant studies
Petri dish freezing tolerance assays, and peat pellet freezing tolerance assays, were carried out following the methods previously described by Xin and Browse [16], using P5CS mutant plants, to study the role of proline in ANE-mediated freezing tolerance. Arabidopsis proline mutant p5cs1.1 (SALK_058000) [85] seeds were a generous gift from Dr. Laszlo Szabados (Institute of Plant Biology, Biological Research Center, Szeged, Hungary).
Transcriptional analysis of proline biosynthesis and degradation genes
The expression of proline biosynthesis genes, P5CS1 and P5CS2, and the proline degradation gene, PRODH (proline dehydrogenase), was studied by quantitative Real-Time PCR. Leaf samples were taken from −2°C treatments. Leaf tissue from five plants constituted a replicate and there were three replications for each treatment. The leaves were flash frozen in liquid nitrogen, ground with a pre-chilled mortar and pestle and stored at −80°C until further use. Total RNA was isolated using RNAqueous® Plant RNA isolation Kit (Ambion Inc. Austin, TX) following the manufacturer’s instruction. Ten micrograms of total RNA were treated with DNAase using the TURBO DNA-free®kit (Ambion Inc. Austin, TX), and first-strand cDNA was synthesized using the Retroscript® Reverse Transcription Kit (Ambion Inc. Austin, TX). Quantitative Real-time PCR, performed to assess the fold change in P5CS1 and P5CS2,PRODH transcript abundance, was carried out in a ‘Step One Plus® Real-Time PCR System’ (Applied Biosystems) using ‘Fast Start Universal SYBR Green Master®’ (Roche Diagnostics, Indianapolis, IN) adopting manufacturer’s instructions. Data were analyzed using ‘Step One software V2.0’ with a ‘Relative Standard Curve’ mode. Primer sequences used for this study are provided in the Additional file 2: Table S1.
Total soluble sugars estimation and sugar mutant studies
Total soluble sugars were estimated by the colorimetric assay developed by Farrar [86]. An Arabidopsis sugar mutant (sfr4) (FR67) [87] was used to study the role of soluble sugar accumulation in ANE-mediated freezing tolerance. The sfr4 mutant used in this study was a kind gift from Drs. Matsuo Uemura and Lenn Thorlby (School of Biological Sciences, University of London, UK). The sfr4 mutant is highly sensitive to freezing temperatures as compared to wild-type plants, due to a low sugar content, which leads to a loss of osmotic responsiveness [40]. A peat pellet freezing tolerance assay was carried out to confirm the role of sugar accumulation in ANE-mediated freezing tolerance. Trypan blue stain (3,3-[(3,3-dimethyl-4,4-biphenylylene) bis (azo)] bis(5-amino-4-hydroxy-2,7-naphthalenedisulfonic acid) tetra sodium salt) was used to visualize the extent of freezing-induced tissue damage in extract-treated and control plants. Damaged tissues stained dark blue, while the viable cells did not stain, due to an intact cell membrane barrier [88].
Plant growth conditions, extract treatments and freezing temperatures were as described in the peat pellet freezing assay, as described in Xin and Browse [16]. At each treatment/temperature combination, five plants per treatment were removed from the incubator, thawed at 4°C for 12 h in the dark, and then returned to the normal growth conditions (16 h:8 h day:night cycle; 22/18°C day/night temperatures at an irradiance of 100 μmol photons m-1 s-1). Two days later, uniform sized leaves, from the second or third whorl, were sampled (by excising at the base of the petiole) and tissue damage was assessed, as described previously by Rate et al. [88]. The area of the damaged tissue was measured using Image-j®image processing and analysis software (Research Services Branch, NIH). For each treatment, at least 15 individual leaves collected from five randomly selected plants (three leaves per plant), were stained and analyzed. Two independent experiments were carried out with similar results.
Metabolite profiling
Global profiling
Global metabolite profiling of freeze-stressed Arabidopsis plants was carried out using a method described by Ward et al. [44], with minor modifications. Total metabolites were extracted from 100 mg of frozen plant tissue using 10 ml methanol (100%), by sonicating for 1 h using an ice/water bath. The extract was decanted into a fresh glass vial and re-extracted with a fresh batch of methanol. The extracts were combined, dried under a stream of nitrogen, and freeze dried to remove residual water. The dried extract was re-suspended in 800 μl deuteriated methanol (Methanol-d4) (Cat no 530530, Sigma Aldrich, Oakville, ON), sonicated, and a 750 μl aliquot was transferred into a 5 mm NMR tube (Wilmad-Lab glass, Buena, NJ) for analysis.
NMR spectra were acquired on a Bruker Avance III 600 MHz NMR spectrometer (Bruker Corporation, East Milton, ON), operating at 600.28 MHz 1H observation frequency, and a temperature of 25±0.2°C. Spectra were obtained with a gradient broadband inverse 5 mm probe (optimized for 1H), auto-tuned, matched and shimmed on each sample, at a flip angle of 30° pulse, followed by a 2.66 s acquisition time and 2 s relaxation delay. Each spectrum consisted of 32 scans of 64 k data points, with a spectral width of 12335.52 Hz. 1H NMR chemical shifts in the spectra were referenced to CD2H peak of methanol at δ 3.31 ppm, and that peak intensity was also used as a reference for comparison of the level of metabolites level between samples. The signals were acquired, processed and analyzed using TopSpin® NMR data acquisition and processing software (Bruker Biospin Ltd, East Milton, ON) integrated with the spectrometer. The experiment was conducted with three biological replicates and the experiment was repeated twice.
Targeted profiling
For targeted metabolite profiling, representative samples used in the NMR experiments above were dried and re-suspended with 20 mM phosphase buffer (pH 7.0), 1 mM NaN3 and 0.49 mM DSS (3-(trimethylsilyl)-1-propanesulfonic acid) in D2O. NMR spectra were acquired on the same Bruker Avance III 600 NMR spectrometer, as described above, using a 5 mm inverse gradient probe set to 25.0±0.1°C. 1D NOESY spectra with pre-saturation on water were acquired with 256 scans, an acquisition time of 2.73s, a sweep width of 20 ppm, a mixing time of 100 ms, and relaxation delay of 5s. All spectra were referenced using the internal DSS standard set to 0 ppm. NMR spectra were processed and binned for the region of d 0–10 ppm (water peak region d 4.7-4.83 ppm excluded) within 0.04 ppm chemical shift window by using the NMR software package NMR Suite 5.0 (Chenomx, Edmonton, Canada). The same software package was used to do targeted profiling. Multivariate data analysis (PCA and PLS-DA) was performed by SIMCA-P+ 12.01.0 software package (Umetrics, Umea, Sweden). The experiment had three biological replicates.
Microarray analysis
RNA extraction and cRNA preparation
Total RNA was isolated using an RNAqueous® Plant RNA isolation kit (Ambion Inc. Austin, TX) following the manufacturer’s instructions. The RNA samples were quantified on a Nanodrop ND1000® (Thermo Scientific, Beverly, MA) and the integrity was checked on an Agilent Bioanalyzer 2100® (Agilent Technologies Canada Inc. Mississauga, ON). CopyRNA was prepared using an Illumina TotalPrep® RNA Amplification kit according to the standard protocol provided by the manufacturer. Briefly, the protocol consisted of first- and second-strand reverse transcription steps, followed by a single, in vitro transcription (IVT) amplification step that incorporated biotin-labeled nucleotides. The synthesized cRNA was later purified and quantitated.
Hybridization and signal acquisition
Equal quantities of purified cRNA were hybridized to GeneChip® Arabidopsis ATH1 Genome Arrays (Affymetrix Inc. Santa Clara, CA). Subsequent steps included washing, blocking, and streptavadin-Cy3:Cy5 staining. Fluorescence emission was quantitatively detected usingthe iScan® System (Illumina Inc. Hayward, CA) with Illumina Beadstudio® software (Illumina Inc. Hayward, CA). Each treatment had three biological replicates.
Microarray data analysis
Statistical analysis
The raw data were analyzed using Flex Array statistical software (Genome Quebec, Montreal, QC) to obtain a measure of gene expression. Background correction, normalization, and summarization of probe-level gene chip data was performed using the RMA (Robust Multi-chip Average) algorithm in Flex Array. The statistical test used was Wright and Simon’s implementation of the Empirical Bayes method [89]. The gene lists were imported from the annotation file, ATH1-121501.na21.annot.csv (Affymetrix Inc. Santa Clara, CA), available in TIGR database (ATH1-121501).
Functional classification of differentially expressed genes
The differentially expressed gene lists were distributed into functional categories using the web based tool, Classification Super-Viewer (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi), Bio-Array Resource for Arabidopsis functional genomics, University of Toronto) based on Gene Ontology classes available on the TAIR database (ftp://ftp.arabidopsis.org/home/tair/Ontologies/GeneOntology). Those IDs falling into classification categories, other than unclassified and classification not yet clear-cut, were removed from these two categories. The tool then normalized the number of genes from the experimental data to the number of genes in each class present on the chip, so that differences were more easily perceived. A class score for normalization was calculated based on the following equation [90].
Furthermore, the input set was bootstrapped one hundred times by sampling the input set (with repeats) and then reclassifying each set generated. The standard deviation for the scores generated from the bootstrap sets was displayed along with the normalized class score. In this way classes represented by small numbers of genes on the chip were easily identified [90].
Pathway analysis
The Pathway Tools Omics Viewer (formerly the Pathway Tools Expression Viewer) (http://gohelle.cirad.fr:1555/expression.html) was used for pathway analysis of genes differentially expressed in the microarray experiment. We followed the standardized AraCyc-defined metabolic pathways catalogue [91], which included 1759 Arabidopsis enzyme genes, to identify genes in each pathway.
Heat map and Venn diagram
Heat mapping was perfromed using the web based tool, Heat-mapper Plus (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi) (Bio-Array Resource for Arabidopsis functional genomics, University of Toronto) and the Venn diagram was generated using the web based tool, Venn Diagram Generator (http://www.pangloss.com/seidel/Protocols/venn.cgi).
Validation of microarray experiment using RT-PCR
A ‘Two-step RT-PCR’ was used for quantifying transcript yield and comparing differential gene expression. Ten micrograms of total RNA were treated with DNase using the TURBO DNA-free®kit (Ambion Inc. Austin, TX), and first-strand cDNA was synthesized using the Retroscript® Reverse Transcription Kit (Ambion Inc. Austin, TX). The cDNA samples were purified with QAIquick® PCR purification Kit (Qiagen Inc. Mississauga, Ontario) and normalized with the Quantum RNA® Universal 18S internal standard (Ambion Inc. Austin, TX). A standard PCR reaction mixture was used and the transcripts were amplified using the following profile: 1 cycle of 94°C for 2 min, 30 cycles of 94°C for 30s, 60°C for 30s, 72°C for 1min, followed by 1 cycle of 72°C for 7 min.
Competing interests
The authors declare no competing interests.
Authors’ contributions
BP, PR conceived the experiment. BP, PR, JZ, CK, MDH, BB planned the study. PR,JZ, XJ, CK conducted experiments. PR, JZ, CK, MDH, BB, ATC, DH, SK, BP analyzed, interpreted data and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Global gene expression of Arabidopsis plants treated with lipophilic fraction of ANE (LPC) during freezing stress and post freezing recovery period. Volcano plot of differential expression pattern during (a) freezing stress and (b) post freezing recovery with ≥1.5 foldchange and P≤0.05 cut off. Scatter plot of log2 signal intensities of three replicate samples from LPC-treated plants during (c) freezing stress and (d) post freezing recovery. Figure S2. Functional categorization of differentially expressed genes. Normalized frequency (Provart and Zhu, 2003) categories of (a) up, or (c) down-regulated genes in LPC-treated plants during freezing. (b&d) functional categorization (b) up or (d) down-regulated genes during freezing according to the actual number of genes. Figure S3. Functional categorization of differentially expressed genes. Normalized frequency (Provart and Zhu, 2003) categories of (a) up, or (c) down-regulated genes in LPC-treated plants during post-freezing recovery period. (b& d) functional categorization (b) up or (d) down-regulated genes according to the actual number of genes during post-freezing recovery period. Figure S4. Heat map of selected differentially expressed genes in LPC treated plants during (FR) freezing stress and (TH) post freezing recovery period. Expression-level change is shown in a color relative to the expression level, as indicated in the color scale bar.
Table S1. Primer sets and the PCR conditions used for gene expression studies. This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/. Table S2. A list of common genes which showed the similar pattern of expression (either up-regulated, or down-regulated) during freezing and post-freezing thawing period. Table S3. List of genes selected for RT-PCR confirmation of microarray results.
Contributor Information
Prasanth Nair, Email: pnair@uoguelph.ca.
Saveetha Kandasamy, Email: saveetha.kandasamy@dal.ca.
Junzeng Zhang, Email: junzeng.Zhang@nrc-cnrc.gc.ca.
Xiuhong Ji, Email: Xiuhong.Ji@nrc-cnrc.gc.ca.
Chris Kirby, Email: chris.kirby@agr.gc.ca.
Bernhard Benkel, Email: bernhard.benkel@dal.ca.
Mark D Hodges, Email: mark.hodges@agr.gc.ca.
Alan T Critchley, Email: alan.critchley@acadian.ca.
David Hiltz, Email: david.hiltz@acadian.ca.
Balakrishnan Prithiviraj, Email: bprithiviraj@dal.ca.
Acknowledgements
BP’s lab is supported by the grants from Natural Sciences and Engineering Research Council of Canada (NSERC), Nova Scotia Department of Agriculture & Marketing (NSDAF) and Acadian Seaplants Limited. Authors are grateful to Dr. Laszlo Szabados (Institute of Plant Biology, Biological Research Center, Szeged, Hungary) for providing P5CS1 mutants and Dr. Matsuo Uemura & Dr. Glenn Thorlby (School of Biological Sciences, University of London, UK) for providing FR67 (sfr4) mutants.
References
- Bray EA, Bailey-Serres J, Weretilnyk E. In: Biochemistry and Molecular Biology of Plants. Gruissem W, Buchannan B, Jones R, editor. Rockville: American Society of Plant Biologists; 2000. Responses to abiotic stresses; pp. 1158–1249. [Google Scholar]
- Fuller MP, Hamed F, Wisniewski M, Glenn DM. Protection of plants from frost using hydrophobic particle film and acrylic polymer. Ann Appl Biol. 2003;143:97–97. [Google Scholar]
- Ilker R, Warring AJ, Lyons JM, Breidenbach RW. The cytological responses of tomato seedling cotyledons to chilling and the influence of membrane modifications upon these responses. Protoplasma. 1976;90:229–252. doi: 10.1007/BF01275677. [DOI] [Google Scholar]
- Horváth I, Van Hasselt PR. Inhibition of chilling-induced photooxidative damage to leaves of Cucumis sativus L. by treatment with amino alcohols. Planta. 1985;164:83–88. doi: 10.1007/BF00391029. [DOI] [PubMed] [Google Scholar]
- Temple WD, Bomke AA. Effects of Kelp (Macrocystis integrifolia and Eklonia maxima) foliar applications on bean crop growth. Plant Soil. 1989;117:85–92. doi: 10.1007/BF02206260. [DOI] [Google Scholar]
- Metting B, Zimmerman WJ, Crouch T, Van-Staden J. In: Introduction of Applied Phycology. Akatsuka I, editor. Hague: SPB Academic Publishing; 1990. Agronomic uses of seaweed and microalgae; pp. 589–627. [Google Scholar]
- Yan J. Influence of plant growth regulators on turfturf grass polar lipid composition, tolerance to drought and salinity stresses, and nutrient efficiency.Ph.D. dissertation. Blacksburg: Virginia Polytechnic Institute and State Univ; 1993. [Google Scholar]
- Featonby-Smith BC, Van-Staden J. The effect of seaweed concentrate on the growth of tomatoes in nematode infested soil. Scient Hort. 1983;20:137–146. doi: 10.1016/0304-4238(83)90134-6. [DOI] [Google Scholar]
- Nabati DA. Response of two grass species to plant growth regulators, fertilizer N, chelated Fe, salinity and water stress. Ph.D. dissertation. Blacksburg: Virginia Polytechnic Institute and State Univ; 1991. [Google Scholar]
- Nabati DA, Schmidt RE, Parrish DJ. Alleviation of salinity stress in Kentucky bluegrass by plant growth regulators and Iron. Crop Sci. 1994;34:198–202. doi: 10.2135/cropsci1994.0011183X003400010035x. [DOI] [Google Scholar]
- Burchett S, Fuller MP, Jellings AJ. Application of seaweed extract improves winter hardiness of winter barley cvIgri. Abstracts – Plant Cell Topics. York Meeting. 1998.
- Wilson S. University of Tasmania Research Report - UT 99/1. Australia: Grape and Wine Research and Development Corporation; 2001. Frost Management in Cool Climate Vineyards; p. 34. [Google Scholar]
- Swaaij ACV, Jacobsen E, Feenstra WJ. Effect of cold hardening, wilting and exogenously applied proline on leaf proline content and frost tolerance of several genotypes of Solanum. Physiol Plant. 1985;64:230–236. doi: 10.1111/j.1399-3054.1985.tb02341.x. [DOI] [Google Scholar]
- Kushad MM, Yelenosky G. Evaluation of polyamine and proline levels during low temperature acclimation of citrus. Plant Physiol. 1987;84:692–695. doi: 10.1104/pp.84.3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörffling K, Dörffling H, Lesselich G, Luck E, Zimmermann C, Melz G, Jurgens HU. Heritable improvement of frost tolerance in winter wheat by in vitro-selection of hydroxyproline-resistant proline overproducing mutants. Euphytica. 1997;93:1–10. doi: 10.1023/A:1002946622376. [DOI] [Google Scholar]
- Xin Z, Browse B. Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad USA. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett. 1999;461:205–210. doi: 10.1016/S0014-5793(99)01451-9. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]
- Rudolph AS, Crowe JH, Crowe LM. Effects of three stabilizing agents – proline, betaine, and trehalose on membrane phospholipids. Arch Biochem Biophys. 1986;245:134–143. doi: 10.1016/0003-9861(86)90197-9. [DOI] [PubMed] [Google Scholar]
- Iyer S, Caplan A. Products of proline catabolism can induce osmotically regulated genes in rice. Plant Physiol. 1998;116:203–211. doi: 10.1104/pp.116.1.203. [DOI] [Google Scholar]
- Saradhi P, Alia P, Arora S, Prasad KVSK. Proline accumulates in plants exposed to UV radiation and protects them against UV induced peroxidation. Biochem Biophys Res Commun. 1995;209:1–5. doi: 10.1006/bbrc.1995.1461. [DOI] [PubMed] [Google Scholar]
- Hare PD, Cress WA. Metabolic implications of stress induced proline accumulation in plants. Plant Growth Regul. 1997;21:79–102. doi: 10.1023/A:1005703923347. [DOI] [Google Scholar]
- Brugière N, Dubois F, Limami AM, Lelandais M, Roux Y, Sangwan RS, Hirel B. Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell. 1999;11:1995–2011. doi: 10.1105/tpc.11.10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayirath P, Benkel B, Hodges DM, Allan-Wojtas P, MacKinnon SL, Critchley AT, Prithiviraj B. Lipophilic components of the brown seaweed, Ascophyllum nodosum, enhance freezing tolerance in Arabidopsis thaliana. Planta. 2009;230:135–147. doi: 10.1007/s00425-009-0920-8. [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow M. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313X.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Pennycooke JC, Jones ML, Stushnoff C. Down-Regulating α-Galactosidase. Enhances freezing tolerance in transgenic petunia. Plant Physiol. 2003;133:901909. doi: 10.1104/pp.103.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra SS, Wolfraim L, Poole RJ, Dhindsa RS. Molecular cloning and relationship to freezing tolerance of cold-acclimation-specific genes of alfalfa. Plant Physiol. 1989;89:375–380. doi: 10.1104/pp.89.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL. Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:187–223. doi: 10.1146/annurev.pp.41.060190.001155. [DOI] [Google Scholar]
- Jones PG, Inouye M. The cold shock response: a hot topic. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- Nishida I, Murata N. Chilling sensitivity in plants and Cyanobacteria: the crucial contribution of membrane lipid. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:541–568. doi: 10.1146/annurev.arplant.47.1.541. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS. Proline biosynthesis and osmoregulation in plants. Plant J. 1993;4:215–223. doi: 10.1046/j.1365-313X.1993.04020215.x. [DOI] [Google Scholar]
- PengZ LQ, Verma DP. Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Gen Genet. 1996;253:334–341. doi: 10.1007/pl00008600. [DOI] [PubMed] [Google Scholar]
- Hare PD, Cress WA, van Staden J. Proline synthesis and degradation: a model system for elucidating stress-related signal transduction. J Exp Bot. 1999;50:413–434. [Google Scholar]
- Wanner LA, Junttila O. Cold induced freezing tolerance in Arabidopsis. Plant Physiol. 1999;120:391–399. doi: 10.1104/pp.120.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T, Nakamura M, Hayashi H, Inatsugi R, Yano R, Nishida I. The leaf-order-dependent enhancement of freezing tolerance in cold-acclimated Arabidopsisrosettes is not correlated with the transcript levels of the cold-inducible transcription factors of CBF/DREB1. Plant Cell Physiol. 2003;44:922–931. doi: 10.1093/pcp/pcg117. [DOI] [PubMed] [Google Scholar]
- Strand A, Foyer CH, Gustafsson P, Gardestrom P, Hurry V. Altering flux through the sucrose biosynthesis pathway in transgenic Arabidopsis thaliana modifies photosynthetic acclimation at low temperatures and the development of freezing tolerance. Plant Cell Environ. 2003;26:523–535. doi: 10.1046/j.1365-3040.2003.00983.x. [DOI] [Google Scholar]
- McKown R, Kuroki G, Warren G. Cold responses of Arabidopsis mutants impaired in freezing tolerance. J Exp Bot. 1996;47:1919–1925. doi: 10.1093/jxb/47.12.1919. [DOI] [Google Scholar]
- Uemura M, Warren G, Steponkus PL. Freezing sensitivity in the sfr4 mutant of Arabidopsis is due to low sugar content and is manifested by loss of osmotic responsiveness. Plant Physiol. 2003;131:1800–1807. doi: 10.1104/pp.102.013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Steponkus PL. Modification of the intracellular sugar content alters the incidence of freeze-induced membrane lesions of protoplasts isolated from Arabidopsis thaliana leaves. Plant Cell Environ. 2003;26:1083–1096. doi: 10.1046/j.1365-3040.2003.01033.x. [DOI] [Google Scholar]
- Klotke J, Kopka J, Gatzke N, Heyer AG. Impact of soluble sugar concentrations on the acquisition of freezing tolerance in accessions of Arabidopsis thaliana with contrasting cold adaptation: Evidence for a role of raffinose in cold acclimation. Plant Cell Environ. 2004;27:1395–1404. doi: 10.1111/j.1365-3040.2004.01242.x. [DOI] [Google Scholar]
- Ristic Z, Ashworth EN. Changes in leaf ultrastructure and carbohydrates in Arabidopsis thaliana L. (Heynh) cv. Columbia during rapid cold acclimation. Protoplasma. 1993;172:111–123. doi: 10.1007/BF01379368. [DOI] [Google Scholar]
- Ward JL, Harris C, Lewis J, Beale MH. Assessment of 1H NMR spectroscopy and multivariate analysis as a technique for metabolite fingerprinting of Arabidopsis thaliana. Phytochemistry. 2003;62:949–957. doi: 10.1016/S0031-9422(02)00705-7. [DOI] [PubMed] [Google Scholar]
- Iba K. Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Ann Rev Plant Biol. 2002;53:225–245. doi: 10.1146/annurev.arplant.53.100201.160729. [DOI] [PubMed] [Google Scholar]
- Bruce TJA, Matthes MC, Napier JA, Pickett JA. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007;173:603–608. doi: 10.1016/j.plantsci.2007.09.002. [DOI] [Google Scholar]
- Jakab G, Ton J, Flors V, Zimmerli L, Métraux JP, Mauch-Mani B. Enhancing arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 2005;139:267–274. doi: 10.1104/pp.105.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R, Nakamura M, Yoneyama T, Nishida I. Starch-related α-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis. Plant Physiol. 2005;138:837–846. doi: 10.1104/pp.104.056374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle C, Vaultier MN, Taconnat L, Renou JP, Kader JC, Zachowski A, Ruelland E. The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. PlantPhysiol. 2005;139:1217–1233. doi: 10.1104/pp.105.068171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amme S, Matros A, Schlesier B, Mock HP. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J Exp Bot. 2006;57:1537–1546. doi: 10.1093/jxb/erj129. [DOI] [PubMed] [Google Scholar]
- Dominguez-Solis JR, He Z, Lima A, Ting J, Buchanan BB, Luan S. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc Natl Acad Sci USA. 2008;105:16386–16391. doi: 10.1073/pnas.0808204105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell VO, Loper JE. The sigma factor RpoS is required for stress tolerance and environmental fitness of Pseudomonas fluorescens Pf-5. Microbiology. 2005;151:3001–3009. doi: 10.1099/mic.0.28077-0. [DOI] [PubMed] [Google Scholar]
- Steponkus PL. Role of the plasma membrane in freezing injury and cold acclimation. Annu Rev Plant Physiol. 1984;35:543–584. doi: 10.1146/annurev.pp.35.060184.002551. [DOI] [Google Scholar]
- Guy C. In: Cold Shock: Response and Adaptation. Inouye M, Yamanaka K, editor. Norfolk: Horizon Press; 2000. Molecular responses of plants to cold shock and cold acclimation; pp. 85–112. [Google Scholar]
- Horvath E, Szalai G, Janda T. Induction of abiotic stress tolerance by salicylic acid signalling. J Plant Growth Reg. 2007;26:290–300. doi: 10.1007/s00344-007-9017-4. [DOI] [Google Scholar]
- Raskin I. Role of salicylic acid in plants. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:439–463. doi: 10.1146/annurev.pp.43.060192.002255. [DOI] [Google Scholar]
- Klessig DF, Durner J, Noad R, Navarre DA, Wendehenne D, Kumar D, Zhou JM, Shah J, Zhang S, Kachroo P, Trifa Y, Pontier D, Lam E, Silva H. Nitric oxide and salicylic acid signaling in plant defense. Proc Natl Acad Sci USA. 2000;97:8849–8855. doi: 10.1073/pnas.97.16.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim VA, Vess A, Scheel D, Rosahl S. The role of salicylic acid and jasmonic acid in pathogen defence. Plant Biol. 2006;8:307–313. doi: 10.1055/s-2006-924025. [DOI] [PubMed] [Google Scholar]
- Tasgín E, Atící Ö, Nalbantoglu B. Effects of salicylic acid and cold on freezing tolerance in winter wheat leaves. Plant Growth Regul. 2003;41:231–236. [Google Scholar]
- Tasgín E, Atící Ö, Nalbantoglu B, Popova LP. Effects of salicylic acid and cold treatments on protein levels and on the activities of antioxidant enzymes in the apoplast of winter wheat leaves. Phytochemistry. 2006;67:710–715. doi: 10.1016/j.phytochem.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Ding CK, Wang CY, Gross KC, Smith DL. Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta. 2002;214:895–901. doi: 10.1007/s00425-001-0698-9. [DOI] [PubMed] [Google Scholar]
- Scott IM, Clarke SM, Wood JE, Mur LAJ. Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 2004;135:1040–1049. doi: 10.1104/pp.104.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempa S, Rozhon W, Samaj J, Erban A, Baluska F, Becker T, Haselmayer J, Schleiff E, Kopka J, Hirt H, Jonak C. A plastid-localized glycogen synthase kinase 3 modulates stress tolerance and carbohydrate metabolism. Plant J. 2007;49:1076–1090. doi: 10.1111/j.1365-313X.2006.03025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004;135:1674–1684. doi: 10.1104/pp.104.040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov LN, Dominguez-Solis JR, Allary AL, Buchanan BB, Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA. 2006;103:9732–9737. doi: 10.1073/pnas.0603329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- Zuther E, Buchel K, Hundertmark M, Stitt M, Hincha DK, Heyer AG. The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett. 2004;576:169–173. doi: 10.1016/j.febslet.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Hincha DK. Differential effects of galactose containing saccharides on mechanical freeze-thaw damage to isolated thylakoid membranes. Cryoletters. 1990;11:437–444. [Google Scholar]
- Pharr DM, Saravitz DM, Carter TE. Galactinol synthase activity and soluble sugars in developing seeds of four soybean genotypes. Plant Physiol. 1987;83:185–189. doi: 10.1104/pp.83.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J. Responses of Plants to Environmental Stresses 1: Chilling, Freezing and High Temperature Stresses. 2. New York: Academic; 1980. pp. 254–258. [Google Scholar]
- Sakai A, Larcher W. Frost survival of plants: Responses and adaptation of freezing Stress. Berlin: Springer; 1987. pp. 41–47. [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL. Cold-acclimation of Arabidopsis thaliana: effect of plasma membrane composition and freeze induced lesions. Plant Physiol. 1995;109:15–30. doi: 10.1104/pp.109.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MS, Green BR. Biochemical and biophysical properties of thylakoid acyl lipids. Biochim Biophys Acta (Biophys) 1991;1060:133–158. doi: 10.1016/S0005-2728(09)91002-7. [DOI] [Google Scholar]
- Hendrickson L, Vlcková A, Selstam E, Huner N, Oquist G, Hurry V. Cold acclimation of the Arabidopsis dgd1 mutant results in recovery from photosystem I-limited photosynthesis. FEBS Lett. 2006;580:4959–4968. doi: 10.1016/j.febslet.2006.07.081. [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses: role of phospholipase D{alpha} in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- Li W, Wang R, Li M, Li L, Wang C, Welti R, Wang X. Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. J Biol Chem. 2008;283:461–468. doi: 10.1074/jbc.M706692200. [DOI] [PubMed] [Google Scholar]
- Bruce BD. The role of lipids in plastid protein transport. Plant Mol Biol. 1998;38:223–246. doi: 10.1023/A:1006094308805. [DOI] [PubMed] [Google Scholar]
- Dormann P, Benning C. Galactolipids rule in seed plants. Trends Plant Sci. 2002;7:112–118. doi: 10.1016/S1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- Dormann P, Balbo I, Benning C. Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science. 1999;284:2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- Sikorska E, Ondrias K, Farkas T. Physical properties of membranes of cold hardened and non hardened winter rape plants. Acta Biol Acad Sci Hung. 1981;32:266–274. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Rayirath P, Khan W, Palanisamy R, MacKinnon SL, Stefanova R, Hankins SD, Critchley AT, Prithiviraj B. Extracts of brown seaweed, Ascophyllum nodosum, induce gibberellic acid (GA3) independent amylase activity in barley. J Plant Growth Regul. 2008;27:270–279. [Google Scholar]
- Lea PJ, Blackwell RD. In: Photosynthesis and Production in a Changing Environment. Hall DO, Scurlock JMO, Bolhar HR, Leegood RC, Long SP, editor. London: Chapman and Hall; 1993. Ammonia assimilation, photorespiration and amino acid biosynthesis; pp. 313–336. [Google Scholar]
- Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E. et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- Farrar JF. In: Photosynthesis and production in a changing environment: A Field and laboratory Manual. Hall DO, Scurlock JMO, Bolha’r-Nordenkampf HR, Leegood RC, Long SP, editor. London: Chapman and Hall; 1993. Carbon Partitioning; pp. 232–246. [Google Scholar]
- Warren G, McKown R, Marin A, Teutonico R. Isolation of mutants affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;111:1011–1019. doi: 10.1104/pp.111.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell. 1999;11:1695–1708. doi: 10.1105/tpc.11.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart N, Zhu T. A Browser-based Functional Classification SuperViewer for Arabidopsis Genomics. Curr Comput Mol Biol. 2003. pp. 271–272.
- Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Global gene expression of Arabidopsis plants treated with lipophilic fraction of ANE (LPC) during freezing stress and post freezing recovery period. Volcano plot of differential expression pattern during (a) freezing stress and (b) post freezing recovery with ≥1.5 foldchange and P≤0.05 cut off. Scatter plot of log2 signal intensities of three replicate samples from LPC-treated plants during (c) freezing stress and (d) post freezing recovery. Figure S2. Functional categorization of differentially expressed genes. Normalized frequency (Provart and Zhu, 2003) categories of (a) up, or (c) down-regulated genes in LPC-treated plants during freezing. (b&d) functional categorization (b) up or (d) down-regulated genes during freezing according to the actual number of genes. Figure S3. Functional categorization of differentially expressed genes. Normalized frequency (Provart and Zhu, 2003) categories of (a) up, or (c) down-regulated genes in LPC-treated plants during post-freezing recovery period. (b& d) functional categorization (b) up or (d) down-regulated genes according to the actual number of genes during post-freezing recovery period. Figure S4. Heat map of selected differentially expressed genes in LPC treated plants during (FR) freezing stress and (TH) post freezing recovery period. Expression-level change is shown in a color relative to the expression level, as indicated in the color scale bar.
Table S1. Primer sets and the PCR conditions used for gene expression studies. This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/. Table S2. A list of common genes which showed the similar pattern of expression (either up-regulated, or down-regulated) during freezing and post-freezing thawing period. Table S3. List of genes selected for RT-PCR confirmation of microarray results.