Abstract
Background
Japanese encephalitis virus (JEV), a mosquito-borne zoonotic pathogen, is the sole etiologic agent of Japanese Encephalitis (JE); a neurotropic killer disease which is one of the major causes of viral encephalitis worldwide with prime public health concern. JE was first reported in the state of West Bengal, India in 1973. Since then it is being reported every year from different districts of the state, though the vaccination has already been done. Therefore, it indicates that there might be either partial coverage of the vaccine or the emergence of mutated/new strain of JEV. Considering this fact, to understand the JEV genotype distribution, we conducted a molecular epidemiological study on a total of 135 serum/cerebrospinal fluid (CSF) samples referred and/or collected from the clinically suspected patients with Acute encephalitis syndrome (AES), admitted in different district hospitals of West Bengal, India, 2010.
Findings
JEV etiology was confirmed in 36/135 (26.6%) and 13/61 (21.3%) 2–15 days’ febrile illness samples from AES cases by analyzing Mac-ELISA followed by RT-PCR test respectively. Phylogenetic analysis based on complete envelope gene sequences of 13 isolates showed the emergence of JEV genotype I (GI), co-circulating with genotype III (GIII).
Conclusion
This study represents the first report of JEV GI with GIII, co-circulating in West Bengal. The efficacy of the vaccine (derived from JEV GIII strain SA-14-14-2) to protect against emerging JEV GI needs careful evaluation. In future, JE outbreak is quite likely in the state, if this vaccine fails to protect sufficiently against GI of JEV.
Keywords: Acute encephalitis syndrome, Japanese encephalitis virus, Genotype I, Genotype III, West Bengal
Background
The mosquito-borne Japanese encephalitis virus (JEV) is an enveloped, positive-sense single-stranded RNA virus, member of the genus Flavivirus under the family Flaviviridae [1]. JEV is the sole etiologic agent of Japanese Encephalitis (JE); a neurotropic killer disease being one of the major causes of viral encephalitis in human. Since the isolation of this virus in Japan in 1935 [2], it has spread worldwide becoming a major public health problem. Worldwide case-fatality rate of JE was recorded to be 30% approximately with 30-50% of survivors developing permanent neurologic deficit/sequelae [3].
Recent studies have shown that the envelope (E) gene is an established phylogenetic marker for JEV, since this region is free from selective pressure that supports obscure long-term evolutionary relationship [4]. Altogether 5 distinct genotypes have been identified among the JEV strains [5] of which genotype III (GIII) is mostly circulated in the Southeast Asian countries, including Japan, South Korea, China, Taiwan, Vietnam, Philippines, and India [6]. However, it was recently documented that GIII is replaced by genotype I (GI) in South Korea, Thailand and China [7]. Though GIII is predominant in India, GI has been introduced in the country recently [7]. In 1973 JE outbreak was first recorded in the districts of Burdwan and Bankura in West Bengal where 700 cases and 300 deaths were reported [8,9]. Thereafter, several JE outbreaks took place in the state [10-12]. Every year sporadic JE cases are being reported indicating its endemicity in this state despite the vaccination programme undertaken by the State Health Department, Government of West Bengal [13]. In addition, the geographic features, environmental factors and socio-economic status of this state also favor JEV transmission [14]. Moreover, the reports of JE incidences in the state are the indications of either partial coverage of the vaccine or the emergence of mutated/new strain of JEV. Genetic variation of JEV circulating in West Bengal has not yet been investigated and hence to ascertain the same a molecular epidemiological study was undertaken.
Materials and Methods
A total of 92 serum and 43 cerebrospinal fluid (CSF) samples were referred and/or collected from 135 clinically suspected pediatric-adolescent (0–20 years old) and adult (≥ 21 years old) individuals with Acute encephalitis syndrome (AES), showing high grade fever (≥39°c) for 2–15 days including any two of the following symptoms, viz. headache, vomiting, stupor, delirium, abnormal movements, presence of kernig’s sign, convulsions, neck rigidity, altered sensorium, unconsciousness admitted in 8 different district hospitals, West Bengal during the period from July to December in 2010 (Figure 1).
Figure 1.
Map of West Bengal showing the location of sample collection areas.
All the samples were tested for IgM antibody against JEV by using IgM antibody-capture (Mac) ELISA kit (National institute of virology, Pune, India), according to the manufacturer’s protocol.
Only 61 JEV IgM negative samples with a history of ≤ 3 days’ illness were screened and 200 μl of each of them were used for virus isolation on C6/36 cell line according to the standard protocol [13]. The tissue culture fluids were collected from the samples producing prominent cytopathic effect (CPE) and subjected for RNA extraction by QIAamp RNA viral kit (Qiagen, GmbH, Hilden, Germany), following the manufacturer’s protocol.
To identify the isolates as JEV, reverse transcription-PCR (RT-PCR) was carried out with the extracted RNA (50 pg to 1μg) by Qiagen one step RT-PCR kit (Qiagen, GmbH, Hilden, Germany), in accordance with the manufacture’s specifications, using 0.6 μM of primer pairs [13] that specific for structural E gene sequence of JEV. The PCR products were separated by electrophoresis on 1% agarose gel, stained with ethidium bromide.
RT-PCR amplicons were purified using the Qiagen gel extraction kit (Qiagen, GmbH, Hilden, Germany), according to the manufacturer’s protocol, followed by direct sequencing using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA), as per the manufacturer’s instructions and the products were analyzed using an automated DNA sequencer, 3130XL Genetic Analyzer (PE Applied Biosystems, Foster city, CA, USA). The 1,500 nucleotides generated complete sequences of the JEV E gene that were edited and corrected using the Finch TV software (http://www.geospiza.com). Multiple sequence alignment and phylogenetic analysis were performed by using CLUSTALW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and MEGA version 5.0 software (http://www.megasofteware. net). The phylogenetic tree was constructed by the neighbor-joining method, tested with Kimura 2-parameter model.
Results
Out of 135 samples, only 36 (26.6%) samples were reactive to JEV specific IgM antibody, of which 23 (63.8%) and 13 (36.1%) samples were CSF and serum respectively. Only 61 of the remaining 99 JEV IgM negative samples having the history of ≤ 3 days of febrile illness were selected and subsequently subjected to tissue culture resulting in 19 samples producing prominent CPE of which only 13 (21.3%) samples were identified as JE positive by RT-PCR method, consisting of 8 (61.5%) from CSF and 5 (38.4%) from serum.
We have a total of 49 (36 IgM + 13 RT-PCR positive) JE cases (36.2%) of which 30 (19 IgM + 11 RT-PCR positive) were pediatric-adolescent (61.2%) and remaining 19 (17 IgM + 2 RT-PCR positive) were found to be adult cases (38.7%). Moreover, the occurrence of JEV infection was recorded during the month of July to December with the maximum number of cases (46.9%) observed in the month of September.
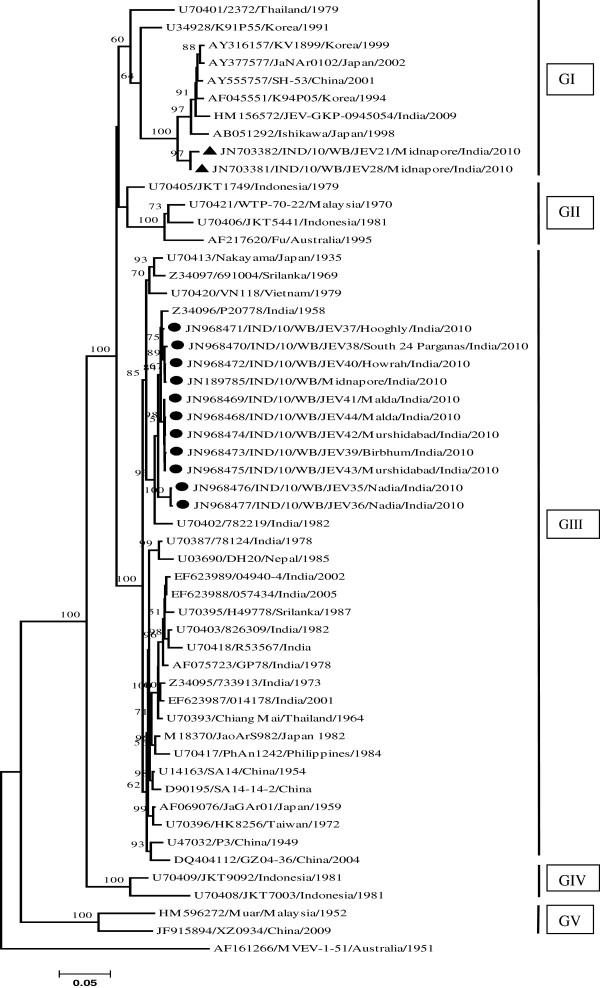
The Figure 2 shows the phylogenetic tree derived from 13 E gene sequences of JEV isolates along with 41 previously published JEV strains, including 12 from India and 29 from worldwide (Table 1). Dendrogram showed 2 E gene sequences of the isolates (GenBank: JN703381, JN703382) belonging to GI and comprising 89%-91% nucleotide (nt) identity with 11 E gene sequences of other isolates (GenBank: JN968468- JN968477, JN189785) belonging to GIII (Figure 2). Moreover, these 2 GI E gene sequences showed 99% nt similarity with each other and were most similar (96%) with Japanese GI strain Ishikawa (GenBank: AB051292), followed by 94%-95% nt similarity with Indian isolate JEV-GKP-0945054 (GenBank: HM156572). Eleven GIII E gene sequences showed 97%-99% nt similarity with each other and 93%-98% nt similarity with other Indian GIII strains, having the highest similarity (97%-98%) with Indian P20778 strain (GenBank: Z34096).
Figure 2.
Phylogenetic analysis of 13 JEV isolates in serum/CSF samples from AES patients, West Bengal. The Neighbor-Joining (NJ) Phylogenetic tree based on complete envelope (E) gene nucleotide sequences of Japanese encephalitis virus (JEV) isolates/strains. The Murray valley encephalitis virus strain (MVEV-1-51) was used as an out group for generating the rooted tree. The robustness of dendrogram was evaluated by 1000 bootstrap pseudo replicates. Bootstrap values (≥50% of replicates) were shown in corresponding nodes. Horizontal branch lengths are proportional to genetic distance and vertical branch lengths have no significance. Each taxon is named systematically by mentioning the accession number, strain name, country of origin and year of isolation. The isolates’ sequences used in this study were marked with filled circle and triangle symbols. Genotypes are indicated on the right. Scale bar indicates nucleotide substitutions per site.
Table 1.
Background information of selected strains/isolates of JEV referenced in this study
| Strains/isolates | Country and Year of isolation | Source | Genotype | GenBank Accession no. |
|---|---|---|---|---|
| Nakayama |
Japan, 1935 |
Human brain |
III |
U70413 |
| JaGAr01 |
Japan, 1959 |
Mosquito |
III |
AF069076 |
| JaoArS982 |
Japan, 1982 |
Mosquito |
III |
M18370 |
| Ishikawa |
Japan, 1998 |
Mosquito |
I |
AB051292 |
| JaNAr0102 |
Japan, 2002 |
Mosquito |
I |
AY377577 |
| Fu |
Australia,1995 |
Human |
II |
AF217620 |
| WTP-70-22 |
Malaysia ,1970 |
Mosquito |
II |
U70421 |
| PhAn1242 |
Philipines,1984 |
Pig serum |
III |
U70417 |
| Muar |
Malaysia, 1952 |
Human |
V |
HM596272 |
| 691004 |
Srilanka, 1969 |
Human |
III |
Z34097 |
|
H49778 |
Srilanka, 1987 |
Human |
III |
U70395 |
| DH20 |
Nepal, 1985 |
Human |
III |
U03690 |
| VN118 |
Vietnam, 1979 |
Mosquito |
III |
U70420 |
| K94P05 |
Korea, 1994 |
Mosquito |
I |
AF045551 |
| KV1899 |
Korea, 1999 |
Pig |
I |
AY316157 |
| K91P55 |
Korea, 1991 |
Mosquito |
I |
U34928 |
| GP78 |
India, 1978 |
Human |
III |
AF075723 |
| 733913 |
India, 1973 |
Human brain |
III |
Z34095 |
|
P20778 |
India, 1958 |
Human |
III |
Z34096 |
|
R53567 |
India, unavailable |
unavailable |
III |
U70418 |
| 782219 |
India, 1982 |
Human |
III |
U70402 |
| 826309 |
India, 1982 |
Human brain |
III |
U70403 |
|
R53567 |
India, unavailable |
unavailable |
III |
U70418 |
| 014178 |
India, 2001 |
Human blood |
III |
EF623987 |
| 04940-4 |
India, 2002 |
Mosquito |
III |
EF623989 |
| 057434 |
India, 2005 |
Human blood |
III |
EF623988 |
| 78124 |
India, 1978 |
Human |
III |
U70387 |
| JEV-GKP-0945054 |
India, 2009 |
Human CSF |
I |
HM156572 |
| P3 |
China, 1949 |
Mosquito |
III |
U47032 |
| SA14 |
China, 1958 |
Mosquito |
III |
U14163 |
| SA14-14-2 |
China, unavailable |
Vaccine |
III |
D90195 |
| GZ04-36 |
China, 2004 |
Mosquito |
III |
DQ404112 |
| XZ0934 |
China, 2009 |
Mosquito |
V |
JF915894 |
| SH-53 |
China, 2001 |
Mosquito |
I |
AY555757 |
| JKT1749 |
Indonesia, 1979 |
Mosquito |
II |
U70405 |
| JKT9092 |
Indonesia, 1981 |
Mosquito |
IV |
U70409 |
| JKT7003 |
Indonesia, 1981 |
Mosquito |
IV |
U70408 |
| JKT5441 |
Indonesia, 1981 |
Mosquito |
II |
U70406 |
| 2372 |
Thailand, 1979 |
Human |
I |
U70401 |
| Chiang Mai |
Thailand, 1964 |
Human |
III |
U70393 |
| HK8256 |
Taiwan, 1972 |
Mosquito |
III |
U03691 |
| IND/10/WB |
India, 2010 |
Human CSF |
III |
JN189785 |
| IND/10/WB/JEV28 |
Midnapore, India, 2010 |
Human CSF |
I |
JN703381 |
| IND/10/WB/JEV21 |
Midnapore, India, 2010 |
Human CSF |
I |
JN703382 |
| IND/10/WB/JEV38 |
South 24 Pgs, West Bengal, India, 2010 |
Human CSF |
III |
JN968470 |
| IND/10/WB/JEV37 |
Hooghly, West Bengal, India, 2010 |
Human serum |
III |
JN968471 |
| IND/10/WB/JEV44 |
Malda, West Bengal, India, 2010 |
Human CSF |
III |
JN968468 |
| IND/10/WB/JEV41 |
Malda, West Bengal, India, 2010 |
Human CSF |
III |
JN968469 |
| IND/10/WB/JEV40 |
Howrah, West Bengal, India, 2010 |
Human CSF |
III |
JN968472 |
| IND/10/WB/JEV39 |
Birbhum, West Bengal, India, 2010 |
Human CSF |
III |
JN968473 |
| IND/10/WB/JEV42 |
Murshidabad, West Bengal, India, 2010 |
Human serum |
III |
JN968474 |
| IND/10/WB/JEV43 |
Murshidabad, West Bengal, India, 2010 |
Human serum |
III |
JN968475 |
| IND/10/WB/JEV35 |
Nadia, West Bengal, India, 2010 |
Human serum |
III |
JN968476 |
| IND/10/WB/JEV36 |
Nadia, West Bengal, India, 2010 |
Human serum |
III |
JN968477 |
| MVEV-1-51 | Australia, 1951 | Human | AF161266 |
All strains/isolates are JEV, except Murray valley encephalitis virus strain (MVE-1-51) used as out group for phylogenetic analysis in this study.
Discussion
JEV infection is considered as a prime issue on public health concern in West Bengal. The present study reveals that 36/135 (26.6%) and 13/61 (21.3%) samples were positive to JE by Mac-ELISA and RT-PCR method respectively. This observation is the proof of JEV infection in recent time and to detect the total number of JE cases, ELISA negative acute samples (from ≤ 3 days’ febrile illness) should be confirmed by RT-PCR test.
JE incidences (61.2% vs. 38.7%) were higher in pediatric-adolescent age group than adult because pediatrics were infected possibly due to lack of immunity and adolescents were directly exposed to the mosquito vector (Culex sp.) bite, as they usually took active part in cultivation in crop-fields where vectors usually breed. In the present study, JE was found to occur in the monsoon period with the maximum number of cases in September when the Culex mosquitoes breed in the paddy fields covered with stagnant rain water.
However, we found that 86 [(99–61)+(61–13)] samples with a history of 2–15 days’ illness were true JE negative possibly due to either mishandling of samples which damaged the IgM antibody/the viral titre or the presence of another etiology responsible for AES.
In our previous reports we have achieved 36 JEV isolates [13] belonging to GIII whose E gene sequences were submitted to NCBI GenBank database like GenBank: JN189782, JN189783, JN189784 and HQ891146 etc. (unpublished data). The present study, therefore, constitutes the first report on E gene based phylogenetic analysis of the JEV isolates from AES cases of West Bengal where JEV GI has emerged very recently, co-circulating with JEV GIII.
Our study reveals that 2 isolates (GenBank: JN703381, JN703382) belonging to GI of JEV from the coastal district Midnapore (22.25°N 87.65°E and 23 meters above sea-level) where this GI might be transmitted from other part of India [7] or by the travellers returning from JE endemic countries, possibly with JEV infection [15]. On the other hand, this district has got many Lakes, swamps, forest and rice fields which provide a wintering and staging grounds for several migratory birds. Such areas are also very suitable for breeding and survival of mosquitoes. In view of these conditions, GI of JEV might have been introduced into West Bengal through migratory birds or cyclonic wind-blown mosquitoes from newer geographic region [16,17]. However, it is still unknown as to how JEV GI has emerged in West Bengal exactly. Therefore, further studies to determine the role of travelers, migratory birds and wind-blown mosquitoes in JEV transmission are required.
The State Health Department of Government of West Bengal undertook the vaccination programme against JE in Midnapore in 2008 [13] using live attenuated JE vaccine derived from GIII strain SA-14-14-2. In this connection, the efficacy of the vaccine to protect against GI of JEV needs careful evaluation. In near future, there is a chance for an impending threat of JEV outbreak in this region/state, if this vaccine fails to protect sufficiently against GI of JEV.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
AS and SCHAT conceived the study, the design, and drafted the manuscript. AS and DT carried out serology and molecular work. AS, DT, SKM, SCHAK and SCHAT contributed to the data analysis and data interpretation. All authors read and approved the final manuscript.
Ethical approval
The study was duly approved by the joint ethical committee of ICMR (Indian Council of Medical Research) virus unit and NICED (National Institute of Cholera and Enteric Diseases), Kolkata, India.
Contributor Information
Arindam Sarkar, Email: arimicro.sarkar@gmail.com.
Debjani Taraphdar, Email: taraphdar.debjani@gmail.com.
Subhra Kanti Mukhopadhyay, Email: skmmicro@gmail.com.
Sekhar Chakrabarti, Email: drsekharchakrabarti@yahoo.co.in.
Shyamalendu Chatterjee, Email: shyamalenduchatterjee@gmail.com.
Acknowledgements
This work was funded by Department of Science and Technology, Govt. of West Bengal, India [Grant No. 396(Sanc.)/ST/P/S&T/9G-27/2007]. The authors express their heartfelt and sincere thanks to the members of DBT centre for bioinformatics, Presidency University, Kolkata for their support during bioinformatics (phylogenetic) analysis. The enthusiastic help obtained from the physicians of District Hospitals in West Bengal, for providing us with the clinically suspected samples for this study, is gratefully acknowledged.
References
- Lindenbach BD, Rice CM. In: Fields Virology. Volume 1 4. Knipe DM, Howley PM, editor. Philadelphia: Lippincott: Williams and Wilkins; 2001. Flaviviridae: the viruses and their replication; pp. 991–1041. [Google Scholar]
- Tanaka M, Aira Y, Igarashi A. Comparative Nucleotide and Amino Acid Sequences of Five Japanese Encephalitis Virus Strains Isolated in Japan and China. Trop Med. 1991;33:15–21. [Google Scholar]
- World Health Organization Japanese encephalitis vaccines-WHO position paper Wkly Epidemiol Rec 200681331–340.16933380 [Google Scholar]
- David TW, Wang LF, Daniels PW, Mackenzie JS. Molecular characterization of the first Australian isolate of Japanese encephalitis virus, the FU strain. J Gen Virol. 2000;81:2471–2480. doi: 10.1099/0022-1317-81-10-2471. [DOI] [PubMed] [Google Scholar]
- Solomon T, Ni H, Beasley DW, Ekkelenkamp M, Cardosa MJ, Barrett AD. Origin and evolution of Japanese encephalitis virus in Southeast Asia. J Virol. 2003;77:3091–3098. doi: 10.1128/JVI.77.5.3091-3098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10(suppl):S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Fulmali PV, Sapkal GN, Athawale S, Gore MM, Mishra AC, Bondre VP. Introduction of Japanese Encephalitis Virus Genotype I, India. Emerg Infect Dis. 2011;17:319–321. doi: 10.3201/eid1702.100815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SN, Rodrigues FM, Seth GP, Tongaonkar SS, Padbidri VS, Gupta NP. Investigations on the outbreak of Japanese encephalitis in Burdwan district, West Bengal. Part II. Serological survey of human population. Indian J Med Res. 1975;63:1472–1477. [PubMed] [Google Scholar]
- Rodrigues FM, Ghosh SN, Banerjee K, Chatterjee AK, Gupta NP. A post-epidemic serological survey of humans in Bankura district, West Bengal, following the epidemic of Japanese encephalitis in 1973. Indian J Med Res. 1975;63:1478–1485. [PubMed] [Google Scholar]
- Rajagopalan PK, Panicker KN. A note on the 1976 epidemic of Japanese encephalitis in Burdwan district West Bengal. Indian J Med Res. 1978;68:3938. [PubMed] [Google Scholar]
- Banerjee K, Sengupta SN, Dandawate CN, Tongaonkar SS, Gupta NP. Virological and serological investigations of an epidemic of encephalitis which occurred at Bankura district, West Bengal. Indian J Med Res. 1976;64:121–130. [PubMed] [Google Scholar]
- Mukhopadhyay BB, Mukherjee B, Bagchi SB, Chakraborty M, Mukherjee KK. An epidemiological investigation of Japanese encephalitis outbreak in Burdwan, District of west Bengal during 1987–1988. Indian J Public Health. 1990;34:107–116. [PubMed] [Google Scholar]
- Sarkar A, Taraphdar D, Mukhopadhyay SK, Chakrabarti S, Chatterjee S. Serological and molecular diagnosis of Japanese encephalitis reveals an increasing public health problem in the state of West Bengal, India. Trans R Soc Trop Med Hyg. 2012;106:15–19. doi: 10.1016/j.trstmh.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Taraphdar D, Mukhopadhyay BB, Kumar M, Mukhopadhyay SK, Chatterjee S. Influence of socio-economic status and environmental factors on serologically diagnosed Japanese encephalitis cases in the state of West Bengal, India during 2005–2010. Health . 2012;4:6–12. doi: 10.4236/health.2012.41002. [DOI] [Google Scholar]
- Centers for Disease Control and Prevention. Japanese Encephalitis in a U.S. Traveler Returning from Thailand, 2004. MMWR weekly. 2005;54:123–125. [PubMed] [Google Scholar]
- Huang JH, Lin TH, Teng HJ, Su CL, Tsai KH, Lu LC, Lin C, Yang CF, Chang SF, Liao TL, Yu SK, Cheng CH, Chang MC, Hu HC, Shu PY. Molecular epidemiology of Japanese encephalitis virus, Taiwan. Emerg Infect Dis. 2010;16:876–878. doi: 10.3201/eid1605.091055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SA, Rochester W. Wind-blown mosquitoes and introduction of Japanese encephalitis into Australia. Emerg Infect Dis. 2001;7:900–903. doi: 10.3201/eid0705.017524. [DOI] [PMC free article] [PubMed] [Google Scholar]