Abstract
Porphyromonas gingivalis, an important periodontal pathogen, is closely associated with inflammatory alveolar bone resorption, and several components of the organism such as lipopolysaccharides have been reported to stimulate production of cytokines that promote inflammatory bone destruction. We investigated the effect of infection with viable P. gingivalis on cytokine production by osteoblasts. Reverse transcription-PCR and real-time PCR analyses revealed that infection with P. gingivalis induced receptor activator of nuclear factor κB (NF-κB) ligand (RANKL) mRNA expression in mouse primary osteoblasts. Production of interleukin-6 was also stimulated; however, osteoprotegerin was not. SB20350 (an inhibitor of p38 mitogen-activated protein kinase), PD98059 (an inhibitor of classic mitogen-activated protein kinase kinase, MEK1/2), wortmannin (an inhibitor of phosphatidylinositol 3 kinase), and carbobenzoxyl-leucinyl-leucinyl-leucinal (an inhibitor of NF-κB) did not prevent the RANKL expression induced by P. gingivalis. Degradation of inhibitor of NF-κB-alpha was not detectable; however, curcumin, an inhibitor of activator protein 1 (AP-1), prevented the RANKL production induced by P. gingivalis infection. Western blot analysis revealed that phosphorylation of c-Jun, a component of AP-1, occurred in the infected cells, and an analysis of c-Fos binding to an oligonucleotide containing an AP-1 consensus site also demonstrated AP-1 activation in infected osteoblasts. Infection with P. gingivalis KDP136, an isogenic deficient mutant of arginine- and lysine-specific cysteine proteinases, did not stimulate RANKL production. These results suggest that P. gingivalis infection induces RANKL expression in osteoblasts through AP-1 signaling pathways and cysteine proteases of the organism are involved in RANKL production.
Periodontitis, an inflammatory disorder of the supporting tissues of the teeth, is one of the most common types of human infection. Alveolar bone resorption, followed by loss of teeth, is clinically the most important issue in periodontitis. A small subset of periodontal bacteria are supposed to play a major part in the pathogenesis of the disease (15, 27, 48), with Porphyromonas gingivalis characterized as a bona fide pathogen in severe forms of adult periodontal diseases (3, 15, 27, 28, 42, 48). P. gingivalis is a gram-negative black pigmented anaerobe that colonizes in periodontal pockets and spreads into deeper tissues, including the connective and bone tissues (15, 18, 27, 28, 30, 42, 48). This pathogen expresses a number of potential virulence factors, such as cysteine proteases named gingipains, as well as fimbriae and lipopolysaccharide (LPS), which may contribute to the pathogenesis of periodontitis (3, 8, 15, 17, 27, 28, 40, 56). Among these, gingipains degrade collagen and fibronectin and inhibit interaction between epithelial cells and the extracellular matrix (8, 17, 25, 46). Gingipains also degrade various cytokines, such as tumor necrosis factor-alpha (TNF-α), interleukin-6 (IL-6), and IL-8, which results in the disturbance of host cytokine network (4, 8, 17, 34, 40). They are classified into two groups, arginine-specific gingipains (Arg-gingipain-A and -B) and lysine-specific gingipain (Lys-gingipain). Fimbriae and their subunit protein, fimbrillin (FimA), are reported to mediate bacterial adhesion to and invasion of human epithelial cells (3, 5, 6, 9, 10, 26, 33, 35, 56). LPS, a major component of the outer membrane of gram-negative bacteria, displays multiple biological and immunological activities through mammalian innate receptors named Toll-like receptors (TLRs) (2). It has been reported that P. gingivalis LPS is a potent stimulator of inflammatory mediators such as IL-1 and prostaglandin E2 (PGE2), which eventually induces bone resorption (15, 16, 31, 32, 50, 57).
Osteoclastic bone resorption involves the differentiation and activation of osteoclasts, i.e., bone-resorbing multinucleate cells differentiated from hemopoietic progenitor cells. The differentiation of osteoclasts requires a recently discovered cytokine known as receptor activator of nuclear factor κB (NF-κB) ligand (RANKL). RANKL is a transmembrane molecule of the TNF ligand superfamily that is expressed in osteoblasts (24, 49, 58), T cells (23), and synoviocytes (51). This molecule is essential for full osteoclastic differentiation from hematopoietic precursor cells into mature multinucleated bone-resorptive osteoclasts in the presence of macrophage colony-stimulating factor (23, 24, 49, 58). On the other hand, osteoclastogenesis is blocked in the presence of osteoprotegerin (OPG) (47, 49), a soluble decoy receptor that inhibits osteoclast formation, function, and survival by preventing the binding of RANKL to its receptor that is present on osteoclast precursors and mature osteoclasts (23, 24, 49). Mice with a null mutation of the OPG gene exhibit severe osteoporosis (47). Therefore, RANKL and OPG are key molecules that act as positive and negative regulators, respectively, in osteoclastogenesis and bone resorption in vivo.
It is known that bacterial infection triggers a wide variety of cellular responses, including the production of proinflammatory cytokines. Previous studies suggested that adhesion and invasion of infected bacteria induce cellular responses (14, 20, 39, 43, 45, 52). Activation of mitogen-activated protein kinase (MAPK) pathways has also shown to be involved in these proinflammatory responses; however, the mechanisms have not been elucidated (12, 14, 39, 45, 52). P. gingivalis can adhere to and invade epithelial and endothelial cells (3, 5, 6, 9, 10, 26, 33, 35, 44, 56), and infection with viable organisms of P. gingivalis induces IL-1, IL-6, IL-8, TNF-α, and monocyte chemoattractant protein 1 (MCP-1) production by human epithelial and endothelial cells (20, 35, 43). We found here that infection with viable P. gingivalis induced RANKL expression in osteoblasts through activator protein 1 (AP-1) signaling. Our results also suggest that the major effectors of RANKL induction by P. gingivalis are gingipains.
MATERIALS AND METHODS
Mice and reagents.
Female ddY mice were obtained from Japan SLC Co. (Hamamatsu, Japan). PGE2, wortmannin, PD98059, SB203580, and curcumin (7) were purchased from Sigma Chemicals (St. Louis, Mo.). Carbobenzoxyl-l-leucinyl-l-leucinyl-l-leucinal (ZLLLal), an inhibitor of NF-κB (41), and the control peptide, carbobenzoxyl-l-leucinyl-l-leucinal (ZLLal), were purchased from Peptide Institute (Osaka, Japan). Rabbit antibodies against total and phosphorylated c-Jun were purchased from Cell Signaling Technology (Beverly, Mass.). Horseradish peroxidase-conjugated anti-rabbit and anti-goat immunoglobulin G (IgG), antibodies against the inhibitor of NF-κB-alpha (IκB-α), and protein A/G-agarose were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Recombinant mouse receptor activator of NF-κB (RANK)/Fc chimera protein (RANK/Fc) and anti-mouse RANKL antibodies were obtained from R&D Systems (Minneapolis, Minn.). Other chemicals were obtained from Sigma Chemicals or Wako Pure Chemicals (Osaka, Japan).
Bacterial strains, culture conditions, and preparation of heat-inactivated bacterial cells.
P. gingivalis ATCC 33277, KDP150 (an FimA-deficient mutant of ATCC 33277, the whole fimA coding region of ATCC 33277 was replaced with a 2.1-kb ermF ermAM DNA block of pKD355 [53]), KDP136 (a triply deficient mutant for three gingipains, Arg-gingipain-A and -B and Lys-gingipain of ATCC 33277) (6, 46), ATCC 53977, and OMZ314 were grown as described previously (46). Fresh bacterial cells from blood agar plates supplemented with hemin (5 μg/ml; Sigma) and menadione (1 μg/ml; Sigma) were inoculated into 5 ml of Trypticase soy broth (BBL Microbiology Systems, Cockeysville, Md.) supplemented with yeast extract (0.1%; BBL), hemin, and menadione. The cultures were incubated anaerobically in an N2-H2-CO2 (80:10:10) atmosphere at 37°C by using an Anaerobic System 1024 (Forma, Marietta, Ohio). Organisms were harvested by centrifugation, washed with phosphate-buffered saline (PBS), and resuspended in alpha minimum essential medium (αMEM) (Gibco-BRL, Grand Island, N.Y.). The numbers of bacteria were determined with a spectrophotometer (at an optical density at 600 nm) based on a standard curve established by colony formation on bacterial plates (33). To prepare heat-inactivated P. gingivalis cells, bacterial suspensions in PBS were heated at 60°C for 30 min, washed with PBS, and resuspended in αMEM.
Culture of primary mouse osteoblasts.
Primary mouse osteoblasts were isolated from newborn mouse calvariae after five routine sequential digestions with 0.1% collagenase (Wako Pure Chemicals) and 0.2% dispase (Godo Shusei, Tokyo, Japan), as described previously (11, 37). Osteoblasts were cultured in αMEM supplemented with 10% fetal calf serum (Gibco-BRL), penicillin G (100 U/ml), and streptomycin (100 μg/ml). Immunochemical staining with osteocalcin and staining for alkaline phosphatase activity demonstrated that >95% of the cells showed an osteoblastic character (11, 37).
For the infection assay, osteoblasts at 80 to 90% confluence were washed three times with serum-free αMEM containing no antibiotics, followed by incubation at 37°C for 1 h in serum-free αMEM containing no antibiotics. Cells were subjected to infection with viable P. gingivalis at a multiplicity of infection (MOI) of 10 to 100. In some experiments, specific protein kinase inhibitors or NF-κB inhibitor were used, in which case they were preincubated at the indicated concentration with the cultures for 1 h to permit penetration into the cells.
Adhesion and invasion assay.
P. gingivalis ATCC 33277 (107 CFU) was washed with sterile PBS and suspended in 1 ml of serum-free αMEM with no antibiotics. The organisms were incubated at 37°C in a CO2 incubator for 1, 3, and 6 h, and the CFU of viable P. gingivalis were determined by using blood agar plates supplemented with hemin and menadione (26, 33).
P. gingivalis was added to osteoblast cultures grown in 24-well culture plates (MOI = 100), and the cultures were incubated at 37°C in 5% CO2. To determine the numbers of adherent and invaded bacteria, the cultures were incubated for 0.5, 1, or 2 h. Unattached bacteria were washed off with PBS, and the osteoblasts were disrupted by extensive pipetting with sterile water and then plated on blood agar plates. For the invasion assay, the attached bacteria were killed by 1 h of incubation with gentamicin (300 μg/ml; Sigma) and metronidazole (200 μg/ml; Sigma). After exposure to the antibiotics, the cells were washed in PBS, the invaded bacteria were released by disruption of the cells in sterile water, and the lysate dilutions were plated on blood agar plates (26).
To visualize the adhesion and invasion of P. gingivalis, osteoblasts grown on a coverglass (18 by 18 mm) were incubated with P. gingivalis (MOI = 100) in serum-free αMEM with no antibiotics at 37°C in 5% CO2. After 1 h of incubation, cells were washed with PBS three times to remove the nonadherent bacteria and fixed with 3% formaldehyde in PBS for 30 min at room temperature. After the cells were washed with PBS, they were incubated with rhodamine-conjugated phalloidin (Wako Pure Chemicals) and SYBR green (Roche Molecular Biochemicals, Mannheim, Germany) to stain for filamentous actin and the nucleus of osteoblasts. P. gingivalis was visualized by SYBR green because this dye bound to the bacterial DNA. The adhesion and invasion of P. gingivalis was observed by using a model MRC1024 laser scanning confocal microscope (Bio-Rad Laboratories, Hercules, Calif.)
RT-PCR assay and real-time PCR analysis.
For reverse transcription-PCR (RT-PCR), total RNA was prepared from the cells by using Trizol reagent (Gibco-BRL), and 2 μg of total RNA was reverse transcribed in the presence of oligo(dT)15 using the SuperScript first-strand synthesis system (Gibco-BRL) according to the manufacturer's instructions. cDNA samples were tested for integrity and the amount of input RNA by RT-PCR for β-actin, an endogenous control. The cDNA samples were subjected to a conventional PCR with cytokine-specific primers (Table 1). The PCR program was 35 cycles at 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min. The PCR products were separated by electrophoresis on a 2% agarose gel and visualized by ethidium bromide staining with UV light illumination. To quantify RANKL mRNA, real-time PCR was performed by using a LightCycler (Roche Molecular Biochemicals) with a SYBR Green reagent. Samples were subjected to 40 cycles of amplification at 95°C for 15 s, followed by 52°C for 20 s and 72°C for 30 s, according to the manufacturer's instructions (Roche Molecular Biochemicals). Each assay was normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA. The normalized data were expressed three ways: (i) the percentage of gene expression in osteoblasts stimulated with 2 μM of PGE2, (ii) the percentage of gene expression in osteoblasts infected with P. gingivalis, and (iii) the fold increase against the mRNA level of unstimulated cells. The primers used are listed in Table 1.
TABLE 1.
Sequence and expected fragment sizes of synthetic oligonucleotides used for RT-PCR
| Target mRNA | Primer sequence | Size (bp) |
|---|---|---|
| RANKL | 5′-TACTTTCGAGCGCAGATGGAT-3′ | 482 |
| 5′-GTACGCTTCCCGATGTTTCAT-3′ | ||
| IL-6 | 5′-ATGAACAACGATGATGCACTTG-3′ | 490 |
| 5′-TAAGTCAGATACCTGACAACAG-3′ | ||
| OPG | 5′-AAACAGCACTGCACAGTGAG-3′ | 489 |
| 5′-TGGTAGGAACAGCAAACCTG-3′ | ||
| β-Actin | 5′-TCCTGTGGCATCCATGAAACT-3′ | 340 |
| 5′-AACGCAGCTCAGTAACAGTC-3′ | ||
| RANKL (real- | 5′-TACTTTCGAGCGCAGATGGAT-3′ | 89 |
| time PCR) | 5′-ACCTGCGTTTTCATGGAGTCT-3′ | |
| IL-6 (real-time | 5′-ATGAACAACGATGATGCACTTG-3′ | 82 |
| PCR) | 5′-TATCCAGTTTGGTAGCATCCAT-3′ | |
| OPG (real-time | 5′-AAACAGCACTGCACAGTGAG-3′ | 108 |
| PCR) | 5′-ACACTGGGCTGCAATACACA-3′ | |
| GAPDH | 5′-AACTACATGGTCTACATGTTCCA-3′ | 63 |
| 5′-CCATTCTCGGCCTTGACTGT-3′ |
Western blot analysis.
Osteoblasts cultured with or without stimulants were washed with ice-cold PBS containing 10 μM of p-tosyl-l-lysine-chloromethyl ketone (TLCK) (Sigma) and dissolved in 50 mM concentration of Tris-HCl (pH 6.8), containing 2% Triton X-100, 10 μM TLCK, 6.25 mM NaF, 12.5 mM β-glycerophosphate, 12.5 mM p-nitrophenyl phosphate, 1.25 mM NaVO3, and a 1% protease inhibitor cocktails (Roche Molecular Biochemicals). The soluble fraction was collected by centrifugation at 16,000 × g for 5 min at 4°C, and the protein concentration was determined by using a BCA protein assay kit (Pierce Chemical, Rockford, Ill.). The cell extract (20 μg of protein) was denatured in sodium dodecyl sulfate (SDS) sample buffer, resolved by SDS-10% polyacrylamide gel electrophoresis (PAGE), and electrotransferred to a polyvinylidine fluoride membrane. The membrane was blocked with SeaBlock blocking buffer (Pirece Chemical) for 1 h and then incubated with primary antibodies (1:500 dilution in PBS containing 0.05% Tween 20 and 20% SeaBlock) overnight at 4°C. After a washing step, horseradish peroxidase-linked anti-rabbit IgG (diluted 1:1,000 in PBS containing 5% skimmed milk) was added to the membrane. Detection of proteins or phosphorylated proteins was performed by using SuperSignal West Dura Extended Duration Substrate (Pierce Chemical).
Ligand receptor precipitation.
Osteoblasts cutlured with or without stimulants were lysed by addition of lysis buffer (50 mM Tris buffer [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 μM TLCK, protease inhibitor cocktails). RANK-immobilized agarose beads were prepared by agitating 20 μg of mouse RANK/Fc with 20 μl of protein A/G-agarose overnight at 4°C. Cell lysates containing 1 mg of protein were precipitated with the RANK-immobilized agarose beads in lysis buffer overnight at 4°C (36). The RANKL-RANK precipitated materials were recovered by boiling in SDS sample buffer and subjected to SDS-PAGE and Western blotting.
AP-1 assay.
P. gingivalis-infected osteoblasts were scraped into ice-cold PBS containing 10 μM TLCK, 6.25 mM NaF, 12.5 mM β-glycerophosphate, 12.5 mM p-nitrophenyl phosphate, and 1.25 mM NaVO3. The nuclear extracts were prepared by using a nuclear extract kit (Active Motif, Carlsbad, Calif.). After centrifugation, the protein concentration in supernatants containing nuclear proteins was determined by using a BCA protein assay kit. AP-1 activity was determined with a Trans-AM AP-1 c-Fos enzyme-linked immunosorbent assay (ELISA) kit (Active Motif). An oligonucleotide containing an AP-1 binding site was attached to a 96-well plate. The active form of AP-1 contained in the nuclear extracts specifically binds to this oligonucleotide and can be revealed by incubation with antibodies by using ELISA with absorbance reading. In the present study, 10-μg portions of nuclear extracts were analyzed for AP-1 binding to the oligonucleotide by using anti-c-Fos antibody according to the manufacturer's instructions. The specificity for the assay was monitored by competition with free wild-type or mutated oligonucleotides according to the manufacturer's instructions.
Statistical analyses.
All data are expressed as the means ± the standard deviation. Statistical analyses were performed by using unpaired Student t test. Multiple comparisons were performed by using one-way analysis of variance and Sheffe's test with STAT View software (SAS Institute, Cary, N.C.).
RESULTS
Expression of RANKL in osteoblasts after P. gingivalis infection.
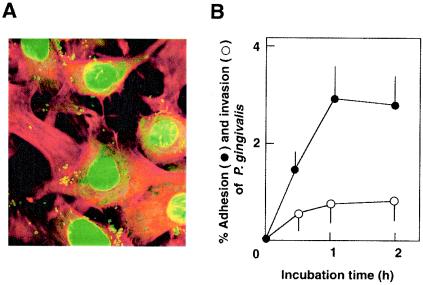
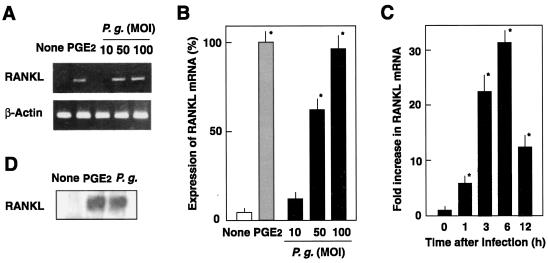
P. gingivalis is known to adhere to and invade mammalian epithelial cells (5, 26, 33, 35, 56). However, little information is available regarding the adhesion to and invasion of osteoblasts by P. gingivalis. Although P. gingivalis is a strict anaerobe, the organisms showed 95% viability in αMEM medium after 1 h of incubation in CO2 incubator (data not shown). A confocal microscopic study showed that P. gingivalis ATCC 33277 adhered to and invaded mouse primary osteoblasts (Fig. 1A). Maximum level of bacterial adhesion at an MOI of 100 was ca. 3%, and the invasion efficiency reached ca. 0.8% (Fig. 1B). Osteoblasts are known to produce RANKL, an osteoclastogenic cytokine, in response to several proinflammatory factors such as PGE2. We investigated whether osteoblasts produce RANKL and other osteotropic cytokines in response to infection with P. gingivalis. RT-PCR revealed an upregulation of RANKL mRNA expression, quantitated by real-time PCR analysis, in mouse primary osteoblasts infected with viable P. gingivalis ATCC 33277 (Fig. 2). The induction of RANKL mRNA expression in osteoblasts exhibited a dose-dependent relationship; a concomitant increase in RANKL mRNA expression, shown as the ratio of the number of P. gingivalis cells in the inoculum to the number of osteoblasts, was increased from 10:1 to 100:1 (Fig. 2B). Also, a time-dependent increase in RANKL mRNA expression is shown in Fig. 2C. The expression of RANKL mRNA in P. gingivalis-infected osteoblasts was 30-fold greater than that in uninfected controls at 6 h, after which the mRNA levels decreased, because the cells were cultured in serum-free medium. Our preliminary study showed that the total amounts of RANKL produced by osteoblasts were below the detection limit of conventional Western blotting. Therefore, we used a ligand precipitation technique to detect RANKL expression of osteoblasts. Western blotting after ligand precipitation demonstrated that osteoblasts stimulated with PGE2 produced RANKL protein with a molecular mass of ca. 40 kDa. Osteoblasts infected with P. gingivalis also produced a detectable amount of RANKL protein (Fig. 2D). We then examined whether other P. gingivalis strains that previously exhibited high virulence in a mouse model (54) induce RANKL responses in osteoblasts. Infection with the virulent strains ATCC 53977 and OMZ314 stimulated the expression of RANKL mRNA in osteoblasts, as strain ATCC 33277 did (data not shown). RT-PCR with primer pairs targeted to OPG, IL-1β, IL-6, or TNF-α revealed that osteoblasts infected with P. gingivalis ATCC 33277 produced IL-6 and OPG (Fig. 3), whereas IL-1β and TNF-α were not detected (data not shown). The mRNA level of OPG in infected osteoblasts was similar to those of control unstimulated osteoblasts.
FIG. 1.
Adhesion to and invasion of osteoblasts by P. gingivalis ATCC 33277. (A) Confocal observation of osteoblasts infected with P. gingivalis ATCC 33277. After 1 h of infection (MOI = 100), osteoblasts were fixed and stained with rhodamine-phalloidin and SYBR green. Filamentous actin and nuclei of osteoblasts visualized as red and green, respectively. P. gingivalis cells were visualized as small green dots. (B) Time course of bacterial adhesion and invasion. The percentage of adhesion or invasion was calculated as follows: (number of CFU adhering to or invading/total number of CFU in inoculum) × 100. Symbols: •, percentage of P. gingivalis released from infected osteoblasts; ○, percentage of invading P. gingivalis. The values shown are the means ± the standard deviation for triplicate assays.
FIG. 2.
Expression of RANKL mRNA by osteoblasts infected with P. gingivalis ATCC 33277. (A) RT-PCR analysis of RANKL. Mouse osteoblasts were cultured in the absence of the stimulant (None) or in the presence of PGE2 (2 μM) for 6 h at 37°C. Other osteoblast cultures were infected with P. gingivalis (P.g.) at MOIs of 10, 50, and 100 for 6 h at 37°C. Total RNA was extracted, reverse transcribed, and subjected to RT-PCR analysis for RANKL and β-actin. (B) Quantification of RANKL by real-time PCR. Cells were cultured in the absence of the stimulant (None) or in the presence of PGE2 for 6 h at 37°C. Other osteoblast cultures were infected with P. gingivalis (P.g.) at MOIs of 10, 50, or 100 for 6 h. RNA was extracted, reverse transcribed, and subjected to real-time PCR. Relative mRNA levels are expressed as percentages of the mRNA level in osteoblasts stimulated with PGE2. (C) Time course of RANKL expression. Osteoblasts were infected with P. gingivalis (MOI = 100) for 1, 3, 6, or 12 h at 37°C. Relative mRNA levels are expressed as the fold increases compared to the RANKL mRNA level in unstimulated cells. The values shown are means ± standard deviations for triplicate assays. An asterisk indicates that the P value was <0.05 in comparison with the RANKL mRNA level in cells cultured in the absence of the stimulant (None). (D) Ligand precipitation of RANKL expressed on the infected osteoblasts. Mouse osteoblasts were cultured in the absence of stimulant (None) or in the presence of PGE2 for 48 h in αMEM containing 10% fetal calf serum. Other cultures of osteoblasts were infected with P. gingivalis (P.g.; MOI = 100) and cultured for 48 h. Cell-associated RANKL was precipitated with RANK-immobilized beads and subjected to SDS-PAGE and Western blotting with anti-RANKL antibodies.
FIG. 3.
Expression of IL-6 and OPG mRNAs by osteoblasts infected with P. gingivalis ATCC 33277. (A) RT-PCR analysis of IL-6 and OPG. Osteoblasts were cultured in the absence of the stimulant (None) or in the presence of PGE2 for 6 h at 37°C. Other osteoblast culture was infected with P. gingivalis (P.g.) at an MOI of 100 for 6 h at 37°C. Total RNA was extracted and subjected to RT-PCR analysis for RANKL and β-actin. (B) Quantification of IL-6 and OPG mRNA by real-time PCR. Relative mRNA levels are expressed as percentages of the mRNA level in osteoblasts stimulated with PGE2 in triplicate assays. An asterisk indicates that the P value was <0.05 in comparison with the RANKL mRNA level in cells cultured in the absence of the stimulant (None).
P. gingivalis infection results in phosphorylation of c-Jun and activation of AP-1.
Various members of the MAPK family may modulate the expression of RANKL in infected osteoblasts. To investigate which MAPK pathway is involved in the expression of RANKL mRNA, we used inhibitors of several MAPKs and NF-κB. Osteoblasts were treated with wortmannin (an inhibitor of phosphatidylinositol 3-kinase, 1 μM), SB20350 (an inhibitor of p38 MAPK, 5 μM), and PD98059 (an inhibitor of MEK1/2, 5 μM) and then stimulated with P. gingivalis for 6 h. However, these inhibitors failed to reduce the RANKL responses in infected cells. Proteasome inhibitor was also used to explore whether activation of NF-κB is involved in the expression of RANKL. Peptide aldehyde ZLLLal reportedly inhibits the proteolytic activity of proteasome and activation of NF-κB, whereas since ZLLal does not inhibit activation of NF-κB; it was therefore used as the control (41). Although the pretreatment of osteoblasts with ZLLLal showed a slight decrease in the expression of RANKL mRNA, no significant difference between the effects of ZLLLal and ZLLal was observed (data not shown). Activation of NF-κB was also measured by the degradation of cytoplasmic IκB-α protein. Western blot analysis showed that no significant degradation of IκB-α occurred in osteoblasts during the 15- to 60-min period after infection with P. gingivalis (data not shown).
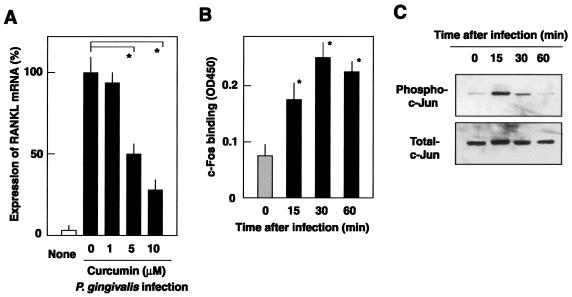
On the other hand, curcumin, a potent inhibitor of c-Jun N-terminal kinase (JNK) and AP-1 activation (7), prevented the upregulation of RANKL expression in infected osteoblasts (Fig. 4A), since the expression of RANKL mRNA was inhibited by 75% with 10 μM curcumin. At this concentration, curcumin did not affect cell viability. To assess the involvement of AP-1 activation in infected cells, AP-1 activity was probed by using an ELISA that detects c-Fos binding to AP-1 consensus oligonucleotide. As shown in Fig. 4B, infection with P. gingivalis appeared to stimulate the c-Fos binding and the binding kinetics suggested that maximal AP-1 activation was reached 30 min after infection. The specificity of this binding was confirmed by adding an excess amount of competing oligonucleotide (data not shown). Since activation of AP-1 is accompanied by activation of JNK and phosphorylation of c-Jun, the latter in P. gingivalis-infected osteoblasts was examined. Western blot analysis revealed that osteoblasts infected with P. gingivalis at an MOI of 100 exhibited an increase in phosphorylation of c-Jun (Fig. 4C), with maximal phosphorylation observed 15 min after infection.
FIG. 4.
Activation of AP-1 in osteoblasts infected with P. gingivalis. (A) Curcumin-mediated inhibition of RANKL mRNA expression in osteoblasts infected with P. gingivalis. Osteoblasts were preincubated for 1 h at 37°C with curcumin at concentrations of 0, 1, 5, and 10 μM and then infected with P. gingivalis ATCC 33277 (MOI = 100) for 6 h at 37°C. RANKL mRNA levels were determined by real-time PCR. The results are expressed as percentages of the RANKL mRNA levels in osteoblasts infected with P. gingivalis in the absence of the inhibitor. Values shown are means ± standard deviations for triplicate assays. An asterisk indicates that the P value was <0.05 in comparison with the RANKL mRNA level in the cells infected with P. gingivalis in the absence of curcumin. (B) AP-1 activation was evaluated by detecting c-Fos binding to the AP-1 consensus oligonucleotide. Osteoblasts were infected with P. gingivalis for 0, 15, 30, and 60 min at an MOI of 100. Nuclear extracts of the infected cells (10 μg) were subjected to Trans-AM AP-1 and c-Fos binding was measured by ELISA. The results are expressed as the mean optical density at 450 nm ± the standard deviation for triplicate assays. An asterisk indicates that the P value was <0.05 in comparison with the c-Fos binding level in the cells at 0 min. (C) Western blot analysis of phosphorylation of c-Jun. Osteoblasts were infected with P. gingivalis for 0, 15, 30, and 60 min at an MOI of 100. Phosphorylation status was analyzed by Western blotting with the antibodies against phospho-c-Jun and total c-Jun.
Involvement of gingipains in the induction of RANKL responses in osteoblasts.
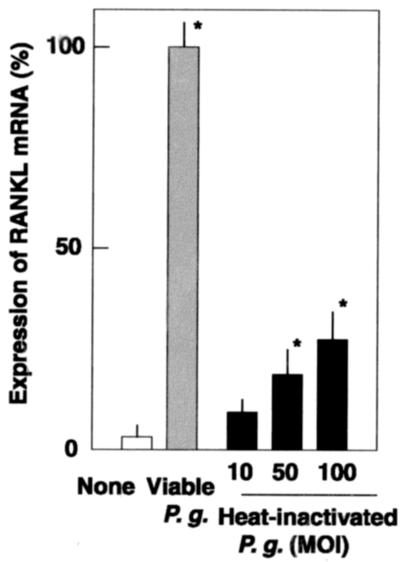
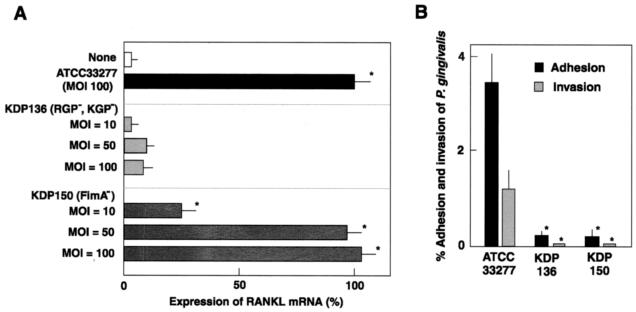
RANKL responses to P. gingivalis infection may result from the binding of surface components such as LPS to the innate receptors, TLRs, on osteoblasts. However, heat-inactivated P. gingivalis was not a potent stimulus of RANKL expression (Fig. 5). Viable P. gingivalis at a ratio of 100:1 routinely induced similar levels of RANKL mRNA expression with PGE2, whereas heat-inactivated P. gingivalis at the same ratio induced only 30% of the mRNA induced by viable P. gingivalis. To investigate P. gingivalis virulence factor(s) on RANKL responses in osteoblasts, we used isogenic mutant strains of P. gingivalis ATCC 33277 to compare the levels of RANKL mRNA expression in osteoblasts (Fig. 6). Although FimA-deficient mutant KDP150 induced RANKL responses similarly to the parent strain ATCC 33277, gingipain-deficient KDP136 failed to induce such expression, even at an MOI of 100. Since both KDP136 and ATCC 33277 produce LPS, the involvement of LPS was likely negligible. In addition, KDP136 and KDP150 have decreased adhesion and invasion frequencies in osteoblasts (Fig. 6B).
FIG. 5.
RANKL mRNA expression in osteoblasts treated with heat-inactivated P. gingivalis. Osteoblasts were cultured in the absence of the stimulant (None) and in the presence of live P. gingivalis ATCC 33277 (MOI of 100) (Viable P.g.) for 6 h at 37°C. Other osteoblast cultures were stimulated with heat-inactivated P. gingivalis ATCC 33277 (heat-inactivated P.g.) at an MOI of 10, 50, or 100 for 6 h. Total RNA was extracted, reverse transcribed, and subjected to real-time PCR. Transcription levels are expressed as percentages of the RANKL mRNA levels in osteoblasts stimulated with viable P. gingivalis. Values shown are means ± standard deviations for triplicate assays. An asterisk indicates that the P value was <0.05 in comparison with the RANKL mRNA level in cells cultured in the absence of the stimulant (None).
FIG. 6.
Infection with gingipain-deficient and FimA-deficient mutant strains of P. gingivalis. (A) Expression of RANKL mRNA in osteoblasts infected with gingipain-deficient and FimA-deficient mutants. Osteoblasts were cultured in the absence of the stimulant (None) for 6 h at 37°C. Other osteoblast cultures were infected with P. gingivalis strains ATCC 33277 (parent strain), KDP136 (deficient in both Arg-gingipains [RGP] and Lys-gingipain [KGP]), or KDP150 (deficient in FimA) for 6 h. Expression of RANKL mRNA was quantified by real-time PCR. The results are expressed as percentages of the RANKL mRNA levels in osteoblasts infected with strain ATCC 33277. Values shown are means ± the standard deviations for triplicate assays. An asterisk indicates that the P value was <0.05 in comparison with the RANKL mRNA level in cells cultured in the absence of the stimulant (None). (B) Adhesion and invasion of gingipain-deficient and FimA-deficient mutants of P. gingivalis. P. gingivalis strains were added to osteoblast cultures grown in 24-well culture plates (MOI = 100). To determine the numbers of adherent and invaded bacteria, the cultures were incubated for 1 h. Unattached bacteria were washed off with PBS, and the osteoblasts were disrupted by extensive pipetting with sterile water and then plated on blood agar plates. For the invasion assay, the attached bacteria were killed by 1 h of incubation with gentamicin and metronidazole. Values shown are means ± the standard deviations for triplicate assays. An asterisk indicates that the P value was <0.05 in comparison with the efficiency of the wild-type strain.
DISCUSSION
The present study demonstrated for the first time that viable P. gingivalis is a potent stimulant for the induction of RANKL production by osteoblasts. RANKL is a cytokine that stimulates the differentiation of osteoclasts and is an activating agent that leads to bone destruction (24, 49, 51, 58). P. gingivalis colonizes in periodontal pockets and spreads into deeper tissues, including the connective tissues (15, 27, 28, 30, 48). This bacterium is also associated with the periapical periodontitis and is isolated from root canal (18). These findings suggest that, in the advanced and destructive periodontitis and periapical pathosis, P. gingivalis may be present in the deeper periodontal tissues, including periodontal ligaments and teeth supporting bone tissues. Therefore, local RANKL production stimulated by P. gingivalis infection would influence alveolar bone destruction in progressive periodontal diseases.
Our RT-PCR findings revealed that P. gingivalis infection induced an upregulation of IL-6 mRNA in addition to RANKL (Fig. 3). IL-6 is involved in osteoclastogenesis, as well as stimulation of osteoclast formation and differentiation (49). However, the mRNA level of OPG, a decoy receptor of RANKL that strongly inhibits osteoclast formation induced by osteotropic factors (47, 49), was not influenced by infection (Fig. 3). In normal bone tissues, osteoblasts control osteoclastogenesis by expressing two functionally conflicting factors, RANKL and OPG. Thus, our findings suggest that infection with P. gingivalis changes the RANKL/OPG expression ratio in osteoblasts and stimulates osteoclastogenesis and bone destruction.
Recent studies have suggested that various bacterial components, such as LPS and peptidoglycans, bind to TLRs and lead to activation of NF-κB signaling pathways, followed by the release of inflammatory cytokines (2). Kikuchi et al. (21) reported that the LPS of gram-negative bacteria induces RANKL gene expression in osteoblasts via TLR4 and suggested that RANKL expression via the TLR pathway plays an important role in the pathogenesis of LPS-mediated bone disorders. Therefore, it may be possible that the LPS of P. gingivalis binds to TLRs and activates related signal pathways, although our experiments showed that heat-inactivated P. gingivalis was a poor stimulator of RANKL in osteoblasts (Fig. 5). Further, the isogenic mutant strain KDP136, which lacks both Arg- and Lys-gingipains but expresses LPS, was unable to induce RANKL expression (Fig. 6).
Sandros et al. (43) reported that infection with viable P. gingivalis strongly induced IL-1, IL-6, IL-8, and TNF-α production in epithelial cells. In that study, heat-inactivated P. gingivalis stimulated an IL-1 response similar to the one obtained for viable bacteria. Since the bacterial adhesion was not diminished by heat treatment, they suggested that P. gingivalis adhesion to epithelial cells was essential for cytokine responses. Other reports showed that infection with viable P. gingivalis induces MCP-1 expression in human epithelial cells (20, 34). In these studies, heat-inactivated bacteria did not induce MCP-1 response, suggesting that invasion of viable P. gingivalis is necessary for chemokine production by epithelial cells. On the other hand, our findings suggest that invasion of P. gingivalis is not related to RANKL production by osteoblasts. It is reported that FimA-deficient mutants of P. gingivalis exhibited decreased levels of adhesion and invasion (34, 35, 56), and we also found that our Fim-A deficent mutant (KDP150) has decreased adhesion and invasion frequencies in osteoblasts (Fig. 6B). However, KDP150 induced RANKL mRNA expression similarly to the parent strain (Fig. 6A), suggesting that bacterial invasion is not essential for RANKL production. P. gingivalis KDP136, which lacks both Arg- and Lys-gingipains, also showed reduced adhesion and invasion frequencies. It is reported that gingipain-deficient mutants of P. gingivalis displayed significant reduction in epithelial invasion (38). Since Arg-gingipain is a processing enzyme for the maturation and translocation of the precursors of FimA in P. gingivalis (8, 19, 25), gingipain-deficient mutants are unable to express mature FimA on the bacterial surface.
To investigate the signaling pathway that regulates RANKL response in osteoblasts infected with P. gingivalis, we examined specific inhibitors of various MAPKs, as well as those of NF-κB and AP-1. Among these, AP-1 activation was related to the RANKL expression. Additional experiments showed that phosphorylation of c-Jun occurred with activation of AP-1 in P. gingivalis-infected osteoblasts (Fig. 4). NF-κB signals did not seem to be involved in RANKL expression, which is consistent with a report showing no obvious NF-κB binding motifs in the promoter regions of the mouse RANKL gene (22). In this regard, RANKL expression in osteoblasts infected with Streptococcus pyogenes is regulated by p38 MAPK pathways (36), whereas upregulation of IL-6 expression in osteoblasts induced by Staphylococcus aureus is regulated by extracellular signal-regulated kinase 1/2 (ERK1/2) pathways (11). Therefore, signals stimulated by P. gingivalis infection may be different from those stimulated by S. aureus or S. pyogenes infections. Such differences in signal pathways may come from the difference in virulence factors of each bacterium.
Watanabe et al. (55) reported that infection with P. gingivalis activated JNK in human gingival epithelial cells and suggested that the activation of JNK is related to bacterial invasion, whereas NF-κB was not activated by P. gingivalis. These features of epithelial cells are similar to our findings in osteoblasts. They also showed that heat-inactivated P. gingivalis organisms did not stimulate JNK activity. Although further study is required, our data and their results are consistent with the concept that the activation of AP-1 is associated with the infection of viable P. gingivalis, since JNK is the upstream regulator of phosphorylation of c-Jun and activation of AP-1 (12).
Our study suggests that gingipains produced by P. gingivalis are likely responsible for the induction of RANKL response in osteoblasts. Recent studies have found that infection with P. gingivalis induces IL-8 and MCP-1 responses in human endothelial cells at the mRNA level (4, 20, 34). At the same time, these reports also suggested that a high level of P. gingivalis inoculum for an extended time period results in significant proteolysis of the secreted chemokines due to gingipains. Furthermore, Hintermann et al. (13) reported that P. gingivalis-infected oral keratinocytes showed proteolysis of focal contact components, i.e., focal adhesion kinase, adherens junction proteins such as catenins, and adhesion signaling molecules such as Src tyrosine kinase. Transmembrane proteins such as integrins also undergo proteolysis upon P. gingivalis infection (4), which is related to the reduced adhesion to the extracellular matrix of the infected cells. It is possible that proteolytic degradation of osteoblast focal adhesion kinase and integrins triggers stress-related responses in infected cells, including AP-1 activation.
The involvement of protease-activated receptors (PARs) is another possibility. Lourbakos et al. (29) showed that oral epithelial cells express PAR-1, -2, and -3, and PAR activation by P. gingivalis Arg-gingipain stimulated IL-6 secretion from the cells. Thee authors also demonstrated that trypsin is able to activate PARs on epithelial cells and suggested that Arg-gingipain activates PARs by cleavage after arginine residue in their extracellular domains. In addition, an RT-PCR study showed that rat primary osteoblasts express PAR-2 (1). Therefore, Arg- and Lys-gingipains of P. gingivalis likely stimulate RANKL expression through PAR signals in osteoblasts. Gingipains are secreted proteins, associated with extracellular vesicles, found on the bacterial cell surface and in the culture supernatant (8, 15, 17, 25, 40). Thus, these molecules would stimulate RANKL expression with or without direct interaction of P. gingivalis cells with osteoblasts. Further studies are needed to elucidate why and how gingipains stimulate RANKL response in osteoblasts, as well as the possible involvement of PARs.
In summary, we found that gingipains play important roles not only in the degradation of various extracellular and intercellular host proteins but also in the modulation of RANKL responses in osteoblasts infected with P. gingivalis.
Acknowledgments
This study was supported by grants-in-aid for scientific research B-13557181, B-15390645, and C-14657544 and the 21st century COE program from the Japan Society for the Promotion of Science.
Editor: J. N. Weiser
REFERENCES
- 1.Abraham, L. A., C. Chinni, A. L. Jenkins, A. Lourbakos, N. Ally, R. N. Pike, and E. J. Mackie. 2000. Expression of protease-activated receptor-2 by osteoblasts. Bone 26:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 3.Amano, A. 2003. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J. Periodontol. 74:90-96. [DOI] [PubMed] [Google Scholar]
- 4.Baba, A., T. Kadowaki, T. Asao, and K. Yamamoto. 2002. Roles for Arg- and Lys-gingipains in the disruption of cytokine responses and loss of viability of human endothelial cells by Porphyromonas gingivalis infection. Biol. Chem. 383:1223-1230. [DOI] [PubMed] [Google Scholar]
- 5.Belton, C. M., K. T. Izutsu, P. C. Goodwin, Y. Park, and R. J. Lamont. 1999. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell. Microbiol. 1:215-223. [DOI] [PubMed] [Google Scholar]
- 6.Chen, T., K. Nakayama, L. Belliveau, and M. J. Duncan. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect. Immun. 69:3048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y. R., and T. H. Tan. 1998. Inhibition of the c-Jun N-terminal kinase (JNK) signaling pathway by curcumin. Oncogene 17:173-178. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, M. A., J. Aduse-Opoku, and M. Rangarajan. 2001. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 12:192-216. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande, R. G., M. B. Khan, and C. A. Genco. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect. Immun. 66:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn, B. R., J. N. Burks, K. N. Seifert, and A. Progulske-Fox. 2000. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol. Lett. 187:139-144. [DOI] [PubMed] [Google Scholar]
- 11.Ellington, J. K., A. Elhofy, K. L. Bost, and M. C. Hudson. 2001. Involvement of mitogen-activated protein kinase pathways in Staphylococcus aureus invasion of normal osteoblasts. Infect. Immun. 69:5235-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrington, T. P., and G. L. Johnson. 1999. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 11:211-218. [DOI] [PubMed] [Google Scholar]
- 13.Hintermann, E., S. K. Haake, U. Christen, A. Sharabi, and V. Quaranta. 2002. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect. Immun. 70:5846-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobbie, S., L. M. Chen, R. J. Davis, and J. E. Galan. 1997. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J. Immunol. 159:5550-5559. [PubMed] [Google Scholar]
- 15.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 16.Iino, Y., and R. M. Hopps. 1984. The bone resorbing activities in tissue culture of lipopolysaccharides from the bacteria Actinobacillus actinomycetemcomitans, Bacteriodes gingivalis and Capnocytophaga ochracea isolated from human mouths. Arch. Oral Biol. 19:59-63. [DOI] [PubMed] [Google Scholar]
- 17.Imamura, T. 2003. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74:111-118. [DOI] [PubMed] [Google Scholar]
- 18.Jung, I. Y., B. K. Choi, K. Y. Kum, B. D. Roh, S. J. Lee, C. Y. Lee, and D. S. Park. 2000. Molecular epidemiology and association of putative pathogens in root canal infection. J. Endodontol. 26:599-604. [DOI] [PubMed] [Google Scholar]
- 19.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 20.Kang, I.-C., and H. K. Kuramitsu. 2002. Induction of monocyte chemoattractant protein-1 by Porphyromonas gingivalis in human endothelial cells. FEMS Immunol. Med. Microbiol. 34:311-317. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, T., T. Matsuguchi, N. Tsuboi, A. Mitani, S. Tanaka, M. Matsuoka, G. Yamamoto, T. Hishikawa, T. Noguchi, and Y. Yoshikai. 2001. Gene expresson of osteoclast differentiation factor is induced by lipopolysaccharide in mouse osteoblasts via Toll-like receptors. J. Immunol. 166:3574-3579. [DOI] [PubMed] [Google Scholar]
- 22.Kitazawa, R., S. Kitazawa, and S. Maeda. 1999. Promoter structure of mouse RANKL/TRANCE/OPGL/ODF gene. Biochem. Biophys. Acta 1445:134-141. [DOI] [PubMed] [Google Scholar]
- 23.Kong, Y. Y., U. Feige, I. Sarosi, B. Bolon, A. Tafuri, S. Morony, C. Capparelli, J. Li, R. Elliott, S. McCabe, T. Wong, G. Campagnuolo, E. Moran, E. R. Bogoch, G. Van, L. T. Nguyen, P. S. Ohashi, D. L. Lacey, E. Fish, W. J. Boyle, and J. M. Penninger. 1999. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature 402:304-309. [DOI] [PubMed] [Google Scholar]
- 24.Kong, Y. Y., H. Yoshida, I. Sarosi, H. I. Tan, E. Timms, C. Capparelli, S. Morony, A. J. Oliveira-dos-Santos, G. Van, A. Itie, W. Khoo, A. Wakeham, C. R. Dunstan, D. L. Lacey, T. W. Mak, W. J. Boyle, and J. M. Penninger. 1999. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397:315-323. [DOI] [PubMed] [Google Scholar]
- 25.Kontani, M., S. Kimura, I. Nakagawa, and S. Hamada. 1997. Adherence of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol. Microbiol. 24:1179-1187. [DOI] [PubMed] [Google Scholar]
- 26.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamont, R. J., and H. F. Jenkinson. 2000. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol. 15:341-349. [DOI] [PubMed] [Google Scholar]
- 29.Lourbakos, A., J. Potempa, J. Travis, M. R. D'Andrea, P. Andrade-Gordon, R. Santulli, E. J. Mackie, and R. N. Pike. 2001. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect. Immun. 69:5121-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manor, A., M. Lehendiger, A. Shiffer, and H. Tovel. 1984. Bacterial invasion of periodontal tissues in advanced periodontitis in humans. J. Periodontol. 55:567-573. [DOI] [PubMed] [Google Scholar]
- 31.Miller, S. J., E. G. Goldstein, M. J. Levine, and E. Hausmann. 1986. Modulation of bone metabolism by two chemically distinct lipopolysaccharide fractions from Bacteroides gingivalis. Infect. Immun. 51:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair, S. P., S. Meghji, M. Wilson, K. Reddi, P. White, and B. Henderson. 1996. Bacterially induced bone destruction: mechanisms and misconceptions. Infect. Immun. 64:2371-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa, I., A. Amano, M. Kuboniwa, T. Nakamura, S. Kawabata, and S. Hamada. 2002. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect. Immun. 70:277-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nassar, H., H. H. Chou, M. Khlgatian, F. C. Gibson III, T. E. Van Dyke, and C. A. Genco. 2002. Role of fimbriae and lysin-specific cysteine proteinase gingipain K in expression of interleukin-8 and monocyte chemoattractant protein in Porphyromonas gingivalis-infected endothelial cells. Infect. Immun. 70:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Njoroge, T., R. J. Genco, H. T. Sojar, N. Hamada, and C. A. Genco. 1997. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okahashi, N., A. Sakurai, I. Nakagawa, T. Fujiwara, S. Kawabata, A. Amano, and S. Hamada. 2003. Infection by Streptococcus pyogenes induces the receptor activator of NF-κB ligand expression in mouse osteoblastic cell. Infect. Immun. 71:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onoe, Y., C. Miyaura, T. Kaminakayashiki, Y. Nagai, K. Noguchi, Q. R. Chen, H. Seo, H. Ohta, S. Nozawa, I. Kudo, and T. Suda. 1996. IL-13 and IL-4 inhibit bone resorption by supressing cyclooxygenase-2-dependent prostaglandin synthesis in osteoblasts. J. Immunol. 156:758-764. [PubMed] [Google Scholar]
- 38.Park, Y., and R. J. Lamont. 1998. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect. Immun. 66:4777-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pieters, J. 2001. Evasion of host cell defense mechanisms by pathogenic bacteria. Curr. Opin. Immunol. 13:37-44. [DOI] [PubMed] [Google Scholar]
- 40.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000 24:153-192. [DOI] [PubMed] [Google Scholar]
- 41.Read, M. A., A. S. Nash, F. W. Luscinskas, V. J. Palombella, T. Maniatis, and T. Collins. 1995. The proteasome pathway is required for cytokine-induced endothelial-leukocyte adhesion molecule expression. Immunity 2:493-506. [DOI] [PubMed] [Google Scholar]
- 42.Saglie, F. R., A. Martany, and P. Camargo. 1988. Intragingival occurrence of Actinobacillus actinomycetemcomitans and Bacteroides gingivalis in active destructive periodontal lesions. J. Periodontol. 59:259-265. [DOI] [PubMed] [Google Scholar]
- 43.Sandros, J., C. Karlsson, D. F. Lappin, P. N. Madianos, D. F. Kinane, and P. N. Papapanou. 2000. Cytokine responses of oral epithelial cells to Porphyromonas gingivalis infection. J. Dent. Res. 79:1808-1814. [DOI] [PubMed] [Google Scholar]
- 44.Sandros, J. P. Papapanou, and G. Dahlen. 1993. Porphyromonas gingivalis invades oral epithelial cells in vitro. J. Periodontal Res. 28:219-226. [DOI] [PubMed] [Google Scholar]
- 45.Sansonetti, P. J. 2001. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks. FEMS Microbiol. Rev. 25:3-14. [DOI] [PubMed] [Google Scholar]
- 46.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 47.Simonet, W. S., D. L. Lacey, C. R. Dunstan, M. Kelly, M. S. Chang, R. Luthy, H. Q. Nguyen, S. Wooden, L. Bennett, T. Boone, G. Shimamoto, M. DeRose, R. Elliott, A. Colonbero, H. L. Tan, G. Trail, J. Sullivan, E. Davy, N. Bucay, L. Renshaw-Gegg, T. M. Huges, D. Hill, W. Pattison, P. Campbell, and J. W. Boyle. 1997. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89:309-319. [DOI] [PubMed] [Google Scholar]
- 48.Slots, J., and M. A. Listgarten. 1988. Bacteroides gingivalis, Bacteroides intermedius, and Actinobacillus actinomycetemcomitans in human periodontal diseases. J. Clin. Periodontol. 15:85-93. [DOI] [PubMed] [Google Scholar]
- 49.Suda, T., N. Takahashi, N. Udagawa, E. Jimi, M. T. Gillespie, and T. J. Martin. 1999. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocrine Rev. 20:345-357. [DOI] [PubMed] [Google Scholar]
- 50.Takada, H., J. Mihara, I. Morisaki, and S. Hamada. 1991. Induction of interleukin-1 and -6 in human gingival fibroblasts cultures stimulated with Bacteroides lipopolysaccharide. Infect. Immun. 59:295-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takayanagi, H., H. Iizuka, T. Juji, T. Nakagawa, A. Yamamoto, T. Miyazaki, Y. Koshihara, H. Oda, K. Nakamura, and S. Tanaka. 2000. Involvement of receptor activator of nuclear factor κB ligand/osteoclast differentiation factor in osteoclastogenesis from synoviocytes in rheumatoid arthritis. Arthritis Rheum. 43:259-269. [DOI] [PubMed] [Google Scholar]
- 52.Tang, P., C. L. Sutherland, M. R. Gold, and B. B. Finlay. 1998. Listeria monocytogenes invasion of epithelial cells requires the MEK-1/ERK-2 mitogen-activated protein kinase pathway. Infect. Immun. 66:1106-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueshima, J., M. Shoji, D. B. Ratnayake, K. Abe, S. Yoshida, K. Ymamoto, and K. Nakayama. 2003. Purification, gene cloning, gene expression, and muants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun. 71:1170-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Winkelhoff, A. J., B. J. Appelmelk, N. Kippuw, and J. Graaff. 1993. K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol. Immunol. 8:259-265. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe, K., O. Yilmaz, S. F. Nakhjiri, C. M. Belton, and R. J. Lamont. 2001. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect. Immun. 69:6731-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaji, Y., T. Kubota, K. Sasaguri, S. Sato, Y. Suzuki, H. Kumada, and T. Umemoto. 1995. Inflammatory cytokine gene expression in human periodontal ligament fibroblasts stimulated with bacterial lipopolysaccharides. Infect. Immun. 63:3576-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuda, H., N. Shima, N. Nakagawa, K. Yamaguchi, M. Kinoshita, S. Mochizuki, A. Tomoyasu, K. Yano, M. Goto, A. Murakami, E. Tsuda, T. Morinaga, K. Higashio, N. Udagawa, N. Takahashi, and T. Suda. 1998. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA 95:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]