SUMMARY
The expanding repertoire of genetically encoded biosensors constructed from variants of Aequorea victoria green fluorescent protein (GFP) enable the imaging of a variety of intracellular biochemical processes. To facilitate the imaging of multiple biosensors in a single cell, we undertook the development of a dimerization-dependent red fluorescent protein (ddRFP) that provides an alternative strategy for biosensor construction. An extensive process of rational engineering and directed protein evolution led to the discovery of a ddRFP with a Kd of 33 μM and a 10-fold increase in fluorescence upon heterodimer formation. We demonstrate that the dimerization-dependent fluorescence of ddRFP can be used for detection of a protein-protein interaction in vitro, imaging of the reversible Ca2+-dependent association of calmodulin and M13 in live cells, and imaging of caspase-3 activity during apoptosis.
INTRODUCTION
Fluorescent protein (FP)-based biosensors that report on biochemical events in live cells typically operate on one of three principles: the modulation of Förster resonance energy transfer (FRET) efficiency between two FPs of different hues (Miyawaki et al., 1997), the modulation of the fluorescent intensity of a single FP through analyte-induced changes in the chromophore environment (Nakai et al., 2001; Nagai et al., 2001), or the fluorogenic reconstitution of a single FP (also known as complementation) from two polypeptide chains (Ghosh et al., 2000; Kerppola, 2009). Each of these design principles is associated with distinct advantages. For example, FRET-based biosensors benefit from being inherently ratiometric in their response, and single FP-based biosensors often provide better signal-to-noise because of larger intensiometric responses. FP complementation uniquely generates an irreversible response and can be applied for detection of interacting proteins in proteome-wide screens because of negligible background signal (Magliery et al., 2005; Remy and Michnick, 2004).
A shortcoming shared by these three design strategies is that they are most commonly implemented with engineered variants of Aequorea victoria green FP (GFP) and only rarely implemented with Anthozoan-derived red FPs (RFP) (Matz et al., 1999). Although there have been reports of orange FP-RFP FRET pairs (Piljic and Schultz, 2008; Ouyang et al., 2010), single RFP-based Ca2+ indicators (Zhao et al., 2011), and complementation of split RFPs (Jach et al., 2006; Fan et al., 2008), such examples are relatively few in number. Among the contributing factors to the discrepancy between the popularity of GFP and RFP variants in biosensing applications is the poor sensitized emission of RFPs when used as FRET acceptors (Ai et al., 2008) and the challenge of engineering circularly permuted RFPs for use in single FP-based biosensors (Carlson et al., 2010).
We reasoned that a new approach, which better harnesses the inherent properties of RFPs, could serve to stimulate development of a broader selection of RFP-based biosensors. One property that differentiates GFP from naturally occurring RFPs is the obligate tetrameric structure of the latter (Baird et al., 2000). For many applications, the tetrameric structure is undesirable, and substantial effort has gone into engineering dimeric and monomeric FPs (Campbell et al., 2002). However, it is also apparent that the oligomeric structure of RFPs helps stabilize the chromophore in a conformation that favors bright fluorescence. This role is apparent when one compares the quantum yields (Φ) of dimeric (i.e., dimer2 and dTomato; both 0.69) and monomeric (i.e., mRFP1 and mCherry; 0.25 and 0.22, respectively) variants of Discosoma species RFP (DsRed) (Matz et al., 1999; Campbell et al., 2002; Shaner et al., 2004). This dimerization-dependent brightness suggested to us a new strategy for the creation of RFP-based biosensors.
Our idea was to develop an alternative biosensing system by engineering the oligomeric structure of RFPs to create a low-affinity heterodimer that exhibits bright red fluorescence in the associated state and dim fluorescence in the dissociated state. To engineer such a system, we expected that there would be three major hurdles to overcome: first, developing a heterodimeric RFP; second, engineering high contrast between the associated and dissociated states; and third, decreasing the affinity such that the dimer partners were not fully associated at intracellular concentrations typically used for FP imaging (~1–50 μM) (Martin et al., 2005). This manuscript describes our efforts to overcome these challenges and produce a fluorogenic RFP heterodimer that has proven useful in a series of representative biosensing applications.
RESULTS AND DISCUSSION
Engineering and Characterization of ddRFP
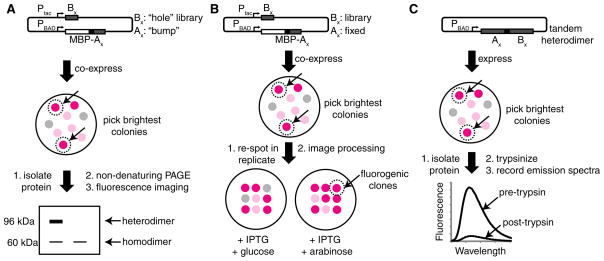
Our strategy for generating a fluorogenic RFP heterodimer was inspired by the well-established practice of introducing interface breaking mutations to produce a monomeric FP from a dimeric precursor (Campbell et al., 2002; Ai et al., 2006). Accordingly, we created a monomeric and dimly fluorescent RFP variant (designated A) by introducing the H162K and A164R substitutions into dTomato (Shaner et al., 2004). We then set out to “rescue” dimer formation and fluorescence by creating a dTomato-derived partner (designated B) with surface modifications that complemented the modifications to A. To achieve this goal, we turned to the first of the three distinct library screening strategies (Figures 1 and 2; Figure S1 available online) that we implemented during this extensive process of molecular evolution (Table S1). For clarity, we append a subscript number to A and B to designate the generation of a given variant. Thus, dTomato-H162K/A164R is designated as A0.1, and the most extensively optimized version of A is designated A1.
Figure 1. Overview of the Screening Procedures Used to Identify Heterodimeric and Fluorogenic RFPs.
(A) Electrophoretic mobility shift screen for heterodimeric pairs of RFPs.
(B) Replicaplating screen for fluorogenic and heterodimeric pairs of RFPs.
(C) Tandem heterodimer proteolysis-based screen. See also Figure S1 and Table S1.
Figure 2. Characterization of Homo- and Heterodimeric Structure by Gel Electrophoresis.
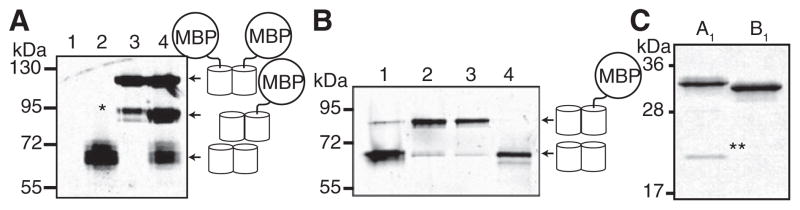
The contrast of each whole image has been adjusted to emphasize weak bands.
(A) Validation of the SDS-PAGE electrophoretic mobility shift screen for pairs of protein variants that exhibit heterodimeric character. Proteins were loaded without boiling and were detected by imaging the red fluorescence. Lane 1, uninduced culture; lane 2, dTomato; lane 3, MBP-dTomato; lane 4, coexpressed dTomato and MBP-dTomato. Asterisk (*) indicates a species resulting from proteolysis of one dTomato-MBP linker.
(B) Red fluorescence image of a gel used for PAGE analysis of four representative variants analyzed during screening (numbered 1–4).
(C) Coomassie-stained gel of purified A1 and B1 variants. Samples were boiled in sample buffer prior to electrophoresis. The double asterisk (**) indicates the 19 kDa product of chromophore hydrolysis routinely observed for RFPs (Gross et al., 2000). The 11 kDa fragment was not observed.
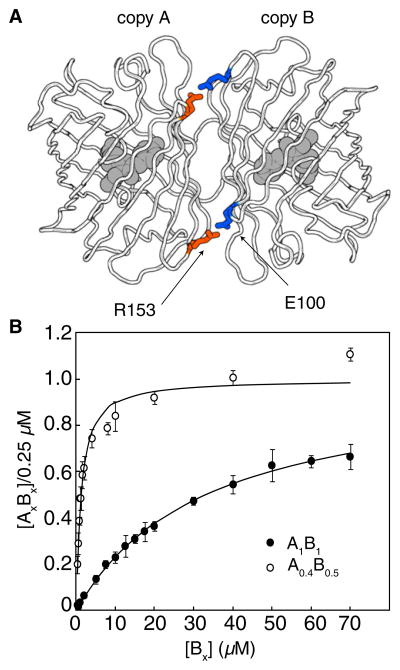
To identify a heterodimeric partner for A0.1, we developed a two-step assay that involved an image-based screen for red fluorescence of Escherichia coli colonies followed by an electrophoretic mobility shift analysis of protein extracts (Figure 1A). A dual-expression plasmid with two different promoters (Ptac and PBAD) was used to express A0.1 as a fusion with the 43 kDa E. coli maltose binding protein (MBP), and a dTomato-R149X/H162X/Y192X (where X = all 20 amino acids) gene library. These three residues were targeted because of their proximity to H162 and A164 across the AC dimer interface of tetrameric DsRed (Yarbrough et al., 2001). Colonies with the brightest red fluorescence were picked and cultured, and the protein extracts were analyzed by polyacrylamide gel electrophoresis (PAGE) under conditions that preserve high-affinity oligomeric interactions of FPs (Baird et al., 2000; Campbell et al., 2002). Heterodimeric proteins migrated slower than B0.1 homodimers during PAGE analysis (Figure 2A). Screening of ~20,000 colonies for fluorescence, and approximately 100 proteins by gel-shift, led to the identification of a pool of 25 variants (B0.1, pool) that exhibited some heterodimeric character. However, even the best of these variants existed as a mixture of the homodimeric and heterodimeric states (Figure 2B). In an effort to engineer variants that existed primarily as heterodimers, we flipped the identity of two residues (R153 and E100) that form a salt-bridge interaction across the interface (Figure 3A and Figure S2). Specifically, we introduced R153E into A0.1 to produce A0.2 and E100R into a B0.1, pool to create B0.2, pool. The R153E mutation had the additional benefit of increasing the amount of soluble protein produced for the A variant. We then created a second-generation library from the template of B0.2, pool by randomizing positions E160 and H162. Screening of this library led to the identification of B0.3, which was equivalent to dTomato-E100R/R149L/E160H/H162F/Y192G and, by PAGE analysis, predominantly formed a heterodimeric complex with A0.2. To further diminish the residual homodimeric character, we removed three residues that contribute to the dTomato interface (F224, L225, and Y226) (Yarbrough et al., 2001) from the C termini of A0.2 and B0.3. The resulting proteins, A0.3 and B0.4, exclusively formed a heterodimer, but exhibited reduced fluorescent brightness.
Figure 3. Engineering Heterodimeric ddRFPs.
(A) Position of the two symmetry-related salt bridges in dTomato.
(B) Saturation-binding curves for A0.4 with B0.5 and A1 with B1. Kd values are 1.1 and 33 μM, respectively. Error bars are ± standard deviation for three independent experiments. See also Figure S2.
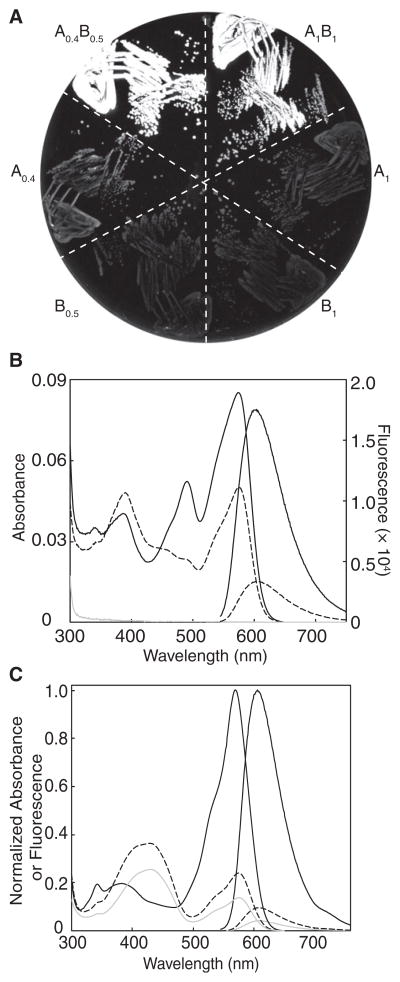
We next turned to a colony-based assay in which replica plating was used to identify noncovalent AB pairs that were brightly fluorescent when both proteins were expressed, but dimly fluorescent when only one protein was expressed (Figure 1B and Figure S1). Briefly, we used the dual-expression plasmid to express one randomly mutated partner under the isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible Ptac promoter, and one genetically fixed partner under the L-arabinose-inducible PBAD promoter. For library screening, transformed E. coli was plated on media permissive for expression of both partners (i.e., supplemented with IPTG and L-arabinose). The brightest colonies were picked and manually arrayed onto two new plates: one with media for expression of both partners and one with media for expression of only the variable partner (i.e., supplemented with IPTG and glucose). The red fluorescence of each plate was digitally imaged and then processed in pairs to determine the fluorogenic contrast for each colony. We performed several rounds of screening in which B0.4-derived variants were randomized and A0.3 was held constant, followed by several rounds with A0.3-derived libraries and B0.5 held constant. This procedure led to our first fluorogenic dimerization-dependent RFP composed of A0.4 and B0.5 (ddRFP-A0.4B0.5). Characterization of ddRFP-A0.4B0.5 revealed that the FP partners exhibited a 5-fold increase in fluorescence upon dimerization both in E. coli (Figure 4A) and in vitro (Figure 4B), and a Kd of 1.1 μM (Figure 3B).
Figure 4. Characterization of ddRFPs.
(A) Fluorescence image of E. coli expressing various proteins discussed in the text. A0.4B0.5 and A1B1 are tandem constructs.
(B) Absorbance and fluorescence emission spectra for A0.4 (dashed line; 10 μM), B0.5 (gray line; 10 μM), and the equimolar mixture of A0.4 and B0.5 (black line; 10 μM each).
(C) Absorbance and fluorescence spectra of A1 (gray line; 20 μM) and tandem ddRFP-A1B1 (black line; 20 μM) before (solid line) and after (dashed line) treatment with trypsin. B1 has baseline absorbance and fluorescence. See also Figure S3.
To further improve the fluorogenic contrast of ddRFP-A0.4B0.5, we developed a third screening procedure that involved the use of proteolyzable tandem heterodimers (Figure 1C) composed of A and B joined by a 23-residue cleavable linker. Libraries of randomly mutated heterodimer genes were expressed in E. coli, and the brightest colonies were picked and cultured. The fluorescence of protein extracts was measured before and after treatment with trypsin, and clones with the greatest contrast were carried on to subsequent rounds. Application of this strategy for several rounds provided only moderate improvements in contrast, so we next attempted to rationally engineer improved contrast.
Because the fluorescence contrast of ddRFP is limited by the residual fluorescence of A in the absence of B (Figure 4B), we reasoned that manipulating the intraprotein interactions of the chromophore in partner A could lead to variants with improved contrast. Accordingly, we introduced the S146A substitution into A0.4 to produce A0.5, which had one less hydrogen bond donor in close proximity to the phenolate oxygen of the A chromophore (Yarbrough et al., 2001). From a library in which positions E144 and A145 of A0.5 were randomized, we picked the clones that were most dimly fluorescent when expressed in the absence of B. The resulting A0.6, pool was fused in tandem to a B0.5-derived gene library in which three residues (Y192, Y194, and H222) in close proximity to residues 144–146 of the dimer partner were randomized (Yarbrough et al., 2001). To decrease the Kd of the heterodimer, we also reverted E100R and installed R153E in the B partner (Figure 3A) such that two favorable Arg-Glu electrostatic interactions were replaced with unfavorable Glu-Glu contacts. The library was screened as tandem heterodimers, and clones with substantially improved contrast were identified. The A and B partners of the clone that exhibited the highest contrast were designated A1 and B1.
Purified A1 and B1 exhibit a 10-fold increase in fluorescence upon dimerization (Figure 4C) and a Kd of 33 μM (Figure 3B). As with ddRFP-A0.4B0.5, interaction with B1 results in an instantaneous increase in the fluorescent brightness of A1. A1 alone has Φ = 0.026 and an extinction coefficient (ε) = 11,800 M−1·cm−1 at pH 7.4, and these values increase to 0.074 and 48,300 M−1·cm−1, respectively, upon formation of a heterodimer with B1. No visible wavelength absorbance or fluorescence could be detected for B1, even after several weeks at 4°C. Furthermore, boiling of purified B1 resulted in none of the backbone hydrolysis that is characteristic of DsRed-type proteins (including A1) with mature chromophores (Figure 2C) (Gross et al., 2000). We observed no limits to the solubility of A1 and B1; expression in E. coli gave yields of 10–80 mg/l. Both A1 and the A1B1 complex exhibit relatively high fluorescence pKas (9.0 and 7.4, respectively), and, thus, their fluorescence is sensitive to physiologically relevant changes in pH (Figures S3A–S3C). Size exclusion chromatography confirmed that both A1 and B1 are monomeric in isolation but form a heterodimer when mixed (Figure S3D). Interestingly, we noted that A1B1 undergoes a reversible reaction with β-mercaptoethanol (possibly a nucleophilic addition to the chromophore; Dong et al., 2008) that results in loss of the red fluorescent state and the formation of a new species that absorbs at 402 nm and emits at 466 nm (Figures S3E and S3F).
The convoluted evolutionary pathway that led to ddRFP-A1B1 reflects the practical challenge of simultaneously optimizing at least five distinct, and possibly conflicting, properties: heterodimeric structure, minimal homodimeric structure, high-contrast fluorogenesis, high brightness in the associated state, and high Kd. Accordingly, ddRFP-A1B1 represents a compromise, but it is the best compromise that we have identified to date.
In Vitro and Live Cell Applications of ddRFP
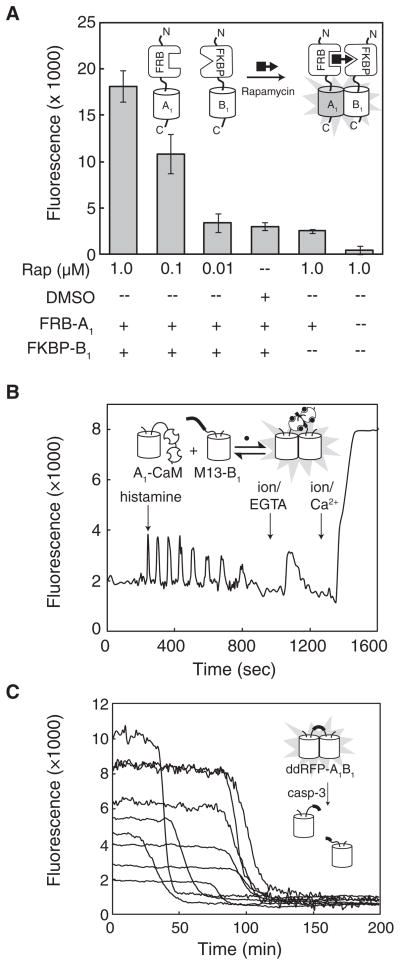
We next investigated the utility of ddRFP-A1B1 in representative in vitro and live cell imaging applications. To determine whether ddRFP-A1B1 could be used to detect protein-protein interactions in vitro, we turned to the rapamycin-dependent interaction of FK506-binding protein (FKBP) and the FKBP-rapamycin-binding domain (FRB) (Chen et al., 1995). We fused A1 and B1 to the C termini of FRB and FKBP with a 43-residue unstructured linker to produce FRB-A1 and FKBP-B1. These fusions were expressed in E. coli, purified, and mixed at a concentration (0.5 μM each) that is well below the Kd for ddRFP-A1B1. As shown in Figure 5A, the addition of rapamycin gave a dose-dependent increase in fluorescence. This result indicates that ddRFP-A1B1 is useful for detection of protein-protein interactions with dissociation constants well below the ddRFP-A1B1 Kd.
Figure 5. Fluorescence Detection of In Vitro Chemically Induced Dimerization and Imaging ddRFP-A1B1 in Live Cells.
(A) Fluorescence of various combinations of FRB-A1 (0.5 mM), FKBP-B1 (0.5 μM), rapamycin, and dimethyl sulfoxide (DMSO) vehicle control. Fluorescence intensity is the mean integrated emission intensities for three independent experiments (± standard deviation) and corrected for background (i.e., assay buffer alone).
(B) Imaging of Ca2+ dynamics with A1-CaM and M13-B1 in HeLa cells. Transfected HeLa cells were treated with histamine, followed by EGTA/ionomycin (abbreviated as ion) and Ca2+/ionomycin.
(C) Imaging of caspase-3 activity with a ddRFP-A1B1 tandem heterodimer with a DEVD substrate in the linker region. Transfected HeLa cells were treated with TNF-α (t = 0) to initiate apoptosis, and red fluorescence was imaged as a function of time.
To determine whether ddRFP-A1B1 could be employed to detect reversible protein-protein interactions in live cells, we borrowed the so-called “split-cameleon” strategy in which the Ca2+-induced interaction of calmodulin (CaM) and the M13 peptide brings two FPs into close proximity (Miyawaki et al., 1999). Accordingly, mammalian expression plasmids were prepared in which A1 was fused to the N terminus of CaM (A1-CaM) and B1 was fused to the C terminus of the M13 peptide (M13-B1). HeLa cells were cotransfected with both plasmids and imaged at 24 hr after transfection. Upon histamine treatment, we observed oscillations in the red fluorescence intensity (Figure 5B) that were consistent with the expected changes in intracellular Ca2+ concentration. Other FP-based Ca2+ indicators based on FRET (Miyawaki et al., 1999) and single FP Ca2+ biosensors (Zhao et al., 2011) exhibit Ca2+ spikes of duration similar to those observed with A1-CaM and M13-B1, indicating that, at least on the timescale of seconds, heterodimer association and dissociation is not a rate-limiting step. In situ calibration of the dynamic range was accomplished by treating cells with ionomycin/EGTA to deplete Ca2+ and ionomycin/Ca2+ to saturate CaM. These experiments revealed that the dynamic range for A1-CaM plus M13-B1 in cells was 5.7 ± 1.3 (N = 16). This dynamic range is approximately half of the in vitro range for A1 and B1 alone, indicating that the partners are approximately 50% associated in the absence of Ca2+ and at the intracellular protein concentrations of these experiments. Accordingly, we estimate that the intracellular concentrations of A1-CaM and M13-B1 in those cells bright enough to be imaged, is approximately 30 μM.
For a third representative application of ddRFP-A1B1, we envisioned a strategy for protease activity sensing in which A1 and B1 were fused in tandem with the well-characterized Asp-Glu-Val-Asp (DEVD) caspase-3 substrate sequence in the linker region (Xu et al., 1998). As in our trypsinolysis assay, cleavage of the substrate should result in a loss of fluorescence as the partners dissociate, provided the concentration is well below the Kd. HeLa cells were transfected with a plasmid harboring the DEVD tandem heterodimer, treated with tumor necrosis factor-α (TNF-α), and imaged through time. We reliably observed a decrease in red fluorescence intensity (dynamic range of 8.6 ± 4.3; N = 9), just prior to the cell shrinkage, and blebbing associated with the end stages of apoptosis (Figure 5C). The rate of the transition from the bright to dim state was comparable to the rate previously observed for a FRET-based biosensor of caspase-3 activity (Ai et al., 2008), indicating that heterodimer association and dissociation is not a rate-limiting step on the relatively slow timescale (minutes) of this experiment. Overall, these results confirm that the affinity of A1 and B1 is sufficiently low to allow protease detection and imply that the tandem heterodimer is sufficiently bright for imaging at intracellular concentrations well below the Kd of 33 μM.
Homology Modeling of ddRFP-A1B1
To gain a better understanding of how the 35 amino acid substitutions in ddRFP-A1B1 (relative to dTomato) contribute to its heterodimeric character and fluorogenesis, we created a homology model of ddRFP-A1B1 (Figure 6). Inspection of this model reveals that A1 and B1 have six and four substitutions, respectively, at positions with side chains directed toward the interior of the β-barrel. In A1, just three of these residues are in close proximity to the chromophore: the conservative I161L substitution and the nonconservative S146A and K163G substitutions that are likely destabilizing the phenolate form of the chromophore because of loss of a hydrogen bond and a favorable electrostatic interaction, respectively. In contrast, the cavity that would normally harbor the chromophore of B1 has been dramatically remodeled with multiple nonconservative substitutions that include K70E, Y120C, I161S, and E215G. The remodeled chromophore environment is consistent with the conclusion, supported by spectroscopic and biochemical evidence, that B1 does not form a chromophore.
Figure 6. Structural Model of ddRFP-A1B1 Based on the AC Dimer of DsRed.
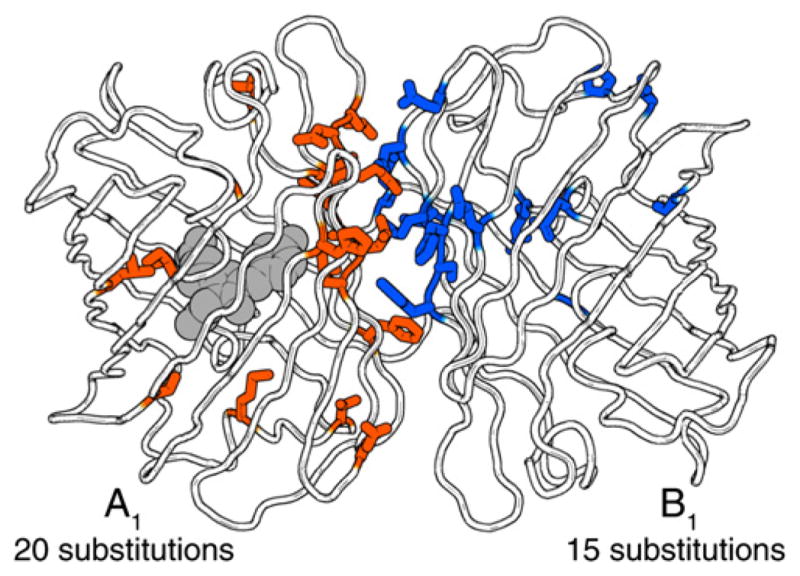
The amino acid side chains for all mutations in ddRFP-A1B1 are shown in stick representation (Yarbrough et al., 2001). Figure was generated with PyMOL (http://www.pymol.org). See also Figure S4.
The homology model of ddRFP-A1B1 also reveals that shape complementarity and electrostatic interactions across the dimer interface (Figure S4C) have been substantially modified relative to dTomato (which preserves the interface of DsRed) (Figure S4D). Indeed, of the 19 substitutions in A1 and B1 with side chains directed to the exterior of the β-barrel, 15 are located in the interface. The homology model suggests that a principal determinant of the heterodimeric character is a new “bump-and-hole” interaction across the dimer interface. In the X-ray crystal structure of DsRed, the large side chain of R149 is positioned close to the small side chain of A164 on the dimer partner (Figure S4B) (Yarbrough et al., 2001). In ddRFP-A1B1, the A164R substitution of A1 is a “bump” that was introduced to destabilize the dTomato homodimer as a result of steric clashes and electrostatic repulsion with R149 of B1. In our model, the “hole” on the surface of B1 is formed by reorientation of the side chain of R149M of B1 such that it is directed away from A164R of A1 and is filling an adjacent cavity created by Y194C of B1 (Figure S4A).
Two factors contribute to the fluorogenesis of ddRFP-A1B1: pKa modulation and Φ modulation. At physiological pH and in the absence of B1, the fully formed chromophore of A1 exists primarily in the protonated and nonfluorescent state because of its pKa of 9.0. Upon interaction with B1, the chromophore environment of A1 is modified such that the pKa is lowered to 7.4 and the equilibrium shifts toward the anionic fluorescent state. A similar mechanism has been proposed for a system in which a single domain antibody (nanobody) modulates the fluorescence of GFP (Kirchhofer et al., 2010). An additional contribution to fluorogenesis is the increase in Φ from 0.026 in A1 to 0.074 in the A1B1 complex. We speculate that, in free A1, the chromophore spends more time in noncoplanar conformations that favor excited-state deactivation by processes other than fluorescence (Shu et al., 2006). Upon formation of the A1B1 complex, a modified chromophore environment stabilizes a coplanar conformation of the chromophore, and Φ increases. Further structural studies should shed light on the molecular details of the interface interactions that contribute to fluorogenesis.
Outlook for ddRFP-A1B1 as a Biosensing Strategy
With the advent of ddRFP-A1B1, a new entry has been added to the short list of strategies (i.e., FRET, insertion in a single FP, and FP complementation) that can be used for creation of FP-based biosensors. We have demonstrated the practicality of ddRFP-A1B1 by using it to detect protein-protein interactions, image a reversible Ca2+-dependent protein-protein interaction, and image protease activity. The primary advantage of ddRFP-A1B1 relative to other strategies is that it provides a general means of creating red intensiometric biosensors with a reversible response. Potential disadvantages include the pH sensitivity and relative low brightness of ddRFP-A1B1, though we note that the brightness is comparable to the commercially available DsRed-monomer (Strongin et al., 2007). Relative to the three well-established biosensing strategies, dimerization of A1 and B1 to form ddRFP-A1B1 is most closely analogous to FP complementation (Ghosh et al., 2000; Kerppola, 2009). A key difference between single FP complementation and ddRFP-A1B1 heterodimerization is that, although the fragments of a split FP are non-fluorescent, A1 retains 10% of the brightness of the complex. This residual fluorescence is likely to complicate efforts to use ddRFP-A1B1 in high-throughput screens for identification of interacting proteins (Jackrel et al., 2010).
Future efforts to improve ddRFP-A1B1 should provide variants with decreased affinity and increased contrast. Until such variants become available, the most appropriate applications of ddRFP-A1B1 are as an alternative to intermolecular FRET and for protease activity sensing with tandem heterodimers. As we have done with the split cameleon-type Ca2+ biosensor and the caspase-3 biosensor, it should be relatively straightforward to convert existing FRET-based biosensors into ddRFP-A1B1-based biosensors. In these contexts, ddRFP-A1B1 will complement the existing repertoire of FP-based biosensing strategies, because it enables creation of spectrally distinct biosensors with an intensiometric and reversible red fluorescent signal.
SIGNIFICANCE
By employing a series of three protein library screening strategies, we have engineered ddRFP-A1B1, a fluorogenic DsRed-derived heterodimer with an associated state that is 10-fold brighter than the dissociated state. The interaction has been engineered to have a Kd of 33 μM, and, therefore, the heterodimers can exist primarily in the dissociated state at cytoplasmic concentrations sufficient for live cell imaging. Homology modeling has revealed that this heterodimeric character results from a new “bump-and-hole” interaction that is not present in the homodimeric parent protein. The utility of ddRFP-A1B1 has been demonstrated in three applications, including detection of a protein-protein interaction in vitro, imaging of a Ca2+-dependent protein-protein interaction in live cells, and imaging of caspase-3-dependent proteolysis during apoptosis.
EXPERIMENTAL PROCEDURES
Library Screening by Gel Shift
A custom bacterial dual-expression plasmid, containing a XhoI/HindIII poly-linker under control of Ptac and an EcoRI/BglII polylinker under control of PBAD, was used for the rescue-of-binding assay. A silent mutation was introduced into the gene encoding maltose-binding protein (MBP) to remove an internal BglII site. The fusion of MBP and dTomato H162K/A164R with an intervening KpnI site was ligated into the PBAD polylinker. Random mutations at sites R149, H162, and Y192 were introduced into a second dTomato gene by overlap extension PCR (Bessette et al., 2003), and the gene product was ligated into the Ptac polylinker. Colonies of E. coli were screened with 535/50 nm excitation and visualization through 600 nm long-pass goggles. Bright colonies were picked into Luria-Bertani (LB) media supplemented with 100 μg/ml ampicillin, 0.1 mM IPTG, and 0.02% L-arabinose and were incubated overnight. Proteins were purified using Nickel-NTA, electrophoresed by SDS-PAGE, and analyzed for in-gel fluorescence using digital imaging with appropriate filters (excitation 535/50 nm; emission 630/60 nm).
Library Screening for Fluorogenesis
Error-prone PCR was performed using Taq polymerase as previously described (Campbell et al., 2002). E. coli was transformed with gene libraries in the Ptac and PBAD polylinker sites of the dual-expression plasmid. Bright colonies were picked and replica plated in a regular grid pattern on one plate supplemented with 1 mM IPTG and 0.2% L-arabinose and on one plate supplemented with 1 mM IPTG and 10 mM glucose. Following overnight incubation at 37°C, the red fluorescence of both plates was imaged, and the ratios of intensities on the L-arabinose versus glucose plates for each colony were determined using image processing macros. Clones with the highest ratios were pooled and used in the next round.
Library Screening with Tandem Heterodimers
Tandem heterodimers were initially constructed by a three-part ligation strategy, which provided a chimera of the form A-linker-B in XhoI/HindIII sites of pBAD/His B, where the linker was a 23-residue sequence that included a KpnI site. Bright colonies were picked and cultured in LB with ampicillin and L-arabinose (0.02%) overnight. Crude protein extracts were treated with trypsin at approximately 10 μg/ml for 30 min, and emission spectra were acquired using a 96-well microplate reader. Contrast ratios were calculated as the integrated emission peak area of the nontrypsinized extract divided by the peak area of the trypsinized extract.
Protein Production and Characterization
To obtain purified proteins, the genes for tandem ddRFP variants and monomeric partners were subcloned into pBAD/His B and were used to transform E. coli. Cultures at an optical density of 0.5–0.7 were induced with 0.02% L-arabinose and allowed to grow for a further 12–16 hr. Soluble proteins were purified from cleared lysates by Nickel-NTA and were dialyzed into 5 mM Tris-Cl and 100 mM NaCl (pH 7.5). pH sensitivity measurements were performed by incubating purified proteins in buffers of desired pH and by acquiring emission spectra with a 96-well microplate reader. Spectra represented in this manuscript were recorded with a DU-800 UV-visible spectrophotometer (Beckman) or a QuantaMaster spectrofluorimeter (Photon Technology International, Inc). The alkaline chromophore denaturation method was used to determine ε values (Ward, 1981). dTomato was used as the reference for Φ determination.
To determine the Kd of the purified recombinant ddRFP partners, an increasing amount of nonfluorescent B1 was mixed with a fixed amount of A1 to generate A1B1 complexes in 5 mM Tris-HCl and 100 mM NaCl (pH 7.4). The integrated fluorescence emission peaks recorded as a function of B1 concentration were used to generate saturation binding curves. Experimental data were fit using a modified Langmuir isotherm to account for ligand depletion.
FKBP-FRB Interaction
The genes encoding FRB and FKBP were fused to A1 and B1, respectively, to generate FRB-A1 and FKBP-B1 fusions. For both constructs, the linker sequence corresponded to residues 95 to 135 of phage λ repressor (accession NP_040628). The fusion proteins were expressed from pBAD/His B and soluble proteins were purified as described for the tandem ddRFP variants. Protein concentrations were determined by the BCA method.
Live Cell Imaging
The A1 and B1 copies in the DEVD tandem heterodimer were linked by the sequence GHGTHSTHSHSSHTASHDEVDGA. The gene sequence encoding the nuclear exclusion sequence (LALKLAGLDIGS) was appended to the 3′ end of the gene (Wen et al., 1995). For the split cameleon-type biosensors, the genes encoding A1 and CaM, as well as M13 and B1, were joined by the sequence GHGTGSTGSGSSTASSEDMA. The genes encoding the tandem heterodimers and both the CaM and M13 fusions were ligated into the XhoI/HindIII sites of pcDNA3.1(+) (Invitrogen). HeLa cells were transfected using the Turbofect transfection reagent (Fermentas) and were imaged on an inverted Nikon Eclipse Ti microscope equipped with a 200W metal halide lamp (PRIOR Lumen) and a QuantEM: 512SC 16-bit cooled CCD camera (Photometrics). For Ca2+ imaging, cells were imaged in HEPES-buffered Hank’s balanced salt solution and were consecutively treated with histamine (25 μM), EGTA (3 mM) with ionomycin (5 μM), and 10 mM CaCl2 with 5 μM ion-omycin. For caspase-3 imaging, apoptosis was initiated by treatment with 100 ng/ml TNF-α.
Homology Modeling
The homology model for ddRFP-A1B1 (Figure 6 and Figure S4) was constructed using the Rosetta fixed backbone design protocol (Kuhlman et al., 2003) with the A and C chains of PDB ID 1G7K (Yarbrough et al., 2001). Regions with high-energy substitutions were energy minimized with localized backbone flexibility, as implemented in Rosetta’s kinematic closure loop modeling protocol (Mandell et al., 2009). Small changes in the backbone conformation were sufficient to allow the side chains to assume a lower energy conformation and avoid steric clashes.
Supplementary Material
Acknowledgments
We thank the University of Alberta MBSU for technical assistance and Andreas Ibraheem for the dual-expression plasmid. Work in the Campbell laboratory is made possible by grants from CIHR and NSERC. S.C.A. is supported by Ph.D. scholarships from NSERC and Alberta Ingenuity. R.E.C. holds a Tier II Canada Research Chair in Bioanalytical Chemistry.
Footnotes
ACCESSION NUMBERS
The GenBank accession numbers for the ddRFP-A1 and ddRFP-B1 gene sequences reported in this article are JN381545 and JN381546, respectively.
Supplemental Information includes four figures and one table and can be found with this article online at doi:10.1016/j.chembiol.2012.01.006.
References
- Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessette PH, Mena MA, Nguyen AW, Daugherty PS. Construction of designed protein libraries using gene assembly mutagenesis. Methods Mol Biol. 2003;231:29–37. doi: 10.1385/1-59259-395-X:29. [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson HJ, Cotton DW, Campbell RE. Circularly permuted monomeric red fluorescent proteins with new termini in the beta-sheet. Protein Sci. 2010;19:1490–1499. doi: 10.1002/pro.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Abulwerdi F, Baldridge A, Kowalik J, Solntsev KM, Tolbert LM. Isomerization in fluorescent protein chromophores involves addition/elimination. J Am Chem Soc. 2008;130:14096–14098. doi: 10.1021/ja803416h. [DOI] [PubMed] [Google Scholar]
- Fan JY, Cui ZQ, Wei HP, Zhang ZP, Zhou YF, Wang YP, Zhang XE. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem Biophys Res Commun. 2008;367:47–53. doi: 10.1016/j.bbrc.2007.12.101. [DOI] [PubMed] [Google Scholar]
- Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J Am Chem Soc. 2000;122:5658–5659. doi: 10.1021/ja994421w. [DOI] [Google Scholar]
- Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jach G, Pesch M, Richter K, Frings S, Uhrig JF. An improved mRFP1 adds red to bimolecular fluorescence complementation. Nat Methods. 2006;3:597–600. doi: 10.1038/nmeth901. [DOI] [PubMed] [Google Scholar]
- Jackrel ME, Cortajarena AL, Liu TY, Regan L. Screening libraries to identify proteins with desired binding activities using a split-GFP reassembly assay. ACS Chem Biol. 2010;5:553–562. doi: 10.1021/cb900272j. [DOI] [PubMed] [Google Scholar]
- Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci USA. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci USA. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Ouyang M, Huang H, Shaner NC, Remacle AG, Shiryaev SA, Strongin AY, Tsien RY, Wang Y. Simultaneous visualization of protumorigenic Src and MT1-MMP activities with fluorescence resonance energy transfer. Cancer Res. 2010;70:2204–2212. doi: 10.1158/0008-5472.CAN-09-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piljic A, Schultz C. Simultaneous recording of multiple cellular events by FRET. ACS Chem Biol. 2008;3:156–160. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick SW. Regulation of apoptosis by the Ft1 protein, a new modulator of protein kinase B/Akt. Mol Cell Biol. 2004;24:1493–1504. doi: 10.1128/MCB.24.4.1493-1504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- Strongin DE, Bevis B, Khuong N, Downing ME, Strack RL, Sundaram K, Glick BS, Keenan RJ. Structural rearrangements near the chromophore influence the maturation speed and brightness of DsRed variants. Protein Eng Des Sel. 2007;20:525–534. doi: 10.1093/protein/gzm046. [DOI] [PubMed] [Google Scholar]
- Ward WW. Properties of the Coelenterate green-fluorescent proteins. In: De Luca M, McElroy DW, editors. Bioluminescence and Chemiluminescence: Basic Chemistry and Analytical applications. New York: Academic Press; 1981. pp. 235–242. [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Xu X, Gerard AL, Huang BC, Anderson DC, Payan DG, Luo Y. Detection of programmed cell death using fluorescence energy transfer. Nucleic Acids Res. 1998;26:2034–2035. doi: 10.1093/nar/26.8.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc Natl Acad Sci USA. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.