Abstract
We identified cytolethal distending toxin and its gene (cdt) in 17 of 340 non-O157 Shiga toxin-producing Escherichia coli (STEC) strains (serotypes O73:H18, O91:H21, O113:H21, and O153:H18), all of which were eae negative. cdt is either chromosomal and homologous to cdt-V (serotypes O73:H18, O91:H21, and O113:H21) or plasmidborne and identical to cdt-III (serotype O153:H18). Among eae-negative STEC, cdt was associated with disease (P = 0.003).
Cytolethal distending toxins (CDTs) are tripartite toxins encoded by three adjacent or slightly overlapping genes (cdtA, cdtB, and cdtC) (15, 19, 20, 22) and found in multiple pathogens (5). All cdt genes are required for arrest of the eukaryotic cell cycle in the G1 or G2 phase (5, 15, 19, 20), which characteristically distends cell morphology and eventually causes cell death (5, 14, 15, 19). Recent studies suggest that within the CDT holotoxin, CdtB is the enzymatically active subunit (14, 15), which damages (through DNase I-like activity) (6, 14) host cell DNA (14), thereby triggering the DNA damage checkpoint response (5) that arrests the cell cycle (5, 6, 14, 15). The CdtA and CdtC polypeptides constitute the heterodimeric subunit (15, 16) that is required for CDT binding to target cells (16) and for intracellular delivery of CdtB (15). Five different Escherichia coli cdt sequences have been reported. Four of the sequences encode CDT-I (22), CDT-II (20), CDT-III (19), and CDT-IV (24) in enteropathogenic E. coli (EPEC), E. coli causing extraintestinal infections, and animal pathogenic E. coli. We recently described the fifth cdt allelic cluster in sorbitol-fermenting (SF) Shiga toxin (Stx)-producing E. coli (STEC) O157:H− strain 493/89 (11); we have designated the fifth sequence (following nomenclature recommendations) (5) cdt-V. CDT-V is produced by most (87%) SF STEC O157:H− isolates and a subset (6%) of STEC O157:H7 isolates (11).
In this study we used PCR to detect all presently known E. coli cdt alleles to determine the frequency and distribution of cdt among non-O157 STEC human isolates. We sequenced cdt genes from STEC isolates of different serotypes, determined their genomic locations, and investigated their expression characteristics through the use of a cell culture assay. The cell culture assay was also applied (in parallel with PCR analyses) to identify potential producers of CDT encoded by an allele or alleles undetectable by the PCR strategies employed.
The frequency and distribution of cdt among non-O157 STEC isolates.
A total of 340 non-O157 STEC (130 eae-positive and 210 eae-negative) isolates belonging to 100 different serotypes (Table 1) and isolated from patients with hemolytic-uremic syndrome (HUS) (n = 66) or uncomplicated diarrhea (n = 206) and from asymptomatic carriers (n = 68) (7, 8) were subjected to PCRs with primers targeting cdt-I, cdt-II, cdt-III, cdt-IV, and cdt-V (Table 2). cdt-I, cdt-II, and cdt-IV were identified in none of these strains (Table 1). A total of 3 and 14 strains tested positive for cdt-III and cdt-V, respectively (Table 1); all others tested negative. The cdt-positive STEC isolates belonged to serotype O153:H18 (cdt-III+ strains) and to serotypes O73:H18, O91:H21, and O113:H21 (cdt-V+ strains) (Table 1). Within these serotypes, all or most isolates possessed cdt (Table 1). Each of the 17 cdt-positive STEC isolates was eae negative (Table 1) and originated from patients (3 of the patients had HUS and 14 had uncomplicated diarrhea) (Table 3). Among the 210 eae-negative STEC isolates investigated, cdt was significantly more frequent in those from patients with HUS (3 of 7) and in those from patients with diarrhea (14 of 138) than in those from asymptomatic carriers (0 of 65) (P = 0.0018 and P = 0.0057, respectively; Fisher's exact test). The difference in the level of association of cdt with HUS versus that with diarrhea was not significant (P = 0.073).
TABLE 1.
Distribution of cdt-I, cdt-II, cdt-III, cdt-IV, and cdt-V alleles among 340 non-O157 STEC strainsa
| Serotype(s)b | Total no. of isolates | No. of isolates PCR positive for:
|
|||||
|---|---|---|---|---|---|---|---|
| eae | cdt-I | cdt-II | cdt-III | cdt-IV | cdt-Vc | ||
| O26:H11/H−d | 40 | 40 | 0 | 0 | 0 | 0 | 0 |
| O103:H2/H− | 23 | 23 | 0 | 0 | 0 | 0 | 0 |
| O111:H8/H− | 14 | 14 | 0 | 0 | 0 | 0 | 0 |
| O145:H28/H25/H− | 23 | 23 | 0 | 0 | 0 | 0 | 0 |
| Otherse | 30 | 30 | 0 | 0 | 0 | 0 | 0 |
| O8:H2/H10/H14/H19/H28/H− | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| O73:H18 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| O91:H14/H− | 14 | 0 | 0 | 0 | 0 | 0 | 0 |
| O91:H21 | 15 | 0 | 0 | 0 | 0 | 0 | 9 |
| O113:H4 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| O113:H21 | 6 | 0 | 0 | 0 | 0 | 0 | 4 |
| O128:H2/H12/H− | 13 | 0 | 0 | 0 | 0 | 0 | 0 |
| O146:H8/H10/H21/H28/H31/ H51/H− | 20 | 0 | 0 | 0 | 0 | 0 | 0 |
| O153:H18 | 3 | 0 | 0 | 0 | 3 | 0 | 0 |
| O174:H2/H8/H21/H− | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| Othersf | 105 | 0 | 0 | 0 | 0 | 0 | 0 |
The strains were isolated at the Institute of Hygiene and Microbiology, University of Würzburg, Würzburg, at the Institute of Hygiene, University Hospital Münster, Münster, and at the Robert Koch Institute, Wernigerode, Germany, between 1996 and 2002 and serotyped as described previously (8, 21).
Only serotypes comprising cdt-positive strains and serogroups or serotypes with more than 10 isolates are listed in the table; the other serotypes are collectively called “Others.”
All 14 strains positive for cdt-V produced amplicons of expected sizes in each of the three PCRs for the detection of cdt-VA, cdt-VB, and cdt-VC (Table 2).
H−, nonmotile.
Serotypes (number of isolates, where more than one was used, in parentheses): O4:H−, O6:H− (2), O8:H2 (4), O11:H2, O25:H− (2), O55:H6, O84:H4/H6/H− (4), O92:H33, O118:H16/H− (3), O119:H2, O121:H10/H19 (2), O129:H−, O136:H−, O156:H−, O165:H−, Orough:H10 (Orough, autoagglutinable strain) (2), ONT:H11 (ONT, O antigen, nontypeable) (2).
Serotypes (number of isolates, where more than one was used, in parentheses): O3:H2/H10 (2), O4:H−, O5:H− (6), O6:H− (5), O15:H21, O22:H8 (6), O23:H19, O31:H−, O40:H8 (5), O41:H−, O62:H−, O68:H4, O74:H−, O75:H8, O76:H19 (8), O78:H− (3), O84:H4, O86:H− (2), O92:H−, O104:H16/H21 (3), O112:H2/H− (4), O115:H10 (2), O136:H2, O152:H4, O176:H−, O178:H19/H− (6), O181:H16/H49 (6), Orough:H2/H11/H19/H21/H45/H− (18), ONT:H2/H4/H10/H14/H18/H19/H21/H32/H− (15).
TABLE 2.
PCR primers, conditions, and control strains used in this study
| Primer | Sequence | Target | PCR conditions (°C, s)a
|
PCR product (bp) | Source or reference | Positive controlb (reference) | ||
|---|---|---|---|---|---|---|---|---|
| Denaturing | Annealing | Extension | ||||||
| cdtI-fc | 5′-TGG TGA GAA TCG GAA CTG-3′ | cdt-IA | 94, 30 | 51, 60 | 72, 60 | 418 | This study | 6468/62 (22) |
| cdtI-rc | 5′-CAT TCC ATC AGG TTT GTC-3′ | |||||||
| cdtII-fd | 5′-AAT CCC TAT CCC TGA ACC-3′ | cdt-IIA | 94, 30 | 52, 60 | 72, 60 | 542 | This study | 9142/88 (20) |
| cdtII-rd | 5′-GTT CTA TTG GCT GTG GTG-3′ | |||||||
| cdtIII-f | 5′-AAACAGGACGGTAATAATGACTAATA-3′ | cdt-III | 94, 30 | 54, 60 | 72, 180e | 2,230 | 4 | 1404 (19) |
| cdtIII-r | 5′-GTGATCTCCTTCCATGAAAATATAGT-3′ | Complete sequence | ||||||
| CDT-IVs | 5′-CCTGATGGTTCAGGAGGCTGGTTC-3′ | cdt-IVB | 94, 60 | 55, 60 | 72, 60e | 350 | 24 | M375/95 (this study) |
| CDT-IVas | 5′-TTGCTCCAGAATCTATACCT-3′ | |||||||
| c338f | 5′-AGC ATT AAA TAA AAG CAC GA-3′ | cdt-VA | 94, 30 | 52, 60 | 72, 60 | 1,329 | 11 | 493/89 (11) |
| c2135r | 5′-TAC TTG CTG TGG TCT GCT AT-3′ | |||||||
| c1309f | 5′-AGC ACC CGC AGT ATC TTT GA-3′ | cdt-VB | 94, 30 | 52, 60 | 72, 60 | 1,363 | 11 | 493/89 (11) |
| c2166r | 5′-AGC CTC TTT TAT CGT CTG GA-3′ | |||||||
| P105 | 5′-GTC AAC GAA CAT TAG ATT AT-3′ | cdt-VC | 94, 30 | 49, 60 | 72, 60 | 748 | 11 | 493/89 (11) |
| c2767r | 5′-ATG GTC ATG CTT TGT TAT AT-3′ | |||||||
| c338f | 5′-AGC ATT AAA TAA AAG CAC GA-3′ | cdt-V | 94, 30 | 51, 60 | 72, 180e | 2,449 | 11 | 493/89 (11) |
| c2767r | 5′-ATG GTC ATG CTT TGT TAT AT-3′ | Complete sequence | ||||||
PCRs were performed with 25-μl volumes as previously described (8) and included 30 cycles followed by a final extension of 5 min at 72°C unless otherwise stated.
The E. coli control strains 6468/62 (serotype O86:H34, allele cdt-I+), 9142/88 (O128:H−, cdt-II+), and 1404 (O78:H?, cdt-III+) were provided by D. A. Scott (University of Maryland School of Medicine, Baltimore, Md.), C. L. Pickett (University of Kentucky Medical Center, Lexington, Ky.) and E. Oswald (Ecole Nationale Veterinaire, Toulouse, France), respectively. E. coli strain M375/95 (O75, cdt-IV+) was isolated from a patient with a urinary tract infection (M. Bielaszewska, unpublished data), and the presence of cdt-IVB was verified by sequencing. E. coli strain C600 was a negative control in all PCRs.
The primers for the detection of cdt-IA were designed from the published cdt-I sequence (GenBank accession number U03293) (22).
The primers for the detection of cdt-IIA were designed from the published cdt-II sequence (GenBank accession number U04208) (20).
The final extension was performed at 72°C for 10 min.
TABLE 3.
Serotypes, disease associations, cdt alleles, and CDT titers of CDT+ STEC strains
| Strain/yr of isolation | Serotype | Disease associationa | cdt alleleb | CDT titer (CHO cells)d |
|---|---|---|---|---|
| 2996/96 | O73:H18 | HUS | cdt-Vc | 2-4 |
| 1745/97 | O91:H21 | D | cdt-V | 4 |
| 3332/99 | O91:H21 | HUS | cdt-V | 4 |
| 6605/00 | O91:H21 | D | cdt-V | 4 |
| 4292/01 | O91:H21 | D | cdt-V | 2-4 |
| 9101/01 | O91:H21 | D | cdt-V | 4 |
| 9108/01 | O91:H21 | D | cdt-V | 2-4 |
| 9282/01 | O91:H21 | D | cdt-Vc | 2-4 |
| 1269/02 | O91:H21 | D | cdt-V | 4-8 |
| 2596/02 | O91:H21 | HUS | cdt-V | 4 |
| 691/99 | O113:H21 | D | cdt-V | 16 |
| 6090/00 | O113:H21 | D | cdt-V | 32-64 |
| 2896/01 | O113:H21 | D | cdt-V | 64 |
| 5249/01 | O113:H21 | D | cdt-Vc | 16-32 |
| 5107/00 | O153:H18 | D | cdt-III | 4-8 |
| 7272/02 | O153:H18 | D | cdt-III | 4-8 |
| 9063/02 | O153:H18 | D | cdt-IIIc | 8 |
HUS, hemolytic uremic syndrome (defined as microangiopathic hemolytic anemia, thrombocytopenia, and renal insufficiency [7]); D, watery diarrhea.
As detected by PCR.
Confirmed by sequence analysis.
The reciprocal of the highest dilution of a culture filtrate that distended 50% of CHO cells after 5 days of incubation. CDT titers or titer ranges obtained for each strain in three independent experiments are shown; no (where only one titer value is given) or only nonsignificant (1 dilution) titer differences were observed at the repeat testings.
cdt sequencing.
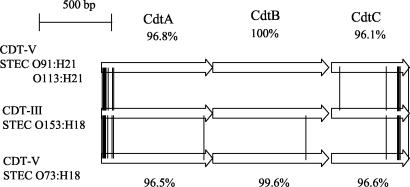
cdt gene clusters were amplified with primer pair c338f and c2767r or primer pair cdtIII-f and cdtIII-r (Table 2) and sequenced using an automated ABI Prism 3100 Avant Genetic Analyzer and an ABI Prism BigDye Terminator Ready Reaction cycle sequencing kit (Perkin-Elmer Applied Biosystems, Weiterstadt, Germany). Sequences were analyzed with DNASIS software (Hitachi Software). Homology searches were performed using the EMBL-GenBank database. cdt clusters from STEC isolates of serotypes O73:H18 (strain 2996/96), O91:H21 (strain 9282/01), O113:H21 (strain 5249/01), and O153:H18 (strain 9063/02) consisted of three open reading frames (ORFs) of 777, 810, and 546 bp each, which encode CdtA, CdtB, and CdtC, respectively. The cdt sequences from STEC O91:H21 strain 9282/01 (GenBank accession number AY365042) and STEC O113:H21 strain 5249/01 (GenBank accession number AY365043) were 100% identical and differed by 1, 2, and 2 nucleotides in their cdtA, cdtB, and cdtC genes, respectively, from the corresponding genes of STEC O73:H18 strain 2996/96 (GenBank accession number AY365045). The sequences of cdtA, cdtB, and cdtC and the deduced amino acid sequences of the corresponding proteins from these three strains were identical or closely related (≥98.8% homology) to those published for cdt-V and CDT-V, respectively, from SF STEC O157:H− strain 493/89 (GenBank accession number AJ508930) (11). In contrast, the sequences of each of the three cdt genes and of the corresponding proteins from STEC O153:H18 strain 9063/02 (GenBank accession number AY365044) were 100% identical to those published for cdt-III and CDT-III, respectively, from necrotoxigenic E. coli (NTEC) O15:H21 strain S5 (GenBank accession number U89305) (19). CDT-III from the STEC O153:H18 isolate differed from CDT-V from STEC O91:H21, O113:H21, and O73:H18 isolates by 15, 15, and 16 amino acid residues, respectively (Fig. 1). With a single exception, the amino acid differences were confined to CdtA and CdtC whereas CdtB proteins were conserved (Fig. 1).
FIG. 1.
Amino acid sequence differences between CDT-III from STEC O153:H18 (strain 9063/02) (accession number AY365044) and CDT-V from STEC O91:H21 (strain 9282/01) (accession number AY365042), O113:H21 (strain 5249/01) (accession number AY365043), and O73:H18 (strain 2996/96) (accession number AY365045). The exchanges in the amino acids are marked by connecting lines between the three CDT ORFs. The amino acid sequence homologies between CDT-III from STEC O153:H18 and CDT-V from STEC O91:H21 and O113:H21 (the latter two share 100% identical CDT-V sequences) are given above the three depicted ORFs. The amino acid sequence homologies between CDT-III from STEC O153:H18 and CDT-V from STEC O73:H18 are given below the three ORFs.
Genomic location of cdt.
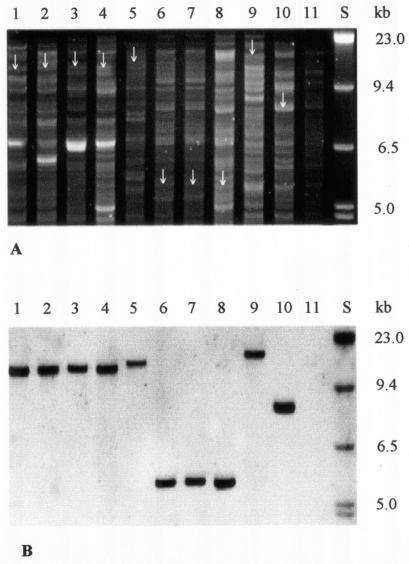
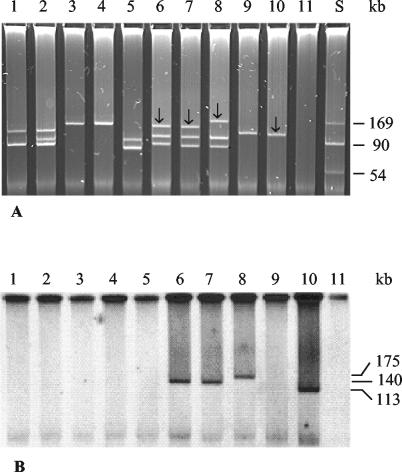
To determine the cdt location in non-O157 STEC isolates, EcoRV-digested genomic DNA and isolated plasmids (23) were electrophoretically separated (Fig. 2A, Fig. 3A) and hybridized with digoxigenin-labeled cdtB probe derived from strain 493/89 by PCR with primers c1309f and c2166r (Table 2). The probe was detected using a DIG luminescent detection kit (Roche Molecular Biochemicals, Mannheim, Germany). SF STEC O157:H− strain 493/89 (which contains cdt-V on the chromosome) (11) and NTEC O78:H? strain 1404 (which harbors cdt-III on a large Vir plasmid) (19) were used as controls. A single fragment of genomic DNA between 5.5 and 18 kb in size hybridized with the cdtB probe in each of five representative STEC isolates harboring cdt-V (Fig. 2B, lanes 1 to 5), in each of the three STEC isolates harboring cdt-III (Fig. 2B, lanes 6 to 8), and in both control strains (Fig. 2B, lanes 9 and 10). In contrast, only large (140- or 175-kb) plasmids of the three cdt-III-positive STEC O153:H18 isolates (Fig. 3B, lanes 6 to 8) and the large plasmid of strain 1404 (Fig. 3B, lane 10) hybridized with the cdtB probe. No signals were elicited from large plasmids of any of the five STEC isolates harboring cdt-V (Fig. 3B, lanes 1 to 5) or of strain 493/89 (Fig. 3B, lane 9). Taken together, these data demonstrate that cdt-V in STEC isolates O73:H18, O91:H21, and O113:H21 is chromosomal whereas cdt-III in STEC O153:H18 is on a large plasmid.
FIG.2.
Agarose gel electrophoresis (A) and hybridization with the cdtB probe (B) of EcoRV-digested genomic DNA from cdt-harboring STEC strains and controls. Lanes S, molecular mass markers (digoxigenin-labeled DNA Molecular Weight Marker II; Roche Biochemicals). In lanes 1 to 8, the following STEC strains (serotypes and cdt alleles are shown in parentheses) are displayed: lane 1, strain 9282/01 (serotype O91:H21, allele cdt-V); lane 2, 2596/02 (O91:H21, cdt-V); lane 3, 5249/01 (O113:H21, cdt-V); lane 4, 2896/01 (O113:H21, cdt-V); lane 5, 2996/96 (O73:H18, cdt-V); lane 6, 9063/02 (O153:H18, cdt-III); lane 7, 7272/02 (O153:H18, cdt-III); lane 8, 5107/00 (O153:H18, cdt-III). Lanes 9 to 11 contain control strains as follows: lane 9, strain 493/89 (SF STEC serotype O157:H−, allele cdt-V); lane 10, 1404 (NTEC O78:H?, cdt-III); lane 11, E. coli C600 (negative control). Arrows in panel A indicate the EcoRV fragments that hybridized with the cdtB probe.
FIG. 3.
Plasmid profiles (A) and plasmid hybridization with the cdtB probe (B) of cdt-harboring STEC strains and controls. Lane S, molecular mass markers (plasmids R27 [169 kb] and R100 [90 kb] [Roche Molecular Biochemicals] and a 54-kb plasmid from strain V517 [17]). In lanes 1 to 8, the following STEC strains (serotypes and cdt alleles are shown in parentheses) are displayed: lane 1, strain 9282/01 (serotype O91:H21, allele cdt-V); lane 2, 2596/02 (O91:H21, cdt-V); lane 3, 5249/01 (O113:H21, cdt-V); lane 4, 2896/01 (O113:H21, cdt-V); lane 5, 2996/96 (O73:H18, cdt-V); lane 6, 9063/02 (O153:H18, cdt-III); lane 7, 7272/02 (O153:H18, cdt-III); lane 8, 5107/00 (O153:H18, cdt-III). Lanes 9 to 11 contain control strains as follows: lane 9, strain 493/89 (SF STEC serotype O157:H−, allele cdt-V); lane 10, 1404 (NTEC O78:H?, cdt-III); lane 11, E. coli C600 (negative control). The arrows in panel A indicate the plasmids that hybridized with the cdtB probe. The sizes of the hybridizing plasmids are given in panel B.
CDT expression.
Using a Chinese hamster ovary (CHO) cell assay (11), each of the 17 cdt-harboring STEC isolates produced CDT, as evidenced by a progressive distension of the cells for up to 5 days after their exposure to culture filtrates of these strains (data not shown). CDT titers ranged from 1:2 to 1:64 (Table 3). No CDT activity was detected by the CHO assay in any of the remaining 323 STEC isolates that were PCR negative for cdt-I, cdt-II, cdt-III, cdt-IV, and cdt-V, suggesting that no additional cdt allele undetectable by the PCR approaches used occurred among the STEC strains investigated. In accordance with a previous report that CHO cells are resistant to Stx (13), none of the 340 STEC isolates caused an Stx-like cytotoxic effect on these cells.
Our data demonstrate that two different cdt alleles with different genomic locations and specific serotype associations encode active CDT in non-O157 STEC strains. The finding that the location of each of the two cdt alleles is restricted to a limited spectrum of serotypes raises questions about the acquisition of these genes. In SF STEC O157:H− strain 493/89, cdt-V is located within the late gene region of bacteriophage P2 (11), suggesting that it might have been acquired by transduction (11). Currently, we seek to determine whether cdt-V in non-O157 STEC strains is also located within a phage genome, to characterize such a phage, and to find out whether the association of cdt-V with several particular STEC serotypes reflects a specific ability of such strains to be infected with a cdt-encoding phage. In addition, the finding of cdt-III on large plasmids of STEC O153:H18 strains is intriguing and prompts efforts to determine whether these plasmids are conjugative plasmids similar to the cdt-III-harboring Vir plasmid of NTEC strains (19) and whether a horizontal transfer through such plasmids contributes to the spread of cdt-III among STEC strains. The association of CDT with particular non-O157 STEC serotypes parallels the situation reported for EPEC (1, 2, 9) and extraintestinal pathogenic E. coli (12) and suggests that (as proposed for the latter two E. coli groups) (2, 9, 12) the particular serotypes might denote specific STEC clones that frequently harbor cdt and produce CDT. Until now, no association of CDT with diarrhea was found in epidemiological studies investigating EPEC (1, 2). Also, the absence of CDT from all eae-positive STEC isolates investigated in this study, including those belonging to the major non-O157 serogroups (serogroups O26, O103, O111, and O145) (Table 1) that frequently cause HUS (7), demonstrates that CDT is not essential for the pathogenicity of eae-positive STEC. However, our finding of a significant association between CDT production by eae-negative STEC isolates and clinical symptoms in patients infected with such strains suggests that CDT might contribute to the pathogenicity of these organisms. In particular, the finding of CDT in STEC strains of serotypes O91:H21 and O113:H21 that can cause serious human diseases including HUS (this study and references 10, 13 and 18) despite their lacking eae (18), an STEC factor associated with virulence and with HUS (3), warrants further investigation of a potential role of CDT in the pathogenesis of diseases caused by these strains. Our data thus add to a growing literature profiling the loci (beyond those of genes encoding Stx or intimin) potentially needed for STEC to cause disease.
Nucleotide accession sequence numbers.
The nucleotide sequences of cdt genes from STEC strains 9282/01 (O91:H21), 5249/01 (O113:H21), 9063/02 (O153:H18), and 2996/96 (O73:H18) have been entered into the GenBank database (accession numbers AY365042 to AY365045, respectively).
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG), no. KA 717/4-1, by a grant from the Bundesministerium für Bildung und Forschung (BMBF) Project Network of Competence Pathogenomics Alliance “Functional Genomic research on enterohaemorrhagic, enteropathogenic and enteroaggregative Escherichia coli”, Project Group “Schmidt/Karch, Universitätsklinikum Münster” (BD numbers 119523 and 207800), and by a grant from Fonds der Chemischen Industrie.
We thank P. I. Tarr (Washington University School of Medicine, St. Louis, Mo.) for critical reading of the manuscript and stimulating discussions, colleagues who provided us with CDT control strains, and M. Hüllsman (Münster) and B. Knüppel (Wernigerode) for skillful technical assistance.
Editor: A. D. O'Brien
REFERENCES
- 1.Albert, M. J., S. M. Faruque, A. S. G. Faruque, K. A. Bettelheim, P. K. B. Neogi, N. A. Bhuian, and J. B. Kaper. 1996. Controlled study of cytolethal distending toxin-producing Escherichia coli infections in Bangladeshi children. J. Clin. Microbiol. 34:717-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansaruzzaman, M., M. J. Albert, S. Nahar, R. Byun, M. Katouli, I. Kuhn, and R. Mölby. 2000. Clonal groups of enteropathogenic Escherichia coli isolated in case-control studies of diarrhoea in Bangladesh. J. Med. Microbiol. 49:177-185. [DOI] [PubMed] [Google Scholar]
- 3.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, B. J. Johnson, and C. L. Gyles. 1999. Association between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, C. G., S. T. Johnson, R. H. Easy, J. L. Campbell, and F. G. Rodgers. 2002. PCR for detection of cdt-III and the relative frequencies of cytolethal distending toxin variant-producing Escherichia coli isolates from humans and cattle. J. Clin. Microbiol. 40:2671-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortes-Bratti, X., T. Frisan, and M. Thelestam. 2001. The cytolethal distending toxins induce DNA damage and cell cycle arrest. Toxicon 39:1729-1736. [DOI] [PubMed] [Google Scholar]
- 6.Elwell, C. A., and L. A. Dreyfus. 2000. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 37:952-963. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich, A. W., M. Bielaszewska, W.-L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich, A. W., J. Borell, M. Bielaszewska, A. Fruth, H. Tschäpe, and H. Karch. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41:2448-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghilardi, A. C. R., T. A. T. Gomes, and L. R. Trabulsi. 2001. Production of cytolethal distending toxin and other virulence characteristics of Escherichia coli strains of serogroup O86. Mem. Inst. Oswaldo Cruz. 96:703-707. [DOI] [PubMed] [Google Scholar]
- 10.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 11.Janka, A., M. Bielaszewska, U. Dobrindt, L. Greune, M. A. Schmidt, and Helge Karch. 2003. The cytolethal distending toxin (cdt) gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect. Immun. 71:3634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spandjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal meningitis in the Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, W. M., and H. Lior. 1988. A new heat-labile cytolethal distending toxin (CLDT) produced by Escherichia coli isolates from clinical material. Microb. Pathog. 4:103-113. [DOI] [PubMed] [Google Scholar]
- 14.Lara-Tejero, M., and J. E. Galan. 2000. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290:354-357. [DOI] [PubMed] [Google Scholar]
- 15.Lara-Tejero, M., and J. E. Galan. 2001. CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infect. Immun. 69:4358-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, R. B., D. C. Hassane, D. L. Cottle, and C. L. Pickett. 2003. Interactions of Campylobacter jejuni cytolethal distending toxin subunits CdtA and CdtC with HeLa cells. Infect. Immun. 71:4883-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macrina, F. L., D. J. Kopecko, K. R. Jones, D. J. Ayers, and S. M. McCowen. 1978. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1:417-420. [DOI] [PubMed] [Google Scholar]
- 18.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga-toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peres, S. P., O. Marches, F. Daigle, J. P. Nougayrede, F. Herault, C. Tasca, J. DeRycke, and E. Oswald. 1997. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol. Microbiol. 24:1095-1107. [DOI] [PubMed] [Google Scholar]
- 20.Pickett, C. L., D. L. Cottle, E. C. Pesci, and G. Bikah. 1994. Cloning, sequencing, and expression of the Escherichia coli cytolethal distending toxin genes. Infect. Immun. 62:1046-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prager, R., U. Strutz, A. Fruth, and H. Tschape. 2003. Subtyping of pathogenic Escherichia coli strains using flagellar H- antigens: serotyping versus fliC polymorphisms. Int. J. Med. Microbiol. 292:477-486. [DOI] [PubMed] [Google Scholar]
- 22.Scott, D. A., and J. B. Kaper. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 62:244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tietze, E., and H. Tschäpe. 1983. Plasmid pattern analysis of natural bacterial isolates and its epidemiological implication. J. Hyg. (Cambridge) 90:475-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toth, I., F. Herault, L. Beutin, and E. Oswald. 2003. Production of cytolethal distending toxins by pathogenic Escherichia coli isolated from human and animal sources: Establishment of the existence of a new cdt variant (type IV). J. Clin. Microbiol. 41:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]