Abstract
Adult mesenchymal progenitor cells have enormous potential for use in regenerative medicine. However, the true identity of the progenitors in vivo and their progeny has not been precisely defined. We hypothesize that cells expressing a smooth muscle α-actin promoter (αSMA) directed Cre transgene represent mesenchymal progenitors of adult bone tissue. By combining complementary colors in combination with transgenes activating at mature stages of the lineage we characterized the phenotype and confirmed the ability of isolated αSMA+ cells to progress from a progenitor to fully mature state. In vivo lineage tracing experiments using a new bone formation model confirmed the osteogenic phenotype of αSMA+ cells. In vitro analysis of the in vivo labeled SMA9+ cells supported their differentiation potential into mesenchymal lineages.
Utilizing a fracture-healing model, αSMA+ cells served as a pool of fibrocartilage and skeletal progenitors. Confirmation of the transition of αSMA+ progenitor cells to mature osteoblasts during fracture healing was assessed by activation of bone specific Col2.3emd transgene. Our findings provide a novel in vivo identification of defined population of mesenchymal progenitor cells with active role in bone remodeling and regeneration.
INTRODUCTION
A full understanding of the recruitment of multipotent progenitors into the skeletal lineage and the factors influencing their differentiation is critical to the development of protocols to modulate bone regeneration and to the design of novel targets for pharmacological intervention. However, a major obstacle has been the unavailability of reproducible methods to identify these progenitors and to track their fate. Initial studies have characterized some phenotypic properties of these cells, but have not provided clear information on their origin.
A perivascular niche has been implicated as a source of mesenchymal progenitors 1, 2. The human bone marrow derived stromal population expressing STRO-1 and CD146 exhibits the ability to form bone in vitro and in a heterotopic bone formation assay in vivo. This population of cells is localized in the proximity of endothelial cells and was positive for αSMA expression 3. Sacchetti et al. defined a population of osteoprogenitors as CD146+ subendothelial cells in bone marrow suggesting that perivascular cells exhibit potential for self-renewal and differentiation supportive of hematopoiesis 4. Crisan et al observed perivascular cells in various organs with the ability to differentiate into multiple mesenchymal lineages 5.
Recently, we described a population of cells expressing smooth muscle actin alpha (αSMA) that have osteogenic potential 6. The αSMAGFP cells are located in perivascular niches, periosteum and sutures, regions that contain osteoprogenitor cells. Moreover,αSMAGFP+ cells derived from primary BMSCs and ADSCs of transgenic mice exhibit osteogenic and adipogenic potential 6. A high proportion of αSMAGFP+ cells express the stem cell markers; stem cell antigen 1 (Sca1) and CD90 but lack expression of the CD117 (c-kit) or CD11b 7–9.
A detailed characterization of progenitor cells requires methods to isolate the cells and trace their transition into different phenotypes. The use of cell surface markers is limited by the fact that progenitor cells may loose markers as they differentiate.
We have generated αSMACreERT2 transgenic mice, which provide the system in which the fate of the progenitors could be traced. We propose to examine the ability of αSMACreERT2 expressing cells as mesenchymal progenitors during bone formation and repair and to determine whether these progenitor cells become committed to specific lineages. The combinatorial approach of using Cre recombinase fused to a modified estrogen receptor ligand binding domains (ERT2) permits the temporal induction by delivery of a tamoxifen and tissue specificity by directing the Cre expression with a lineage specific promoter 10. When crossed with Ai9 reporter mice, recombination can be detected by expression of red fluorescence (TdTomato) 11. This system allows for the spatial and temporal activation of Cre labeling and identifying the labeled population, and tracing the lineage progenies of the labeled cells for prolonged period after the tamoxifen treatment has been terminated. Here we utilized the visual markers for transgene expression and the Cre/loxP recombination system for lineage tracing using a fragment of the αSMA promoter 6. We have utilized murine models harboring real-time visual transgenes in combination with cell surface markers and Cre/loxP technology as a powerful way of characterizing mesenchymal progenitors.
These approaches have defined a mesenchymal progenitor cell in vivo that actively participates in the physiological process of bone remodeling and during the regenerative processes of fracture healing.
MATERIALS AND METHODS
Generation of transgenic mice and animal studies
Detailed description of generation of αSMACreERT2 mice is outlined in the Suppl. Methods. The generation and characterization of αSMAGFP, Col2.3cyan, Col2a1cyan and Col2.3emd mice has been published 6, 12, 13. Mice transgenic for AP2cyan and αSMAcherry have been recently developed 14. The αSMACreERT2/Ai9 dual transgenic mice mice (term SMA9 will be used) were generated by breeding αSMACreERT2 male mice and Ai9 female mice that were obtained from Jackson labs (stock # 007909). The Ai9 mice harbor a targeted mutation of the Gt(ROSA)26Sor locus with a loxP-flanked STOP cassette preventing transcription of a CAG promoter-driven red fluorescent protein variant (tdTomato). For in vivo lineage tracing studies 4–5 weeks old mice were treated with tamoxifen at the dose of 75 μg/g of weight. Mice were treated twice in the interval of 24 hours and sacrificed 2 days or 17 days later. The SMA9 untreated mice and αSMACreERT2 negative/Ai9 treated mice were evaluated as controls.
In vitro studies
Primary BMSC were prepared as previously described 15. Following cell separation using fluorescent activated cell sorting (FACS), cells were plated at the density of 10,000 cells/cm2 in 24 well plates. Cells were induced to osteogenesis using DMEM/10%FCS medium supplemented with 50μg/ml ascorbic and 8mM β-glycerol phosphate for two weeks, while adipogenesis was induced media supplemented with 1μM Insulin and 0.5μM rosiglitasone for 7 days. To evaluate chondrogenesis sorted SMA9+ and SMA9− population were seeded as a 20μl spot containing 5×104 cells. After 2hr, αMEM/10%FCS was added and cells were for 7 days, placed under chondrogenic induction (serum free media supplemented with 50μg/ml ascorbic acid, 10ng/ml TGFβ3, 100nM dexamethasone, 40μg/ml L-proline, sodium pyruvate, ITS+ 1 and cultured in 5% oxygen). Methods evaluating osteogenesis, adipogenesis and chondrogenesis in vitro are presented within Suppl. Methods section.
Histology
Bone samples for histology were fixed for 3–5 days in 10% formalin (Sigma) and decalcified in 15%EDTA for 4–7 days depending on the age of animal, placed in sucrose overnight and embedded in cryomedium (Thermo scientific). Soft tissues were fixed overnight and after 24 hours in sucrose they were embedded and sectioned. Sections of 5μm were obtained using a Leica cryostat and tape transfer system (Japanese tape). Images were obtained by appropriate filter cubes optimized for GFP variants (Chroma) using a Zeiss Observer.Z1 microscope. Images were obtained in grey scale, pseudocolored and composite images assembled. To obtain a full size image of bone, images were scanned at high power and then stitched into a composite. Following fluorescent imaging sections were stained with hematoxyllin and imaged. Cells embedded within the bone matrix were counted within the trabecular area using sections counterstained with DAPI to visualize the population of osteocytes.
Flow cytometry
FACS analysis and cell sorting of αSMAcherry+, and αSMACreERT2/Ai9 cells were performed using a BD FACSAria I (BD Biosciences. San Jose, CA, USA) equipped with five lasers and 18 fluorescence detectors. Flushed bone marrow cells, and enzymatically digested (collagenase 0.2%, hyalouronidase 0.2% in 0.25 trypsin) periosteal layer cells were used. Erythrocytes were lysed by hypotonic shock. Sorting gates were defined using cells from non-transgenic and transgenic mice bearing individual transgenes. Single cell suspensions were prepared in staining medium (2% FCS/HBSS/HEPES). For phenotype characterization we used commercially available antibodies (Ab) as described in detail in Supp. Material.
RESULTS
αSMA+ cells exhibit characteristics of mesenchymal progenitor cells
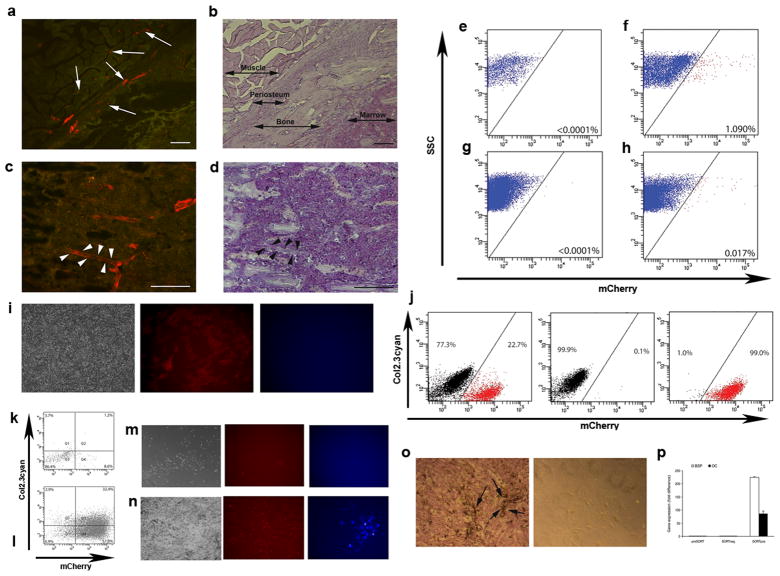
To evaluate the localization of cells expressing αSMA we analyzed mice expressing cherry FP under the control of αSMA promoter. In bone marrow and periosteum we observed αSMAcherry+ cells in association with microvasculature. In addition to perivascular location, we observed αSMAcherry+ fibroblastic shaped cells in periosteum (figure 1a–d, see suppl. figure 1a for whole bone section). FACS analyses of freshly isolated cells indicated that the αSMA transgene is active in 0.02–0.03% of cells in bone marrow and in around 1% of scraped cells from periosteum (figure 1e–h). The identified population in bone marrow can be expanded in vitro by day 7 (figure 1i). To assess the ability to differentiate into osteogenic lineage αSMAcherry+ and αSMAcherry− population were sorted from a dual transgenic mouse harboring the osteoblast specific reporter, Col2.3cyan (figure 1j). These cells were then replated and grown under lineage inductive conditions. We confirmed that αSMAcherry+ cells differentiated into mature osteoblasts and expressed Col2.3cyan on day 7 (figure 1l) and day 14 (figure 1n). In contrast, αSMAcherry− cells did not generate colonies or express osteogenic marker (figure 1k, m). The profile of cells derived from αSMAcherry+ population was confirmed by alkaline phosphatase expression, mineralization and the expression of bonesialoprotein and osteocalcin (figure 1o–p). Similar experiments driving sorted cells into adipogenesis and chondrogenesis, showed that the αSMAcherry+ population has the ability to differentiate into mature adipocytes and chondrocytes. Adipogenesis was detected by staining for oil-red-o and expression of AP2cyan and mRNA for adipsin and adiponectin (suppl. figure 1b), while chondrogenic differentiation was analyzed by alcian blue staining and aggrecan mRNA expression (data not shown).
Figure 1. αSMAcherry expressing cells display osteoprogenitor potential.
(a–d) The expression of αSMAcherry is localized to the perivascular area of capillaries within periosteum and muscle (a–b, see arrows) and bone marrow (c–d, arrowheads).
(e–h) FACS analysis for αSMAcherry expression in samples derived from enzymatically digested periosteum (e–f) or whole bone marrow from transgenic (g–h) or non-transgenic mice (e, g). Proportion of αSMAcherry expressing cells is indicated in the right lower corner of the plots (four αSMAcherry mice were analyzed individually and the respresentative values are shown).
(i–j) Primary BMSC derived from αSMAcherry/Col2.3cyan mice were grown for 7 days and imaged under transmitted (left), red (middle) and blue (right) epifluorescence. αSMAcherry expression was present in fibroblastic shaped cells while no expression of Col2.3cyan was detected (i). αSMAcherry-expressing cells (j, left panel, 23% of cells) were collected by FACS sorting; reanalysis of the sorted population purity is shown for αSMAcherry− and αSMAcherry+ cells (j, middle and left panels).
(k–l) Following sorting, cells were replated and induced to osteogenesis and FACS analyzed on day 7. αSMAcherry− cells did not activate Col2.3cyan (k) while α-SMAcherry+ showed differentiation into Col2.3cyan+ cells (l). Col2.3cyan was detected at day 14 in population of differentiated α-SMAcherry+ cells (n), with no expression in α-SMAcherry− population (m).
(o–p) Alkaline phosphatase expression and mineralization (arrows) was detected inαSMAcherry+ cells in contrast to αSMAcherry− cells (o). Real-time PCR shows increased expression of BSP and OC after osteogenic induction of α-SMAcherry+ cells (p).
Lineage tracing identifies progenitor population during normal skeletal remodeling
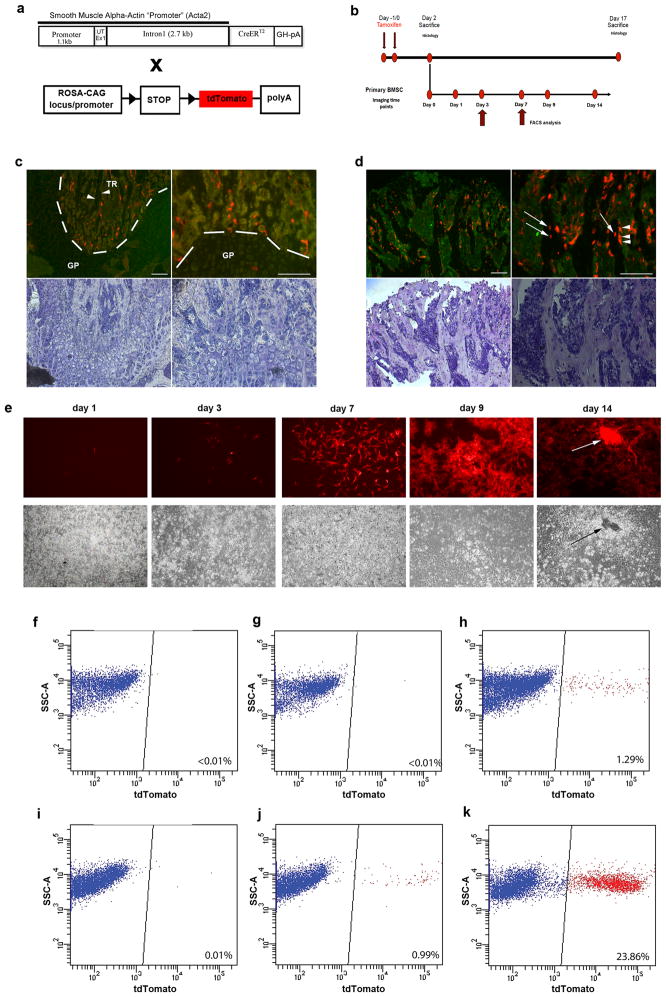
In order to determine whether αSMA promoter can identify cells with progenitor activity in vivo, we generated αSMACreERT2 transgenic mice and we characterized its expression by crossing it with the Ai9 reporter transgenic line to generate αSMACreERT2/Ai9 (SMA9) mice (figure 2a). In these mice the expression of the Cre molecule is activated under the control of the αSMA promoter upon treatment with tamoxifen. This allows the tracking of developmental progression of αSMA expressing cells by activating the expression of the tdTomato visual reporter through Cre-mediated recombination. Mice were treated with tamoxifen and analyzed at days 2 and 17 after treatment (figure 2b–d). Two days after treatment SMA9+ cells defined a small population of cells present in primary spongiosa (figure 2c) or within the periosteum (suppl. figure 2a). No cells expressing SMA9 reporter were detected within the bone matrix (figure 2c). Interestingly, 17 days after the tamoxifen activation we found numerous osteoblast and osteocytes labeled by the transgene. Image analysis revealed that 23.4±3.2% of bone matrix embedded cells were SMA9+ at this time point. These observations confirm the ability of the αSMA directed Cre to target osteoprogenitor cells in vivo (figure 2d). We have confirmed that the αSMACreERT2 activity was ligand dependent in adult bone and other tissues. In the absence of tamoxifen we did not detect leaky expression of tdTomato in bone tissue (suppl. figure 2a) However, a significant leak of the Cre expression was observed in tissues with a strong endogenous activity of αSMA gene like bladder, and aorta (suppl. figure 2b).
Figure 2. Cell lineage tracing experiments following in vivo transgene activation.
(a) TheαSMACreERT2 transgenic construct harboring sequence of αSMA regulatory elements. These mice were crossed with a Cre recombinase reporter strain Ai9. In dual expressing SMA9 mice tamoxifen-induced Cre recombinase activates expression of tdTomato.
(b) Diagram of experimental approach for the cell lineage tracing experiments. Mice were sacrificed on day 2 and 17 following tamoxifen treatment and evaluated by histological analysis. Primary BMSC were established from mice treated with tamoxifen and sacrificed on day 2 after treatment.
(c–d) Histological analysis of SMA9 recombination identifies SMA9+ cells within the trabecular area (TR) but not inside the bone matrix (arrowheads). No expression is observed in the growth plate (GP) chondrocytes. Seventeen days after the initial evaluation (day 17) we could track the SMA9+ cells to osteoblasts lining trabecular bone (d, arrowheads) and ostecytes embedded within bone (arrows). Shown are representative images of 6–7 mice per group analyzed at each time point.
(e) Epifluorescent imaging of primary BMSC derived from SMA9 mice treated with tamoxifen. Once labeled in vivo, SMA9+ cells expanded during the first week in culture formed nodules and underwent mineralization (see arrow).
(f–k) FACS analysis of primary BMSC (days 3 (f–h) and 7 (j–k)) derived from SMA9+ labeled cells following tamoxifen injection in vivo was performed onαSMACreERT2negative/Ai9 mice treated with tamoxifen (f, i), and SMA9 mice without treatment (g, j) served as a controls. In vivo treatment labeled SMA9+ cells account for about 1% of cells on day 3 and close to 25% on day 7 of the culture (h, k).
To further evaluate the distribution of labeled cells generated after αSMACre activation, we analyzed primary cultures at days 3 and 7 by flow cytometry. We did not detect expression of tdTomato in cells derived from αSMACrenegative/Ai9/Tx treated mice (figure 2f, i) at days 3 or 7, or in cells derived from SMA9 untreated mice on day 3 in culture (figure 2g). However, an appreciable activation of Cre-directed recombination was observed in SMA9 treated mice by day 3 (1–2%) and by day 7 (23–27%) (figure 2h, k). A small proportion of cells showed a spontaneous Cre activation in primary cultures derived from SMA9 non-treated mice (figure 2j) by day 7. The spontaneous activation of SMACreERT2 transgene in vitro is due the increase of SMA promoter activity and the presence of number of serum response elements in the proximal sequence of SMA promoter 16.
In addition, primary cultures derived from untreated SMA9 mice, subsequently treated in vitro by 4-OH tamoxifen, showed expression of tdTomato in a similar proportion of cells (suppl. figure 2c). This confirms our in vivo observation that we are selectively targeting a progenitor population that exhibits ability to differentiate into mature osteoblasts in vivo and in vitro.
Differentiation potential of SMA9 labeled progenitor population
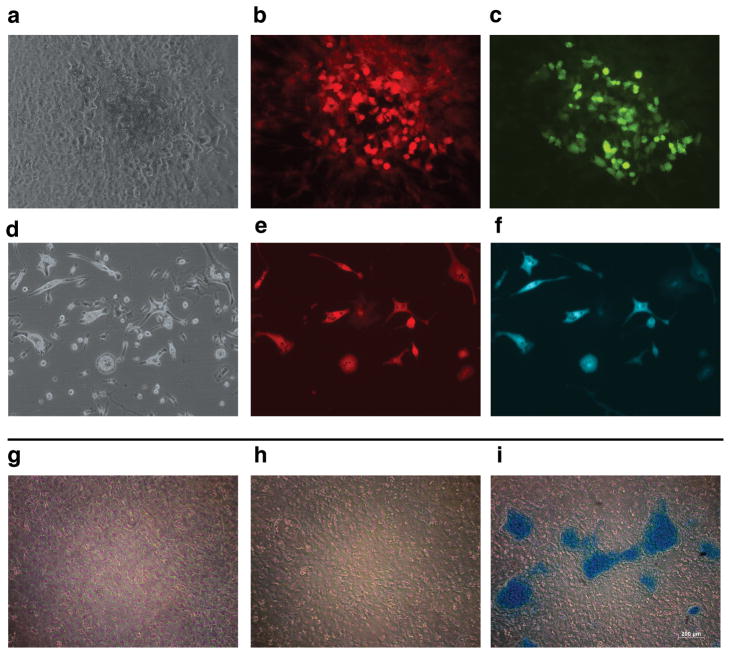
To test the progenitor ability of the in vivo labeled SMA9 cells we established primary cultures from SMA9 mice that were treated with tamoxifen for 2 days (figure 2b). Following one day in culture, individual SMA9+ cells were evident, and over time they expanded in number forming colonies during the first 7 days of culture (figure 2e). After induction of osteogenic differentiation SMA9+ cells formed multilayered nodules with the ability to mineralize. To substantiate these results, we have utilized primary BMSC cultures derived from SMA9/AP2Cyan and SMA9/Col2.3emd mice treated with tamoxifen. Two days after treatment, cultures were established and after reaching confluence were induced to adipogenesis and osteogenesis. We have observed numerous SMA+ cells differentiating into adipocytes or osteoblasts as detected by the expression of AP2cyan (figure 3b–c), or Col2.3emd respectively (figure 3e–f). To evaluate chondrogenic differentiation SMA9 mice were utilized, and cells we sorted into SMA9+ and SMA9− populations on day 7 of culture. Following differentiation we observed the formation of alcian blue positive nodules in the cultures derived from SMA9+ cells (figure 3i). Cultures of unsorted or SMA9− cells did not form alcian blue positive nodules (figure 3g–h) even after longer period in culture. The SMA9+ population shows also enhanced mRNA expression of chondrogenic markers Col2a1 and aggrecan (data not shown).
Figure 3. Multilineage potential of SMA9 progenitor cells.
(a–f) Primary BMSC were established from SMA9/Col2.3emd and SMA9/AP2cyan mice treated with tamoxifen for 2 days. Cells were then grown to confluence and induced to osteogenic or adipogenic differentiation respectively. Osteogenic differentiation was confirmed by dual expression of red-labeled SMA9 (b), and green-labeled Col2.3emd osteoblast lineage cells (c), (phase is shown in a). Adipogenic differentiation was confirmed by dual expression of red-labeled SMA9 (e), and blue labeled AP2cyan adipocytes (f), (phase is shown in d).
(g–i) To evaluate chondrogenesis cells derived from SMA9 mice cells were cultured for 7 days, and then sorted into SMA9+ and SMA9− and placed under chondrogenic conditions for 9 days. Formation of chondrogenic nodules was detected using alcian blue staining in sorted SMA9+ cells (i), in contrast to SMA− (h) and unsorted BMSC (g).
Our data clearly shows that the SMA9 transgenic constructs provided a bona-fide identification of mesenchymal progenitor cells and should allow further characterization of this population of cells using markers currently proposed as signature molecules for stem cells.
Multiple mesenchymal stem cell markers are present on SMA9 labeled cells
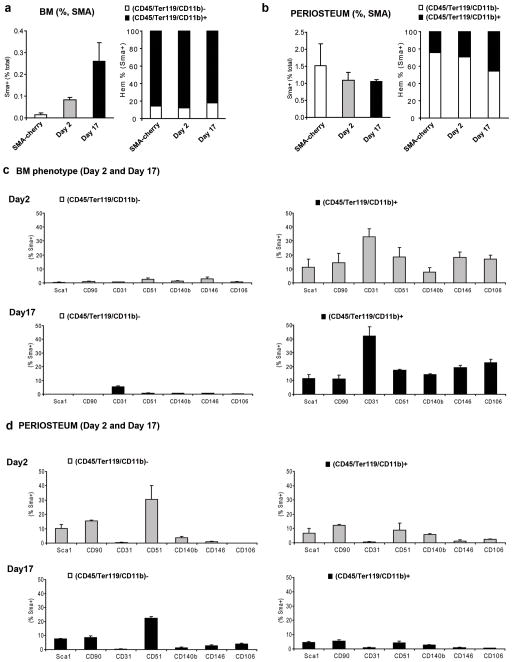
In addition to the histological assessment, we followed the percentage of SMA9+ labeled cells in vivo at days 2 and 17 after tamoxifen treatment by flow cytometry. We found an increase in the percentage of cells expressing tdTomato reporter in the bone marrow up to 0.3% at day 17, compared to the 0.02% observed for the expression of αSMAcherry in basal conditions (figure 4a). In contrast, the percentage of SMA9+ cells isolated from the periosteal layer was approximately 1% and was maintained along a time-course evaluation (figure 4b, and suppl figure 3a). We used multicolor FACS analysis to test if the reporter signal correlated to markers of the hematopoietic lineage. To evaluate hematopoietic markers we used a combination of antibodies for CD45, Ter119 and CD11b. Interestingly, most of the SMA9+ cells present in the bone marrow cells expressed markers characteristic of hematopoietic lineage, whereas the majority of the SMA9+ periosteal cells were (CD45/Ter119/CD11b)− (figures 4a–b). Among the (CD45/Ter119/CD11b)− SMA9+ bone marrow population, approximately one third of the cells were CD45+Ter119−CD11b− (not shown). We also analyzed gated SMA9+ cells for the co-expression of cell surface markers associated with mesenchymal progenitor cells and found that SMA9+ population maintained a stable expression pattern when analyzed at day 2 and 17 after transgene induction (figures 4c–d, suppl figure 3b). Bone marrow cells that are tdTomato+ and (CD45/Ter119/CD11b)− were mostly negative for the included mesenchymal lineage markers, except for CD31 that was weakly expressed on approximately 5% cells on day 17(figure 4c).
Figure 4. Distinct phenotype of αSMACre labeled cells from the bone marrow compartment and periosteum.
(a–b) Left-side graphs present FACS analysis of the percentage of αSMAcherry and SMA9 labeled cells (SMA9+) (2 and 17 days following tamoxifen injection) within the bone marrow (a) and among periosteal cells (b). Right-side graphs present relative proportion of (CD45/Ter119/Cd11b)+ vs. (CD45/Ter119/Cd11b)− populations among αSMAcherry and SMA9+ cells. Each group included 2–3 independent samples pooled from the respective tissue of 2–3 mice. To determine the percentage of SMA9+ cells at least 2.5×106 cells for bone marrow and 0.25×106 for periosteal cells were analyzed. Gates were set in accord to the control populations from tamoxifen injected αSMACrenegative/Ai9 mice. Mice without tamoxifen treatment, carrying the transgene SMA9 served for the Cre-leakage control and contained a percentage of tdTomato+ cells comparable to the control mice (not shown).
(c–d). Phenotyping of SMA9+ cells 2 and 17 days following transgene activation. Graphs represent the percentage of the population expressing respective mesenchymal cell markers within (CD45/Ter119/Cd11b)+ vs. (CD45/Ter119/Cd11b)− compartments of the SMA9+ cells. FACS analysis was performed by gating SMA9+ cells and plotting them using markers for hematopoietic lineage (CD45/Ter119/CD11b) vs respective proposed mesenchymal lineage marker (Sca1, CD90, CD31, CD51 Cd140b, CD146 or CD106) within the bone marrow (c) and periosteal cell populations (d). Values represent mean±SD of the percentage of respective population from 2–3 independent samples pooled from 2–3 mice.
Percentages of hematopoietic marker positive SMA9+ bone marrow cells expressing mesenchymal markers vary between 10–30%, with CD31 being the most prominently expressed (figure 4c). This percentage of CD31+ cells was much lower (<0.1%) in αSMAcherry labeled cells (data not shown), indicating induction of CD31 expression after tamoxifen activation of Cre mediated recombination.
The periosteal SMA9+ cells had a different phenotype, mostly lacking expression of hematopoietic markers but strongly expressing Sca1 and CD90 on approximately 20% of the population and weakly expressing CD51 on approximately a third of the population at day 2 after transgene induction (figure 4d and suppl. figure 3b).
The in vitro system allows for the expansion of the progenitor population, but comes with a number of variables including serum composition and selective expansion of the adherent population of cells. Therefore, we evaluated the expression of cell surface markers that are used to characterize mesenchymal progenitor cells in vitro. Following our in vivo indication that majority of SMA9+ bone marrow cells are positive for the hematopoietic lineage markers we have evaluated the expression of SMA9+ cells for the presence of Sca1, CD51, CD140b, CD31 and CD146 in the context of the and CD45/Ter119 expression. During the first 3 days in culture SMA9+ cells lose the expression of CD45/Ter119, which is present in about 2% of cells on day 3 and 6% on day 7 (suppl. fig. 4). The expression of Sca1 and CD51 was observed by 30–40% of cells on day 3 and increased up to 50–60% of cells on day 7, while CD140b was expressed at much lower levels (suppl. fig. 4). We did not observe the expression of CD31 or CD146 in primary stromal cell cultures at either time point (not shown).
Origin of mesenhymal lineages during fracture repair
To determine whether SMA9 expressing cells participate in the regenerative processes including generation of chondrocytes and osteoblasts we utilized a tibia fracture model. The experimental design included treatment with tamoxifen, on the day before and on the day of fracture. The aim was to label the population of αSMA expressing cells and then trace their progeny during the fracture healing process.
During the first week following injury expansion of SMA9+ cells within bone marrow and periosteum was observed (figure 5a). Interestingly, we detected the presence of SMA9+ expressing cells in newly formed chondrogenic areas of the fracture callus, and areas of the new bone formation (figure 5b). We correlated the presence of these osteoblasts with the dynamic formation of new bone through injection of calcein dye one day before sacrificing the mice. The osteoblasts derived from SMA9+ progenitor cells showed intense deposition of new bone, indicated by the correlation between calcein label (green) and tdTomato (red) positive cells (figure 5b). These results clearly indicated that the labeled populations present within the periosteum and bone marrow actively participate in all the aspects of fibrochondral ossification.
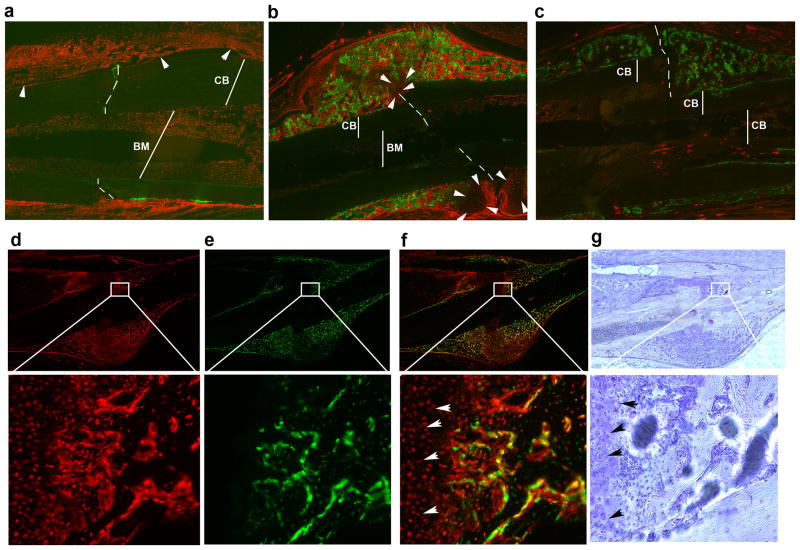
Figure 5. Mesenchymal progenitor cell lineage tracing during fracture healing.
(a–b) Osteogenesis during fracture healing is examined at one and two weeks following fracture. The expression of SMA9 is detected in the periosteal layer of cells (a, arrowheads point to the expanded SMA9+ cells within periosteum) located in the proximity to the fracture site (indicated by perforated lines; CB-cortical bone, BM-bone marrow). One week after fracture SMA9+ cells comprise most of the periosteal layer around fracture site. Bone marrow is also filled with SMA9+ cells. Two weeks after the fracture new cartilaginous callus formation is observed (b, arrowheads), while new bone formation is detected by deposition of calcein (green). Osteogenic lineage cells labeled by the SMA9 reporter (red, tdTomato+) persist in proximity to the large and intermingled calcein labeled areas within the callus.
(c) Two week post fracture callus of a SMA9 control mouse (tamoxifen non treated) showing extensive calcein labeling but minimal reporter leakage.
(d–e) The same experimental design was completed in a SMA9/Col2.3emd model. The expression of SMA9+ cells (d, red,) co-localizes with the expression of osteoblast specific marker Col2.3emd (e, green), as indicated by the presence of yellow color (overlay image, f). To provide better orientation within the area of callus same sections were stained by hematoxylin (g).
To track the population of SMA9+ cells into mature osteoblast we bred a bone specific transgene into the SMA9 mice. The expression of SMA9+ cells (red-tdTomato) co-localized to Col2.3emd expressing mature osteoblasts (green, SMA9/2.3GFP) (figure 5d–g). This finding is direct evidence for terminal differentiation of SMA9+ cells into mature osteoblasts in vivo. A representative image of a control fractured bone from SMA9 mouse not injected with tamoxifen is shown in figure 5c. No major leak of spontaneous Cre expression was detected following fracture of SMA9 untreated mice.
The majority of the cartilaginous callus cells were SMA9+ indicating differentiation into chondrocytes (figure 5b and 5f–g arrowheads). In addition to the morphological characterization of chondrocyte we have introduced a chondrocyte lineage directed reporter (Cyan) using a Col2a1 transgene. One week following bone fracture in SMA9/Col2a1 mice we have observed a population of dual SMA9/Col2a1Cyan expressing cells indicating that the SMA9+ population labeled prior to fracture healing can gave rise to Col2a1 expressing cells (suppl. figure 5).
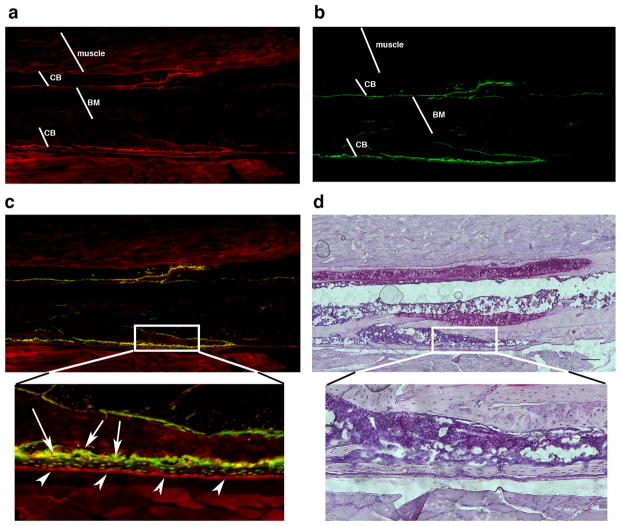
To define the contribution of the SMA9 mesenchymal progenitor cells to newly formed bone following completion of the fracture healing process we assessed the 6-week post-fracture time point. Mice transgenic for SMA9/Col2.3emd were fractured and cell-lineage traced following callus remodeling (figure 6). The contribution of the SMA9+ cell was detected by persistence of the dual SMA9+/Col2.3emd labeled cells (osteoblasts and osteocytes) within the new bone 6 weeks into the repair process (figure 6a–d, arrows indicate numerous dual SMA9/Col2.3emd expressing osteoblasts and osteocytes).
Figure 6. SMA9+ cells contribute to new bone formation following fracture healing.
(a–c) Osteogenesis following fracture healing and callus remodeling is examined at 6 weeks following fractures. The expression of SMA9 (a), Col2.3emd (b) and the overlayed image was evaluated for the presence of the dual expressing mature osteoblast lineage cells (c). Hematoxylin staining of the same section has been completed following epifluorescence imaging (d). The expression of SMA9 is detected in the periosteal layer of cells (a, c, see arrowheads point to the expanded SMA9+ cells within periosteum), within the osteoblast layer (c, indicated by arrows), among osteocytes and within the muscle. High magnification (20x) clearly identifies the presence of numerous cells that express both SMA9 and Col2.3emd markers (c, see yellow cells on the bone surface and within the bone matrix). BM- bone marrow, and CB-cortical bone. For this experiment unilateral fractures were performed in 3 mice.
DISCUSSION
Skeletal stem cell, as a term, has been proposed for the population of mesechymal progenitor cells residing within bone marrow compartment 17, 18. The most accepted markers to define human MSCs are STRO-1, recognizing a milieu of cell surface glycoproteins, and MCAM/CD146, a marker that has been associated with subendothelial perivascular cells 5, 19. Recently, the expression of nestin driven reporters has been associated with a population of bone marrow cells that can differentiate into mesenchymal lineages 20. Although useful to isolate cells from bone marrow and primary cell cultures, these markers do not allow for lineage tracing of the progeny of the mesenchymal stem cell since they are only transiently expressed at certain stages of differentiation.
The origin of the mature osteoblast remains elusive. We hypothesize that the αSMA transgene expression could be a marker for a cell that can give rise not only to osteoblasts but also to other mesenchymal lineage cells. We have addressed this by using an αSMACreERT2 transgene aiming to confirm the progenitor potential of αSMA expressing perivascular cell in vivo. We were able to trace the progeny of the perivascular cell into the mature osteoblasts and osteocytes, the final stage of skeletal lineage differentiation. These data indicate that during the process of bone formation and remodeling the αSMA transgene identifies skeletal stem cells.
These observations prompted us to define the surface markers in the cells identified by αSMAcherry as well as SMA9 expression. A high proportion of αSMAcherry+ and SMA9+ cells, expressed markers associated with hematopoietic cells including CD45. The exclusiveness of CD45 as pan-hematopoietic marker has been recently challenged, indicating that mesenchymal progenitor cells can express markers currently used to define hematopoietic lineage 21,22. Skeletal stem cell interactions with endothelial cells are critical for early invasion of vasculature during embryonic development and for the translocation of skeletal stem cells into the newly formed bone marrow cavity 23. However, recent literature has proposed alternative definitions for the identity of the mesechymal progenitor cells regarding their expression of endothelial cell markers. Medici et al. described cells with endothelial phenotype exhibiting the ability to differentiate into mesenchymal cell lineages 24. Other report, using a co-culture model of human embryonic stem cells with mouse OP9 stromal cells, defined a common precursor for endothelial and mesenchymal cell lineages, termed mesenchymoangioblast 25. These studies cannot be strictly compared as they are derived from different experimental models. However, they provide evidence for a close relationship between mesenchymal and endothelial lineage cells. In our study we have observed the expression of αSMAcherry in proximity to endothelial cells, while very few or none αSMAcherry+ co-expressed CD31 marker. Nevertheless, when the progeny of the SMA9 cells was traced in vivo we observed a weak co-expression of CD31 in approximately 5% of the (CD45/Ter119/CD11b)−, SMA9+ cells. The proportion of CD31+/SMA9+ cells represents about 30% of the cells positive for markers of hematopoietic lineage. These findings suggest the possibility that SMA9 labeled progenitor cells, located in the perivascular region, have the ability to mature into cells with an endothelial phenotype. In addition, within SMA9+ bone marrow population that are positive for hematopoietic markers we observed approximately 20% of CD146+/SMA9+ cells, while there were very few or none CD146 expressing cells among periosteal layer or in vitro expanded SMA9+ cells. Future studies have to be aimed to define these populations using a combination of approaches and multiple markers for both mesenchymal and endothelial lineages.
To test functionally the proposed hypothesis, the osteogenic potential of SMA9 labeled cells was tested in a bone injury model. Our data indicate that the majority of the callus cells, including chondrocytes and osteoblasts, are derived from SMA9 expressing cells. Interestingly, the expansion of the SMA9 cells occurred both in periosteum and within bone marrow, and resulted into SMA9+ cells with mature bone cell phenotypes contributing to the fracture healing process. Our work provides direct in vitro and in vivo evidence that the use of SMA9 labeled cells should allow the selective identification and isolation of progenitor cells that participate actively in the fracture healing process. In combination with visual markers of mature lineages, we could identify pathways important for lineage determination that could provide therapeutic targets for the enhancement of the regenerative processes.
Supplementary Material
Acknowledgments
Supported by: This work has been supported by NIH/NIAMS AR059315-01 and AR055607-01 grants to I.K., NIH/NHLBI 1RC1HL100569-01 to H.L.A. and NIH/NIAMS AR043457 to D.W.R. SP and DR have been supported by Croatian Science Foundation.
Footnotes
Author Contribution
Danka Grcevic: conception and design, collection and assembly of data, manuscript writing, final approval of manuscript
Slavica Pejda: collection and assembly of data, data analysis and interpretation, manuscript writing
Brya Matthews: collection and assembly of data, final approval of manuscript
Dario Repic: collection and assembly of data, final approval of manuscript
Liping Wang: collection and assembly of data, final approval of manuscript
Haitao Li: collection and assembly of data, final approval of manuscript
Mark S. Kronenberg; collection and assembly of data, final approval of manuscript, manuscript writing
Xi Jiang: collection and assembly of data, data analysis and interpretation
Peter Maye: data analysis and interpretation, manuscript writing
Douglas J. Adams: collection and assembly of data, final approval of manuscript
David W. Rowe: provision of study materials, financial support, manuscript writing
Hector L. Aguila: data analysis and interpretation, financial support, manuscript writing, final approval of manuscript.
Ivo Kalajzic: conception and design, collection and assembly of data, data analysis and interpretation, financial support, manuscript writing, final approval of manuscript.
References
- 1.Bianco P, Riminucci M, Gronthos S, et al. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- 2.Gronthos S, Zannettino AC, Hay SJ, et al. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J Cell Sci. 2003;116:1827–1835. doi: 10.1242/jcs.00369. [DOI] [PubMed] [Google Scholar]
- 3.Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res. 2003;18:696–704. doi: 10.1359/jbmr.2003.18.4.696. [DOI] [PubMed] [Google Scholar]
- 4.Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 5.Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Kalajzic Z, Li H, Wang LP, et al. Use of an alpha-smooth muscle actin GFP reporter to identify an osteoprogenitor population. Bone. 2008;43:501–510. doi: 10.1016/j.bone.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 8.Van Vlasselaer P, Falla N, Snoeck H, et al. Characterization and purification of osteogenic cells from murine bone marrow by two-color cell sorting using anti-Sca-1 monoclonal antibody and wheat germ agglutinin. Blood. 1994;84:753–763. [PubMed] [Google Scholar]
- 9.Welm BE, Tepera SB, Venezia T, et al. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 10.Feil R, Wagner J, Metzger D, et al. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 11.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokota T, Kawakami Y, Nagai Y, et al. Bone marrow lacks a transplantable progenitor for smooth muscle type alpha-actin-expressing cells. Stem Cells. 2006;24:13–22. doi: 10.1634/stemcells.2004-0346. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Toole BP, Dealy CN, et al. Hyaluronan in limb morphogenesis. Dev Biol. 2007;305:411–420. doi: 10.1016/j.ydbio.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kronenberg M, Jiang X, Kalajzic I, et al. Adipocyte Directed Expression of GFP in Transgenic Mice. J Bone Miner Res. 2004;(Suppl 1):Abstract. [Google Scholar]
- 15.Kalajzic I, Kalajzic Z, Kaliterna M, et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 16.Lee TC, Chow KL, Fang P, et al. Activation of skeletal alpha-actin gene transcription: the cooperative formation of serum response factor-binding complexes over positive cis-acting promoter serum response elements displaces a negative-acting nuclear factor enriched in replicating myoblasts and nonmyogenic cells. Mol Cell Biol. 1991;11:5090–5100. doi: 10.1128/mcb.11.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 19.Gronthos S, Graves SE, Ohta S, et al. The STRO-1+ fraction of adult human bone marrow contains the osteogenic precursors. Blood. 1994;84:4164–4173. [PubMed] [Google Scholar]
- 20.Mendez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers I, Yamanaka N, Bielecki R, et al. Identification and analysis of in vitro cultured CD45-positive cells capable of multi-lineage differentiation. Exp Cell Res. 2007;313:1839–1852. doi: 10.1016/j.yexcr.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 22.Schipani E, Kronenberg HM. StemBook [Internet] Cambridge (MA): Harvard Stem Cell Institute; 2008–2009. Jan 31, Adult mesenchymal stem cells. 2008. [PubMed] [Google Scholar]
- 23.Maes C, Kobayashi T, Selig MK, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medici D, Shore EM, Lounev VY, et al. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16:1400–1406. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vodyanik MA, Yu J, Zhang X, et al. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell. 7:718–729. doi: 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.