Abstract
Alternative cleavage and polyadenylation (APA) leads to mRNA isoforms with different coding sequences (CDS) and/or 3′ untranslated regions (3′UTRs). Using 3′ Region Extraction And Deep Sequencing (3′READS), a method which addresses the internal priming and oligo(A) tail issues that commonly plague polyA site (pA) identification, we comprehensively mapped pAs in the mouse genome, thoroughly annotating 3′ ends of genes and revealing over five thousand pAs (~8% of total) flanked by A-rich sequences, which have hitherto been overlooked. About 79% of mRNA genes and 66% of long non-coding RNA (lncRNA) genes have APA; but these two gene types have distinct usage patterns for pAs in introns and upstream exons. Promoter-distal pAs become relatively more abundant during embryonic development and cell differentiation, a trend affecting pAs in both 3′-most exons and upstream regions. Upregulated isoforms generally have stronger pAs, suggesting global modulation of the 3′ end processing activity in development and differentiation.
Keywords: cleavage and polyadenylation, splicing, mouse, mRNA, lncRNA, development, differentiation
Introduction
Cleavage and polyadenylation of nascent RNA is essential for maturation of almost all eukaryotic mRNAs, and is coupled to termination of transcription 1. The cleavage and polyadenylation site, or polyA site (pA), is defined by surrounding cis elements 2, 3, including upstream ones, such as UGUA, AAUAAA or its variants (also known as the polyadenylation signal or PAS), and U-rich elements, as well as downstream ones, such as U-rich and GU-rich elements. The 3′ end processing machinery, composed of ~20 core factors and many associated factors 4, is responsible for the cleavage and polyadenylation reaction.
Over half of the human mRNA genes have been found to have multiple pAs, leading to mRNA isoforms containing different coding sequences (CDS) and/or variable 3′ untranslated regions (3′UTRs) 5. Alternative cleavage and polyadenylation (APA) can play a significant role in mRNA metabolism by controlling the length of 3′UTR and its encoding cis elements 6, 7. Dynamic regulation of 3′UTR by APA has been reported in different tissue types 8, 9, cell proliferation/differentiation and development 10, 11, cancer cell transformation 12, 13, and response to extracellular stimuli 14. By contrast, pAs in introns and upstream exons have not been fully studied at the genomic level. In addition, to what extent APA regulates long non-coding RNAs (lncRNAs), which are increasingly found to play important roles in the cell 15, 16, is largely unknown.
Identification of pAs typically relies on the cDNA sequence corresponding to the poly(A) tail, which is generated by oligo(dT)-based reverse transcription 17, 18. However, oligo(dT) can also prime at internal A-rich sequences, which are completely converted to As in the final sequence, becoming indistinguishable from the sequence derived from the real poly(A) tail 19. This problem, commonly known as the ‘internal priming’ issue, is usually addressed computationally by eliminating putative pAs mapped to genomic A-rich regions. However, this approach not only does not guarantee full elimination of false positives caused by internal priming, but can also discard real pAs. In addition, RNAs in the cell can have oligo(A) tails that are synthesized by noncanonical poly(A) polymerases, such as those involved in exosome-mediated RNA decay 20. While the length distribution of the oligo(A) tail is not yet clear, a recent study showed that the RNA species bound by yeast Trf4, a poly(A) polymerase in the TRAMP complex involved in nuclear RNA surveillance, have the median A-tail length of 5 nucleotides (nt)21. However, as shown by the study, about 10% of the population have an oligo(A) tail between 10 and 14 nt, making them potential targets for oligo(dT) priming.
Here we present a high-throughput method, named 3′ Region Extraction And Deep Sequencing (3′READS), to map pAs and quantitatively measure APA isoform expression. This method completely addresses the issue of internal priming and minimizes the complication of oligo(A) tail. Using 3′READS, we systematically mapped pAs in the mouse genome, and found APA isoforms in ~79% of mRNA and ~66% of lncRNA genes. This method enabled us to uncover over five thousand real pAs located in genomic A-rich regions, which have hitherto been overlooked, underscoring the necessity to address the internal priming issue for comprehensive pA analysis. Using 3′READS, we studied APA in development and differentiation, and found overall relative downregulation of isoforms using promoter-proximal pAs, including those in the 3′-most exon as well as in introns and upstream exons. Expression and cis element analyses indicated that this general trend of transcript lengthening is likely due to global modulation of the 3′ end processing activity.
Results
Development of 3′ Region Extraction And Deep Sequencing (3′READS)
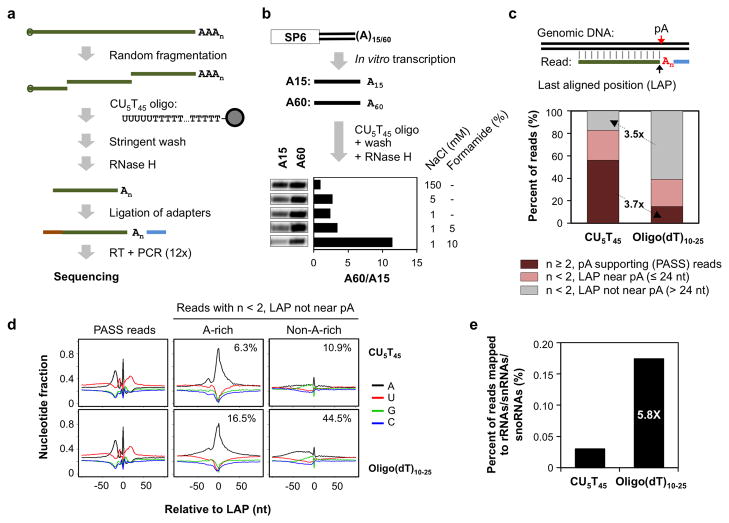
We wanted to develop a deep sequencing method which can address issues that commonly plague pA identification, namely internal priming and oligo(A) tail, as described in the Introduction. We reasoned that the former could be solved by eliminating the use of oligo(dT) in reverse transcription and sequencing, and the latter could be alleviated by using a condition that maximally distinguishes RNAs with long A-tails from those with short A-tails. To this end, we developed a method named 3′ Region Extraction And Deep Sequencing (3′READS), as illustrated in Figure 1a (see Methods for detail). Briefly, after fragmentation of RNA, we captured poly(A)-containing RNA fragments onto magnetic beads coated with a chimeric oligonucleotide (oligo), which contained 45 thymidines (Ts) at the 5′ portion and 5 uridines (Us) at the 3′ portion, dubbed CU5T45. We developed an experimental condition which enriched RNAs with 60 terminal As by ~12-fold as compared to those with 15 As (Figure 1b). RNase H digestion was used to release RNA from the beads and to remove most of the As of the poly(A) tail. Eluted RNA was ligated to 5′ and 3′ adapters, followed by reverse transcription, PCR amplification, and deep sequencing (see Supplementary Table 1 for adapter and primer sequences). The resulting reads were aligned to the genome, and those with at least 2 non-genomic As at the 3′ end were considered as PolyA Site Supporting (PASS) reads, and were used for pA site analysis (Figure 1c).
Figure 1. Mapping pAs by 3′READS.
(a) Schematic of the 3′READS method. See Methods for detail. (b) Optimization of washing condition to enrich RNAs with long poly(A) tails. Radioactively labeled A15 and A60 RNAs were synthesized by in vitro transcription using SP6 RNA polymerase. The X-ray film image shows the eluted RNA after RNase H digestion. The A60/A15 ratio indicates the difference in amount between eluted A60 and A15 RNAs. (c) Reads generated by 3′READS using the CU5T45 oligo or oligo(dT)10–25 (See Methods for detail). Top, schematic showing alignment of a read to genomic DNA. The last aligned position (LAP) and the putative pA are indicated by arrows. Bottom, distribution of three types of reads: 1) reads with ≥ 2 As immediately downstream of the LAP, which were used for pA identification and were called polyA site supporting (PASS) reads; 2) reads with <2 As immediately downstream of the LAP, and the LAP is near a pA (≤ 24 nt); 3) same as 2) except that the LAP is not near a pA (> 24 nt). (d) Nucleotide profiles around the LAP (set to position 0), as illustrated in (c). Top panels are reads generated by CU5T45 and bottom ones by oligo(dT)10–25. Left, PASS reads; middle and right, reads with <2 As immediately downstream of the LAP and the LAP is not near a pA, i.e., type 3 in (c). Reads whose LAP is flanked by A-rich sequences (middle) or non-A-rich sequences (right) areshown. The percent of total reads is shown in each graph. An A-rich sequence is defined as ≥6 consecutive As or ≥7 As in a 10 nt window in the −10 to +10 nt region around the LAP. (e) Percent of PASS reads assigned to rRNA, snoRNA, and snRNA genes for data generated by CU5T45 or oligo(dT)10–25. The ratio of the values is indicated.
Using mouse reference RNA (cell line mix 1 in Supplementary Table 2), we found that 56% of the reads generated from 3′READS were PASS reads (Figure 1c). As expected, the nucleotide profile of the genomic region around the last aligned position (LAP) of these reads (Figure 1c) was similar to that of pAs reported before 5, indicating that PASS reads are suitable for pA mapping. About 27% of all reads were also aligned near pAs but had no or 1 non-genomic A (Figure 1c). Presumably, the poly(A) tail sequence of the RNA fragments for these reads had been completely digested by RNase H. The remaining 17% of the reads were distributed along transcripts (see Supplementary Figure 1 for an example). About one third of them (6% of total) had the LAP flanked by A-rich sequences (Figure 1d, middle), whereas the rest (11% of total) did not (Figure 1d, right). Conceivably, the former reads were generated because of binding of RNA with internal A-rich sequences to the CU5T45 oligo, whereas the latter ones might come from degraded RNAs with oligo(A) tails.
For comparison, we used also a regular oligo(dT) column commonly used for poly(A)+ RNA selection, which contained oligo(dT)10–25. This column led to far fewer PASS reads (3.7-fold) and more reads mapped to A-rich or other regions (3.5-fold, Figures 1c and 1d, lower panels), supporting the effectiveness of using CU5T45 in distinguishing poly(A) tails from internal A-rich sequences. Importantly, since reads containing no additional As after alignment were not used for pA identification, the issue of “internal priming” essentially did not exist. In addition, since the 5 Us in the CU5T45 oligo can protect some As from digestion by RNase H due to the RNA:RNA base-pairing, the eluted RNAs were more likely to have terminal As than those eluted from oligo(dT)10–25-coated beads (Figure 1c), making the resultant reads more usable for pA analysis.
To further evaluate the performance of 3′READS, we examined PASS reads mapped to rRNAs, snoRNAs and snRNAs, which are not polyadenylated. Reads mapped to these RNAs would either be due to internal A-rich sequences or the oligo(A) tail produced during their maturation or degradation 20. As shown in Figure 1e, the CU5T45 oligo generated much fewer (5.8-fold) PASS reads mapped to rRNAs/snoRNAs/snRNAs as compared to oligo(dT)10–25.
We next compared 3′READS with several deep sequencing methods recently developed for pA mapping that employed oligo(dT) in reverse transcription, such as PolyA-seq 22 and PAS-seq 23. As shown in Supplementary Figure 2a, 3′READS generated >10-fold fewer reads aligned to rRNAs/snoRNAs/snRNAs, indicating that 3′READS can significant mitigate false positives caused by internal A-rich sequences and oligo(A) tails. Notably, we found over half of the original pAs (not filtered for internal priming) mapped by PolyA-seq were surrounded by A-rich sequences (Supplementary Figure 2b), with the overall nucleotide profile of these sites similar to that of the sites for non-PASS reads in this study (Figure 1d, middle panels). This indicates that internal priming is a serious issue for methods using oligo(dT) for reverse transcription. We also compared our data with those of 3P-seq, which does not use oligo(dT) for reverse transcription. As shown in Supplementary Figure 2c, 3′READS gave rise to 54% more usable reads for pA mapping than 3P-seq 24. This is presumably due to the stringent washing condition and/or fewer sample processing steps used in 3′READS (see Supplementary Discussion).
Using replicate samples, we found that 3′READS had good reproducibility, with r (Pearson correlation) ≥ 0.95 between replicates (Supplementary Figure 3a). As expected, genes expressed at low levels had higher variations than those expressed at high levels (Supplementary Figure 3a). When different isoforms of a gene were combined, the 3′READS data had a good correlation with that of RNA-seq, with r (Pearson correlation) = 0.89 (Supplementary Figure 3b), indicating that 3′READS data is quantitative. Taken together, our data indicate that 3′READS is an accurate and efficient method for quantitative analysis of APA isoforms and gene expression.
Comprehensive mapping of pAs in the mouse genome
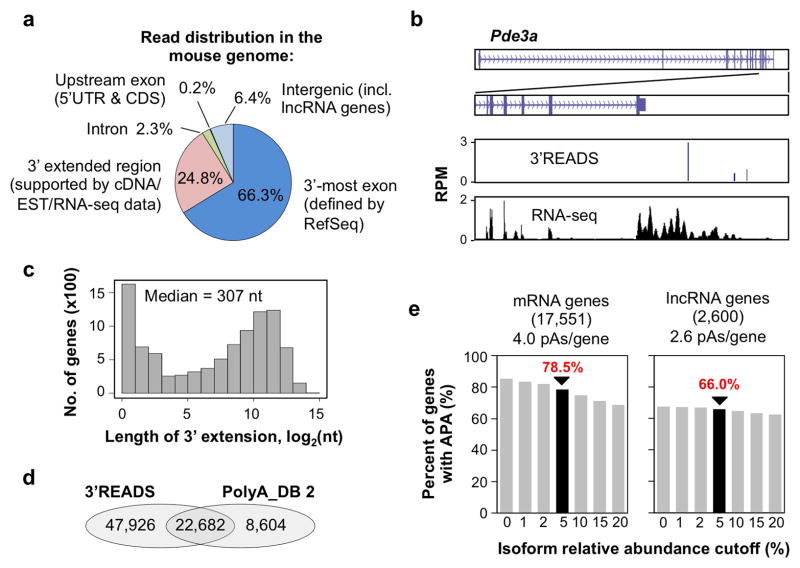
Using 3′READS, we set out to comprehensively map pAs in mouse which, despite its central role as a model for mammalian biology, has poor pA annotations compared to human 17. We used RNA samples from 1) male and female whole bodies, 2) embryos at 11, 15, and 17 days, 3) brain and testis tissues at different postnatal stages, and 4) over 11 cell lines, yielding ~54 million PASS reads in total (Supplementary Table 2). We found ~25% of the PASS reads were aligned to regions downstream of RefSeq-supported 3′ ends, indicating incomplete gene annotation by the RefSeq database (Figures 2a and 2b). To address this issue, we used cDNA/EST sequences from NCBI, and strand-specific RNA-seq reads from the ENCODE project 25 to connect the pAs mapped by 3′READS to RefSeq-defined genic regions (see Methods for detail). This step resulted in extension of the 3′ end for 9,612 genes with the median extension length of 307 nt (Figure 2c). Our 3′READS data significantly expanded pAs currently annotated for mouse in the PolyA_DB 2 database by more than 2.5-fold (Figure 2d). Consistent with our previous results 5, we found 42% of the pAs were associated with AAUAAA, 15% with AUUAAA, 22% with variants of A[A/U]UAAA, and 21% were not associated with any prominent PAS in the −40 to −1 nt region (Supplementary Figure 4a).
Figure 2. Mouse pAs identified by 3′READS.
(a) Distribution of PASS reads in the mouse genome (data from all the samples are included). (b) An example gene (Pde3a) showing PASS reads from 3′READS and RNA-seq reads (ENCODE project) used to assign pAs to the gene. (c) Histogram of the length of 3′ end extension for RefSeq mRNA genes (9,612 genes with extension > 0 nt). The median is indicated. (d) Venn diagram comparing pAs in the PolyA_DB 2 database with those identified in this study. (e) Percent of mRNA or lncRNA genes considered to have APA at different isoform relative abundance cutoffs. Numbers of mRNA and lncRNA genes analyzed are indicated.
Overall, we examined 17,551 mRNA genes and 2,600 lncRNA genes in the mouse genome. When the relative abundance of an APA isoform was required to be above 5% in at least one sample, 78.5% of mRNA genes and 66.0% of lncRNA genes were found to have APA (Figure 2e). On average, we found 4.0 pAs per mRNA gene, and 2.6 pAs per lncRNA gene (Figure 2e and see Supplementary Figure 4b for histogram of number of pAs per gene). Data simulation indicated that our mouse pA collection for mRNA genes was near saturation with the RNA samples used in the study (Supplementary Figure 4c). Overall, the pAs in mRNA and lncRNA genes were surrounded by similar cis elements (Supplementary Figure 5 and Supplementary Table 3).
pAs in A-rich regions
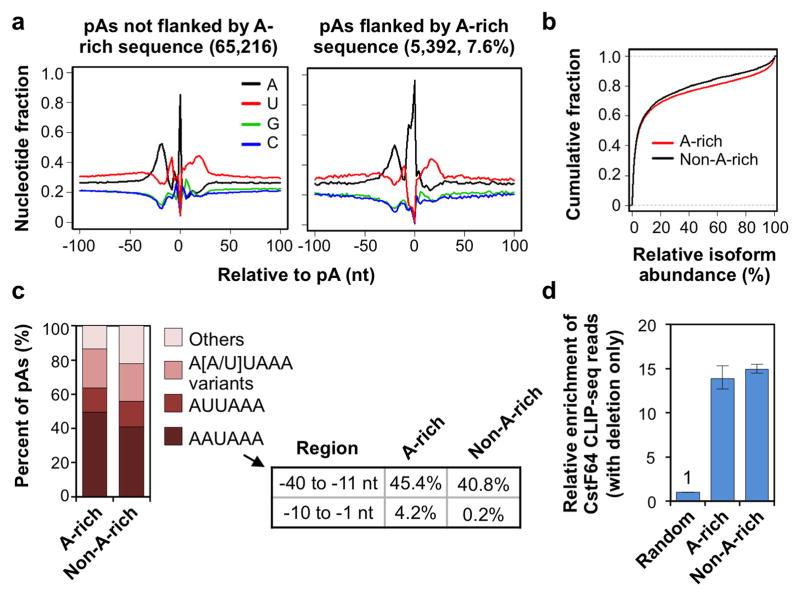
Interestingly, 5,392 identified pAs (7.6% of total) were surrounded by genomic A-rich sequences, which would have been filtered out as internal priming candidates if a method employing oligo(dT) in reverse transcription had been used 26. Except for the A-rich sequences around the cleavage site, these pAs, named A-rich pAs for simplicity, had similar upstream A-rich and downstream U-rich peaks around the cleavage site to regular pAs (Figure 3a). This is in stark contrast to the internal A-rich sequences that led to non-PASS reads (Figure 1d). Notably, transcripts using A-rich pAs were generally more abundant than those using non-A-rich pAs (Figure 3b), and A-rich pAs were more likely to be associated with AAUAAA than non-A-rich pAs (Figure 3c): ~50% of the A-rich pAs had AAUAAA in the −40 to −1 nt region as compared to 41% for the non-A-rich pAs.
Figure 3. Comparison of pAs flanked by A-rich or non-A-rich sequences.
(a) Nucleotide profile around the pAs identified in this study. Left, pAs not flanked by A-rich sequences; right, pAs flanked by A-rich sequences. (b) Relative abundance of isoforms using pAs flanked by A-rich sequences or other pAs (non-A-rich). The cumulative fraction curves based on all genes analyzed in this study and on all samples combined. (c) PAS distribution in the −40 to −1 nt region for A-rich and non-A-rich pAs. The frequencies of occurrence of AAUAAA in −40 to −11 nt and −10 to −1 nt regions for A-rich and non-A-rich pAs are shown in a table. (d) Enrichment of CstF64 CLIP-seq reads around A-rich or non-A-rich pAs relative to randomly selected gene regions. Error bars represent the 90% confidence interval derived from bootstrapping (1,000 ×) of data.
We next wanted to validate the A-rich pAs identified by 3′READS. We reasoned that the surrounding regions of real pAs should have stronger binding of cleavage and polyadenylation factors than random regions in the gene. To this end, we carried out cross-linking immunoprecipitation and high-throughput sequencing (CLIP-seq) using C2C12 cells and an antibody against the core cleavage and polyadenylation factor CstF64 (Supplementary Figure 6 and see Methods for detail). As shown in Figure 3d, A-rich and non-rich pAs had similar CstF64 bindings in their surrounding regions and both types had significantly stronger (>13-fold) CstF64 association than did randomly selected regions in genes. This result further confirms that the A-rich pAs we identified here are genuine sites.
Alternative pAs in mRNA and lnRNA genes
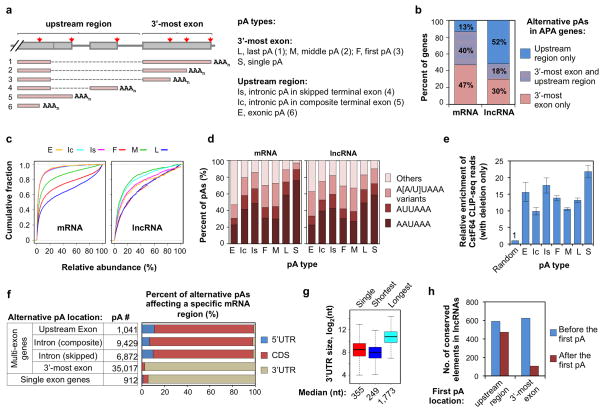
We next examined alternative pAs in the mouse genome. pAs can be located in the 3′-most exon or upstream regions (Figure 4a). pAs in the former group were further divided into the “single” type when there was only one pA in the 3′-most exon, or the “first”, “middle” and “last” types, according to their relative locations (Figure 4a). pAs in upstream regions were grouped into the “intronic” type, if there was RefSeq evidence indicating that the pA could be removed by splicing, or the “exonic” type otherwise. As we did previously 27, intronic pAs were further separated into two sub-groups: intronic pAs in skipped terminal exons or composite terminal exons (Figure 4a).
Figure 4. APA of mouse mRNA and lncRNA genes.
(a) Schematic of pA types. The full and short names for different pA types are indicated. The type number in parenthesis corresponds to the isoform number shown in the graph. Dotted lines indicate splicing. (b) Distribution of alternative pAs in different regions of mRNA or lncRNA genes. The p-value for the difference in distribution between mRNA and lnRNA genes is 0 (Chi-squared test). (c) Relative abundance of APA isoforms using different types of pA. The cumulative fraction curve is based on all genes analyzed in this study and on all samples combined. (d) Frequency of various PAS types for different types of pAs in mRNA and lncRNA genes. (e) Enrichment of CstF64 CLIP-seq reads around different types of pAs relative to randomly selected gene regions. Error bars represent the 90% confidence interval derived from bootstrapping (1,000 x) of data. (f) mRNA regions affected by alternative pAs. pAs were grouped based on gene type (multi-exon or single exon) and pA location. mRNA regions were separated into 5′UTR, CDS and 3′UTR. For intronic pAs, the mRNA region affected was defined by the exon immediately upstream of the pA. (g) Distribution of 3′UTR length for genes without alternative pAs in the 3′UTR (single), and genes with APA in the 3′UTR. For the latter, the shortest and longest isoforms were used for analysis. (h) APA regulates conserved elements in lncRNAs. Conserved elements are based on 30 mammalian species (see Methods for detail). The numbers of the conserved elements upstream or downstream of the first pA were calculated. In total, 599 and 391 lncRNA genes have the first pA in the upstream region and 3′-most exon, respectively. Only isoforms with relative expression level > 20% were analyzed.
We found mRNA genes were more likely to have alternative pAs in the 3′-most exon, whereas lncRNA genes were more likely to have pAs in upstream regions (Figure 4b): 70% of lncRNA genes with APA had intronic or upstream exonic pAs compared to 53% for mRNA genes with APA. This notion was further supported by relative expression levels of different APA isoforms (Figure 4c): for mRNA genes, APA isoforms using 3′-most exon pAs were expressed at much higher levels than those using upstream region pAs, whereas the difference between these isoform types was much smaller for lncRNA genes. The PAS usage pattern for different pA types in lncRNA genes was similar to that for mRNA genes (Figure 4d). For example, the single and last pAs were more likely to be associated with AAUAAA than other types. Confirming the overall validity of identified pAs, all types of pAs had significantly stronger CstF64 binding than randomly selected regions (Figure 4e). However, pAs in different locations appeared to have distinct interactions with CstF64: single pAs in genes had the highest CstF64 binding, and pAs in composite terminal exons and in the middle of 3′-most exons had the lowest binding. Future analyses are needed to address underlying mechanisms for variation of CstF64 binding.
As shown in Figure 4f, about one third of all alternative pAs in multi-exon mRNA genes were in upstream regions, most of which (>97%) led to isoforms with different CDS. APA in the 3′-most exon on average resulted in ~7-fold difference in 3′UTR length between the shortest and longest isoforms (medians of 249 nt and 1,773 nt for these isoforms, respectively, Figure 4g). Therefore, APA can significantly impact the proteome and mRNA metabolism in the cell. To understand the significance of APA for lncRNAs, we examined pA locations relative to conserved elements of lncRNAs, assuming these elements are important for lncRNA functions. We found that ~45% of the conserved elements were downstream of the first pA when the site was located in an intron/upstream exon, or ~15% when it was in the 3′-most exon (Figure 4h), suggesting that APA can play a significant role in regulation of lncRNA functions.
Regulation of APA in development and differentiation
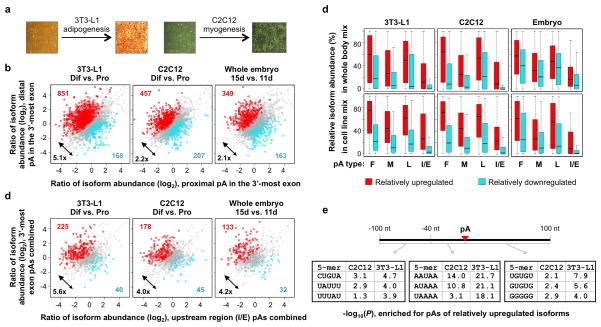
We previously reported progressive lengthening of 3′UTRs in mouse development and in cell differentiation by surveying about 1,000 genes using microarray data 10. With 3′READS we now can have more systematic analysis of APA. To this end, we induced differentiation of C2C12 and 3T3-L1 cells, which represent myogenesis and adipogenesis, respectively (Figure 5a). In addition, we compared whole embryos at 11 and 15 embryonic days. We first examined APA in the 3′-most exon (Figure 5b). Consistent with our previous findings, genes having relatively upregulated distal pA isoforms significantly outnumbered those having relatively upregulated proximal pA isoforms in 3T3-L1 differentiation, C2C12 differentiation, and embryonic development (by 5.1-, 2.2-, and 2.1-fold, respectively). In addition, the number of APA events consistently regulated in these processes was significantly greater than that of events oppositely regulated (Supplementary Figure 7a). However, distinct APA events in each sample set could be clearly discerned. An example of consistent APA regulation in cell differentiation is shown in Supplementary Figure 8a. This result indicates general 3′UTR lengthening in development and differentiation.
Figure 5. General transcript lengthening in cell differentiation and embryonic development.
(a) Schematic showing differentiation of 3T3-L1 and C2C12 cells. 3T3-L1 cells were stained with the Oil Red O (ORO). (b) Regulation of alternative pAs in the 3′-most exon. The number of genes with significantly upregulated distal pA isoforms (red dots) and the number of genes with significantly upregulated proximal pA isoforms (cyan dots) are indicated in each graph. The ratio of the numbers (upregulated vs. downregulated) is also indicated to show the general trend of regulation. Significantly regulated isoforms are those with p-value <0.05 (Fisher’s Exact test) and abundance change >5%. Only the two most abundant isoforms for each gene were analyzed. (c) Regulation of alternative pAs in upstream regions. As in (b), except that upstream region pA isoforms were compared with 3′-most exon isoforms. All upstream region pA isoforms were grouped together and so were the 3′-most exon isoforms. (d) Isoforms using strong pAs tend to be relatively upregulated in differentiation and development. Isoform relative abundance in whole body mix (top panels) and in in cell line mix (bottom panels, cell line mix 1 in Supplementary Table 2) for those upregulated (UP) and downregulated (DN) in differentiation and development. Regulated isoforms are those with p-value < 0.05 (Fisher’s Exact test) and abundance change > 5%, compared to all other isoforms of the same gene. Differentiation of 3T3-L1 and C2C12 cells and embryonic development are shown. Upstream region pAs are also shown as I/E (intron/upstream exon) pAs. (e) Top 5-mers consistently enriched for regions around the pAs of upregulated isoforms in differentiation of C2C12 and 3T3-L1 cells. P-value was based on the Fisher’s Exact test comparing pAs of upregulated isoforms with those of downregulated ones. Three regions surrounding the pA were analyzed, i.e., −100 to −41 nt, −40 to −1 nt, and +1 to +100 nt.
We next examined alternative pAs in upstream regions. We first grouped together all isoforms using intron/upstream exon pAs for each gene and compared their change of abundance with that of isoforms using 3′-most exon pAs, which were also grouped together (Figure 5c). Strikingly, we found that more genes had relatively upregulated 3′-most exon pA isoforms than had relatively upregulated intron/upstream exon pA isoforms, by 5.6-, 4.0-, and 4.2-fold for 3T3-L1 differentiation, C2C12 differentiation, and embryonic development, respectively. Like APA in the 3′-most exon, both commonly and distinctly regulated APA events in these sample sets could be identified (Supplementary Figure 7b). An example of consistent APA regulation in cell differentiation is shown in Supplementary Figure 8b. Together with the data of APA in 3′-most exons, this result indicates that isoforms using promoter-distal pAs are generally upregulated in development and differentiation, regardless of intron/exon locations.
We next wanted to address whether the isoforms regulated in development and differentiation have common features other than their pA locations. We first examined isoform abundance in the whole body mix and cell line mix samples. As shown in Figure 5d, isoforms relatively upregulated in development and differentiation tend to have higher expression levels in these samples than those relatively downregulated, regardless of their pA locations. This suggests that isoforms with strong pAs are more likely to be upregulated than those with weak pAs. Consistent with this hypothesis, we found that 5-mers known to enhance cleavage and polyadenylation were enriched for regions around the pAs of upregulated isoforms (Figure 5e), including AAUAAA in the −40 to −1 nt region, UGUA and U-rich elements in the −100 to −41 nt region, and UGUG elements in the +1 to +100 nt downstream region. Consistently, upregulated isoforms were more likely to have AAUAAA compared to other PAS types than downregulated isoforms (Supplementary Figure 9). Thus, we conclude that pA strength is a significant parameter in determining APA regulation in development and differentiation.
Discussion
A number of deep sequencing methods for pA analysis have recently been reported 23, 24, 28–32. However, most of these methods utilize the oligo(dT) sequence for reverse transcription, opening the possibility of internal priming. For example, more than half of the original pAs mapped by PolyA-seq are flanked by A-rich sequences (Supplementary Figure 2b), underscoring the severity of this problem. Direct RNA sequencing using the Helicos system 28 does not require reverse transcription, but this method can also be affected by internal A-rich sequences because of using oligo(dT) to fill the poly(A) tail region before sequencing. The key issue with internal priming is that it is impossible to determine whether the unaligned As in reads come from the real poly(A) tail or the oligo(dT) sequence in primer. We found that even under a stringent condition, such as the one used in 3′READS, RNAs with internal A-rich sequences can still bind oligo(dT)(Figure 1c). In addition, RNA fragments not from genomic A-rich regions can also bind oligo(dT)(Figure 1d), presumably due to oligo(A) tails. Surprisingly, these two types of RNA species can account for ~17% and ~60% of the total reads generated from CU5T45 oligo and oligo(dT)10–25, respectively. Thus, for pA mapping, it is critically important 1) not to use oligo(dT) for priming in reverse transcription and 2) to use unaligned As in reads for quality control. On the other hand, because 3′READS is not affected by the internal priming issue, we have been able to uncover nearly 8% of all mouse pAs that are located in genomic A-rich regions; they would have been removed by normal criteria to address internal priming 26.
We found a global trend of upregulation of isoforms using promoter-distal pAs in development and differentiation. The regulation of alternative pAs in the 3′-most exon is consistent with the result we previously reported using microarray data 33, 34, and is in line with the notation that proliferating cells generally express short 3′UTR isoforms 11. The regulation of intron/upstream exon pAs in development and differentiation, however, has never been reported mainly due to technical limitations in using microarrays. Our result indicates that both intron/upstream exon pAs and 3′-most exon pAs follow the same global trend of regulation, leading to transcript lengthening. A number of mechanisms may contribute to this phenomenon. First, our cis element analysis indicated that, compared to downregulated isoforms, upregulated ones tend to have pAs with enhancing elements for pA usage, such as upstream AAUAAA, UGUA, and U-rich elements and downstream GU-rich elements. This suggests that the 3′ end processing activity is weakening in development and differentiation. This is consistent with our previous result showing downregulation of mRNA expression in development and differentiation for many genes involved in 3′ end processing 34. Importantly, this result readily explains why not all genes have 3′UTR lengthening in these processes: it is the strength of pA rather than its location that is the primary determinant for regulation. Second, a recent study by the Gideon Dreyfuss lab reported a significant role of U1 snRNP in regulation of transcript length 35. Modulation of the activity of U1 snRNP relative to that of 3′ end processing in development and differentiation can contribute to more usage of promoter-distal pAs. The detailed mechanisms need to be elucidated in the future, so are the biological implications of this phenomenon.
Online Methods
Cell culture, tissue harvest and RNA samples
Mouse cell lines Tib75, CMT93, B16, F9, and C2C12 were cultured in DMEM with 10% fetal bovine serum (FBS) and NIH3T3, 3T3-L1 and MC3T3-E1 cells were cultured in DMEM with 10% fetal calf serum (FCS). Differentiation of C2C12 and 3T3-L1 cells was carried out as previously described 10, 36. Differentiated C2C12 and 3T3-L1 cells correspond to 4 days and 8 days after initiation of differentiation, respectively. Mouse whole body tissue RNA sample was purchased from SABiosciences and cell line mix 1 sample was purchased from Agilent. All mouse embryos and pups used in this study were derived from mating of FVB females and males. To obtain embryos, pregnant females were sacrificed by CO2 asphyxiation at 11, 15 and 17 days of pregnancy. Embryos were carefully dissected free of decidual and extraembryonic tissues. Postnatal pups were sacrificed at 3 weeks, 6 weeks and 9 weeks after birth. The whole brain and testes cleared from tunica albuginea and the seminiferous tubules were removed. All tissue samples were flash-frozen in liquid nitrogen. All animal work was conducted according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at UMDNJ-New Jersey Medical School. Total RNA from cells/tissues was isolated using Trizol (Invitrogen) or the Qiagen RNeasy kit. RNA samples were checked for integrity by Agilent Bioanalyzer using the RNA pico6000 kit (Agilent Technologies). RNA samples with the RNA integrity number (RIN) above 8.0 were used for subsequent processing.
Plasmids
Constructs expressing transcripts containing 15 or 60 terminal As, named pALL-A15 and pALL-p60, respectively, were obtained from Dr. Lance Ford (Bioo Scientific). RNAs were made by in vitro transcription using SP6 RNA polymerase.
3′READS
Total RNA was subjected to 1 round of poly(A) selection using the Poly(A)Purist™ MAG kit (Ambion) according to manufacturer’s protocol, followed by fragmentation using Ambion’s RNA fragmentation kit at 70°C for 5 min. Poly(A)-containing RNA fragments were isolated using the CU5T45 oligo (Sigma) which were bound to the MyOne streptavidin C1 beads (Invitrogen) through biotin at its 5′ end. The oligo(dT)10–25-coated beads were from the Poly(A)Purist MAG kit. Binding of RNA with CU5T45 oligo-coated beads was carried out at room temperature for 1 hr in the binding buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA), followed by washing with the low salt buffer (10 mM Tris-HCl pH 7.5, 1 mM NaCl, 1 mM EDTA, 10% Formamide). RNA bound to the CU5T45 oligo was digested with RNase H (5 U in 50 μl reaction volume) at 37°C for 1 hr, which also eluted RNA from the beads. Eluted RNA fragments were purified by Phenol: Chloroform extraction and Ethanol precipitation, followed by phosphorylation of the 5′ end with T4 kinase (NEB). Phosphorylated RNA was then purified by the RNeasy kit (Qiagen) and was sequentially ligated to a 5′-adenylated 3′ adapter with the truncated T4 RNA ligase II (Bioo Scientific) and to a 5′ adapter with T4 RNA ligase I (NEB). The resultant RNA was reverse-transcribed to cDNA with Superscript III (Invitrogen), and the cDNA was amplified by 12 cycles of PCR with Phusion high fidelity polymerase (NEB). We designed adapter sequences so that the RNA fragments can be sequenced from the 5′ end (forward sequencing) or from the 3′ end (reverse sequencing). Adapter sequences and primer sequences are listed in Supplementary Table 2. cDNA libraries were sequenced on an Illumina Genome Analyzer GAIIx (1×72 nt). RNA-seq was carried out using essentially the same protocol except that 1) the 3′ end region extraction step using the CU5T45 oligo was omitted and 2) fragmented RNA was dephosphorylated at the 3′ end with shrimp alkaline phosphatase (Roche) before ligation with adapters.
CLIP-seq
Our CLIP-seq method was largely based on the protocol used by Wang et al. 37 with some minor modifications. Briefly, three 15-cm dishes of C2C12 cells were UV-irradiated using Stratalinker (Stratagene) at 254 nm with 2000 × 100 μJ/cm2 and then lysed on ice. The lysate was treated with 20 U/ml RNase T1 (Fermentas) at 25°C for 10 min, followed by centrifugation at 14, 000 × g for 5 min at 4°C. The supernatant was incubated for 2 hours at 4°C with mouse anti-CstF64 monoclonal antibody (gift from Dr. Clint MacDonald, Texas Tech University) conjugated to magnetic Protein G Dynabeads (Invitrogen). Co-immunoprecipitated RNA fragments were dephosphorylated by calf intestinal alkaline phosphatase (NEB) and end-labeled with [γ-32P]-ATP by T4 polynucleotide kinase (NEB). The RNA-protein complex was then resolved with Bis-Tris buffered SDS-PAGE, and transferred to nitrocellulose membrane, which was then exposed to X-ray film. The 85–150 kDa region was cut out, and the RNA from the cut-out membrane was isolated. cDNA library preparation was based on the protocol for Illumina’s Small RNA v1.5 kit. The libraries were sequenced on an Illumina Genome Analyzer GAIIx (1×70 nt).
Identification of pA
For forward sequencing, the reads were aligned to the reference genome (mm9) and exon-exon junction database by Bowtie 38 using the first 25 nt as seed, allowing up to 2 mismatches. The exon-exon junction database contained all possible exon-exon junction sequences in the genome 9. Aligned reads were scored from 5′ to 3′ using the scheme: +1 for match and −2 for mismatch. The position in a read with the maximum score was considered as the last aligned position (LAP). The best hit for each read was chosen, and was considered uniquely mapped if its score was greater than the second best hit by at least 5. If a read contained ≥ 2 non-genomic As immediately after the LAP, we considered the read as a polyA site supporting (PASS) read and the cleavage site is immediately downstream of the LAP. For data from reverse sequencing, we first trimmed the 5′ region of read, including the first 4 random nucleotides and subsequent continuous Ts. We then aligned the reads to the reference genome and exon-exon junction database by Bowtie using the first 36 nt, allowing up to 2 mismatches. For uniquely aligned reads, we compared the trimmed Ts with reference genome and exon-exon junction sequences. The reads with at least 2 non-genomic Ts are PASS reads. Since each pA can have multiple cleavage positions in a small window 5, we merged cleavage positions into pAs: we first clustered together cleavage positions located within 24 nt from one another. If a cluster size was ≤24 nt, the position with the greatest number of PASS reads was used as the representative position for the pA. If a cluster was > 24 nt, we first identified the cleavage site with the greatest number of PASS reads and re-clustered reads located > 24 nt from the position. This process was repeated until all pAs in the cluster were defined. To reduce false positives, we required a real pA to have 1) PASS reads from more than one sample, and 2) ≥2 distinct PASS reads (defined by the number of As and the 4 random Ns) and ≥5% of all PASS reads for the same gene in at least one sample.
Extension of the 3′ end of genes
We used cDNA, EST and directional paired-end RNA-seq data from the ENCODE project 25 to extend the 3′ ends defined by RefSeq. An extended region is between the 3′ end defined by RefSeq and pAs mapped by 3′READS and is covered by cDNA/EST sequences or RNA-seq reads without a gap greater than 40 nt. We also required that the 3′UTR extension does not exceed the transcription start site of the downstream gene. For genes located in an intron of another gene, the 3′ end extension was required not to go beyond the 3′SS of the intron.
pAs flanked by A-rich sequences
An A-rich sequence around a pA was defined as ≥6 consecutive As or ≥7 As in a 10 nt window in the −10 to +10 nt region around the pA. pAs associated with A-rich sequences are typically filtered because they can be derived from internal priming when a primer containing oligo(dT) is used in reverse transcription 17.
APA analysis
The expression level of each APA isoform was indicated by the Reads Per Million (RPM) value, which was calculated as the total number of PASS reads normalized to per million total uniquely mapped PASS reads for the sample. For analysis of APA regulation, the Fisher’s Exact test was used to examine whether the abundance of an APA isoform compared to that of other isoforms was significantly different between two comparing samples.
lncRNA genes
lncRNAs were based on noncoding genes annotated in the RefSeq and Ensemble databases, excluding rRNAs, microRNAs, snoRNAs, snRNAs, and tRNAs, and those overlapping with mRNA genes on the same strand. We required lncRNAs to be longer than 200 nt. Conserved elements were obtained from the UCSC table browser (Euarchontoglires Conserved Elements for mm9)39 and were mapped to exonic regions of lncRNAs.
Identification of PAS
As previously described 5, we first selected the hexamer with the highest occurrence in the −40 to −1 nt region upstream of all pAs. Once a hexamer (PAS) was identified, all associated pAs were removed and the remaining pAs were searched for the next most prominent hexamer. This process was repeated until the top 10 most prominent PAS hexamers were identified.
Cis element analysis
We studied cis elements in four regions around the pA, i.e., −100 to −41 nt, −40 to −1 nt, +1 to +40 nt, and +41 to +100 nt. As previously described 40, for each region, we calculated Zoe for each hexamer to reflect the difference between observed and expected occurrences:
, where No(H) is the observed occurrence of hexamer H, Ne(H) is the expected occurrence based on the 1st-order Markov Chain model of the region, and voe(H) is the variance of No(H)−Ne(H) 40.
CLIP-seq data analysis
CLIP-seq reads were aligned to the mouse genome (mm9) using the program novoalign (http://www.novocraft.com/). Identical reads in a sample were counted only once. The reads with deletion(s), which are caused by skipping of reverse transcriptase at the UV crosslinked nucleotide 41, were used for analysis and the genome location corresponding to the deletion was used to indicate the read location. The enrichment score was the ratio of read density within 40 nt around the pA to that in randomly selected regions of the same size. A bootstrap resampling method was used to get the 90% confidence interval as described before 33. The Z score for hexamers was calculated by Z = N(H)/SD(H), where N(H) is the occurrence of hexamer H, and SD(H) is the standard deviation of N(H).
Supplementary Material
Acknowledgments
We thank Yun Zhao and Anita Antes for technical assistance, Victor Jin for help with 3T3-L1 differentiation, Clint MacDonald (Texas Tech University) for the anti-CstF64 antibody and Sam Gunderson (Rutgers University) for helpful discussions. This work was funded by NIH grants (GM084089 and DK094207) to BT.
Footnotes
Author Contributions
Conceived and designed the experiments: MH, ZJ, BT. Performed the experiments: MH, DZ, WL (Luo), BY. Analyzed the data: ZJ, WL (Li), JYP. Contributed reagents/materials: GY. Wrote the paper: MH, ZJ, BT.
Competing Financial Interests
US Patent application PCT/US2012/052122 based on this work is pending.
References
- 1.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 2.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian B, Graber JH. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA. 2011 doi: 10.1002/wrna.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. WIREs RNA. 2011;2:23–31. doi: 10.1002/wrna.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh P, et al. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer research. 2009;69:9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen LL, Carmichael GG. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA. 2010;1:2–21. doi: 10.1002/wrna.5. [DOI] [PubMed] [Google Scholar]
- 16.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JY, Yeh I, Park JY, Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007;35:D165–168. doi: 10.1093/nar/gkl870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brockman JM, et al. PACdb: PolyA Cleavage Site and 3′-UTR Database. Bioinformatics. 2005;21:3691–3693. doi: 10.1093/bioinformatics/bti589. [DOI] [PubMed] [Google Scholar]
- 19.Nam DK, et al. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci U S A. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt MJ, Norbury CJ. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip Rev RNA. 2010;1:142–151. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- 21.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. Embo J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shepard PJ, et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. Rna. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium TEP. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JY, Park JY, Tian B. Identification of mRNA polyadenylation sites in genomes using cDNA sequences, expressed sequence tags, and Trace. Methods Mol Biol. 2008;419:23–37. doi: 10.1007/978-1-59745-033-1_2. [DOI] [PubMed] [Google Scholar]
- 27.Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangone M, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Y, et al. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011 doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fox-Walsh K, Davis-Turak J, Zhou Y, Li H, Fu XD. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Genomics. 2011;98:266–271. doi: 10.1016/j.ygeno.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon OK, Brem RB. Noncanonical transcript forms in yeast and their regulation during environmental stress. Rna. 2010;16:1256–1267. doi: 10.1261/rna.2038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji Z, et al. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol. 2011;7:534. doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Tollervey J, Briese M, Turner D, Ule J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods. 2009;48:287–293. doi: 10.1016/j.ymeth.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.