Abstract
Chromatin immunoprecipitation (ChIP) assays have contributed greatly to our understanding of the role of histone modifications in gene regulation. However, a major limitation is that they do not permit analysis with single cell resolution thus confounding analyses of heterogeneous cell populations. Herein we present a new method which permits visualization of histone modifications of single genomic loci with single-cell resolution in formaldehyde-fixed paraffin-embedded tissue sections based on combined use of In Situ Hybridization (ISH) and Proximity Ligation Assays (PLA). Using this method we show that H3K4dime of the MYH11 locus is restricted to the smooth muscle cell (SMC) lineage in human and mouse tissue sections, and that the mark persists even in phenotypically modulated SMC within atherosclerotic lesions that show no detectable expression of SMC marker genes. This new methodology has promise for broad applications in the study of epigenetic mechanisms in complex multicellular tissues in development and disease.
Keywords: Proximity Ligation Assay, Epigenetics, Cell lineage, Differentiation, Smooth, muscle cells, atherosclerosis
Introduction
Epigenetics is defined as a heritable code other than the genomic sequence and include histone post-translational modifications, DNA methylation and ATP-dependent chromatin remodeling1–3. Differentiation from embryonic stem cells (ESCs) to multiple differentiated cell types requires establishment of spatiotemporal expression of gene patterns4, 5. As one example out of many6–9, we have previously shown that a SMC-specific epigenetic repertoire of histone modifications are acquired during development of SMC from ESC, including H3K4dime of SMC marker genes such as ACTA2 and MYH11, the latter being the most specific marker of SMC lineage10–12. Importantly, results showthat H3K4dime of these genes is restricted to SMCs and absent in non-SMCs. Moreover, H3K4dime enrichment of these genes persists when cultured SMCs are induced to undergo phenotypic switching to a less differentiated state11, a process dependent on activation of the ESC pluripotency factor KLF4 both in vivo13 and in cultured cells11. These results suggest that H3K4dime of the MYH11 and other SMC marker gene loci represent stable epigenetic markers of SMC lineage. However, a major limitation in studies thus far is that data are derived nearly exclusively from studies in cultured SMC which are poorly differentiated since cell culture systems do not recapitulate complex environmental cues that regulate SMC differentiation in vivo14, 15 under normal as well as in pathological conditions. Moreover, it is impossible to obtain chromatin exclusively derived from SMCs from tissue sources, since all SMC-containing tissues contain multiple other cell types. Conversely, all “non-SMC” tissues contain large numbers of SMCs since all tissues are vascularized. As such, conventional ChIP16, 17 analyses cannot be used to rigorously test if H3K4dime of SMC gene loci are an exclusive epigenetic signature of SMC lineage in vivo. Indeed, a major general limitation of ChIP assays is that they do not permit analysis of histone modifications at a given gene locus within individual cells thus confounding interpretation of analyses of heterogeneous cell populations, and precluding direct assessment of the role of specific histone modifications within individual cell types within complex multicellular tissues including disease specimens. For example, ChIP analyses on a tumor biopsy or atherosclerotic tissue specimen, represents a composite signal derived from the many different cell types present within that tissue sample. Although one may perform ChIP assays on a given cell population obtained by cell sorting, such analyses of course results in loss of critical information regarding the spatial orientation of cells within tissues, and may be subject to epigenetic changes that occur during the tissue dissociation and/or the sorting procedures.
Here, we describe a new method that permits visualization of histone modifications at a single genomic locus in human and mouse formalin-fixed paraffin-embedded tissue sections that is an equivalent of a “single-cell ChIP” assay by combination of ISH and PLA methods. Moreover, using this new methodology, we demonstrate that H3K4dime of the MYH11 gene locus is a highly specific marker of SMC lineage in vivo that persists even in phenotypically modulated SMC within atherosclerotic lesions that lack detectable expression of endogenous SMC marker genes.
Results
Our overall strategy made use of the Proximity Ligation Assay (PLA)18–20, to detect proximity between a biotin-labeled probe targeting the MYH11 promoter and H3K4dime at this locus (Fig. 1a). PLA is widely performed for detection of protein/protein interaction or protein post-translation modifications in both cultured cells and tissue sections20, 21 but to our knowledge has not been used for assessing histone modifications at specific gene loci. We sought to combine PLA with ISH methods using two primary antibodies targeting: 1) histone modifications; and 2) biotin residues included in a probe annealing to the genomic locus of interest. The feasibility of the approach was first assessed by estimating intermolecular distances between DNA and histone tails to ascertain the compatibility with the PLA range of detection of approximately 40 nm (Supplementary Fig. 1). The major methodological steps included the following (Fig. 1b): 1) immunostaining of human or mouse formalin-fixed paraffin embedded tissue sections with antibodies to SMC or non-SMC markers; 2) ISH with a biotinylated DNA probe targeting the MYH11 locus; 3) PLA including incubation with anti-biotin (rabbit) and H3K4dime (mouse) primary antibodies. We validated that nuclear detection of H3K4dime was preserved following the ISH procedure (Supplementary Fig. 2) and mapped H3K4dime enrichment of the MYH11 promoter to identify boundaries for our ISH probe (Supplementary Fig. 3). Moreover, for validation of our ISH procedure, we used a 5-TAMRA dUTP labeled Y chromosome probe in human samples from male patients, and we observed similar hybridization efficiencies of the Y chromosome in SMCs and non-SMCs cell types (Supplementary Fig. 4).
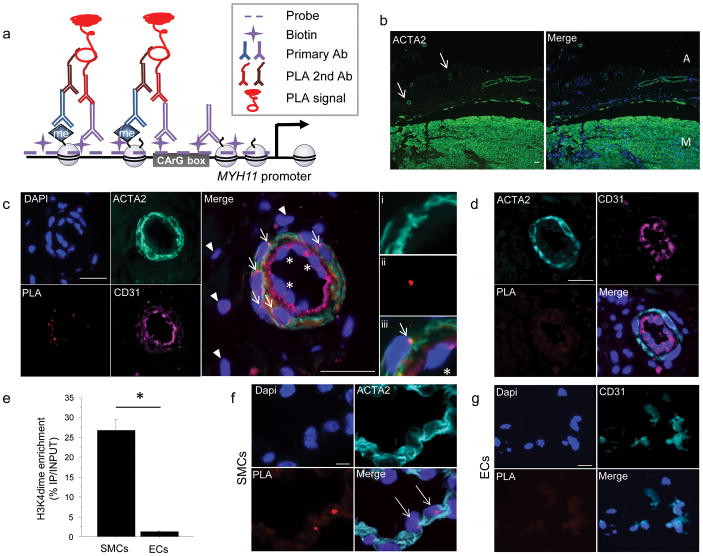
Figure 1. ISH-PLA: a new method of detection of histone modificationsat a single genomic locus in tissuesections.
(a) Schematic of the combined in situ hybridization (ISH) and Proximity Ligation Assay (PLA) method for detecting H3K4dime of the MYH11 promoter. (b) Workflow of the ISH-PLA procedures. (c) Immunostaining of 5 μm-thick sections of human carotid artery for ACTA2 and DAPI (diamidinophenyindole). Two distinct layers are identified: the media (M) and the adventitia (A). Small vessels consisting of ACTA2+ SMCs are visualized within the adventitia (arrows). Scale bar = 100 μm. (d) ISH-PLA in adventitial small arteries (n = 5). PLA amplification (PLA+) is visualized as a red spot localized within nuclei. MYH11 H3K4dime PLA+ signal is restricted to ACTA2+ medial SMCs (arrows) and is absent from CD31+ ECs (stars) as well as adventitial fibroblasts (arrow heads). (i–iii): High magnification with i) ACTA2, ii) PLA and iii) merge.Scale bar = 50 μm. (e)ISH-PLA negative control using an empty vector probe in adventitial vessel of human carotid artery sections. A total absence of PLA amplification demonstrates that hybridization of the biotin-labeled MYH11 probe is required for ISH-PLA amplification. Scale bar = 50 μm (f) Conventional ChIP assays showing enrichment of H3K4dime of MYH11 in SMCs but not in ECs. Mean ± s.d.; n = 3; *P < 0.05. (g) ISH-PLA in SMCs (n = 3) and ECs (n = 3) in vitro. MYH11 H3K4dime PLA amplification is restricted to SMCs.Scale bars = 10 μm.
We performed ISH-PLA detection of H3K4dime at the MYH11 locus in human coronaryarteries since these are highly relevant to atheroscleroticdisease, and these vessels contain three distinct cell layers: 1) the intima consisting of endothelial cells (ECs); 2) the media composed primarily of Smooth muscle α-actin+ (ACTA2+) SMCs; and 3) the adventitia mainly composed of fibroblasts negative for SMC markers but with abundant small blood vessels (Fig. 1c). We initially focused on small arteries within the adventitia of coronary arteries which have a well-defined SMC layer. MYH11 H3K4dime PLA+ signal (i.e. visualized by red spots within the nucleus) was observed exclusively within ACTA2+ medial SMCs (Fig. 1d) whereas adventitial cells and ECs (CD31+) were negative. An empty biotinylated vector was used as a negative control and showed no PLA amplification in SMCs and non-SMCs demonstrating that the MYH11 probewasrequired for PLA amplification (Fig. 1e). To further validate our ISH-PLA method, we compared results of ISH-PLA and ChIP assays in cultured cells (Fig. 1f,g and Supplementary Fig. 5). These results showed complete concordance in that H3K4dime enrichment of the MYH11 locus was observed exclusively in SMCs by both ChIP and ISH-PLA analyses.
To provide a model system for testing the stability of the MYH11 H3K4dime mark in phenotypically modulated SMCs in vivo, we developed a SMC lineage tracing system by crossing Myh11-CreERT2 mice22 with ROSA26 STOP-floxeYFP+/+ mice(SMC-eYFP+/+ mice)(Fig. 2a). Tamoxifen treatment of mice from age six to eight weeks induced high efficiency eYFP expression exclusively in SMCs in all tissues examined (Fig. 2b,c and Supplementary Fig. 6). In contrast, no detectable expression of eYFP was observed without tamoxifen treatment (Fig. 2b) or in SMC-eYFP−/− mice treated with tamoxifen (Fig. 2d), whereas >95% of medial cells within SMC-eYPF+/+ mice were YFP+ (Fig. 2e) indicating high efficacy of SMC specific recombination. Moreover, a substantial fraction of eYFP+ SMCs within large arteries (i.e. aorta) of these SMC-eYFP+/+ mice were Myh11 H3K4dime PLA+ (Fig. 2e). In contrast, non-SMCs including ECs were Myh11 H3K4dime PLA−. No PLA signal was seen in negative controls using an empty cloning vector as probe in place of the MYH11 in situ probe (Fig. 2f). We estimated that ISH-PLA has an equivalent efficiency (66%) compared with regular FISH procedures(64%) (Supplementary Figs.4 and 7). Moreover, we found comparable efficiency using human and mouse tissue sections.
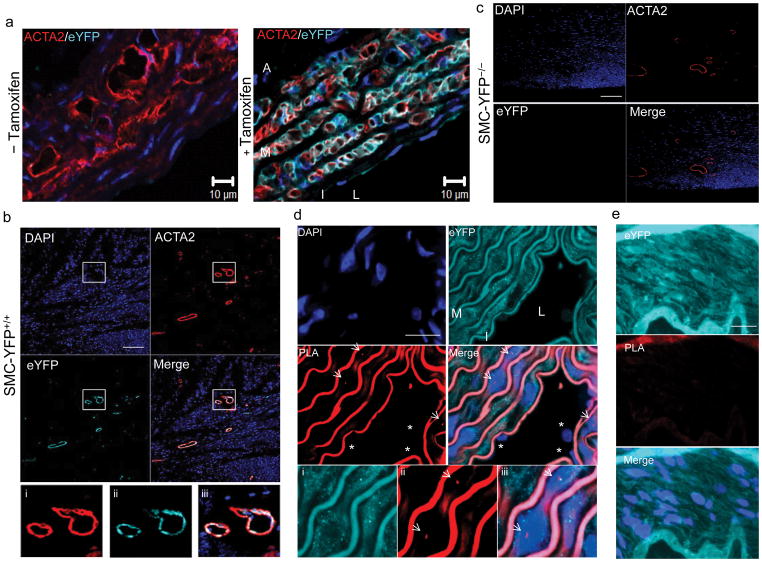
Figure 2. Validation of ISH-PLA using SMC lineage tracing mouse model.
(a) SMC lineage tracing was done by crossing Myh11-CreERT2 transgenic mice with ROSA26 STOPfloxeYFP+/+ mice and treating mice with tamoxifen between six and eight weeks of age thereby providing SMC-specific and permanent lineage tagging of SMCswith eYFP(n =5).(b) Immunostaining of the aorta of SMC-eYFP+/+ mice for eYFP and ACTA2 with (bottom image) and without (top image) tamoxifen injections. eYFP expression was observed only after tamoxifen treatmentand was exclusively observed in SMCs within the media (M), compared with the intima (I) and the adventitia (A) negative for eYFP staining. L: lumen. Scale bar = 10 μm. (c) Assessment of eYFP expression in heart tissue sections of SMC-eYFP+/+ mice by immunostaining for eYFP (cyan), ACTA2 (red) and Dapi (blue). eYFP expression is strictly restricted to ACTA2+ cells. (d) SMC-eYFP−/−mice present a complete lack of eYFP expression in ACTA2+ cells in heart tissue sections. Scale bar (c–d) = 100 μm. e. Results of ISH-PLA in aortas from SMC-eYFP+/+ mice showing that Myh11 H3K4dime PLA positivity was restricted to eYFP+ medial SMCs(arrows)(media: M). No Myh11 H3K4dime PLA signal was detected in ECs (stars) (intima: I). L: lumen. i–iii show eYFP+ PLA+ SMCs at higher magnification with i) eYFP, ii) PLA and iii) merge. Scale bar = 10 μm. f. Negative control wherein ISH was done using an empty biotinylated vector in SMC-eYFP+/+ mice. No Myh11 H3K4dime PLA+ cells were identified.Scale bar = 10 μm.
To test whether ISH-PLA can be readily adapted to examination of additional gene loci or histone modifications, we performed ISH-PLA experiments using a probe targeting the Vascular Endothelial-Cadherin (VEC) promoter (CDH5). CDH5 is a specific marker of the EC lineage23, and enrichment of H3K4dime is found on this promoter specifically in cultured ECs based on conventional ChIP assays11. Results of analyses of mouse and human tissue sections of healthy vessels showed specific CDH5 H3K4dime PLA+ staining of ECs. In contrast, SMCs (medial ACTA2+ human, eYFP+ in SMC-eYFP+/+ mouse) were strictly PLA− (Supplementary Fig. 8). We also performed PLA analysis of H4 acetylation of SM22α(Tagln), a gene selectively expressed in SMC within healthy adult blood vessels24, 25. Consistent with expectations, Tagln H4 acetylation PLA+ staining was restricted to SMC (Supplementary Fig. 9).
Multiple non-SMC including ECs, fibroblasts and ESCs in culture show marked enrichment of H3K27trime of the MYH11 gene locus based on ChIP assays, consistent with its transcriptional silencing in these cells11. Thus, to further validate our PLAmethod in vivo, we evaluated not only H3K4dime but also H3K27trime of the MYH11 locus in human carotid arteries (Fig. 3a). Consistent with PLA results in small arteries within the adventitia (Fig. 1d), and ChIP results in cultured cells (Supplementary Fig. 5), MYH11 H3K4dime PLA+ cells were restricted to ACTA2+ SMCs within the media (Fig. 3b) whereas adventitial fibroblasts and endothelial cells were negative (Fig. 3b and Supplementary Fig. 10). We did not observe any PLA amplification when we used a non-relevant probe (Fig. 3c). Medial SMC were negative for H3K27trime of the MYH11 locus, whereas ECs (CD31+ cells) (Fig. 3d) and adventitial fibroblasts (ACTA2− cells) (data not shown) were positive for this silencing mark based on PLA analyses. Consistent with these results in human coronary arteries, PLA analyses of human brain sections (Fig. 3e and Supplementary Fig. 11) showed exclusive MYH11 H3K4dime PLA labeling of SMC whereas neuronal cells were MYH11 H3K4dime PLA− but MYH11 H3K27trimePLA+. These results provide compelling evidence of the reliability of our ISH-PLA method to specifically detect histone modifications at specific gene loci in single cells in human and mouse tissue sections and are consistent with results of our previous studies using conventional ChIP assays on cultured cells11. However, the preceding studies are the first to identify a cell-type and locus-specific histone modification in cells in vivo within intact tissue sections within a complex multicellular tissue specimen. Furthermore, we provide evidence that H3K4dime of the MYH11 gene locus represents a unique and specific epigenetic signature of cells of the SMC lineage in vivo. Finally, we show that our PLA methodology is readily adaptable to multiple gene loci and histone modifications.
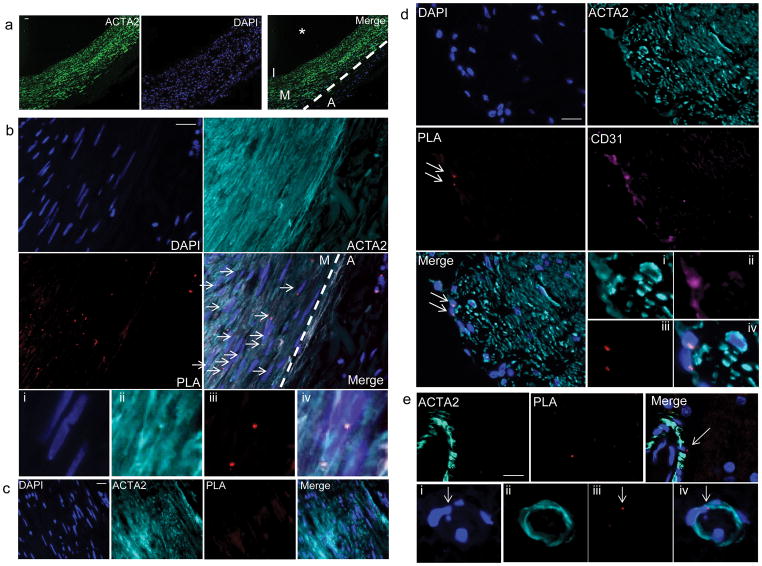
Figure 3. Visualization of H3K4dime on the MYH11 promoter in SMCs in situ in histological sections of humancarotidarteries.
(a) Immunostaining of human carotid artery sections illustrating the media (M) consisting primarily of ACTA2+SMCs, as well as the adventitia (A), intima (I)and the vessel lumen (*)(n= 5). Scale bar = 100 μm. (b) Results of MYH11 H3K4dime PLA in human carotid artery sections with DAPI (blue) and ACTA2 (cyan). PLA signal (red) was exclusively observed in ACTA2+ medial SMCs (arrows). None of the ACTA2− cells within the adventitia were MYH11 H3K4dime PLA+. Scale bar = 50 μm. Higher magnification from b with i) DAPI (blue), ii) ACTA2 (cyan), iii) PLA (red), and iv) merged image. (c) Negative control where ISH was done with an empty biotinylated vector followed by PLA with biotin and H3K4dime antibodies. Scale bar = 50 μm. (d) Detection of H3K27trime on the MYH11 promoter in human carotid artery sections. Results showed a positive MYH11 H3K27trime PLA signal in CD31+ ECs (arrows). In contrast, ACTA2+ cells were MYH11 H3K27trime PLA−. Scale bar = 50 μm. i–iv) Higher magnification images with: i) ACTA2 (cyan), ii) CD31 (purple), iii) PLA (red), and iv) merged, DAPI (blue). (e) MYH11 H3K4dime PLA analysis of human brain tissues stained with ACTA2 (cyan). SMCs in brain vessels are MYH11 H3K4dime PLA+ (arrow). Scale bar = 10 μm. i–iv) Small capillary within brain tissue with DAPI (i), ACTA2 (ii), PLA (iii) and merged image (iv).
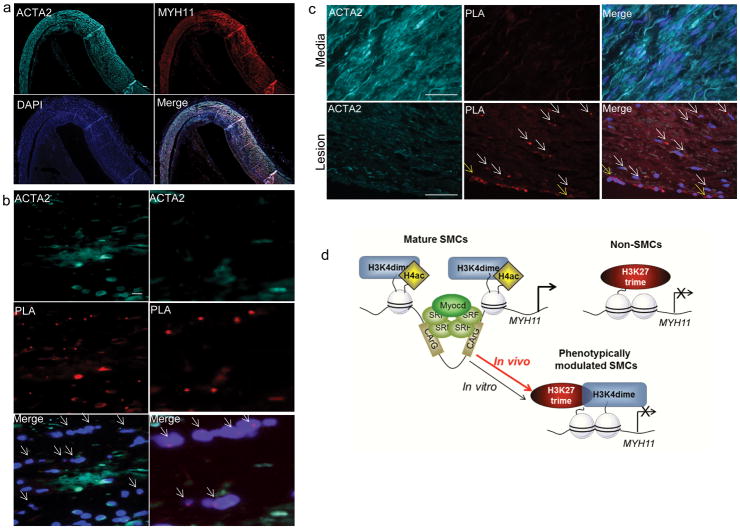
Mature SMCs retain remarkable plasticity and can undergo phenotypic switching characterized by markedly decreased SMC marker gene expression and increased proliferation and migration in pathological environments such as atherosclerosis14, 26, 27. However, because phenotypically modulated SMCs down-regulate expression of SMC markers (including ACTA2 and MYH11) there are major ambiguities as to which cells within lesions are derived from SMCs28. Previous studies demonstrated that PDGF-BB-induced phenotypic switching of cultured SMCs was associated with marked reductions in SMC marker expression and reduced H4 acetylation but no changes in H3K4dime enrichment of the ACTA2 or MYH11 promoters11. We extended these studies herein by showing that induction of SMC phenotypic switching with the pro-atherogenic oxidized phospholipid, 1-(palmitoyl)-2-(5-oxovaleroyl)-sn-glycero-3-phosphatidylcholine (POVPC), had similar effects, including reduced MYH11 expression but no change in MYH11 H3K4dime enrichment (Fig. 4a–b). To determine if Myh11 H3K4dime persists during SMC phenotypic switching in vivo, we generated advanced atherosclerotic lesions by crossing our SMC-eYFP+/+ mice with ApoE−/− mice and fed them a Western diet for 18 weeks. Remarkably, we identified large numbers of phenotypically modulated SMCs within lesions which show no detectible expression of MYH11 (Fig. 4c) or ACTA2 (Fig. 4d) and are identifiable as being of SMC origin only through detection of the SMC-specific eYFP lineage tracing gene product. Importantly, our SMC lineage tracing model is conditionally activated through an ERT2-Cre system allowing us to permanently lineage tag mature SMCs that were expressing MYH11 during the tamoxifen treatment period (i.e. 6–8 weeks of age). These cells and their progeny constitutively express eYFP, allowing us to lineage trace these cells independent of the expression of traditional SMC markers which can be down-regulated. In contrast, any SMC derived from a non-SMC source at a later time point, will not express eYFP. Remarkably, results of our studies with this highly rigorous SMC lineage tracing system showed that >95% of SMC-derived cells within atherosclerotic lesions within brachiocephalic arteries of SMC-eYFP+/+ ApoE−/− ice were not detectable as being of SMC origin based on ACTA2 (Fig. 4f) or MYH11 immunostaining (Fig. 4c). Moreover, of major significance, we showed that the majority of these cells were Myh11 H3K4dime PLA+ (Fig. 4f). Taken together, the preceding results not only establish the SMC specificity of H3K4dime of the MYH11 gene locus, but also show that this epigenetic mark is stable in vivo even in SMC that are not identifiable as being of SMC origin based on detection of endogenous marker genes. Moreover, results indicate that one may use detection of MYH11 H3K4dime by ISH-PLA as an epigenetic lineage tracing system for assessing the role of SMC in the pathogenesis of a number of major human diseases in which SMC phenotypic switching is postulated to play an important role including atherosclerosis, cancer, asthma, and hypertension. Indeed, consistent with this possibility, we found that a large fraction of cells within advanced atherosclerotic lesions within human coronary arteries do not express SMC marker genes such as ACTA2 and MYH11 (Fig. 5a and Supplementary Fig. 12) but are MYH11 H3K4dime PLA+ (Fig. 5b). In addition, we detected MYH11 H3K27trime PLA+ cells only in ACTA2− cells within the lesions but not in medial ACTA2+ SMCs (Fig. 5c). In contrast, MYH11 H4ac was restricted to medial ACTA2+ SMCs and was absent within lesion cells (Supplementary Fig. 13). Fig 5d presents a summary model of histone modifications in differentiated mature SMC versus phenotypically modulated SMC based on observations in vivo in the present studies and our previous studies in cultured SMCs11.
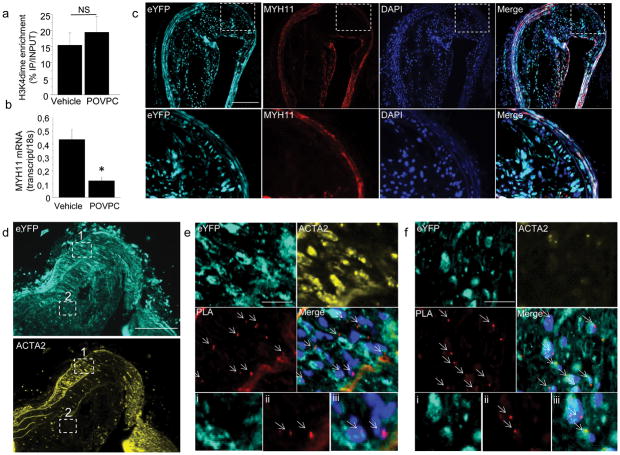
Figure 4. H3K4dime on the MYH11 promoter persists during phenotypic switching in vivo in SMC lineage tracing mice developing atherosclerosis.
(a) ChIP assay performed in cultured SMCs treated with vehicle (DMSO) or POVPC (10 μg/mL, 24h) showing similar H3K4dime enrichment on the MYH11. Mean s.d.; n = 3. (b) mRNA quantification of MYH11 showing POVPC-induced decrease in MYH11 mRNA level compared with vehicle. Mean ± s.d; n = 3; *P <0.01. (c)Identification of modulated SMCs in SMC-eYFP+/+ ApoE−/− mice fed with Western diet for 18 weeks. Brachiocephalic arteries (BCA) sections of these mice were stained with eYFP (cyan) and MYH11 (red). eYFP+ SMCs were identified within the media where they co-express MYH11 and within the atherosclerotic lesion where they lose expression of MYH11. Scale bar = 100 μm. (d) BCA sections of SMC-eYFP+/+ ApoE−/− mice were stained with eYFP (cyan) and ACTA2 (yellow) and were analyzed for MYH11 H3K4dime PLA.Scale bar = 100 μm.(e). Image corresponding to region 1 boxed in (d) Medial SMCs are ACTA2+ eYFP+ and Myh11 H3K4dime PLA+ (arrows). Higher magnification with eYFP (i), PLA (ii) and merged image with Dapi (iii). (f)Image corresponding to region 2 boxed in (d). Within the atherosclerotic lesion, a substantial fraction of eYFP+ACTA2− cells is Myh11 H3K4dime PLA+ indicating cells of SMC origin not identifiable based on detection of endogenous SMC markers (arrows). Scale bar (e–f) = 10 μm. Higher magnification with eYFP (i), PLA (ii) and merged image with Dapi (iii).
Figure 5. Identification of epigenetic regulation of phenotypically modulated SMCs within human coronary atherosclerotic lesions by ISH/PLA.
(a) Immunostaining of 5 μm-thick sections of human coronary arteries illustrates loss of SM marker genes expression within the atherosclerotic lesion: ACTA2 (cyan), MYH11 (red) and Dapi (blue). Scale bar = 100μm. (b) Immunostaining was combined with MYH11 H3K4dime ISH/PLA with ACTA2 (cyan), PLA (red) and Dapi (blue). A large fraction of ACTA2− lesion cells are positive for MYH11 H3K4dime PLA (arrows), suggesting that these cells are of SMC origin. Higher magnification on the right panels. Scale bar = 10 μm. (c) MYH11 H3K27trime ISH-PLA in human coronary atherosclerotic lesions. Medial SMCs are strictly MYH11 H3K27trime PLA− whereas lesion SMCs (white arrows) and ECs (yellow arrows) are MYH11 H3K27trime PLA+. The lower panels are the higher magnification of the middle panels. Scale bar = 50 μm. (d) Cartoon summarizing the epigenetic regulation on the MYH11 promoter in mature SMCs, phenotypically modulated SMCs and non-SMCs in vivo.
Discussion
Herein we develop and validate a new method for visualizing histone modifications at a single genomic locus within single cells in histological sections, and using it provide novel evidence that H3K4dime of the MYH11 gene locus is: 1) a highly specific marker of SMC lineage in vivo; and 2) this epigenetic mark persists even in SMC that have undergone phenotypic transitions to a less differentiated state wherein the cell is unidentifiable as being of SMC origin except using genetic lineage tracing methodologies. We believe these observations are highly important for the following reasons:
First, although several groups29, 30 have employed PLA to detect protein-nucleic acid interactions in vitro, our studies are the first to our knowledge to describe a method that permits detection of specific histone modifications at a given gene locus in single cells in fixed histological specimens which we believe will have tremendous utility for investigating the role of epigenetic changes in development and disease. Indeed, there is a large body of research that have investigated the role of epigenetic regulation and chromatin signature during stem cell differentiation or development of specific types of cancer31. Nevertheless, the technical limitations of ChIP assays described previously do not allow the in vivo characterization of such epigenetic signatures except in the study of hematopoietic stem cells32 or leukemia33 in which cell sorting can be performed without extensive tissue dissociation prior to ChIP analyses. Thus, we believe that ISH-PLA will be a powerful tool for the investigation of epigenetic regulation in solid tumors in which it is virtually impossible to ascertain if a given epigenetic change is present within tumor or stromal cells. Similarly, our ISH-PLA method will have tremendous utility in investigating the role of specific histone modifications at selective gene loci during development of complex multicellular tissues during development. Moreover, the fact that the method can be applied to paraformaldehyde fixed paraffin embedded tissue specimens, the standard protocol for archiving human biopsy specimens, will allow unprecedented studies of the role of epigenetic changes in the pathogenesis of human diseases using existing tissue banks. Finally, we believe that our ISH-PLA method may be adapted for the detection and visualization of DNA methylation or transcription factor binding on specific promoters with single-cell resolution which will further expand our ability to investigate transcriptional regulation of a given gene locus within its native context within complex multicellular tissues in vivo.
Second, our results are the first to show persistence of a cell specific epigenetic signature when cells undergo phenotypic transitions to a less differentiated or altered phenotypic state in vivo, wherein the cell origin cannot be identified by detection of endogenous cell selective marker genes. Evidence of appearance and maintenance of cell-specific epigenetic signature (other than the SMC lineage) have been previously described but with the limitation that results were based on studies in cultured cells under conditions that fail to recapitulate complex cell-cell, cell-matrix, and other critical environmental cues that exist in vivo34, 35. In addition, to having important implications for lineage tracing of cells in complex tissues including human biopsy specimens, these observations may also have critical implications for the propagation of epigenetic mechanisms that control cell lineage memoryand cell specific bookmarking during cell mitosis, normal development, and disease progression34, 36. However, it remains to be determined whether persistence of H3K4diMe of SMC gene loci is important in the ability of SMC to undergo reversible phenotypic switching during repair of vascular injury. As such, our studies have provided no evidence as to whether this histone modification plays a causal role in controlling SMC lineage and phenotype and further mechanistic studies are needed in this important area but at present are not possible due to the lack of an approach to selectively remove this mark exclusively at SMC gene loci.
Third, of major significance, our results indicate that numerous studies of atherosclerosis that relied on identification of intimal SMC based on detection of ACTA2 have grossly under-estimated the frequency of SMCs within lesions, and likely failed to detect possible transition of these cells to alternative phenotypes that may be critical in the pathogenesis of the disease. Until now, lineage tracing of lesion cells in vivo in human studies has been restricted to studying the possible role of hematopoietic cells within lesions based on analyses of cross-gender bone marrow transplant specimens combined with Y chromosome detection by in situ hybridization37. However, these approaches are highly limited due to such samples being extremely rare, the approach does not allow rigorous lineage tracing of non-hematopoietic cells, and results are confounded by development of transplant atherosclerosis which has a different etiology than normal atherosclerosis. Of major interest, we believe that ISH-PLA will greatly advance studies of human atherosclerosis by facilitating novel studies of the mechanisms and factors that regulate phenotypic transitions in SMC within atherosclerotic lesions and their possible functional roles in plaque development, progression and end-stage events leading to clinical complications such as myocardial infarction or stroke.
Methods
Human tissues
Coronary and carotid arteries specimens from patients (n =10) were collected during Coronary artery bypass graft (CABG) and carotid endarterectomy surgery as well as during autopsy. These specimens were processed, fixed in paraformaldehyde and paraffin-embedded blocks were cut into 5 μm sections. Human temporal lobe sections were from autopsy cases from the University of Virginia, Department of Pathology. Brain tissue was immersion-fixed in 10% zinc-buffered formalin, and embedded in paraffin using standard procedures. The institutional review board at University of Virginia approved the use of all autopsy specimens.
Mice
Animal protocols were reviewed and approved by the University of Virginia Animal Care and Use Committee. Myh11-CreERT2 mice were generously provided by Stefan Offermanns (Max Planck Institute, Germany)22; ROSA26 STOP flox eYFP and ApoE−/− were obtained from Jackson Laboratories. All mouse strains have been backcrossed to a C57BL/6J for greater than six generations. Myh11-CreERT2 mice were genotyped by PCR as previously described22. ROSA26 STOP flox eYFP mice were genotyped using the following primers: oIMR0316 5′-GGAGCGGGAGAAATGGATATG-3′, oIMR0872 5′-AAGTTCATCTGCACCACCG-3′, oIMR1416 5′-TCCTTGAAGAAGATGGTGCG-3′, oIMR3621 5′-CGTGATCTGCAACTCCAGTC-3′. Cre Recombinase was activated with a series of ten 1 mg tamoxifen intraperitoneal injections (Sigma Aldricht) from six to eight weeks of age for a total of 10 mg of tamoxifen/25 g mouse. Mice were fed a high-fat (Western type) diet containing 21% milk fat and 0.15% cholesterol (Harlan Teklad) for 18 weeks starting at eight weeks of age. Mice were euthanized by CO2 asphyxiation then perfused via the left ventricle with 5 mL PBS followed by 10 mL 4% paraformaldehyde and an additional 5 mL PBS. Brachiocephalic arteries and various organs were carefully dissected and fixed for 30 min in 4% paraformaldehyde prior to embedding in paraffin.
Cell culture and harvesting
Mouse SMCs were cultured in growth medium (DMEM/F12, Gibco) supplemented with Fetal Bovine Serum (10%), L-glutamine (1.6mM, Gibco) and Penicillin-Streptomycin (100U/mL, Gibco). Cells were starved for three days in serum-free medium and treated with 10μg/mL POV-PC (1-(palmitoyl)-2-(5-oxovaleroyl)-sn-glycero-3-phosphatidylcholine, Cayman Chemical). Human coronary SMCs, human coronary ECs, U937 (human monocyte) and RAW264.7 (human macrophage) cell lines were purchased from Lonza and ATCC. Cells were grown in corresponding medium in the presence of serum to confluence. For ISH/PLA, SMCs and ECs were harvested and fixed in 10% phosphate buffered formalin (PBF, Fisher). After formalin fixation, cells were pelleted and suspended in 1% agarose (Sigma). Cell pellets were processed and paraffin-embedded blocks were cut into 5μm sections.
Immunofluorescent staining
Sections were de-paraffinized and rehydrated in xylene and ethanol series. After antigen retrieval (antigen retrieval solution, Vector), section were blocked with Fish Skin Gelatin/PBS (6 g/L) containing 10% of goat or horse serum for one hour at room temperature. Endogenous mouse IgG in mouse tissue sections was blocked by incubation with unconjugated Fab Fragment Goat anti-mouse IgG (H+L) (Jackson ImmunoResarch Labs) for 1h at room temperature. Slides were incubated with the following antibodies: mouse monoclonal SM α-actin-FITC (ACTA2) (4.4 μg/mL, clone 1A4, Sigma Aldrich), rat monoclonal SM Myosin Heavy Chain (MYH11) (1 μg/mL, clone KM3669, Kamiya Biomedical Company), goat polyclonal anti-PECAM-1 (CD31) (2 μg/mL, # sc-1506, Santa Cruz), mouse monoclonal dimeH3K4 (4 μg/mL, clone CMA303, Millipore) and goat polyclonal anti-GFP (4 μg/mL, # ab6673, Abcam) for detection of eYFP. The secondary antibodies were donkey anti-rat conjugated to Alexa 555 (5 μg/mL, Abcam), donkey anti-goat conjugated to Alexa-647 (4 μg/mL, Invitrogen), donkey anti-mouse conjugated to Alexa-555 (4μg/mL, Invitrogen).
Probe preparation & hybridization
Human MYH11, mouse Myh11, human CDH5, mouse Cdh5 and mouse Tagln biotin-labeled probes were generated. The proximal 2kb of these promoters were amplified by PCR (see primers in Supplementary Table). PCR products were cloned into pCR2.1 vector for amplification (TOPO cloning kit, Invitrogen). Probes were generated by Nick Translation (Roche) using biotin-14-dATP (Invitrogen). Nick translation efficiency was assessed by migration on 1% agarose gel. Labeled-DNA probes (40 ng/slide) undergo denaturation in Hybridization Buffer (2× SSC, 50% high grade formamide, 10% dextran sulfate, 1μg of human or mouse Cot-1 DNA) for 5 min at 80°C. Directly after immunostaining, slides were dehydrated in ethanol series and incubated in 1mM EDTA (pH 8.0) for 20 min. Then, samples were incubated with pepsin (0.5%) in buffer (0.05M Tris, 2mM CaCl2, 0.01M EDTA, 0.01M NaCl) at 37°C for 20 min, as previously described37. Hybridization mixture containing biotinylated probes or 5-TAMRA-dUTP labeled Y chromosome probe (Clone RP11–88F4, Empire Genomics) was applied on sections. Sections were incubated at 80°C for 5 min, followed by 16–24h incubation at 37°C. Hybridization was followed by multiple washes in 2× SSC, 0.1% NP-40 buffer.
Proximity Ligation assay (PLA)
PLA was performed directly after ISH following manufacturer’s instructions (Olink). After blocking, sections were incubated with mouse H3K4dime (5 μg/mL, clone CMA303 Millipore), mouse H3K27trime (5 μg/mL, # ab6002, Abcam), mouse H4ac (5 μg/mL, clone 3HH4–4C10, Millipore) and rabbit Biotin (5 μg/mL, # ab53494, Abcam) antibodies overnight at 4°C, followed by incubation with secondary antibodies conjugated with PLA probe at 37°C for 1h as recommended by manufacturers. Then, ligation and amplification were performed (Duolink detection kit Orange, 555nm). Finally, mounting medium with DAPI was used.
Image acquisition and analysis
Images were acquired with Olympus BX41 fitted with a Q imaging Retiga 2000R camera. Image acquisition was performed with the Q Capture Pro software (Media Cybernetics & QImaging Inc). Settings were fixed at the beginning of both acquisition and analysis steps and were unchanged. Brightness and contrast were lightly adjusted after merging. Image analysis was performed with Image J. Confocal images were acquired using a Zeiss LSM700 scanning confocal microscope with 405nm, 488nm, 555nm, and 637nm solid state lasers. Analysis of confocal images was completed using Zeiss Zen 2009 software.
Chromatin Immunoprecipitation
ChIP was performed on cultured cells as previously described11. Cells were fixed with 1% paraformaldehyde for 10 min at room temperature. Cross-link was stopped by addition of 125 mM glycine for 10 min. The cross-linked chromatin was sonicated to shear chromatin into fragments of 200–600 base pairs. The sheared chromatin was immunoprecipitated with 2 μg of dimeH3K4 antibody (clone CMA303, Millipore), while negative control was incubated with mouse IgG and input DNA without antibody, and immune complexes were recovered with magnetic beads (Millipore).
Quantitative PCR
Total RNA was prepared from cultured cells using Trizol (Invitrogen) according to the manufacturer’s protocol. One microgram of RNA was used for reverse transcription with iScript cDNA synthesis kit (BioRad). Real-time PCR was performed (iQ SYBR Green Supermix, Biorad) on cDNA or DNA extracted after ChIP using primers listed in Supplementary Table 1.
Statistics
Values are expressed as means ± s.d. For ChIP, three independent experiments were performed. Each experiment was repeated in triplicate. Comparison between groups was tested using ANOVA test. A value of P<0.05 was considered significant.
Supplementary Material
Acknowledgments
Fundings
We would like to thank M.E. McCanna and R.S. Tripathi for their knowledge and technical expertise. We thank J.W. Mandell (Department of pathology, University of Virginia) for providing us human brain sections. This work was supported by National Institutes of Health grants R01 HL57323, R01 HL098538, and R01 HL087867 (to G.K. Owens). D. Gomez is supported by the American Heart Association Postdoctoral Fellowship 11POST7760009. L.S Shankman is funded by a predoctoral American Heart Association Fellowship 11PRE17008. A.T Nguyen is funded by a postdoctoral American Heart Association Fellowship 12POST11630032. The authors declare no affiliations, funding or financial holdings that might be perceived as affecting the objectivity of this paper.
Abbreviations
- SMC
Smooth Muscle Cell
- ESC
Embryonic Stem Cell
- PDGF-BB
Platelet Derived Growth Factor-BB
- ChIP
Chromatin Immunoprecipitation
- PLA
Proximity Ligation Assay
- ISH
In situ Hybridization
Footnotes
Author Contributions
G.K. Owens supervised this study; D. Gomez and G.K. Owens conceived the in situ hybridization-PLA strategies, designed studies and wrote the paper; D. Gomez generated labeled DNA probes, performed immunostaining and all ISH-PLA experiments and analyzed data; D. Gomez performed in vitro experiments, ChIP and qPCR; L.S. Shankman generated Myh11-CreERT2 ROSA Flox STOP eYFP+/+ mice; L.S. Shankman and A.T. Nguyen performed immunostaining on mouse sections; D. Gomez, L.S. Shankman and A.T. Nguyen performed image acquisition and analysis.
Reference List
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 6.Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol. 2011;11:478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- 7.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 9.Shechter D, et al. Analysis of histones in Xenopus laevis. I A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions. J Biol Chem. 2009;284:1064–1074. doi: 10.1074/jbc.M807273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manabe I, Owens GK. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ Res. 2001;88:1127–1134. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- 11.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circ Res. 1994;75:803–812. doi: 10.1161/01.res.75.5.803. [DOI] [PubMed] [Google Scholar]
- 13.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative binding of KLF4, pELK-1, and HDAC2 to a G/C repressor element in the SM22alpha promoter mediates transcriptional silencing during SMC phenotypic switching in vivo. Circ Res. 2012;111:685–696. doi: 10.1161/CIRCRESAHA.112.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 15.Alexander MR, Owens GK. Epigenetic Control of Smooth Muscle Cell Differentiation and Phenotypic Switchingin Vascular Development and Disease. Annu Rev Physiol. 2011 doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- 16.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nat Protoc. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 17.Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- 18.Soderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 19.Lievens S, Tavernier J. Single protein complex visualization: seeing is believing. Nat Methods. 2006;3:971–972. doi: 10.1038/nmeth1206-971. [DOI] [PubMed] [Google Scholar]
- 20.Rantala JK, et al. SHARPIN is an endogenous inhibitor of beta1-integrin activation. Nat Cell Biol. 2011;13:1315–1324. doi: 10.1038/ncb2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brobeil A, et al. PTPIP51 is phosphorylated by Lyn and c-Src kinases lacking dephosphorylation by PTP1B in acute myeloid leukemia. Leuk Res. 2011;35:1367–1375. doi: 10.1016/j.leukres.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Wirth A, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 23.Lampugnani MG, et al. A novel endothelial-specific membrane protein is a marker of cell-cell contacts. J Cell Biol. 1992;118:1511–1522. doi: 10.1083/jcb.118.6.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Miano JM, Mercer B, Olson EN. Expression of the SM22alpha promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regan CP, Adam PJ, Madsen CS, Owens GK. Molecular mechanisms of decreased smooth muscle differentiation marker expression after vascular injury. J Clin Invest. 2000;106:1139–1147. doi: 10.1172/JCI10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med. 2000;6:389–395. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D. Signaling vascular morphogenesis and maintenance. Science. 1997;277:48–50. doi: 10.1126/science.277.5322.48. [DOI] [PubMed] [Google Scholar]
- 28.Gomez D, Owens GK. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovasc Res. 2012 doi: 10.1093/cvr/cvs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weibrecht I, et al. Visualising individual sequence-specific protein-DNA interactions in situ. N Biotechnol. 2011 doi: 10.1016/j.nbt.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Gustafsdottir SM, et al. In vitro analysis of DNA-protein interactions by proximity ligation. Proc Natl Acad Sci U S A. 2007;104:3067–3072. doi: 10.1073/pnas.0611229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 35.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 36.Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- 37.Caplice NM, et al. Smooth muscle cells in human coronary atherosclerosis can originate from cells administered at marrow transplantation. Proc Natl Acad Sci U S A. 2003;100:4754–4759. doi: 10.1073/pnas.0730743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.