Abstract
Background and purpose
Intracerebral hemorrhage (ICH) is the acute manifestation of a progressive disease of the cerebral small vessels. The severity of this disease appears to influence not only risk of ICH, but also the size of the hematoma. As the burden of high blood pressure(BP)-related alleles is associated with both hypertension-related end-organ damage and risk of ICH, we sought to determine if this burden influences ICH baseline hematoma volume (BHV).
Methods
Prospective study in subjects of European descent with supratentorial ICH who underwent genome-wide genotyping. Forty-two single nucleotide polymorphisms (SNPs) associated with high BP were identified from a publicly available database. A genetic risk score was constructed based on these SNPs. The score was utilized as the independent variable in univariate and multivariate regression models for admission ICH volume and poor clinical outcome (modified Rankin Scale 3–6).
Results
A total of 323 ICH cases were enrolled in the study (135 deep and 188 lobar intracranial hematomas). The BP-based genetic risk score was associated with both BHV and poor clinical outcome specifically in deep ICH. In multivariate regression analyses, each additional standard deviation of the score increased mean deep ICH volume by 28% (or 2.7-mL increase, beta=0.28, standard error=0.11, p=0.009) and risk of poor clinical outcome by 71% (odds ratio=1.71, 95% confidence interval 1.05–2.80, p=0.03).
Conclusion
Increasing numbers of high BP-related alleles are associated with mean BHV and poor clinical outcome after an ICH. These findings suggest that the small vessel vasculopathy responsible for the occurrence of the hemorrhage could also influence its volume.
INTRODUCTION
The volume of intraparenchymal bleeding is the strongest predictor of clinical outcome in intracerebral hemorrhage (ICH).1 In a model proposed by Fisher, the volume of the ICH is influenced by the severity of the small vessel disease that initially caused the hemorrhage.2 According to this model, the final volume of the hematoma depends on the additional bleeding contributed by the rupture of adjacent, previously diseased small vessels located in the periphery of the artery that originated the ICH. Importantly, the small vessel disease referred to by this model is thought to differ by location: while hypertensive vasculopathy has been found in deep ICH,3 amyloid angiopathy is the predominant finding in lobar ICH.4
The aforementioned hypothesis provides an appropriate framework to describe the role of genetic variation within the apolipoprotein E gene in the occurrence of ICH. Genetic variants at this locus cause familial and sporadic forms of cerebral amyloid angyopathy.5 Accordingly, the apolipopretein E epsilon-2 allele has been shown to be associated with both increased risk6 and larger volume of the hematoma in ICH.7 These associations were described specifically for hemorrhages occurring in lobar regions of the brain, in line with the histopathological variation by topography described above. The objective of the present study was to explore whether a similar model applies to hypertensive vasculopathy and volume of deep ICH.
Genetic risk scores (GRSs) express in a single measure the burden of risk alleles for a specific outcome carried by a given individual. Higher numbers of a blood pressure(BP)-based GRS are associated with higher BP measured through direct observation.8 Accordingly, we have previously found that a BP-based GRS is associated with risk of ICH in deep supratentorial structures.9 And a similar GRS has been shown to correlate with left ventricular hypertrophy, a measure of end-organ damage due to long lasting hypertension.10 Of note, animal studies show that the presence of hypertension-related end-organ damage in one organ correlates with tissue damage in other organs.11 Based on these findings, we aimed to evaluate whether the aggregate burden of high BP-related alleles, as measured by a BP-based GRS, influences the volume of ICH at presentation. We hypothesize that in subjects with deep ICH, higher numbers of this GRS will be associated larger intracranial hematomas due to a more severe underlying hypertensive vasculopathy.
METHODS
Study design and patients
We utilized a cohort design to assess the relationship between high BP-associated risk alleles (or single nucleotide polymorphisms, SNPs) and ICH baseline hematoma volume (BHV) and risk of poor functional outcome at 90 days (defined as modified Rankin Scale [mRS] scores 3–6). Subjects were recruited through the Genetics of Cerebral Hemorrhage on Anticoagulation (GOCHA) study.12 All individuals underwent genome-wide genotyping as part of an ongoing genome-wide association study of ICH. Participating sites included Massachusetts General Hospital, Beth Israel Deaconess Medical Center, Mayo Clinic Jacksonville, and the Universities of Michigan, Virginia, Florida at Jacksonville and Washington. The study was approved by the institutional review board or ethics committee of participating institutions. All participants or their legal proxies provided written consent.
Case ascertainment
Ascertainment of ICH cases was performed by stroke neurologists who were blinded to genotype data. Enrolled subjects were primary acute ICH cases that presented to the participating institutions, were >55 years of age and had confirmation of primary ICH through computed tomography (CT). Exclusion criteria included warfarin treatment, trauma, brain tumor, hemorrhagic transformation of a cerebral infarction, vascular malformation, or any other cause of secondary ICH. A detailed clinical history with relevant covariate information was obtained for each subject, including admission systolic and diastolic BP reading. At each participating institution, BP management was implemented according to the American Heart Association Guidelines.13 Patients (or their caregivers) were interviewed via telephone at 3 months post-ICH to assess their functional outcome using the mRS.
Imaging analysis
ICH location was assigned based on admission CT by neurologists who were blinded to genotype and clinical data. ICH exclusively involving the thalamus, basal ganglia, internal capsule and deep periventricular white matter was defined as deep ICH. ICH originating at the cortex and cortical–subcortical junction was defined as lobar ICH. Hemorrhages involving both territories were defined as mixed ICH and were excluded from the current analysis. Admission ICH hemorrhage volumes were centrally assessed at the Massachusetts General Hospital (the coordinating center). Admission ICH hemorrhage volumes were measured using Alice (PAREXEL International Corporation) and Analyze 9.0 (Mayo Clinic, Rochester, MN) software using previously described methods.14 The distribution of BHVs across participating institutions was assessed by means of analysis of variance, and no significant differences were observed (p=0.45 and p=0.08 for deep and lobar BHVs, respectively). Intraventricular bleeding was not included in volume calculations.
Exposure ascertainment
Peripheral whole blood was collected at each participating institution at the time of consent. Blood samples were subsequently shipped to the Massachusetts General Hospital, the coordinating center, and genotyping was carried out at the Broad Institute (Cambridge, MA). Genotyping was performed using Illumina (San Diego, CA, USA) HumanHap 610 Quad. Stringent quality control procedures were implemented following standard protocols.15 IMPUTE2 software16 was utilized to impute unobserved SNPs based on reference panels from HapMap and the 1000-genomes project.
Statistical analysis
Selection of SNPs associated with BP
SNPs associated with BP p<1×10−7 were selected from the National Human Genome Research Institute GWAS catalog. To ensure that the results of this study reflect independent genetic effects, SNPs in each chromosome were pruned to exclude variants in high linkage disequilibrium (r2 > 0.5).
Population stratification
Principal components analysis was implemented to account for population structure.17 Principal components were initially applied to identify and remove population outliers. Subsequently, principal component 1 and 2 were added as covariates to the regression models that were fit to test each hypothesis.
Genetic association analysis for individual variants
BHV was natural log-transformed to approximate normality. Single-SNP genetic association testing with BHV was completed using linear regression, assuming additive effects for each risk allele present, and including age, gender and principal components 1 and 2 in the model.
Genetic risk score
The main exposure of interest in the present study was the burden of risk alleles for increased BP, as expressed by a weighted GRS. A weighted GRS is the sum of the products of the risk allele count (0, 1 or 2) at each locus multiplied by the reported effect (obtained from the National Human Genome Research Institute GWAS catalog) of that risk allele on BP. For SNPs that had minor alleles associated with decreased BP, the major allele was used as the reference when constructing the score in order to assure common directionality of effects.
Association analysis for the GRS
Univariate and multivariate linear regression analysis was implemented to model the change in mean BHV and mean admission systolic BP. Multivariate logistic regression was utilized to model the change in mean log-odds of poor functional outcome. The GRS Z-score was calculated for each subject (the subject’s value of the GRS minus the sample’s mean GRS, divided by the sample’s standard [SD] deviation for the GRS) and entered as a linear predictor in the regression models. In this setting, the obtained regression coefficient can be interpreted as the change BHV per 1 SD increase in the GRS. Multivariate models included factors known or suspected to influence BHV: age, sex, pre-ICH treatment with aspirin and pre-ICH history of hypertension.18,19 To assess influence of outlier observations, ICH volumes >4 SDs from the mean were dropped in sensitivity analysis. The effect of including time from symptom onset to CT scan to the model was also explored in sensitivity analysis.
Statistical significance was considered to be Bonferroni-corrected p<0.001 for single-SNP association testing (42 tests), and p<0.05 for GRS analyses, all tests being two-sided. All statistical analyses were performed in SAS 9.2 (SAS Institute, Cary, NC USA).
RESULTS
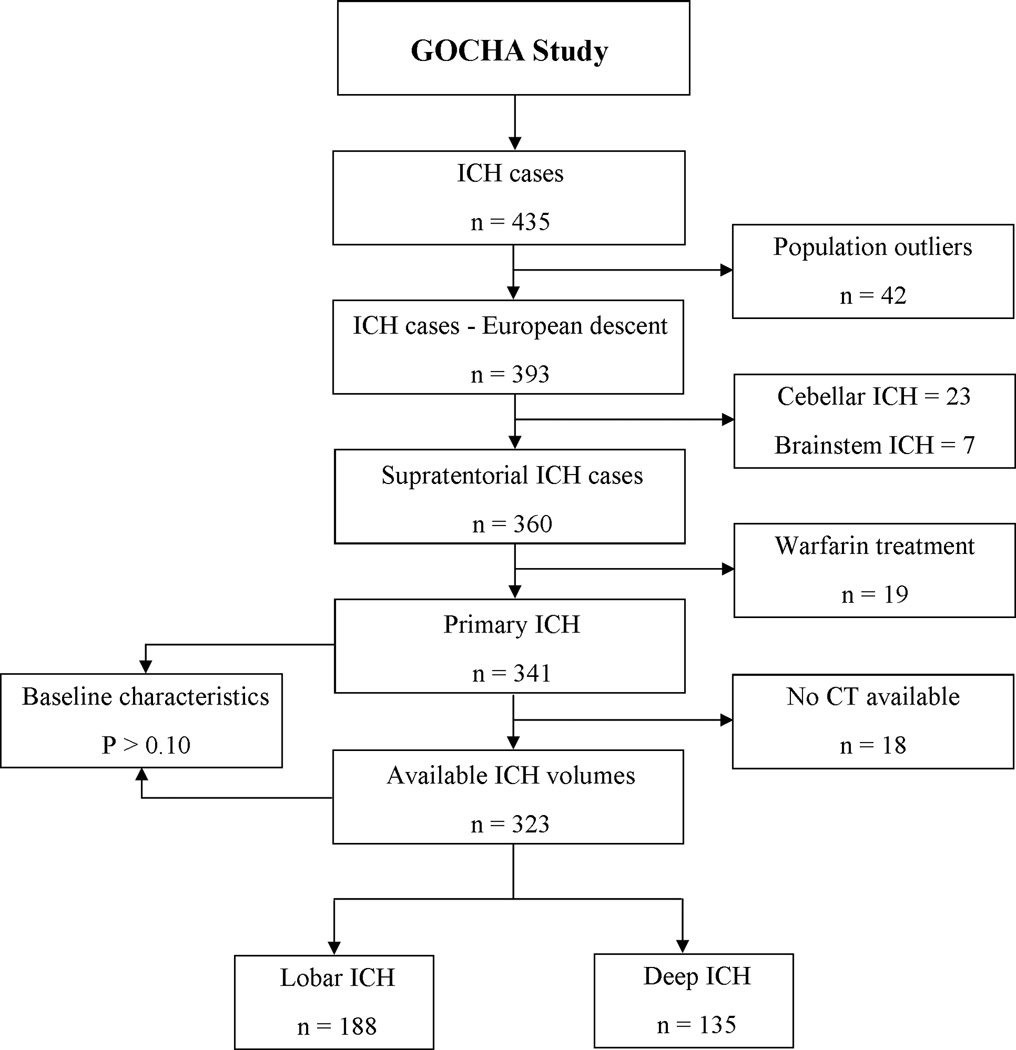
Three hundred and forty one subjects met this inclusion criteria during the study period (Figure 1). Of these, 18 had no available CT data. No significant differences were observed when comparing the full cohort with the final sample that had complete data. Of the 323 subjects available for analysis, 135 (48%) had deep and 188 (52%) lobar hemorrhages (Table 1).
Figure 1. Study flowchart.
Complete cohort of eligible cases (n=341), including subjects with missing data for admission computed tomography, was compared with the cohort of cases with complete data (n=323). Tested characteristics included age, sex, vascular risk factors and pre-ICH treatment with aspirin.
Table 1.
Cohort characteristics.
| Covariate | All ICH n = 323 |
Deep ICH n = 135 |
Lobar ICH n = 188 |
|||
|---|---|---|---|---|---|---|
| Age, mean (SD) | 74 | (10) | 73 | (10) | 75 | (11) |
| Sex (female), n (%) | 150 | (46) | 53 | (41) | 96 | (51) |
| Hx of hypertension, n (%) | 236 | (73) | 108 | (83) | 122 | (65) |
| Hx of diabetes mellitus, n (%) | 65 | (20) | 36 | (28) | 29 | (16) |
| Hx of hypercholesterolemia, n (%) | 120 | (37) | 52 | (40) | 66 | (35) |
| Hx of coronary artery disease, n (%) | 58 | (21) | 38 | (29) | 29 | (16) |
| Previous ischemic stroke, n (%) | 29 | (9) | 14 | (11) | 14 | (7) |
| Pre-ICH aspirin Tx, n (%) | 167 | (52) | 68 | (52) | 97 | (52) |
| Admission systolic BP, mean (SD) | 173 | (34) | 176 | (36) | 93 | (21) |
| Admission diastolic BP, mean (SD) | 91 | (20) | 169 | (31) | 88 | (19) |
| Baseline ICH volume, median (IQR) | 23 | (51) | 15 | (34) | 38 | (64) |
| Baseline IVH volume, median (IQR) | 8 | (28) | 15 | (38) | 5 | (9) |
| Intraventricular extension, n (%) | 147 | (46) | 71 | (55) | 71 | (38) |
| ICH volume 0 – 30 ml, n (%) | 179 | (55) | 92 | (71) | 82 | (44) |
| ICH volume 30 – 60 ml, n (%) | 58 | (18) | 13 | (14) | 40 | (21) |
| ICH volume > 60 ml, n (%) | 86 | (27) | 20 | (15) | 65 | (35) |
| 90-day mRS = 0, n (%) | 31 | (11) | 12 | (11) | 19 | (10) |
| = 1 | 32 | (11) | 10 | (9) | 20 | (11) |
| = 2 | 24 | (9) | 6 | (6) | 18 | (10) |
| = 3 | 30 | (11) | 13 | (12) | 17 | (9) |
| = 4 | 31 | (11) | 17 | (16) | 14 | (7) |
| = 5 | 21 | (8) | 8 | (7) | 12 | (6) |
| = 6 | 111 | (39) | 43 | (40) | 65 | (35) |
Abbreviations: ICH = Intrecerebral hemorrhage, SD = Standard deviation, Hx = History, Tx = Treatment, IQR = Interquartile range, IVH = Intraventricular hemorrhage, mRS = Modified rankin, ml = milliliter.
A total of 42 SNPs were selected to build the BP-based GRS (Supplemental Table 1). There were 157 entries related to BP or hypertension in the Catalog of Published Genome-Wide Association Studies available through the National Human Genome Research Institute. Of these, 86 corresponded to repeated entries (i.e., referred to genetic variants identified in more than one study). Of the remaining 71 SNPs, 58 had been reported to be associated with BP levels at p-values of < 1×E-7; 3 of these had no available risk allele. After genotype pruning based on linkage disequilibrium (r2<0.5), 42 SNPs remained to be utilized in the score, all with minor allele frequencies >5%. Eighteen and 22 of these variants were directly genotyped in Illumina HumanHap 610 Quad array; the remaining variants were inferred through imputation. The maximum number of BP-associated alleles that a given individual in the study could have was 84. Mean (±SD) numbers of high-BP-associated alleles were 41 (±4) and mean GRS was 11.4 (±0.9). None of these SNPs, when assessed individually, was associated with BHV (Supplemental Table 2).
The burden of BP-related alleles was associated with mean BHV in deep ICH. In univariate analysis, each additional SD of the BP-based GRS was associated with a 29% increase (or 3.2 mL, β=0.29, SE=0.10, p=0.005) in mean BHV for ICH located in deep regions (Figure 2). This association between deep ICH BHV and the GRS remained significant, and the estimates unchanged, in multivariate analysis (28% or 3.1 mL increase, β=0.28, SE=0.11, p=0.009, Table 2). No association was found when considering all (deep and lobar combined) or lobar ICH in neither univariate (Figure 2) nor multivariate analysis (Table 2).). Sensitivity analyses aimed to explore the exclusion of outlier observations and the inclusion of time from symptom onset to CT scan did not significantly modify these results (data not shown).
Figure 2. Univariate analysis: Natural log - Admission ICH volume as a function of the BP-based Genetic Risk Score.
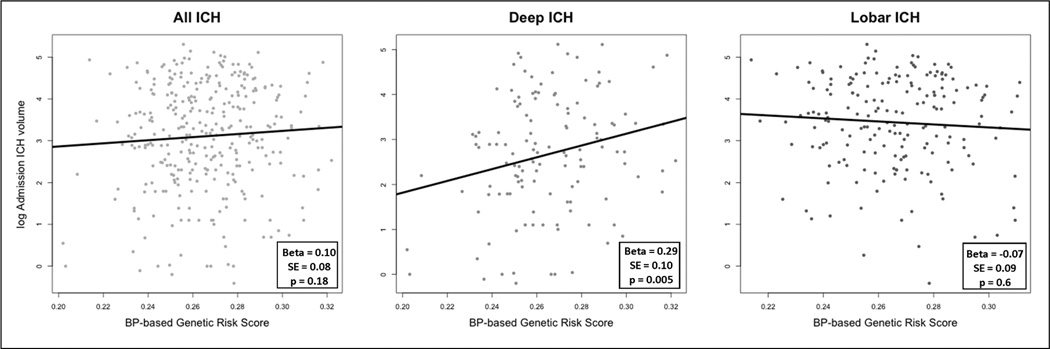
Scatter plot of log-ICH admission volume as a function of the BP-based weighted genetic risk score. Left panel shows deep and lobar ICH combined; central and right panels correspond to deep and lobar hemorrhages, respectively. Each panel also presents the regression line obtained after fitting each model.
Table 2.
Multivariate linear regression results – Dependent variable: Natural log-transformed admission ICH volume.
| All ICH n = 323 |
Deep ICH n = 135 |
Lobar ICH n = 188 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Covariate | Regression coefficient |
SE | p | Regression coefficient |
SE | p | Regression coefficient |
SE | p |
| BP - Based GRS | 0.10 | 0.07 | 0.15 | 0.28 | 0.11 | 0.009 | −0.08 | 0.09 | 0.40 |
| Age | −0.01 | 0.01 | 0.28 | −0.01 | 0.01 | 0.58 | −0.005 | 0.01 | 0.59 |
| Sex (male) | 0.03 | 0.15 | 0.85 | 0.26 | 0.24 | 0.27 | −0.14 | 0.18 | 0.44 |
| Pre-ICH Antiplatelet Tx | 0.17 | 0.15 | 0.24 | 0.14 | 0.24 | 0.57 | 0.21 | 0.18 | 0.24 |
| Hx of Hypertension | −0.17 | 0.17 | 0.31 | −0.15 | 0.31 | 0.63 | −0.15 | 0.19 | 0.42 |
| ICH location | 0.91 | 0.15 | <0.0001 | - | - | - | - | - | - |
Abbreviations: ICH = Intracerebral Hemorrhage, SE = Standard error, BP = Blood pressure, GRS = Genetic risk score, Tx = Treatment, Hx = History.
The burden of BP-related alleles was also associated with poor clinical outcome after ICH. Ninety-day mortality was 111 (39%), 43 (40%) and 65 (35%) for all, deep and lobar ICH, respectively. Poor outcome (mRS 3–6) was observed in 193 (60%), 81 (60%) and 108 (57%) subjects with all, deep and lobar ICH, respectively. As expected given the effect of the GRS on deep BHV, in multivariate logistic regression analysis each additional SD of the score was associated with a 71% increase (OR=1.71, 95%CI 1.05–2.80, p=0.03) in risk of having a poor clinical outcome (Table 3). Sensitivity analysis excluding outlier observations did not significantly modify these results (data not shown).
Table 3.
Multivariate logistic regression results – Dependent variable: Poor functional outcome (mRS 3–6).
| All ICH n = 323 |
Deep ICH n = 135 |
Lobar ICH n = 188 |
||||
|---|---|---|---|---|---|---|
| Covariate | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p |
| BP - Based GRS | 1.39 (1.04 – 0.87) | 0.03 | 1.71 (1.05 – 2.80) | 0.03 | 1.07 (0.75 – 1.52) | 0.72 |
| Age | 1.02 (1.02 – 1.08) | 0.002 | 1.08 (1.02 – 1.14) | 0.01 | 1.06 (1.02 – 1.09) | 0.002 |
| Sex (male) | 0.71 (0.38 – 1.33) | 0.29 | 1.11 (0.38 – 3.28) | 0.84 | 0.90 (0.44 – 1.81) | 0.76 |
| Pre-ICH Antiplatelet Tx | 1.23 (0.66 – 2.31) | 0.52 | 1.33 (0.47 – 3.81) | 0.59 | 1.32 (0.65 – 2.69) | 0.45 |
| Hx of Hypertension | 1.30 (0.65 – 2.58) | 0.46 | 2.79 (0.74 – 10.49) | 0.13 | 1.31 (0.62 – 2.74) | 0.48 |
| ICH location (lobar) | 0.76 (0.40 – 1.45) | 0.40 | - | - | - | - |
Abbreviations: mRS (modified Rankin Scale), ICH = Intracerebral Hemorrhage, OR = Odds ratio, CI = Confidence interval BP = Blood pressure, GRS = Genetic risk score, Tx = Treatment, Hx = History.
The BP-based GRS was also associated with poor clinical outcome in all ICH (deep and lobar combined). Each additional SD of the score was associated with a 39% increase (OR=1.39, 95%CI 1.04–2.87, p=0.03) in risk of having a poor clinical outcome (table 3). This effect, however, appears to be driven by the embedded effect of the GRS on deep hemorrhages. Supporting this notion, the point estimate of the odds ratio for poor clinical outcome in all ICH was 1.39, versus 1.71 in deep ICH. Further, the analysis for all ICH achieved the same association strength as that obtained for deep ICH (p=0.03), with twice the sample size.
DISCUSSION
Our results demonstrate that the burden of BP-related risk alleles, as measured through a GRS, is associated with increased BHV in ICH. This association is subtype-specific for hematomas occurring in deep structures of the brain. The GRS, likely through its effect on hematoma volume, also predicted poor clinical outcome, defined as severe disability or death 90 days after the bleeding episode, in deep and all (deep and lobar combined) ICH. These results are in line with previous evidence that show that the underlying biology of ICH varies with location. ICH occurring in deep locations of the brain has been primarily attributed to the effects on the cerebral vessels of long-standing hypertension,3 while a substantial proportion of lobar ICH appears to arise in the setting of amyloid angiopathy.4
The present results extend our understanding of the role of blood pressure in ICH. In an expanded set of cases and controls, we have previously demonstrated that the burden of BP-related genetic variants increases risk of deep hemispheric, but not lobar, ICH.9 These findings point to a mechanism that could mediate the observed effect: the same small vessel vasculopathy that caused the intracranial bleeding could also be responsible for the severity of the hematoma. This model proposes that severer stages of the intracranial vasculopathy—hypertensive in this case—lead to vessels more prone to rupture in the vicinity of the ICH, thus generating a cascading effect that would results in larger hematomas. As mentioned in the introduction, this paradigm has been put forward to explain the dual effect of the epsilon 2 allele at the apolipoprotein E locus on both risk6 and hematoma volume7 of ICH. The fundamental role of amyloid angiopathy and common genetic variation at the apolipoprotein E locus in the occurrence of lobar ICH also explains why the present study yielded a null result for lobar ICH, despite evidence showing that high BP also plays a role in risk of ICH in this location.20
Another mechanism that could mediate the effect of the burden of BP-related alleles on admission ICH volume involves BP levels during the bleeding episode. Higher BP levels after admission for ICH have been associated with increased risk of hematoma expansion, and, conversely, aggressive BP management is associated with a decreased risk of hematoma expansion.19,21 Higher number of BP-associated alleles could lead to worse and, possibly, more difficult-to-manage hypertension status. In the present study no association was observed between the BP-based GRS and admission systolic BP (Supplemental table 3). This result, however, is limited by the fact that only one BP measurement per subject was available for analysis. Further studies should evaluate the effect of BP-based GRS on patients’ complete BP profile, obtained through repeated BP measurements during the hospital admission.
In addition to contributing to understand the biology underlying ICH volume and expansion, these data have important implications for risk prediction. Given the limited impact of acute treatment in ICH, it is paramount to identify subjects at the highest risk of sustaining an ICH, as well as those at the highest risk of suffering severe bleedings. This would open the possibility of implementing aggressive preventive strategies in high-risk individuals. Genetic variants can aid in this goal, as these data are available from birth, long before hypertension is diagnosed, are constant over time, and are not subject to misclassification, and can be collected quickly, cheaply and painlessly. Importantly, this same approach could be applied to other risk factors and intermediates, and combined genetic data on common variants for these intermediates could be used to build increasingly precise risk prediction models.
This study has a number of limitations. First, due to the difficulty inherent in collecting ICH cases with both genotype data and quantification of BHV, we are not able to provide replication of these results in an independent dataset. It should be emphasized, however, that the weights utilized in the score were obtained from previous, independent genome-wide analyses, rather than from within our study population. Second, selection bias could be present in the form of survival bias. Patients identified in the setting of the implemented study design would be those who survived the onset of an ICH, thus reaching the hospital and allowing for their enrollment. As has been shown recently, however, simulation results suggest that the effect on risk estimates introduced in this setting would be relatively small. 22 Third, clinical status at three months was carried out by telephone interview; this could have introduced some misclassification in this outcome. Previous studies have shown, however, that telephone ascertainment of the mRS has acceptable reliability.23 Finally, the results of this study were obtained in a population from European ancestry. Further analyses would be needed to extrapolate our conclusions to populations of different ethnic background.
Supplementary Material
SUMMARY.
Considered individually, no single BP-related SNP is associated with ICH admission volume. This is expected, given the small effect on BP exerted by each of these genetic variants when considered in isolation. The aggregate burden of these risk alleles, on the other hand, is associated with mean hematoma volume upon admission in deep ICH, and poor functional outcome at 90 days in both all (deep and lobar) and deep ICH.
ACKNOWLEDGMENTS
FUNDING AND SUPPORT
All funding entities had no involvement in study design, data collection, analysis, and interpretation, writing of the report and in the decision to submit the paper for publication.
Genetics Of Cerebral Hemorrhage on Anticoagulation: This study was funded by NIH-NINDS grants R01NS059727, the Keane Stroke Genetics Research Fund, the Edward and Maybeth Sonn Research Fund, by the University of Michigan General Clinical Research Center (M01 RR000042) and by a grant from the National Center for Research Resources.
DECIPHER Study: This study is funded by NIH-NINDS grant 5U54NS057405 (DECIPHER).
Dr. Goldstein was supported by the NIH - K23NS059774. Drs. Biffi and Anderson were supported in part by the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research (0775010N). Dr. Brouwers was supported by the NIH – NINDS SPOTRIAS fellowship grant P50NS051343.
DISCLOSURES
S.M.G. received a research grant the NIH; received honoraria from Medtronic and Pfizer; and is a consultant/on the advisory board of Hoffman-La Roche, Janssen Alzheimer Immunotherapy, and Bristol-Myers Squibb Company.
J.N.G. received a research grant from the NINDS and is a consultant/on the advisory board of CSL Behring.
J.R. received a research grant from the NIH and the AHA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Search Terms: Intracerebral Hemorrhage, ICH volume, Hypertension, Common Genetic Variants, Genetic Risk Score
REFERENCES
- 1.Broderick JP, Brott TG, Duldner JE, Tomsick T, Huster G. Volume of Intracerebral Hemorrhage. A Powerful and Easy-to-Use Predictor of 30-Day Mortality. Stroke. 1993;24:987–993. doi: 10.1161/01.str.24.7.987. [DOI] [PubMed] [Google Scholar]
- 2.Fisher CM. Pathological observations in hypertensive cerebral hemorrhage. J. Neuropathol. Exp. Neurol. 1971;30:536–550. doi: 10.1097/00005072-197107000-00015. [DOI] [PubMed] [Google Scholar]
- 3.Fisher CM. Lacunar strokes and infarcts: a review. Neurology. 1982;32:871–876. doi: 10.1212/wnl.32.8.871. [DOI] [PubMed] [Google Scholar]
- 4.Vinters HV. Cerebral amyloid angiopathy. A critical review. Stroke. 1987;18:311–324. doi: 10.1161/01.str.18.2.311. [DOI] [PubMed] [Google Scholar]
- 5.Zhang-Nunes SX, Maat-Schieman MLC, van Duinen SG, Roos RAC, Frosch MP, Greenberg SM. The cerebral beta-amyloid angiopathies: hereditary and sporadic. Brain Pathol. 2006;16:30–39. doi: 10.1111/j.1750-3639.2006.tb00559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biffi A, Sonni A, Anderson CD, Kissela B, Jagiella JM, Schmidt H, et al. Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann. Neurol. 2010;68:934–943. doi: 10.1002/ana.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biffi A, Anderson CD, Jagiella JM, Schmidt H, Kissela B, Hansen BM, et al. J. APOE genotype and extent of bleeding and outcome in lobar intracerebral haemorrhage: a genetic association study. Lancet Neurol. 2011;10:702–709. doi: 10.1016/S1474-4422(11)70148-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taal HR, Verwoert GC, Demirkan A, Janssens ACJW, Rice K, Ehret G, et al. Genome-wide profiling of blood pressure in adults and children. Hypertension. 2012;59:241–247. doi: 10.1161/HYPERTENSIONAHA.111.179481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falcone GJ, Biffi A, Devan W, Jagiella JM, Schmidt H, Kissela K, et al. Burden of risk alleles for Hypertension Increases Risk of Intracerebral Hemorrhage. Stroke. 2012 doi: 10.1161/STROKEAHA.112.659755. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luft FC, Mervaala E, Müller DN, Gross V, Schmidt F, Park JK, et al. Hypertension-Induced End-Organ Damage A New Transgenic Approach to an Old Problem. Hypertension. 1999;33:212–218. doi: 10.1161/01.hyp.33.1.212. [DOI] [PubMed] [Google Scholar]
- 12.The Genes for Cerebral Hemorrhage on Anticoagulation (GOCHA) Collaborative Group. Exploiting common genetic variation to make anticoagulation safer. Stroke. 2009;40:S64–S66. doi: 10.1161/STROKEAHA.108.533190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broderick J, Connolly S, Feldmann E, Hanley D, Kase C, Krieger D, et al. Guidelines for the management of spontaneous intracerebral hemorrhage in adults: 2007 update: a guideline from the American Heart Association/American Stroke Association Stroke Council, High Blood Pressure Research Council, and the Quality of Care and Outcomes in Research Interdisciplinary Working Group. Stroke. 2007;38:2001–2023. doi: 10.1161/STROKEAHA.107.183689. [DOI] [PubMed] [Google Scholar]
- 14.Rosand J, Eckman MH, Knudsen KA, Singer DE, Greenberg SM. The effect of warfarin and intensity of anticoagulation on outcome of intracerebral hemorrhage. Arch. Intern. Med. 2004;164:880–884. doi: 10.1001/archinte.164.8.880. [DOI] [PubMed] [Google Scholar]
- 15.Weale ME. Quality control for genome-wide association studies. Methods Mol. Biol. 2010;628:341–372. doi: 10.1007/978-1-60327-367-1_19. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiology. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Wasay M, Yousuf A, Lal D, Awan S. Predictors of the intracerebral hemorrhage volume in hypertensive patients. Cerebrovasc Dis Extra. 2011;1:1–5. doi: 10.1159/000323270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson CS, Huang Y, Wang JG, Arima H, Neal B, Peng B, et al. Intensive blood pressure reduction in acute cerebral haemorrhage trial (INTERACT): a randomised pilot trial. The Lancet Neurology. 2008;7:391–399. doi: 10.1016/S1474-4422(08)70069-3. [DOI] [PubMed] [Google Scholar]
- 20.Jackson CA, Sudlow CLM. Is hypertension a more frequent risk factor for deep than for lobar supratentorial intracerebral haemorrhage? J Neurol Neurosurg Psychiatry. 2006;77:1244–1252. doi: 10.1136/jnnp.2006.089292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson CS, Huang Y, Arima H, Heeley E, Skulina C, Parsons MW, et al. Effects of Early Intensive Blood Pressure-Lowering Treatment on the Growth of Hematoma and Perihematomal Edema in Acute Intracerebral Hemorrhage The Intensive Blood Pressure Reduction in Acute Cerebral Haemorrhage Trial (INTERACT) Stroke. 2010;41:307–312. doi: 10.1161/STROKEAHA.109.561795. [DOI] [PubMed] [Google Scholar]
- 22.Anderson CD, Nalls MA, Biffi A, Rost NS, Greenberg SM, Singleton AB, et al. The effect of survival bias on case-control genetic association studies of highly lethal diseases. Circ Cardiovasc Genet. 2011;4:188–196. doi: 10.1161/CIRCGENETICS.110.957928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen PM, Visser NA, Dorhout Mees SM, Klijn CJM, Algra A, Rinkel GJE. Comparison of telephone and face-to-face assessment of the modified Rankin Scale. Cerebrovasc. Dis. 2010;29:137–139. doi: 10.1159/000262309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.