Abstract
Purpose of review
The microbiome continues to demonstrate an important role in immune and metabolic programming. This review will focus on the mechanistic implications of recent findings for diabetes pathogenesis and treatment.
Recent findings
Multiple techniques are developing to specify the microbiome. At the same time, new insights have emerged into local interactions of microbial products with human development. New findings demonstrate that key bacteria and their products result in the programming of diabetes-modulating Th17 and regulatory T lymphocytes within and outside the intestine. The role of the bacterial metagenome in programming human metabolism has also revealed new insights. In turn, these findings suggest a framework in which the microbiome may be modified to change the course of diabetes.
Summary
The microbiome is a key regulator of metabolism and immunity. Specific bacteria and their secreted products are now known to program Th17 and regulatory T-cell development, which may change the course of diabetes. Bacterial genomics are demonstrating important, modifiable roles of bacterial gene products in metabolism. Further understanding of this symbiotic relationship will provide new avenues for intervention in diabetes.
Keywords: diabetes, immunity, metabolism, microbiome, T regulatory cells, Th17
INTRODUCTION
Type 1 diabetes (T1D) develops from the complex interplay of genetic and environmental factors in a stochastic manner. How the environment modifies disease risk and progression has been a long-term subject of investigation as environmental modification represents an attractive approach to change the course of this disease. These investigations have suggested environmental programming begins at birth as early-life exposures modulate T1D outcomes [1,2]. With regard to these early-life events, the pioneering and well regarded BABYDIAB study group recently reported, for instance, that birth by cesarean section appeared to increase the risk for T1D by shortening the duration between the first detection of anti-β cell-specific autoantibodies and diabetes onset [3]. In the immediate postpartum period, breastfeeding has also been ascribed a protective function, although whether through constituents of the breast milk or avoidance of formula remains a matter of debate. The recently reported early results from the nutritional Trial to Reduce Insulin-dependent diabetes in the Genetically at-Risk (TRIGR), in which infants at genetic risk were randomized to casein hydrosylate formula and/or breastfeeding versus cow’s milk formula, also support the role of early exposures as it indicated that hypoallergenic nutrition reduces the development of β-cell autoimmunity within the first 10 years of life [4▪].
The classic concept of the role of the environment in diabetes is as a trigger that initiates autoimmunity. When viewed through the immunologic lens, a triggering event is expected to lead directly to activation of autoreactive lymphocytes and production of autoantibodies. In this context, it is difficult to explain how a trigger early in life requires decade or more before autoimmunity becomes evident, even when autoimmunity is defined as autoantibody production prior to evident β-cell destruction. One possibility is that environmental signals early in life shape the developing immune system over time. One critical early signal with potential for longstanding effects on immune homeostasis is the development of the (intestinal) microbiome. On the basis of emerging evidence of the role of the microbiome in immune development and homeostasis, alterations in the microbiome may be a new mechanism by which the risks from early environmental insults are propagated over an individual’s lifetime.
In this review, we will examine new insights into the development of the intestinal microbiome, its role in programming immune regulation, and the emerging definitions of diabetes-promoting and diabetes-protecting microbiota. We will also briefly consider what is known on the role of the microbiome in metabolism and development of type 2 diabetes (T2D) and present opportunities for future interventions that will derive from these insights.
UNCOVERING THE ROLE OF THE MICROBIOME IN HEALTH AND DISEASE
The intestinal microbiome most likely influences the risk of diabetes either through the specific types of bacteria that are present and their effect on cellular and immune development or through expression of bacterial gene products that shape metabolism or produce novel epitopes from ingested food. Defining the role of the microbiome remains more complex than defining the human genome as bacteria outnumber human cells 10 : 1 and their collective genome outsizes the human genome by a factor of 100 [5]. Large scale, collaborative research efforts, exploiting novel molecular, and genetic technologies, have been launched, including the Human Microbiome Project of the National Institutes of Health and comparable European Union sponsored initiatives [5].
Defining human microbiome–disease interactions is dependent on our ability to characterize microbes and their functions by culture-independent techniques, a process that presents unique challenges at each stage, as outlined in Table 1. Pyrosequencing, as exemplified by the 454 platform and related technologies, offers the advantage of long sequence reads that lead to accurate classification of bacterial taxonomy. However, these techniques may be limited in the number of sequences generated and may thus incompletely specify the microbiome. Additionally, replicates from the same sample may not generate the same bacterial taxa. Currently, very few microbiome studies perform replicates. Including publications from 2009 (total of 441 articles) in five major microbial ecology journals, replicate samples were employed in only 30% of studies [6]. Alternatively to pyrosequencing, Illumina sequencing-by-synthesis approaches generate a significantly larger number of reads; however, the short-read length often limits bacterial taxonomic classification, although it is being applied and refined for use in this field. Ultimately, techniques that combine adequate read length with massive numbers of unique sequences may be needed to adequately specify the microbiome.
Table 1.
Considerations in microbiome study design
| Process | Question for investigator | Considerations |
|---|---|---|
| Sample collection | Will fecal samples be an adequate surrogate for the bacterial populations of interest? | Most studies use fecal samples, although there may be important differences between adherent mucosal bacterial communities and microorganisms detected in feces. |
| How many samples are representative of an individual’s microbiome? | ||
| DNA amplification | What gene target should be employed (i.e., 16S rRNA vs cpn60 UT)? | A resource for investigators considering cpn60 or other chaperonin proteins for their microbiome studies is a database organized by several Canadian research organizations available at www.cpndb.ca/cpnDB/home.php. |
| Which primer set within the gene should I select? | Nine variable regions exist within the 16S gene that can be selected. Commonly used targets include V2, V4, and V6. | |
| High-throughput sequencing | What read length and sampling depth do I need to address the goal of my project? | Illumina (paired-end 101 bp reads) and 454 pyrosequencing (>400 bp reads) show relative consistency with each other. However, some studies suggest the majority of Illumina reads cannot currently be assigned a genus due to shorter lengths and higher error rates above 60 bp. |
| What sequencing platform should I select (454 pyrosequencing versus Illumina)? | Study-to-study data comparisons are only possible when the same sequencing platforms are used. | |
| Data analysis | How should I filter out my low-quality reads? | Phylogenetic analysis employs a phylogenetic tree to connect sequences and is useful when asking how much evolutionary history is unique to a particular sample. Taxon-based analysis looks at how many different ‘species’ are in a sample and treats all assigned species as if they were phylogenetically equivalent. |
| Should I perform taxon-based or phylogenetic analyses? | Multiple programs for filtering and analysis are available. The database and algorithms must be considered for each. | |
| How will I define OTU? |
Analysis of the microbiome requires acquisition of samples, amplification of a target gene with both adequate conserved regions across all bacteria and variable regions for taxonomic classification, sequencing of the amplified target gene, and quality control of sequence data with assignment of sequences to specific bacteria. Potential pitfalls and considerations at each stage are presented. bp, base pairs; cpn, chaperonin; OTU, operational taxonomic unit; UT, universal target.
Identifying bacteria from target genes other than 16S rRNA may allow bacterial identification from shorter products, which would facilitate Illumina-type approaches. Chaperonin-60 (cpn60) universal target amplicons represents one such target that may provide short lengths (549–567 base pairs), greater taxonomic resolution, and relatively uniform distribution of variability across the entire length of the target. A comparison of the 16S rRNA versus cpn60 target for analysis of vaginal microbial communities demonstrated a greater number of operational taxonomic units identified by the cpn60 target, suggesting improved species resolution with this approach [7].
Beyond specifying an individual’s bacterial classes, identification of the metatranscriptome specifies the function of these bacterial communities via isolation of actively transcribed genes. In a recent study utilizing this approach in 10 individuals, species varied between individuals, although tremendous functional overlap existed, especially with regard to the presence of microorganisms responsible for fermentation of polysaccharides and dietary fiber [8▪]. Thus, different bacterial types may have the same local function. Combination of the representation of bacterial species diversity with their function is becoming a new way to define the host–microbiome interaction-denoted enterotypes [9▪▪]. The presence of a limited number of human enterotypes defined to date suggests the possibility of stable ‘nodes’ for host–microbiome symbiosis. Whether diabetes risk can be stratified according to enterotypes remains to be determined.
In addition to choosing the optimal technique, the field is also challenged by determining the optimal intestinal location for study. Fecal samples are most readily available, but studies comparing fecal and tissue samples from the same person detect significant variability between the two sources, questioning the accuracy of fecal samples as surrogates for the intestinal tissue microbiome [10,11]. Because T1D may be programmed by events in the mesenteric and pancreatic lymph nodes close to the small intestine, intestinal tissue biopsy samples may be a more appropriate source for diabetes-related microbiome analyses.
THE MICROBIOME IN IMMUNE SYSTEM DEVELOPMENT, AUTOIMMUNITY, AND TYPE 1 DIABETES
The integrated development of the gut and pancreas and the role of the gut as the body’s largest immune organ make the mucosal immune system a logical site for diabetes initiating and preventing interactions. The role of the microbiome as a determinant of T1D has long been supported by the widely held belief that germ-free, diabetes-prone nonobese diabetic (NOD) mice have a higher incidence of T1D than the specific-pathogen free (SPF) colony counterparts. At least two studies now indicate that this is not strictly true and that diabetes in the germ-free setting may not exceed maximal rates in SPF colonies, although penetrance may be more consistently high [12▪,13▪]. Acceleration in germ-free environments suggested that the predominant role of bacteria is to protect against T1D. Indeed, several bacterial strains have been associated with protection from diabetes in both murine and rat diabetes models, including protective roles for Bacillus cereus and Lactobacillus johnsonii [12▪,14]. However, in more complex settings, it is highly likely that bacteria play both diabetes promoting and protective roles.
In mice, specific members of the microbiota named segmented filamentous bacteria (SFB), a nonculturable Clostridia-related species, can induce a functional and balanced intestinal host response [15]. Nevertheless, the autoimmunity-promoting role of this single bacterium has also been the most well characterized of the constituents of the microbiome. In the NOD-derived arthritogenic model, the K/BxN mouse, colonization with SFB is sufficient to lead to the arthritogenic phenotype by producing Th17 cells that support arthritogenic antibody production through germinal center reaction [16▪].
Although these SFB appear to promote Th17-dependent arthritis development, a recent study indicates that the presence of detectable SFB colonization is correlated with reduced diabetes incidence in an otherwise diverse microbial community [17▪▪]. Although insulitis was not attenuated, progression to diabetes was limited, at least in female mice. Whether other microbial species influence initiation of insulitis and why sex differences exist in this phenomenon remain important areas for future research. SFB are important inducers of interleukin (IL) 17-producing immune cells residing within the gut. The role of these cells in T1D is unresolved but the development of Th17 cells may limit the development of more diabetogenic Th1 cells. Other studies have indicated that the development of Th17 cells promoted by SFB is antigen independent, which questions how these induced cells could specifically protect pancreatic islets while still permitting or fostering other types of autoimmunity [18]. However, there may also be a role for antigen specificity and non-SFB bacteria in the development of an important subset of RORγt+ cells, double positive Foxp3+RORγt+ cells [18]. These cells express both Th17 and Foxp3+ regulatory T-cell (Treg) determining transcription factors, and it has been suggested that some Treg may partially express effector cell programs in order to successfully protect the microbe-exposed host [19]. Overall, the complex interaction of the microbiota and antigen-specific T cells may in part explain how different autoimmune disorders cosegregate in the same kindred. For example, SFB may induce arthritogenic and encephalitogenic Th17 cells that protect from diabetes, whereas introduction of additional bacterial species may produce additional regulatory cells that further modify the course of these diseases.
Commensals are clearly important in the induction of adequate numbers of immune regulatory cells and, in at least one instance, a mechanism has emerged. The important human commensal, Bacteroides fragilis, directs the development of Treg in the intestine. Treg are critical for immune homeostasis and functional defects in these cells have been found in the pancreatic-draining lymph nodes of patients with T1D [20]. A specific immunomodulatory molecule of B. fragilis, polysaccharide A (PSA), mediates the conversion of CD4+ T cells to IL-10-producing Treg and actively suppresses Th17-cell development in the intestinal tract [21▪]. Interestingly, unlike pathogens that trigger inflammatory responses through Toll-like receptors (TLR) to clear infections, colonization by B. fragilis is promoted by mucosal tolerance induction through TLR2 activation of Treg [22▪]. In addition, microbiota from the Firmicutes phylum such as murine Clostridium species and human Lactobacillus strains are now known to induce Treg in the large intestine as well as in other organs and promote systemic immune homeostasis [23▪▪,24]. It is important to note that Treg induction by microbiota is regulated differently in the small and large intestine. For example, whereas probiotic components have successfully been implemented in the promotion of Treg in the large intestine, commensal bacterial DNA in the small intestine can suppress Treg development through TLR9-mediated activation of lamina propria dendritic cells [25].
The role of TLR’s in the microbiome–immune system interaction fulfills the expectation of a role for pattern recognition receptors in this process. However, despite a clear role for TLR2/9 in Treg-inducing pathways, deficiency in multiple individual TLRs has not been found to modify T1D. Nonetheless, deficiency of MyD88, the common TLR adaptor protein, prevents diabetes through a microbiome-dependent process [26]. Other recent studies have indicated that MyD88 deficiency leads to loss of the protective space normally present between the intestinal epithelium and the bulk of fecal bacteria. This process is dependent on production of the protein RegIIIγ, a C-type lectin antibacterial against Gram-positive bacteria [27▪]. Thus, RegIIIγ deficiency leads to increased numbers of Gram-positive bacteria including SFB, which may explain the protective role of MyD88 deficiency in NOD mice although SFB were not detected in the initial report of MyD88−/− NOD mice. How the role of this process, along with bacteria/epithelium interactions that may elucidate the mechanisms behind concepts such as the role of the ‘leaky’ epithelium in diabetes risk, remain for further investigation [28]. An additional critical implication of the study by Wen et al. [26] is that a gene knockout that was expected to alter diabetes by disrupting innate immunity directly exerted its diabetes-protective effect through the microbiome. Whether other models targeting immune function in NOD mice prevent diabetes by common microbial alterations is an urgent area in need of reconciliation with our understanding of immune defects in T1D.
Although the mechanisms of autoimmunity protection and promotion by bacterial colonization are emerging in animal models of diabetes, translation of these findings to prediction and prevention of human disease awaits further investigation. To date, a single study has reported microbiome analysis in children from Finland’s Diabetes Prediction and Prevention study in which children are defined as high risk for diabetes based on Human Leukocyte Antigen (HLA)-DQ genotype. Children in the study by Giongo et al. [29▪] were followed for seroconversion to more than two autoantibodies as the definition of autoimmunity, and all developed T1D. Those children who seroconverted showed loss of bacterial diversity over time with single species dominating in the Bacteroidetes and Firmicutes phyla. Whether this loss of diversity plays a specific pathologic role or is a correlate of other changes in the microbiome requires further investigation. Defining the molecular pathways that protect or promote diabetes is an outstanding opportunity for advancing disease prevention and is the subject of ongoing international T1D trials.
INTERACTION BETWEEN MICROBIOME, METABOLISM, AND TYPE 2 DIABETES
Although the microbiome can modulate immune system development, it also plays an important role in the interaction with ingested nutrients. It is an old fact that microbial constituents of the gut break down numerous energy substrates. However, ingested bacteria are now known to induce significant changes in several metabolic pathways, most prominently those related to carbohydrate metabolism [30▪▪]. Thus, the microbiome may program the whole body metabolism. A recent gut microbiome metagenomics analysis showed a higher proportion of butyrate-producing and mucin-degrading bacteria in controls compared with patients with T1D [31▪]. Disruptions in metabolism may lead to chronic inflammation and both drive autoimmunity (T1D) and produce metabolic disease states including T2D [32].
Attempts to elucidate the interrelationships between dietary intake, intestinal microbial flora, and host metabolism, a field of study often referred to as metabolomic profiling, is a compelling new area of investigation. Randomization of lean and obese individuals to either a 2400 or 3400 k calorie per day diet for a 3-day period, with simultaneous serial stool monitoring and 16S rRNA pyrosequencing, demonstrated rapid changes in the two major gut phyla. Specifically, a 20% increase in Firmicutes and a reciprocal decrease in Bacteroidetes corresponded with an increased energy harvest of approximately150 k calorie in lean individuals. Overfeeding of lean individuals also correlated with a greater fractional decrease in stool energy loss [33▪].
Compositional changes in the microbiota of individuals with T2D have also been observed, including reductions in the amount of Firmicutes, when compared with a control group of nondiabetic adults. Furthermore, the ratios of Bacteroidetes-to- Firmicutes correlated positively with plasma glucose concentration, although not with BMI [34▪]. Obese individuals have an altered gut microbiota when compared with lean controls that is characterized by a reduced number of Bacteroidetes [35], although not all studies have demonstrated this [36], and some studies in mice have suggested that microbiota perturbations associated with obesity are age dependent [37]. In both T2D and obesity, these alterations on an organism level likely correspond to modifications in host energy metabolism.
The mechanism of interaction between diet and gut flora may now be understood as microbial modifications of nutrient absorption that occur with alterations in dietary substrates and that process may be driven by the presence of polysaccharide utilization loci (PUL) contained within the genome of the main intestinal bacterial taxa. These PULs, which encode the machinery responsible for degradation and utilization of extracellular polysaccharides, have been best studied in the Bacteroidetes phylum. The exact gene content of this locus differs within the Bacteroidetes phylum, which phenotypically translates into bacteria with varied abilities to metabolize fructose-based dietary polysaccharides [38▪▪]. Those that can effectively utilize fructans are ultimately able to ‘outcompete’ other bacterial community members with further proliferation. Individual differences in glucose metabolism may in fact be a product of these species-specific PULs and the functional role they have in polysaccharide breakdown.
CONCLUSION
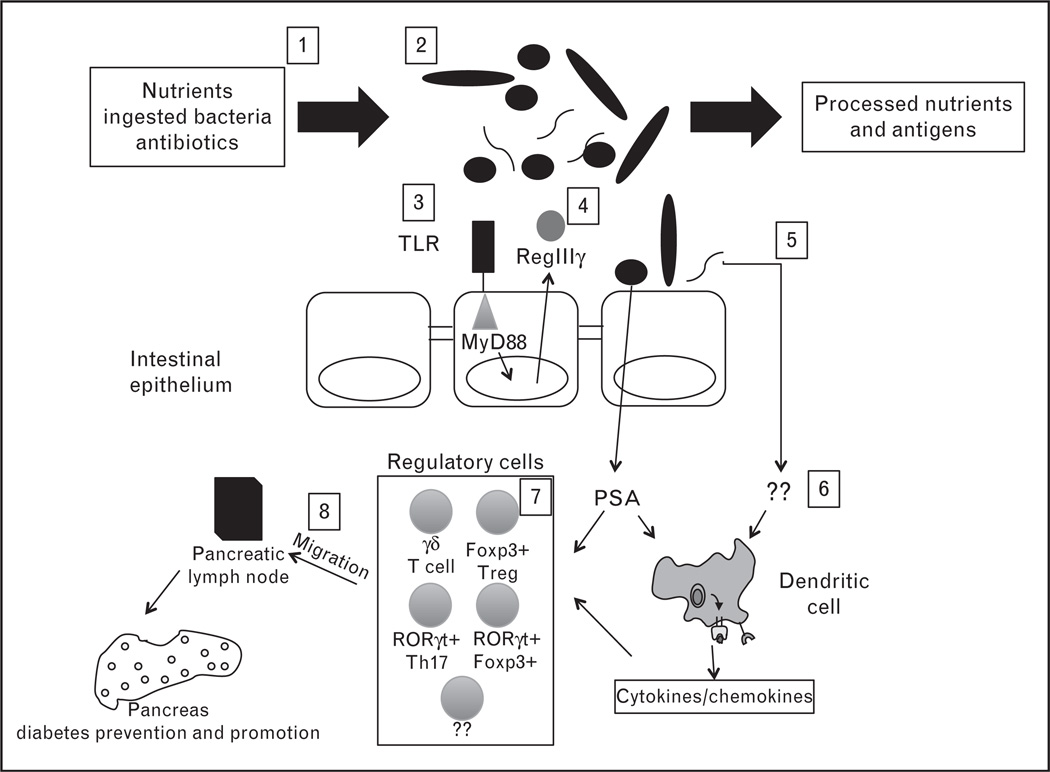
Emerging data on the intestinal microbiome reveals important symbiotic relationships that program human metabolism and immune system development. Human genetics and development also shape the microbiome resulting in a stable interaction in the healthy individual. The first studies on humans with diabetes suggest that this relationship may be perturbed during diabetes progression. An overall model for a series of immune–microbiome interactions in the context of diabetes is presented in Fig. 1.
FIGURE 1.
The microbiome is programmed by numerous environmental factors. 1, selected nutrients and ingested agents determine which bacteria are most likely to thrive. Nutrient availability also influences bacterial gene expression, which may modify overall human metabolism; 2, the diversity of the gut microbiome challenges the immune system to adapt and develop; 3, this interaction is mediated in part by Toll-like receptors (TLRs) including TLR2/9 and their common signaling adaptor MyD88. 4, MyD88 normally induces RegIIIγ, a protein which limits Gram-positive bacterial association with the epithelium; 5, this protection may break down because of inflammation or epithelial injury; 6, accumulation of regulatory T cell (Treg) is regulated differently in the small and large intestine and operates either through direct effects of the microbiota on regulatory cells or indirectly through intermediates such as dendritic cells. 7, overall these factors induce a diversity of cells within the lamina propria. Some, such as CD4+Foxp3+ Treg, RORgt+Th17, or newly described RORgt+Foxp3+ double positive cells, may have diabetes preventing potential; and 8, autoimmune diabetes is initiated in the pancreatic lymph node so these cells induced in the lamina propria likely migrate by an unknown mechanism to the pancreas and pancreatic node in which they exert their protective functions. Conversely, activated diabetogenic cells may migrate to the intestine where they could be regulated. How the interaction of bacteria with locally induced specialized lymphocytes leads to diabetes modulation remains for future mechanistic dissection.
Many questions remain in order to translate this new information to clinical impact for persons with diabetes. With respect to immune system development, it is not yet clear how the same bacterial species may promote some autoimmune diseases, like arthritis, while protecting from others, like diabetes. We also are not yet able to specifically modulate the microbiome. Recent studies suggest we can foster outgrowth or suppression of certain bacteria. Additionally, seemingly radical approaches such as fecal transplant are also gaining attention with some reported success in treating recurrent Clostridium difficile. We expect that defining the microbiome and its function will reveal multiple new opportunities to intervene in the progression of diabetes while simultaneously refining our understanding of immune and metabolic development in this new context. Although study of the microbiome presently remains specialized, further technologic progress is also likely to make microbiome specification a common and required aspect of future diabetes studies.
KEY POINTS.
The microbiome impacts human metabolism and immune system development.
Microbes within the gut and interacting with the intestinal epithelium can induce specific T-cell subsets including Th17 and Treg lineages.
The microbiome adapts its genome (the human metagenome) to ingested nutrients, and thereby modulates human metabolism and energy intake.
Understanding type 1 and type 2 diabetes pathogenesis will require understanding the role of the microbiome.
Acknowledgements
None.
Footnotes
Conflicts of interest
We apologize to the numerous authors whose contributions we were not able to cite for space limitations. J.R.K is supported by T32HD068256. J.H.W is supported by NIH K08HD061607, the Vanderbilt CTSA grant UL1 RR024975–01 from NCRR/NIH (to JHW), and the Vanderbilt University Medical Center’s Digestive Disease Research Center sponsored by NIH grant P30DK058404. DJM is supported by NIH K08DK090146, by a Vanderbilt DRTC Pilot and Feasibility Grant (5P60DK20593–33), and by the Turner-Hazinski award from the Vanderbilt Department of Pediatrics.
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (p. 142).
- 1.Stene LC, Ronningen KS, Undlien DE, Joner G. Does the relative risk for type 1 diabetes conferred by HLA-DQ, INS, and PTPN22 polymorphisms vary with maternal age, birth weight, or cesarean section? Pediatr Diabetes. 2011;12:91–94. doi: 10.1111/j.1399-5448.2010.00669.x. [DOI] [PubMed] [Google Scholar]
- 2.D’Angeli MA, Merzon E, Valbuena LF, et al. Environmental factors associated with childhood-onset type 1 diabetes mellitus: an exploration of the hygiene and overload hypotheses. Arch Pediatr Adolesc Med. 2010;164:732–738. doi: 10.1001/archpediatrics.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifacio E, Warncke K, Winkler C, et al. Cesarean section and interferon-induced helicase gene polymorphisms combine to increase childhood type 1 diabetes risk. Diabetes. 2011;60:3300–3306. doi: 10.2337/db11-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knip M, Virtanen SM, Seppa K, et al. Dietary intervention in infancy and later signs of beta-cell autoimmunity. N Engl J Med. 2010;363:1900–1908. doi: 10.1056/NEJMoa1004809. This study further supports a role of dietary interventions in modulating autoimmunity. Future specification of the microbiome in this study may be an important stop in understanding microbiome–T1D interactions.
- 5.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prosser JI. Replicate or lie. Environ Microbiol. 2010;12:1806–1810. doi: 10.1111/j.1462-2920.2010.02201.x. [DOI] [PubMed] [Google Scholar]
- 7.Schellenberg J, Links MG, Hill JE, et al. Pyrosequencing of the chaperonin-60 universal target as a tool for determining microbial community composition. Appl Environ Microbiol. 2009;75:2889–2898. doi: 10.1128/AEM.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gosalbes MJ, Durban A, Pignatelli M, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6:e17447. doi: 10.1371/journal.pone.0017447. This study examines the human microbiome from a functional level by applying a metatranscriptomic approach whereby mRNA is extracted from fecal samples to categorize microbial gene expression within the GI tract. This represents an important extension from genome analysis to functional approaches.
- 9. Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. This study represents a new approach to classifying the human microbiome in terms of species and functions. Whether ‘enterotype’ will eventually be recognized as the HLA of the microbiome remains to be seen. However, integrative classification strategies offer a strong opportunity to reveal host–bacteria–disease interactions by accounting for multiple factors involved in these complex interactions.
- 10.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoetendal EG, von Wright A, Vilpponen-Salmela T, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. King C, Sarvetnick N. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One. 2011;6:e17049. doi: 10.1371/journal.pone.0017049. See Ref. [12▪].
- 13. Alam C, Bittoun E, Bhagwat D, et al. Effects of a germ-free environment on gut immune regulation and diabetes progression in nonobese diabetic (NOD) mice. Diabetologia. 2011;54:1398–1406. doi: 10.1007/s00125-011-2097-5. These two studies [12▪,13▪] revisit the notion of acceleration of diabetes in germfree NOD mice. These studies confirm that not all bacteria are likely to be disease protective and invite further mechanistic dissection of the hygiene hypothesis.
- 14.Valladares R, Sankar D, Li N, et al. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One. 2010;5:e10507. doi: 10.1371/journal.pone.0010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 16. Wu HJ, Ivanov II, Darce J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. This study is an important counterpoint to [17▪▪] below as it shows that segmented filamentous bacteria permit certain autoimmune conditions. How the outcome of SFB-immune interactions determines disease versus protection is an important area for development.
- 17. Kriegel MA, Sefik E, Hill JA, et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. This study describes a key diabetes-protective bacterial species. Understanding the mechanism of this protection and how residence of these SFB are modulated in the gut is a key area for advancing diabetes prediction and therapy.
- 18.Lochner M, Berard M, Sawa S, et al. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J Immunol. 2011;186:1531–1537. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- 19.Wohlfert E, Belkaid Y. Plasticity of T reg at infected sites. Mucosal Immunol. 2010;3:213–215. doi: 10.1038/mi.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferraro A, Socci C, Stabilini A, et al. Expansion of Th17 cells and functional defects in T regulatory cells are key features of the pancreatic lymph nodes in patients with type 1 diabetes. Diabetes. 2011;60:2903–2913. doi: 10.2337/db11-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. Round et al. present the first identification of a bacterial product that induces Treg development. B. fragilis PSA was found to induce IL-10 producing Treg.
- 22. Round JL, Lee SM, Li J, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. Induction of intestinal Treg is thought to be orchestrated primarily by CD103+ dendritic cells residing in the gut-associated lymphoid tissues. This study suggests that B. fragilis-derived PSA induces Treg directly through TLR2-mediated signaling to suppress Th17 cell responses, and therefore promote colonization by B. fragilis. This represents the first identification of a bacterial protein that modulates host immunity and may lead to pharmacologic approaches to do the same.
- 23. Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. This study demonstrated that colonization of germ-free mice with a cocktail of Clostridium species promotes Treg induction and activity in the colon but not the small intestine. Clostridium-mediated Treg accumulation was observed at extra-intestinal sites as well and resulted in resistance to experimental autoimmunity.
- 24.Livingston M, Loach D, Wilson M, et al. Gut commensal Lactobacillus reuteri 100-23 stimulates an immunoregulatory response. Immunol Cell Biol. 2010;88:99–102. doi: 10.1038/icb.2009.71. [DOI] [PubMed] [Google Scholar]
- 25.Hall JA, Bouladoux N, Sun CM, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. This study extends the model of innate immunity–commensal interactions by providing the first evidence of a host defense molecule that limits bacterial–gut interactions.
- 28.Visser JT, Lammers K, Hoogendijk A, et al. Restoration of impaired intestinal barrier function by the hydrolysed casein diet contributes to the prevention of type 1 diabetes in the diabetes-prone biobreeding rat. Diabetologia. 2010;53:2621–2628. doi: 10.1007/s00125-010-1903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. This study provides the first description of the microbiome in persons genetically at risk for T1D that progress to autoantibody positivity. The small sample size awaits confirmation and extension.
- 30. McNulty NP, Yatsunenko T, Hsiao A, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002701. 106ra106. This study shows that ingestion of probiotic yogurt did not significantly change the arrangement of the host gut microbiota, but did alter metabolic pathways predominantly those related to carbohydrate processing.
- 31. Brown CT, Davis-Richardson AG, Giongo A, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. This study used a metagenomic approach to identify that patients with type 1 diabetes have a lower proportion of butyrate producing, and therefore mucin promoting bacteria, which may increase gut permeability.
- 32.Huang X, Moore DJ, Ketchum RJ, et al. Resolving the conundrum of islet transplantation by linking metabolic dysregulation, inflammation, and immune regulation. Endocr Rev. 2008;29:603–630. doi: 10.1210/er.2008-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. This study demonstrates how modest alterations in caloric consumption induce rapid changes in the gut microbiota and subsequent nutrient absorption and energy harvest in obese and lean individuals.
- 34. Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from nondiabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. This is one of the first descriptions of the gut microbiome in individuals with type 2 diabetes.
- 35.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 36.Schwiertz A, Taras D, Schafer K, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 37.Murphy EF, Cotter PD, Healy S, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 38. Sonnenburg ED, Zheng H, Joglekar P, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. This study illustrates the role of polysaccharide utilization loci (PUL) within microbial genomes as a mechanism by which bacterial communities respond to dietary modifications through the specificity of PUL expression. This study represents a key step to understanding the rules of symbiosis that we may ultimately manipulate for clinical gains.