Abstract
We investigated the impacts of sarA and agr on fnbA expression and fibronectin-binding capacity in Staphylococcus aureus in vitro and in experimental endocarditis. Although sarA up-regulated and agr down-regulated both fnbA expression and fibronectin binding in vitro and in vivo, fnbA expression was positively regulated in the absence of both global regulators. Thus, additional regulatory loci contribute to fnbA regulation and fibronectin-binding capacities in S. aureus.
Staphylococcus aureus is the most common cause of endovascular infections (2). The capacity of S. aureus to cause human diseases involves a variety of cell surface-associated and extracellular virulence factors (5, 7, 11, 15). Two fibronectin-binding proteins (FnBPA and FnBPB), have been ascribed multiple functions, including cell-specific binding (e.g., epithelial and endothelial cells), invasion and persistence within such cells, and triggering of host cell apoptosis (1, 14, 21, 26). Additionally, FnBPA has been shown to be involved in adherence to damaged heart valves (23). Moreover, FnBPs expressed on the S. aureus surface may be degraded by extracellular proteases (17, 18), suggesting that such enzymes participate in the transition of S. aureus cells from an adhesive to invasive phenotype.
Classically, the expression of FnBPs and the synthesis of extracellular proteases are controlled in vitro by at least two global regulatory loci: the accessory gene regulator (agr) and the staphylococcal accessory regulator (sarA) in S. aureus (3, 22, 24). There is a complex interaction between sarA and agr to coordinately regulate S. aureus virulence factor expression, including selected adhesins and extracellular proteases (7, 9, 22). In the present study, we have characterized the impacts of the sarA and agr loci upon fnbA expression, fibronectin-binding capacity, and protease activity in a set of isogenic S. aureus Newman strains in vitro and in an experimental rabbit endocarditis model.
fnbA promoter expression in vitro.
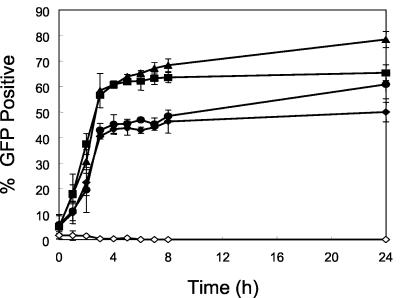
The S. aureus strains and plasmids used in this study are listed in Table 1 (strain Newman is agr type 1). Flow cytometry (FACScalibur; Becton-Dickinson, San Jose, Calif.) was utilized for quantification of fnbA promoter expression, employing a promoter-green fluorescent protein (GFP) reporter fusion, as previously described (28, 30). As expected, fnbA promoter expression was maximal during exponential growth of the parental strain and then plateaued (Fig. 1). In addition, the anticipated positive and negative regulatory effects of sarA and agr, respectively, on fnbA promoter expression were observed (Fig. 1) (3, 24, 29). Interestingly, the percentage of fnbA-expressing cells in the sarA agr double mutant paralleled that of the sarA single knockout mutant during exponential and early postexponential growth phases but increased to near-parental levels in late stationary growth phase (Fig. 1). These data suggest that environmental cues (e.g., low pH, nutrient limitation) or other regulatory loci contribute to fnbA expression during the stationary growth phase in the absence of sarA and agr in vitro (e.g., sae) (27).
TABLE 1.
S. aureus strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| Newman | Wild type | 29 |
| ALC637 | Newman, sarA::Tn917LTV1 | 29 |
| ALC355 | Newman Δagr::tetM | 29 |
| ALC638 | Newman Δagr::tetM sarA::Tn917LTV1 | 29 |
| ALC1829 | Newman with recombinant pALC1484 | This work |
| ALC1827 | Newman with recombinant pALC1484 containing the fnbA promoter | 29 |
| ALC1825 | ALC637 with recombinant pALC1484 containing the fnbA promoter | 29 |
| ALC1835 | ALC355 with recombinant pLAC1484 containing the fnbA promoter | 29 |
| ALC1838 | ALC638 with recombinant pALC1484 containing the fnbA promoter | 29 |
| ALC1645 | RN6390 with Δspa::Etbr mutation | 30 |
| Plasmid | ||
| pALC1484 | pSK236 with a promoterless gfpuvr gene | 29 |
FIG. 1.
Expression of the fnbA promoter in the parental and mutant strains in vitro. The percentage of GFP-positive S. aureus cells during 24 h of incubation in vitro is shown for the fnbA::gfpuvr parental strain (▪), agr mutant (▴), sarA mutant (⧫), sarA agr double mutant (•), and promoterless gfpuvr strain (◊).
Northern blot analysis of fnbA transcription.
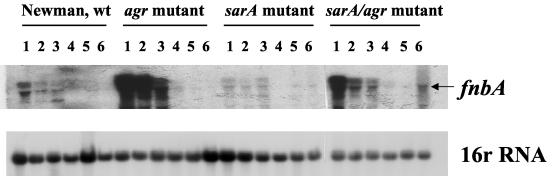
RNA isolation and Northern blot analysis were performed as described previously (29). The transcription of fnbA in the parental strain was maximal during mid-log phase (Fig. 2). As expected, in the agr mutant, there was substantial up-regulation in fnbA transcription during the late log phase, while fnbA transcription in the sarA mutant was markedly reduced compared to that in the parental strain (Fig. 2) (24, 29). It is noteworthy that the level of fnbA transcription in the sarA agr double mutant was between the levels of the single agr and sarA mutants. Interestingly, we also observed a bimodal increase in fnbA transcription in the double mutant, with the first peak occurring during the mid-log phase and a smaller but noticeable peak occurring during the late stationary phase (overnight culture). Therefore, these in vitro transcriptional data concurred with those of the GFP reporter gene fusion data sets above.
FIG. 2.
Northern blots of fnbA transcripts from the wild-type (wt) Newman strain and its isogenic agr, sarA, and sarA agr double mutants. RNAs were harvested from mid-log phase (lanes 1 and 2), late log phase (lane 3), early stationary phase (lane 4), and late stationary phase (lanes 5 and 6). We have included transcription of 16S rRNA as a loading control.
Protease activity in vitro.
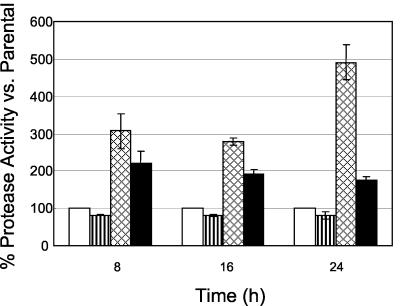
To quantify overall protease activity, a microplate assay kit (Molecular Probes, Eugene, Oreg.) was utilized as previously described (4). Protease activity was slightly decreased in the agr mutant (∼0.8-fold) but significantly increased in the sarA mutant (three- to fivefold; P < 0.05) compared to the parental strain (Fig. 3). These data are consistent with the documented repression of protease production by sarA (17, 18). The sarA agr double mutant had a quantitative protease phenotype that was intermediate between those of the sarA and agr single mutants. No protease activity was observed from any study strain after 4 h of incubation (data not shown).
FIG. 3.
Protease activity in S. aureus fnbA::gfpuvr in the parental strain (□) and agr (▥), sarA (▩), and sarA agr (▪) mutants during 24 h of incubation in vitro. The total protease activity in sarA and/or agr mutants is shown as the percent activity relative to that of the corresponding parent strain, which was normalized at 100%.
To evaluate the effect of the global protease inhibitor, α2-macroglobulin (Boehringer-Mannheim, San Diego, Calif.), the above experiments were repeated with cultures preexposed to the inhibitor (range, 0.4 to 1.6 U/ml). Protease activity was inhibited (>80% for all study strains) in the presence of α2-macroglobulin (1.2 U/ml) (data not shown).
Fibronectin adherence in vitro.
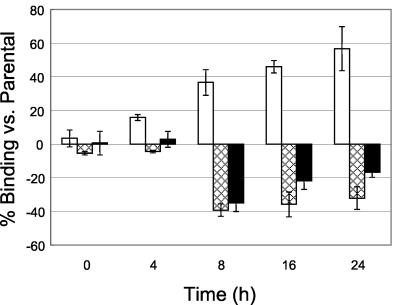
To quantitate correlation between fnbA expression and fibronectin adherence phenotypes, we evaluated the fibronectin-binding capacities of S. aureus by direct binding to immobilized human fibronectin as previously described (17, 19). It is noteworthy that fibronectin-binding properties of the set of strains paralleled the individual fnbA promoter expression profiles of the strains in vitro (Fig. 4). For example, fibronectin binding was higher in the agr mutant than in the parental strain, but it was lower in the sarA mutant (binding of the parental strain to fibronectin reached ∼5% of the inoculum after 24 h of incubation). In addition, the fibronectin-binding capacity of the sarA agr double mutant exceeded that of the sarA mutant during the stationary growth phase (16 to 24 h).
FIG. 4.
Adherence of S. aureus Newman fnbA::gfpuvr in agr (□), sarA (▩), and sarA agr (▪) mutants to fibronectin versus the parental strain in vitro. The results are presented as the mean percentage (± standard deviation [error bar]) fibronectin binding compared to that of the parental strain.
Since the production of proteases is down-regulated in the agr mutant but up-regulated in the sarA mutant and since FnBPs can be degraded by such proteases, it was conceivable that alterations in fibronectin-binding phenotypes might be related to variations in their individual protease production profiles. To test this hypothesis, all study strains were preexposed to α2-macroglobulin (1.2 U/ml), and the fibronectin adherence properties were then determined. For all study strains grown in the presence of α2-macroglobulin, there was a ≥30% increase in fibronectin-binding activities throughout the growth cycle compared to strains grown in the absence of the inhibitor. These data suggest that extracellular protease production contributes modestly to the overall fibronectin-binding capacity of S. aureus strains. However, the fact that all strains were equally affected by protease inhibition and only to a modest extent indicates that the predominant mechanisms dictating phenotypic fibronectin binding probably occur at the level of fnbA transcription.
Experimental rabbit endocarditis model.
Recent studies have demonstrated that S. aureus virulence gene regulation profiles defined in vitro are often not precisely mirrored in vivo (6, 8, 28, 30). These data imply that host environmental cues play a major role in the activation of key S. aureus virulence genes. Thus, we sought to correlate fnbA promoter activation profiles defined in vitro with those delineated in experimental rabbit endocarditis. A well-characterized rabbit endocarditis model was used in these studies as previously described (28, 30).
(i) Microbiologic evaluation.
S. aureus densities achieved in vegetations were significantly higher than in kidney and spleen for all study constructs at 48 h postinfection (P < 0.05) (Table 2). In addition, all the mutants had lower target tissue bacterial densities than the parental strain, although these differences did not reach statistical significance. By comparison, S. aureus densities in vegetations were comparable in all study strains at 8 h postinfection (data not shown).
TABLE 2.
S. aureus fnbA::gfpuvr Newman parental strain and its isogenic agr and/or sarA mutant densities in target tissues of animals with endocarditis challenged with 107 CFU/animal
| Strain (no. of rabbits) | Log10 CFU/g of tissue (mean ± SD) |
||
|---|---|---|---|
| Vegetation | Kidney | Spleen | |
| Parental (10) | 8.62 ± 0.32 | 7.67 ± 0.42a | 6.62 ± 0.56a |
| agr mutant (12) | 8.30 ± 0.37 | 7.06 ± 0.80a | 6.11 ± 0.55a |
| sarA mutant (10) | 7.95 ± 0.88 | 6.72 ± 1.19a | 5.62 ± 0.75a |
| sarA agr double mutant (10) | 7.87 ± 0.52 | 6.66 ± 0.87a | 5.89 ± 0.46a |
Significantly different from the value obtained for vegetation (P < 0.05).
(ii) fnbA promoter expression in vivo.
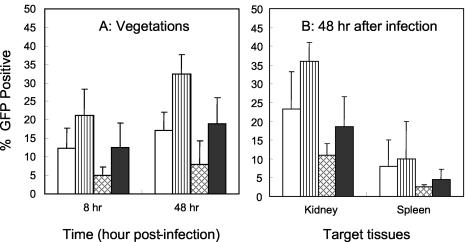
Flow cytometry and a protein A-based immunodetection system were used for detection of fnbA promoter expression in the endocarditis model as previously detailed (28, 30). Interestingly, fnbA promoter expression profiles defined in vivo in all target tissues for the various constructs paralleled in vitro fnbA expression profiles (Fig. 5). Compared to the parental cells, we observed increased fnbA expression in the agr mutant and decreased fnbA expression in the sarA mutant, with expression in the double mutant being greater than that of the sarA mutant in all target tissues (Fig. 5) (P < 0.05 for the parent versus the agr mutant in vegetations at 48 h). As in prior studies (28, 30), there were target tissue-specific differences in gene expression, with maximal fnbA expression seen in vegetations and kidneys, with reduced expression in the spleen. It is interesting that the extent of GFP expression for the various constructs paralleled the percentage of GFP expression in the target tissues studied (data not shown).
FIG. 5.
S. aureus fnbA promoter expression in vegetation, kidney, and spleen during the course of experimental endocarditis. (A) Percent GFP-positive S. aureus cells in vegetations at 8 and 48 h infection. (B) Percent GFP-positive S. aureus cells in kidney and spleen at 48 h infection. Symbols: □, parental strain; ▥, agr mutant; ▩, sarA mutant; ▪, sarA agr double mutant.
(iii) Fibronectin adherence ex vivo.
To determine the relative ability of the study strains (obtained directly from vegetations) to adhere to fibronectin, a modification of the above in vitro adherence assay was performed. Briefly, S. aureus cells (∼5 × 103 CFU based on anticipated vegetation densities) from each vegetation sample (at 24 h after infection [107 CFU/animal]) were directly assessed ex vivo for fibronectin-binding capacity by the in vitro assay detailed above. The in vivo fnbA expression profiles noted above roughly paralleled the fibronectin-binding capacities of the various constructs isolated directly from cardiac vegetations (data not shown). For example, compared to parental cells, the agr mutant cells adhered slightly more to fibronectin, but the sarA mutant adhered significantly less to fibronectin (P < 0.05). Remarkably, the sarA agr double mutant adhered to fibronectin to a higher extent than the sarA single mutant (P < 0.05 for sarA single mutant versus parental strain), mirroring the in vitro and in vivo (intravegetation) profiles of fnbA expression.
Several interesting observations emanated from this investigation. As expected, fnbA promoter expression in vitro was maximal during exponential growth of the parental strain and then plateaued. In addition, using single knockout mutants, the anticipated positive and negative regulatory effects of sarA and agr, respectively, on fnbA promoter expression in vitro were confirmed (3, 24, 29). Surprisingly, the extent of fnbA promoter expression in the sarA agr double mutant paralleled that of the sarA single knockout mutant during exponential and early postexponential growth phases but increased to near-parental levels in late stationary growth phase. Similarly, utilizing Northern blot analysis, Blevins et al. (3) noted fnbA transcription in sarA agr double mutants to be at or above the levels observed for the sarA single mutants in two clinical S. aureus strains. Collectively, these data suggest that during the stationary growth phase, environmental cues (e.g., low pH, nutrient limitation) or other regulatory loci that influence fnbA expression contribute to fnbA regulation in the absence of sarA and agr in vitro (e.g., the sae regulon seems to be required for fnbA activation in the Newman strain [27]). Further, delineation of the regulatory functions of the growing family of sarA homologs may also yield relevant information in this context (10, 12, 20, 25).
To correlate the above differences in fnbA expression profiles in vitro with a key functional phenotype, we compared the temporal fibronectin-binding capacities of this set of strains. It is noteworthy that the growth phase-related fibronectin-binding properties of this set of strains in our solid-phase assay paralleled their individual fnbA promoter expression profiles in vitro (results which are consistent with those reported by Blevins et al. [3], who utilized a liquid-phase fibronectin-binding assay). As with our sarA agr double mutant, in six of the seven strains Blevins et al. (3) studied (including the Newman strain), the fibronectin-binding capacity of the sarA agr double mutant exceeded that of the sarA single mutant by as much as twofold. Importantly, fnbA promoter expression profiles and fibronectin-binding phenotypes defined in vitro for the various constructs in the current study roughly paralleled the profiles and phenotypes defined in vivo in all target tissues in early and well-established infections.
It has been reported that sarA and agr mediate their effects on fnbA at the transcriptional level; this same regulatory pattern has been demonstrated in the Newman strain by Western blot analysis (29). These data indicate that the fibronectin-binding activities of S. aureus strains are at least partially due to direct regulation of fnbA transcription by sarA and agr. However, the overall S. aureus fibronectin-binding capacity is likely multifactorial, including extracellular protease production as well as production of a cadre of other FnBPs (e.g., FnbB, Ebh [host extracellular matrix binding protein homologue], and Emp [extracellular matrix protein binding protein]) (13, 16). Taken together, it is likely that the collective fibronectin-binding capacity of a given S. aureus strain reflects a composite of the activation and regulation of these various loci. Future studies will be required to evaluate the in vitro and in vivo expression paradigms of these other FnBPs, using animal models and gene reporter systems similar to those used in this study, to determine their relative contributions to net fibronectin binding in vitro and in vivo.
Acknowledgments
This work was supported in part by grants from the American Heart Association to Y.-Q.X. (0265054Y) and the National Institutes of Health to A.S.B. (AI-39108), A.L.C. (AI-47441), and M.R.Y. (AI-48031 and RR-13004).
We thank Yin Li for excellent technical assistance. We thank Battouli Said-Salim (Newark, N.J.) for agr genotyping of the Newman strain.
Editor: F. C. Fang
REFERENCES
- 1.Ahmed, S., S. Meghji, R. J. Williams, B. Henderson, J. H. Brock, and S. P. Nair. 2001. Staphylococcus aureus fibronectin binding proteins are essential for internalization by osteoblasts but do not account for differences in intracellular levels of bacteria. Infect. Immun. 69:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 3.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boonacker, E., and C. J. Van Noorden. 2001. Enzyme cytochemical techniques for metabolic mapping in living cells, with special reference to proteolysis. J. Histochem. Cytochem. 49:1473-1486. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, A. L., A. S. Bayer, J. Peter, and J. I. Ward. 1988. Surface proteins of Staphylococcus aureus. Rev. Infect. Dis. 10(Suppl. 2):S351-S355. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, A. L., K. J. Eberhardt, E. Chung, M. R. Yeaman, P. M. Sullam, M. Ramos, and A. S. Bayer. 1994. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 94:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., C. C. Nast, and A. S. Bayer. 1998. Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infect. Immun. 66:5988-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., and P. Ying. 1994. Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J. Bacteriol. 176:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, A. L., and G. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 1:1825-1842. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, S. R., L. G. Harris, R. G. Richards, and S. J. Foster. 2002. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 70:6680-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flock, J. I., G. Froman, K. Jonsson, B. Guss, C. Signas, B. Nilsson, G. Raucci, M. Hook, T. Wadstrom, and M. Lindberg. 1987. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 6:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, T. J., and D. McDevitt. 1994. Surface-associated proteins of Staphylococcus aureus: their possible roles in virulence. FEMS Microbiol. Lett. 118:199-205. [DOI] [PubMed] [Google Scholar]
- 16.Hussain, M., K. Becker, C. von Eiff, J. Schrenzel, G. Peters, and M. Herrmann. 2001. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 183:6778-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kupferwasser, L. I., M. R. Yeaman, C. C. Shapiro, C. C. Nast, P. M. Sullam, S. G. Filler, and A. S. Bayer. 1999. Acetylsalicylic acid reduces vegetation bacterial density, hematogenous bacterial dissemination, and frequency of embolic events in experimental Staphylococcus aureus endocarditis through antiplatelet and antibacterial effects. Circulation 99:2791-2797. [DOI] [PubMed] [Google Scholar]
- 20.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mongodin, E., O. Bajolet, J. Cutrona, N. Bonnet, F. Dupuit, E. Puchelle, and S. de Bentzmann. 2002. Fibronectin-binding proteins of Staphylococcus aureus are involved in adherence to human airway epithelium. Infect. Immun. 70:620-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 23.Que, Y. A., P. Francois, J. A. Haefliger, J. M. Entenza, P. Vaudaux, and P. Moreillon. 2001. Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect. Immun. 69:6296-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saravia-Otten, P., H. P. Muller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha, B., P. Francois, Y. A. Que, M. Hussain, C. Heilmann, P. Moreillon, D. Lew, K. H. Krause, G. Peters, and M. Herrmann. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinhuber, A., C. Goerke, M. G. Bayer, G. Doring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wamel, W., Y. Q. Xiong, A. S. Bayer, M. R. Yeaman, C. C. Nast, and A. L. Cheung. 2002. Regulation of Staphylococcus aureus type 5 capsular polysaccharides by agr and sarA in vitro and in an experimental endocarditis model. Microb. Pathog. 33:73-79. [DOI] [PubMed] [Google Scholar]
- 29.Wolz, C., P. Pohlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 30.Xiong, Y. Q., W. Van Wamel, C. C. Nast, M. R. Yeaman, A. L. Cheung, and A. S. Bayer. 2002. Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J. Infect. Dis. 186:668-677. [DOI] [PubMed] [Google Scholar]