Abstract
The Burkholderia cepacia epidemic strain marker (BCESM) is a useful epidemiological marker for virulent B. cenocepacia strains that infect patients with cystic fibrosis. However, there was no evidence that the original marker, identified by random amplified polymorphic DNA fingerprinting, contributed to pathogenicity. Here we demonstrate that the BCESM is part of a novel genomic island encoding genes linked to both virulence and metabolism. The BCESM was present on a 31.7-kb low-GC-content island that encoded 35 predicted coding sequences (CDSs): an N-acyl homoserine lactone (AHL) synthase gene (cciI) and corresponding transcriptional regulator (cciR), representing the first time cell signaling genes have been found on a genomic island; fatty acid biosynthesis genes; an IS66 family transposase; transcriptional regulator CDSs; amino acid metabolism genes; and a group of hypothetical genes. Mutagenesis of the AHL synthase, amidase (amiI), and porin (opcI) genes on the island was carried out. Testing of the isogenic mutants in a rat model of chronic lung infection demonstrated that the amidase played a role in persistence, while the AHL synthase and porin were both involved in virulence. The island, designated the B. cenocepacia island (cci), is the first genomic island to be defined in the B. cepacia complex and its discovery validates the original epidemiological correlation of the BCESM with virulent CF strains. The features of the cci, which overlap both pathogenicity and metabolism, expand the concept of bacterial pathogenicity islands and illustrate the diversity of accessory functions that can be acquired by lateral gene transfer in bacteria.
The Burkholderia cepacia complex (BCC) is a group of closely related species which inhabit diverse niches from the natural environment (30) to human opportunistic infection (21). Individuals with cystic fibrosis (CF) are particularly prone to BCC infection, with patient-to-patient spread (21) and high rates of morbidity and mortality having been documented (28). Originally known as the single species B. cepacia, these bacteria have been recently reclassified into distinct genetic species or genomovars and currently comprise eight new species and one genomovar (6, 21): B. cepacia (which will remain as the formal name for genomovar I, as it contains the species type strain); B. multivorans (genomovar II); B. cenocepacia, the new name for genomovar III (45); B. stabilis (genomovar IV); B. vietnamiensis (genomovar V); B. cepacia genomovar VI, which has not been formally named; B. ambifaria (genomovar VII); B. anthina (genomovar VIII [44]), and B. pyrrocinia (genomovar IX; 44). All these genomovars or species have been recovered from patients with CF infection (6, 21, 44, 45); however, the relative virulence and pathogenicity of each BCC species has not been fully determined.
The prevalence of infection in several CF populations has shown that B. cenocepacia is the most predominant BCC pathogen, causing on average 67% of cases of BCC infection (21). B. multivorans is the second most prevalent species, causing between 5 and 37% of cases dependent on the CF population examined, and the remaining genomovars constitute around 5% (21). In addition to its dominance as a CF pathogen, B. cenocepacia has also been associated with the majority of well-documented cases of patient-to-patient spread (11, 25, 45). Epidemiological data have demonstrated that it is highly virulent and capable of replacing infection with B. multivorans (28) and causing significant mortality among both CF patients (21) and those which have undergone lung transplantation (2). These data from cases of clinical infection suggest that B. cenocepacia may encode more-potent or larger numbers of virulence factors; yet, despite these advances in our understanding of the taxonomy and epidemiology of the complex, knowledge of specific virulence mechanisms is not extensive. Virulence factors characterized in B. cenocepacia strains and known to play a role in vivo include epithelial cell and mucin binding mediated by the cable pilus (35); a quorum sensing pathway mediated by classical LuxRI homologs, CepRI (17); iron acquisition and the production of siderophores (37); and cellular invasion and intracellular survival mechanisms (29), including a type III secretion system (42). Many of these virulence factors have homologs in other pathogens and none appear to be unique to B. cenocepacia.
The B. cepacia epidemic strain marker is a unique DNA region originally identified in strains of “B. cepacia” which had spread from patient-to-patient in CF (27). It has been widely applied in infection control as a clinical risk marker (21, 28, 40). The B. cepacia epidemic strain marker (BCESM) was identified during random amplified polymorphic DNA (RAPD) typing of “B. cepacia” isolates as a conserved amplification product in otherwise-distinct strain fingerprints (27). The 1.4-kb BCESM DNA encoded a single CDS for a putative negative transcriptional regulator designated esmR (27). Subsequent analysis demonstrated that the marker was exclusive to strains of B. cepacia genomovar III (22), now known as B. cenocepacia (45). Phylogenetic analysis of the recA gene of B. cenocepacia indicated that there were at least two distinct strain lineages within the species, III-A and III-B (22). Epidemiological studies have demonstrated that the BCESM is present in more than 77% of III-A CF strains but is absent in the majority of III-B CF strains (19, 40). The BCESM does not appear to be an absolute marker for the ability of B. cenocepacia strains to cause CF infection and is also not associated with all strains which infect multiple patients (18). However, in CF populations where BCESM positive strains are prevalent, they have been associated with well-documented transmissibility, virulence, and mortality (28, 40). In addition, the major B. cenocepacia clone which infects the majority of CF patients in Canada and the United Kingdom, known as the ET12 lineage (15) or cable pilus strain (41), is unique in being the only B. cenocepacia strain identified to encode both the cable pilus virulence factor and BCESM (21). However, unlike the cable pilus, no virulence function has been associated with the BCESM, despite its correlation to clinically problematic strains. Here we report further characterization of the B. cepacia epidemic strain marker locus using DNA sequence from the B. cenocepacia genome project and gene mutagenesis to determine the role of BCESM encoded genes during infection. Evidence that the BCESM is part of a novel B. cenocepacia genomic island is described.
MATERIALS AND METHODS
Bacterial isolates, culture, strain typing, and general reagents.
The bacterial strains and plasmids used are shown in Table 1. Genome sequence data were obtained for B. cenocepacia strain J2315 and all genetic manipulation was performed in the clonally identical B. cenocepacia strain, K56-2 (Table 1) (25). Other BCC strains were drawn from the Cardiff University collection or previously published studies and cultured as described (7, 22, 24, 25, 28, 40). For genetic selection, B. cenocepacia strain K56-2 was plated onto Tryptone Soya Agar containing antibiotics as needed: trimethoprim (200 μg/ml) or tetracycline (300 μg/ml). Escherichia coli cloning strains were grown and selected for with antibiotics as needed (36). Genomic DNA was extracted as previously described (23), and BCC taxonomic identification was performed by analysis of the recA gene (22). Isolates were typed using RAPD analysis as the primary screen (24) and pulsed-field gel electrophoresis (PFGE) followed by macrorestriction with SpeI as the secondary strain identification method (25, 46). All chemicals were obtained from Sigma-Aldrich Company Ltd., Poole, Dorset, United Kingdom, unless otherwise stated. Restriction enzymes were purchased from Promega Corporation Inc. (Southampton, United Kingdom) and molecular size markers from MBI Fermentas, Inc. (distributed by Helena Biosciences, Sunderland, United Kingdom).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Feature(s) | Reference or source |
|---|---|---|
| Strains | ||
| B. cenocepacia J2315 | CF strain; type strain for species and genome sequence strain | 25, 45 |
| B. cenocepacia K56-2 | CF strain and experimental strain used for construction of site-directed gene mutants and infection studies | 17, 37 |
| B. cenocepacia CEP54B | CF strain representative of Cleveland epidemic strain type 17 | 25 |
| B. cenocepacia CEP54A | Derivative of CF strain CEP54B which lacks BCESM region | This study |
| BCC strains | Selected from published strain panels and Cardiff University collection to be representative of all current genomovars | 7, 22, 25, 40 |
| E. coli JM109 | Standard cloning strain | 36 |
| K56-2cciI | AHL synthase knockout mutant disrupted at single BsmI site (position 235 in 654-bp CDS) | This study |
| K56-2amiI | Amidase knockout mutant disrupted at single ClaI site (position 233 in 1,401-bp CDS) | This study |
| K56-2opcI | Porin knockout mutant disrupted at single BamHI site (position 734 in 1,001-bp CDS) | This study |
| Plasmids | ||
| pGEM-T | PCR product cloning vector | Promega Corporation Inc. |
| pUC-TP | 1.1-kb trimethoprim resistance cassette | 37 |
| pJQ200SK | Gentamicin-resistant, sacB, Mob+, P15A replicon | 32 |
| pEGM105-Tc | pJQ2000SK with tetracycline resistance cassette inserted into BglI site within Gmr gene; suicide vector used for allelic exchange mutagenesis | Kindly provided by G. Mendrano |
| pCciI-Tp | pEGM105-Tc carrying AHL synthase gene interrupted at BsmI site with Tp cassette | This study |
| pAmiI-Tp | pEGM105-Tc carrying amidase gene interupted at ClaI site with Tp cassette | This study |
| pOpcI-Tp | pEGM105-Tc carrying porin gene interupted at BamHI with Tp cassette | This study |
Nucleotide sequence and bioinformatic analysis.
B. cenocepacia strain J2315 DNA sequence data were produced by the Pathogen Sequencing Group at the Sanger Institute, Hinxton, Cambridge, United Kingdom, and can be obtained from http://www.sanger.ac.uk/Projects/B_cenocepacia/. Bioinformatic analysis and annotation was performed using the software Artemis (34) and the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/index.html). Protein family information was obtained from the Protein Families databases of Alignments and HMMs (PFAM; http://www.sanger.ac.uk/Software/Pfam/index.shtml) and the Clusters of Orthologous Groups database (http://www.ncbi.nlm.nih.gov/COG/). Predicted coding sequences were designated numerically with the prefix BCAM (an abbreviation for B. cenocepacia annotation medium, because the sequence data were from the medium-sized chromosome). Restriction mapping, dot-plot analysis and preparation of figures were performed using DS Gene software (Accelerys Ltd., Cambridge, United Kingdom).
PCR and probe labeling.
PCR was performed using Taq DNA polymerase (Qiagen, Crawley, United Kingdom) under the conditions described by the manufacturer with 20 ng of template DNA and 10 pmol of each primer added to a standard 25-μl reaction mixture (22). Thermal cycling was performed on a Flexigene Thermal Cycler (Techne Ltd., Newcastle, United Kingdom) and conditions for all primer sets were as follows: an initial cycle of 94°C for 5 min followed by 30 cycles of 94°C for 30 s, annealing at the appropriate temperature (Table 2 or noted below) for 30 s, and extension at 72°C for 60 s, with a final 5-min extension at 72°C. Southern hybridization probes were prepared by PCR in the presence of the nonradioactive label digoxigenin (DIG)-UTP as described by the manufacturer (Roche Diagnostics Ltd., Lewes, United Kingdom). Products were analyzed and sized by agarose gel electrophoresis as described previously (36).
TABLE 2.
Gene probes used to map the cci by Southern hybridization
| Gene probe name | Putative gene product | PCR primer sequence (5′ to 3′)
|
PCR annealing temp (°C) | Product size (bp) | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| BCAM0236 | Arsenic resistance protein | GGT TCC TGC TGC TGT ACG | TCC TTG CGC TCG CTA TAC | 54 | 574 |
| cciI | AHL synthase | TTG ATC GCG GTC GTA CGA | ACC GCA ACG CTG GTA AAG | 50 | 319 |
| BCAM0248 | Putative IS transposase | GCC GGA AAT CGG AGA AAC | CGT GAT GCT ACC CAT CCA | 53 | 512 |
| BCESM | Original RAPD marker spanning the esmR gene | CCA CGG ACG TGA CTA ACA | CGT CCA TCC GAA CAC GAT | 62 | 1,418 |
| amiI | Putative amidase | GCT TTC TTG CGC GCT ACA | ATG CAC CAC GAT CTT TGC | 52 | 1,821 |
| opcI | Probable porin protein | GCC TTT GTC GGA CTG AGC | GTC TGT GCG TTT CGA GAG | 54 | 612 |
| BCAM0278 | Putative stress protein | GAT CGA CGT CAC CGA AAA | GAT CGC GAG CTT CTT CTG | 52 | 289 |
| BCAM0280 | Hypothetical protein | CGT CGA GGA TGA ACT GGA | CCT TAC CGG ACA ACG TGA | 52 | 317 |
BCC genomic array preparation.
Genomic DNAs from 241 genetically distinct BCC strains were immobilized onto replicate nylon membranes for use in Southern hybridization analysis. Species represented on the array were as follows: B. cepacia genomovar I (23 strains), B. multivorans (53 strains), B. cenocepacia (88 strains), B. stabilis (12 strains), B. vietnamiensis (21 strains), B. cepacia genomovar VI (2 strains), B. ambifaria (19 strains), B. anthina (10 strains), and B. pyrrocinia (13 strains). Genomic DNA from each strain (at a concentration of approximately 150 ng/μl) was aliquoted into 384 well-plates (ABgene Ltd., Epsom, United Kingdom) and spotted onto replicate positively charged nylon membranes (Roche Diagnostics Ltd.), in a two-by-two pattern for each sample, using a Genomic Solutions Flexys robotic microarrayer. Membranes were placed in denaturing solution (0.5 M NaOH) for 5 min, neutralized in 1.5 M NaCl-1.5 M Tris-HCl (pH 8.0) for 5 min, and then washed for 2 min in 2× sodium chloride-sodium citrate buffer (SSC) (36). DNA was fixed to the membranes by exposure to UV light (360 nm) for 3 min.
Southern hybridization analysis.
DNA separated by standard agarose gel electrophoresis or PFGE was transferred to positively charged nylon membranes using a conventional Southern transfer method (36); membranes were then denatured, neutralized and washed as described above for the genomic DNA arrays. All membranes were prehybridized for 2 h at 50°C in EasyHyb buffer (Roche Diagnostics Ltd.) prior to addition of 5 to 10 ng of heat-denatured DIG-labeled PCR probe (Table 2); the probes were then allowed to hybridize overnight at the same temperature. Washing was performed at high stringency with a final 10-min wash in 0.1× SSC containing 0.1% sodium dodecyl sulfate at a temperature of 68°C (36). Chemiluminescent detection of DIG probes was performed using the substrate CSPD in accordance with the manufacturer's protocols (Roche Diagnostics Ltd.) and membranes exposed to autoradiography film (Sigma-Aldrich Ltd.) at 37°C for up to 18 h until clear signals were observable. Control hybridizations to the genomic DNA arrays were performed using a 16S rRNA gene probe (22) amplified from pooled DNA representative of the BCC.
Site-directed gene mutagenesis.
PCR probes designed from the B. cenocepacia J2315 sequence data were used to clone each target for subsequent gene mutagenesis. The AHL synthase gene, cciI, was amplified with a primer pair encoding terminal SmaI sites, 5′-CCCGGGACGCGCTTGATA-3′ and 5′-CCCGGGAAGCGGTGTTTG-3′, which produced the predicted 748-bp product at an annealing temperature of 56°C. The porin gene, opcI, was also amplified with SmaI-encoding PCR primers, 5′-CCCGGGCGATCGATTCAA-3′ and 5′-CCGGGGAGCTTCCAGTCC-3′, which produced the expected product of 988 bp at an annealing temperature of 56°C. The amidase gene was amplified using the primers shown in Table 2. All PCR products were then cloned into pGEM-T as described by the manufacturer (Promega Corporation Inc.). Single restriction sites were identified in the cciI, amiI, and opcI CDSs (BsmI, ClaI, and BamHI, respectively [Table 1]). PCR primers encoding each of these restriction sites as overhanging DNA were designed to amplify the trimethoprim resistance cassette of pUC-TP (Table 1) and the resulting 1.1-kb trimethoprim cassette PCR products cloned into pGEM-T. The appropriate trimethoprim cassette constructs were digested with BsmI, ClaI, and BamHI, respectively, and cloned into the corresponding restriction sites of the cciI, amiI, and opcI pGEM-T constructs. The resulting clones were analyzed by PCR with the appropriate primers shown in Table 2 to identify each correctly interrupted gene construct. Mobilizable suicide constructs were then constructed in pEGM105-Tc, a derivative of plasmid pJQ2000SK (32) encoding tetracycline resistance that was kindly provided by G. Mendrano, Texas A&M University, College Station (Table 1). The cciI and opcI mutated constructs were digested with SmaI and cloned into SmaI-linearized pEGM105-Tc, generating suicide constructs pCciI-Tp and pOpcI-Tp (Table 1). The amiI pGEM-T construct insert was amplified by PCR and the product blunted by incubation with Klenow polymerase as described by the manufacturer (Promega Corporation Inc.) and also cloned into SmaI-digested pEGM105-Tc to generate plasmid pAmiI-Tp (Table 1). Each suicide mutagenesis construct was then introduced into B. cenocepacia strain K56-2 by conjugal transfer in a tri-parental mating with the helper plasmid pRK2013 as previously described (17). Mutants which had undergone allelic exchange were selected by plating on trimethoprim containing media and evaluated for correct double-crossover homologous recombination by PCR with the appropriate primer set (Table 2), PFGE analysis, and conventional Southern hybridization restriction fragment length polymorphism (RFLP) analysis (36).
Agar bead infection model.
Groups of 15 male Sprague-Dawley rats (150 to 170 g; Charles River Canada, Inc.) were tracheostomized under anesthesia and inoculated with approximately 105 CFU of K56-2 or the mutant strains (Table 1) embedded in agar beads as previously described (5, 37). On days 1 and 14 postinfection (p.i.), the lungs from five animals per group were removed aseptically and homogenized (Polytron Homogenizer; Brinkmann Instruments, Westbury, N.Y.) in 3 ml of phosphate-buffered saline, serially diluted and plated on tryptic soy agar and B. cepacia selective agar (13), and incubated at 37°C to determine the number of bacteria present in the lung. Lung homogenates from the mutant strains were also plated on medium with trimethoprim (100 μg/ml) to confirm that the mutations were stable throughout the course of the infections. Lungs from five additional animals per group were examined for quantitative histopathological changes as previously described (39) with the following modifications. The lung sections were scanned using an Epson 1650 scanner. Areas of inflammation, characterized by cellular infiltration consisting of predominantly polymorphonuclear leukocytes and inflammatory exuviate, were identified and digitized with Scion Image software and reported as the percentage of the total area of the lung section that was covered by inflammatory exudate. To determine the ability of the amidase mutant to persist in the lungs, groups of 15 rats were each infected with approximately 5 × 105 CFU of K56-2 or K56-2amiI, and on days 1, 14, and 28 p.i. lungs were removed from groups of animals and quantitative bacteriology analysis was performed as described above.
RESULTS
Stability of the BCESM genomic region.
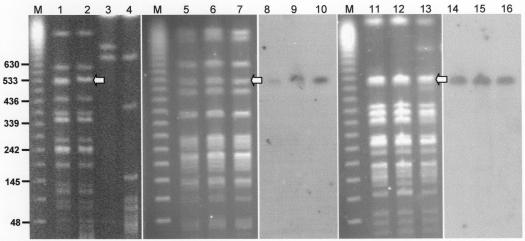
Epidemiological studies had demonstrated that BCESM was not possessed by all B. cenocepacia strains (19, 22), suggesting it may be part of an unstable genomic region. In vitro characterization of a B. cenocepacia III-B CF strain, CEP54 (RAPD type 17, the Cleveland epidemic lineage [25]) indicated that instability of the marker may occur. One hundred colonies from the primary culture of strain CEP54 were analyzed by BCESM colony PCR, and 28 were found to be negative for the marker. Single positive and negative clones were purified and subcultured. PCR testing demonstrated that the BCESM remained stable in the positive derivative, CEP54B (the suffix B indicates that BCESM is present), during subsequent culture passage. PFGE analysis of CEP54B and the negative clone CEP54A (the suffix A indicates that BCESM absent) demonstrated that a complex genomic rearrangement had occurred in strain CEP54A resulting in loss of BCESM encoding DNA (Fig. 1, lanes 1 to 4).
FIG. 1.
PFGE mapping of the BCESM in CF strains from patients with chronic infection. PFGE macrorestriction and Southern hybridization results of B. cenocepacia CF strains are shown. Lanes are as follows: M, molecular size markers with relevant fragment sizes (in kilobases) indicated on the left of the figure; 1 and 2, CEP54A and CEP54B, respectively, digested with SpeI; 3 and 4, CEP54A and CEP54B, respectively, digested with SwaI; 5, 6, and 7, strain type 2 sequential isolates recovered from patient A, after 1, 2, and 3 years of chronic infection (SpeI digests); 8, 9, and 10, corresponding Southern blot of lanes 5 to 7 hybridized to the BCESM; 11, 12, and 13, strain type 4 isolates recovered from patient B, after 1, 3, and 5 years of infection (SpeI digests); 14, 15, and 16, Southern blot of lanes 11 to 13 hybridized to the BCESM. The arrow indicates the conserved 503-kb SpeI fragment to which the BCESM hybridized in all strains.
To determine if the BCESM was stable during chronic CF infection we analyzed sequential isolates from five patients with B. cenocepacia infection of strain type 1, 2 (2 patients), 4, and 6, which all encode the BCESM (27, 28). The PFGE fingerprints of the strains were stable (for up to 11 years in the case of strain type 6; data not shown), with BCESM hybridizing genomic fragment remaining intact throughout chronic lung infection. PFGE and hybridization data for strain types 2 and 4, over a 3- and 5-year period of infection, respectively, are shown in Fig. 1 (lanes 5 to 16). Strain type 2 is the same type as the genome project strain B. cenocepacia J2315 (25), and the 503-kb SpeI fragment to which the BCESM hybridized (Fig. 1B) was the same as that predicted from the genome sequence data. The same isogenic 503-kb SpeI fragment hybridized to the BCESM in strain types 1 and 4 (Fig. 1), and our experimental strain K56-2 (also strain type 2 [25]). These observations from clinical CF infection demonstrate that the BCESM region is stable during infection and conserved on a macrorestriction scale, and they suggest that the loss of the marker observed in B. cenocepacia III-B strain CEP54A was rare.
Bioinformatic analysis of BCESM genomic locus.
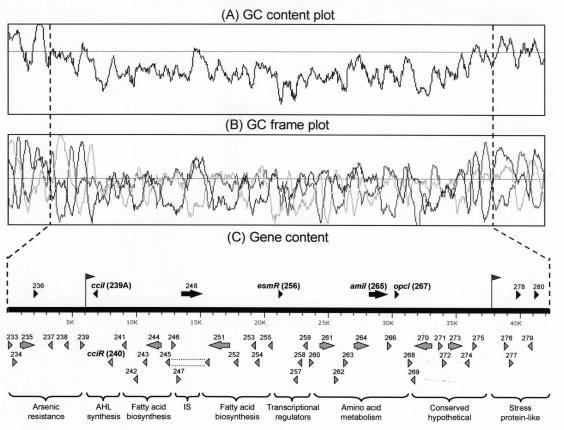
Two approaches were used to map the gene content of the DNA surrounding the original BCESM: nucleotide sequence data from the B. cenocepacia genome project and Southern hybridization using PCR probes designed from the sequence data (see below). Hybridization of BCESM probe to PFGE-separated replicons of B. cenocepacia strains demonstrated that the marker was encoded on the second-largest chromosome of the multireplicon genome (data not shown); this observation was corroborated by the B. cenocepacia genome sequence data. Bioinformatic analysis of the BCESM encoding region of B. cenocepacia J2315 was immediately striking, revealing that the original BCESM (27) was part of a large low-GC-content genomic island (Fig. 2A). The island was approximately 34 kb with a GC content of 61.7%, compared with the second replicon mean of 67.3% (Fig. 2A) and the overall B. cenocepacia genome mean of 66.9%. The reading frame position-specific GC usage was also very different from the rest of the genome (Fig. 2B) indicating that the DNA may have been recently acquired by B. cenocepacia from a different host organism (16). Because of this novel finding and the historical association of the BCESM with B. cenocepacia, the region was designated as the B. cenocepacia island (cci).
FIG. 2.
Bioinformatic analysis of the B. cenocepacia island. (A and B) GC content plots and GC frame plots are shown, respectively; GC percentage was calculated over a window size of 500 bases, and the mean GC content of the second chromosome, 67.3%, is indicated by the line across each panel. (C) The gene content of the DNA in the BCESM region is shown, with each predicted CDS designated by its BCAM number or designated name. Genes mapped by Southern hybridization are shown as black arrows above the sequence bar (size in kilobases indicated on bar); other CDSs are indicated below the sequence bar as grey arrows. The predicted functions of each gene cluster are shown below the brackets. The position of the 13-bp direct imperfect repeats which flank the cci are shown by the arrows linked to the sequence bar.
Preliminary annotation of 43 kb of sequence spanning the cci revealed the presence of 47 predicted CDSs (formal annotation will be completed and published by the Sanger Institute). Thirty-five CDSs were within the low-GC-content portion of the cci (BCAM0239A to BCAM0275), and 12 CDSs were outside this region (BCAM0233 to BCAM0239A and BCAM0276 to BCAM0280 [Fig. 2C]). Dot plot analysis of the genomic region indicated the presence of 13-bp imperfect direct repeats which flanked the island (Fig. 2C). The downstream repeat, 5′-TTTCTGCTCAGGC-3′, was within CDS BCAM0239, and the upstream repeat, 5′-TTTCTGCCCAGGC-3′, was within the intergenic region between BCAM0275 and BCAM0276 (the single mismatched base is underlined). The size of the cci between these repeats was 31.7 kb, smaller than initially predicted from the GC content plots (Fig. 2A and B) and the derived island possessed an even lower GC content of 60.8%. Neither repeat sequence occurred elsewhere in the B. cenocepacia genome and a BLAST search of the DNA databases for short nearly exact sequence matches demonstrated only one perfect prokaryotic match to the downstream repeat which was within the flagellin gene of E. coli serotype O128:H2 (4). However, both repeat sequences occurred widely within eukaryotic organisms.
Four functional gene clusters and one hypothetical gene set were encoded by the cci. Genes with homologies to known genes included: (i) autoinducer synthesis loci, with an N-acyl homoserine lactone (AHL) synthase gene and corresponding transcriptional regulator, which were designated as cciI (BCAM0239A) and cciR (BCAM0240), respectively (Fig. 2C), to distinguish them from the characterized CepIR autoinducer loci of B. cenocepacia (17); (ii) a cluster of genes with homologies to fatty acid biosynthesis genes (BCAM0241 to BCAM0246 and BCAM0251 BCAM0255) that contained a copy of an IS66 family transposon (BCAM0247 and BCAM0248; originally described in the Agrobacterium tumefaciens Ti plasmid) inserted into BCAM0245, a putative decarboxylase; (iii) a set of putative transcriptional regulator loci including the previously described esmR gene (BCAM0256 to BCAM0259); and (iv) a cluster of genes associated with amino acid transport and metabolism (BCAM0260 to BCAM0268 [Fig. 2C]). In addition to these gene clusters with putative functions, the cci also encoded a cluster of seven CDSs with homology to other conserved or hypothetical genes with no currently known functions (BCAM0269 to BCAM0275 [Fig. 2C]). The cci was flanked by a downstream cluster of arsenic and antibiotic resistance-associated genes (BCAM0233 to BCAM0239) and upstream conserved genes with homologies to known stress proteins (BCAM0276 to BCAM0280 [Fig. 2C]).
Southern hybridization mapping of the cci.
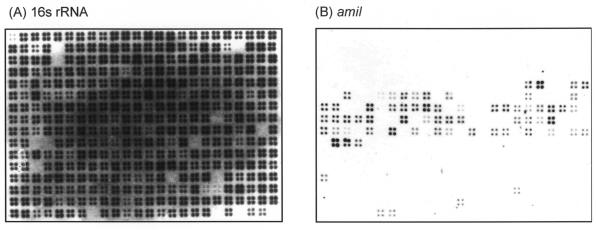
Hybridization of selected cci genes against a collection of 241 genetically distinct BCC strains representative of each genomovar/species was used to determine the extent and prevalence of the cci. PCR hybridization probes were designed from the B. cenocepacia J2315 genome sequence and the following genes were targeted for mapping (Table 2). BCAM0236, BCAM0278, and BCAM0279 were selected because they were outside the low GC island (Fig. 2C) and in the cases of BCAM0236 and BCAM0278, demonstrated good homology to known genes (Table 2). Within the cci, the BCESM (27), the AHL synthase, cciI, and the IS66 family transposase, BCAM0248, were selected (Table 2). In addition, a putative amidase (related to pfam01425), designated amiI, and a predicted porin, designated opcI, related to the OmpC family of gram-negative porins (pfam 00267 [Table 2]) were also selected. PCR probes labeled with digoxigenin were hybridized to the genomic DNA arrays as shown in Fig. 3 and the prevalence of each selected gene is shown in Table 3.
FIG. 3.
Southern hybridization to BCC genomic DNA arrays. (A) Control hybridization to pooled 16S rRNA gene probes. (B) Specific hybridization of the amiI gene probe to strains of B. cenocepacia encoding the BCESM DNA.
TABLE 3.
Prevalence of cci genes mapped by Southern hybridization with the BCC genomic DNA arrays
| Gene probe | No. of strains that hybridized to the indicated BCC genomovar
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| I (n = 23) | B. multivorans (n = 53) | B. cenocepacia (n = 88) | B. stabilis (n = 12) | B. vietnamiensis (n = 21) | VI (n = 2) | B. ambifaria (n = 19) | B. anthina (n = 10) | B. pyrrocinia (n = 13) | |
| Arsenic resistance gene (BCAM0236) | 4 | 13 | 46 | 3 | 3 | 0 | 8 | 0 | 0 |
| AHL synthase (cciI) | 0 | 0 | 48a | 0 | 0 | 0 | 0 | 0 | 0 |
| IS transposase (BCAM0248) | 1 | 0 | 6 | 0 | 0 | 0 | 0 | 2 | 3 |
| BCESM (esmR; BCAM0256) | 0 | 0 | 48a | 0 | 0 | 0 | 0 | 0 | 0 |
| Amidase (amiI) | 0 | 0 | 48a | 0 | 0 | 0 | 0 | 0 | 0 |
| Porin (opcI) | 0 | 0 | 48a | 0 | 0 | 0 | 0 | 0 | 0 |
| Stress protein (BCAM0278) | 0 | 0 | 20 | 1 | 0 | 0 | 7 | 0 | 0 |
| Hypothetical gene (BCAM0280) | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 |
AHL synthase, amidase, and porin gene probes hybridized to exactly the sames strains as the BCESM probe across the array.
The BCESM probe hybridized exclusively to B. cenocepacia strains, corroborating previous findings (25, 27). The BCESM was present in 55% (48 strains) of the B. cenocepacia strains represented on the genomic DNA arrays (Table 3). The cciI, amiI, and opcI genes which were within the cci, all demonstrated the same prevalence as the original BCESM, hybridizing exclusively to B. cenocepacia strains (Table 3). In addition, all the latter probes hybridized to exactly the same strains, suggesting that their distribution was linked. Hybridization of these probes to PFGE-separated DNA also demonstrated that they colocalized to the same genomic region as the BCESM in B. cenocepacia (data not shown). The only gene within the island which did not demonstrate an identical distribution to the BCESM was the IS66 family transposase. It was present in a few genomovar I, B. anthina, and B. pyrrocinia strains, as well as within the B. cenocepacia genome strain J2315 and clonal ET12 strains (including K56-2 [25]). The downstream arsenic resistance gene probe, BCAM0236 (Table 2), hybridized to five other BCC species in addition to B. cenocepacia (Table 3), with only B. cepacia genomovar VI, B. anthina, and B. pyrrocinia not possessing homologous genes. Probes directed to the upstream flank of the cci produced a different distribution profile. The stress protein-like probe, BCAM0278, hybridized only to B. cenocepacia subgroup III-A strains (18 of 31 III-A strains on the array). The hypothetical gene probe, BCAM0280, detected homologs in B. cenocepacia, B. stabilis, and B. ambifaria (Table 3). Overall, the hybridization mapping indicated that gene probes outside the low GC cci were not closely linked in their distribution and that the cciI, amiI, and opcI which were always associated with an intact cci, were conserved across all B. cenocepacia strains possessing the original BCESM.
Mutagenesis of cci genes.
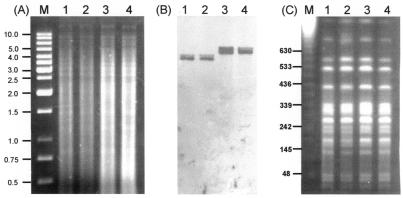
To establish a functional role for the cci in infection, directed mutagenesis of the AHL synthase, amidase, and porin genes was carried out. These genes were selected because they possessed predicted nonregulatory functions, were part of a putative functional cluster of genes (Fig. 2) and were known from the hybridization data to be carried on the cci in all BCESM positive strains. In addition, the CepIR quorum-sensing system of B. cenocepacia had already been shown to play a critical role in virulence factor production (17), and hence the role of the additional CciIR was deserving of evaluation. Knockout mutants were created by homologous recombination with gene constructs interrupted by a trimethoprim cassette inserted at single restriction sites within each CDS as shown in Table 1. Mutants arising from double-crossover homologous recombination were screened by PCR, Southern hybridization and PFGE to confirm they possessed the correct genotype. Southern hybridization analysis of all three mutants probed with the amiI gene (Table 2) is shown in Fig. 4A and B. The amidase mutant, K56-2amiI, demonstrated the correct 1.1-kb increase in RFLP profile expected from the directed mutagenesis; the porin mutant also showed the same increase as it was encoded on the same restriction fragment as the amidase gene. Southern hybridization with the cciI and opcI probes (Table 2) also indicated that each of the latter mutants possessed the correct genotype (data not shown). Finally to confirm that no additional genomic rearrangements had occurred during the mutagenesis procedure, PFGE analysis was performed and demonstrated that all three mutants were isogenic in macrorestriction fingerprint to their parental strain, K56-2 (Fig. 4C). In vitro, the cciI, amiI, and opcI mutants did not demonstrate any altered growth patterns or alterations in observable phenotype when basic growth kinetics were examined. In addition, the genotype of each mutant remained stable when passaged for 10 generations without selection for trimethoprim.
FIG. 4.
Confirmation of the B. cenocepacia K56-2cciI, K56-2amiI, and K56-2opcI mutants. (A and B) Conventional SalI RFLP mapping of the amidase gene mutation. (A) Lanes are as follows: M, 1-kb molecular size marker; 1, K56-2; 2, K56-2cciI; 3, K56-2amiI; and 4, K56-2opcI. (B) Corresponding Southern hybridization probed with the amidase gene probe (Table 2). (C) PFGE SpeI analysis of the following strains: lane 1, K56-2 parent; lane 2, K56-2cciI; lane 3, K56-2amiI; and lane 4, K56-2opcI. PFGE molecular size markers were run in lane M, and the sizes of relevant bands are indicated.
Infection studies on cci mutants.
To establish if cci-encoded genes played a role during infection, the ability of the cciI, amiI, and opcI mutants to establish chronic respiratory infection was compared to the parent strain using the rat agar bead infection model (5, 37). Lungs were removed on day 1 p.i. to determine the ability of the strains to establish an infection. Similar numbers of bacteria were recovered from the lung on day one indicating that the mutants were able to establish an infection (Table 4). Quantitative bacteriology was also performed on day 14 p.i. Approximately 1 log fewer bacteria were recovered from lungs infected with the amidase mutant compared to lungs infected with K56-2 (Table 4). To determine if functional amidase was required for persistence in the lung, a second experiment was performed and quantitative bacteriology analysis was performed on days 1, 14, and 28 p.i. On day 14 p.i. approximately 1-log fewer bacteria were again recovered from the lungs of animals infected with the amidase mutant compared to K56-2 (Table 4; P < 0.05, t test for unpaired observations). A 1-log difference in the number of bacteria recovered was also observed on day 28 p.i. although the difference in this case was not quite statistically significant (data not shown). These data suggest that amidase does play a role in the ability of K56-2 to persist in the lung but that it is not an essential requirement for persistence.
TABLE 4.
Ability of B. cenocepacia cci mutants to establish persistent respiratory infections
| Strain Expt. 1 | Mean log CFU/ml/per lung ± SDa at day:
|
|
|---|---|---|
| 1 p.i. | 14 p.i. | |
| Expt 1 | ||
| K56-2 | 6.1 ± 1.4 | 5.3 ± 0.9b |
| K56-2cciI | 6.0 ± 1.2 | 4.7 ± 1.2 |
| K56-2amiI | 6.8 ± 0.3 | 4.2 ± 0.8 |
| K56-2opcI | 5.6 ± 0.6 | 5.4 ± 0.4 |
| Expt 2 | ||
| K56-2 | 6.7 ± 0.2c | 5.1 ± 0.5 |
| K56-2amiI | 5.6 ± 1.3c | 4.2 ± 0.4d |
Five animals per group unless indicated otherwise.
Six animals per group.
Four animals per group.
Significantly different from result for K56-2 (P < 0.05; t test for unpaired observations).
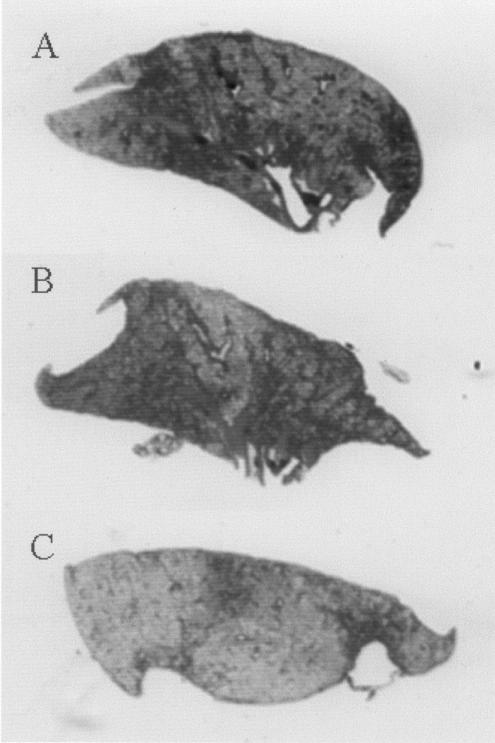
The effect of the mutations on virulence of K56-2 was also determined by performing quantitative histopathology analysis on lungs on day 14 p.i. Mutations in the AHL synthase and porin genes resulted in significantly less histopathological changes in the lungs (Table 5). Scans of representative lung sections from each group of animals are shown in Fig. 5. The inflammation in animals infected with these strains was approximately 50% less than that observed in animals infected with K56-2, despite similar numbers of bacteria being present in the lungs (Table 4). These data indicate that the AHL synthase and porin genes contribute significantly to the virulence of B. cenocepacia. The amidase mutant did not demonstrate any alteration in lung histopathology when compared to the parental strain K56-2. The amiI gene and opcI are very close to one another on the island, with only CDS BCAM0266 intervening (Fig. 2). The contrasting in vivo growth (Table 4) and inflammatory (Table 5) phenotypes of the knockout mutants of each gene indicate that no polar effects were associated with directed mutagenesis of the amidase and porin loci.
TABLE 5.
Comparison of lung histopathology in animals infected with B. cenocepacia mutant and parent strains
| Strain | % Inflammationa (mean ± SD) | P |
|---|---|---|
| K56-2 | 28.2 ± 0.2 | |
| K56-2cciI | 14.0 ± 5.7 | <0.05 |
| K56-2amiI | 23.8 ± 8.3 | NSb |
| K56-2opcI | 13.6 ± 4.8 | <0.05 |
Five animals per group.
NS, not significant.
FIG. 5.
Scans of representative lung sections from animals infected with K56-2 (A), K56-2cciI (B), or K56-2opcI (C). Regions of darker staining are indicative of areas of inflammation. The inflammation calculated for the sections in panels A to C are 28, 13, and 11%, respectively.
DISCUSSION
Our study has demonstrated that the BCESM is part of a novel genomic island encoding genes which play a significant role in the virulence of B. cenocepacia. The BCESM was originally identified by RAPD analysis (27), a strain typing method which we have found particularly useful for typing and molecular epidemiological analysis of several bacterial species (23, 24, 40). In contrast, other investigators have considered RAPD strain fingerprinting methods to lack reproducibility and to be of limited epidemiological use (43). The discovery of the BCESM by RAPD analysis (27), its use as a marker of clinical risk during B. cenocepacia CF infection (28, 40), and the fact that we have now shown that it forms part of a putative pathogenicity island, demonstrates the broad utility of PCR-mediated strain typing methods such as RAPD analysis. These PCR fingerprinting methods may be used for both typing and as differential genomic display methods able to identify useful strain specific markers as shown by this work.
In comparison to other known pathogenicity islands (12), the B. cenocepacia cci possesses most of the features of such horizontally acquired genomic regions. It has an atypical GC content and a large size, possesses flanking direct repeats, has associated insertion (IS) elements, is epidemiologically linked to virulent strains, and most importantly encodes multiple genes—at least three of which we have demonstrated to play a role during chronic lung infection. However, its functional gene content is very unusual in comparison to classical gram-negative pathogenicity islands which often encode type III secretion systems, adhesins, and toxins (12). Of the genes encoded by the cci, only those associated with quorum sensing (cciI and cciR [Fig. 2]) constitute members of genetic systems previously implicated in bacterial virulence (17, 38). As far as we are aware, although it is known that quorum-sensing genes may regulate virulence factors genes encoded on pathogenicity islands (12), the cci is the first pathogenicity island to actually encode classical LuxRI homologs. IS elements and transcriptional regulators are present on many pathogenicity islands (12) and are also present on the cci, although their role in virulence remains undetermined. The IS66 family IS sequence (BCAM0248 [Fig. 2]) in the cci may also be a recent IS into the element, as it was only found in B. cenocepacia strains of the same epidemic lineage, ET12 (15, 21, 41), as the genome strain J2315 (Table 3). The other cci gene clusters associated with fatty acid metabolism, amino acid transport, and metabolism or of a conserved hypothetical nature (Fig. 2) are unusual and not found on other pathogenicity islands. Overall, the cci may be considered to constitute a genomic island which has features of both pathogenicity and metabolic islands (12).
It was already known that the BCESM DNA was encoded by transmissible strains (27) associated with high mortality in patients with CF (28); these data clearly link the presence of the B. cenocepacia cci to virulent B. cenocepacia strains. However, the cci was also absent in significant numbers of B. cenocepacia strains which were all associated with CF infection. Detailed clinical outcome data from infected CF patients need to be gathered to determine if cci encoding strains possess significantly greater virulence than strains which lack the island. Our animal infection model results with cci isogenic mutants suggests that the cci does function to enhance the virulence of B. cenocepacia. The high-prevalence data from this and previous studies (19, 22, 27, 40) suggest that B. cenocepacia strains of recA phylogenetic subgroup III-A may have been the original strains to acquire the cci. The cci appears very stable and intact in B. cenocepacia III-A strains, remaining present throughout chronic lung infection and with only one case of in vitro instability detected in B. cenocepacia III-B strain CEP54A (Fig. 1). A recent controversy in BCC research is the distinction between environmental and clinical strains, and the use of these bacteria as agents of biological control or bioremediation (30). The cci does not provide grounds for distinction between clinical and environmental strains as it was present in four B. cenocepacia environmental strains, including strain M36, a U.S.-registered biocontrol strain which has been withdrawn from commercial use by the manufacturer (26). The virulence role of the cci described herein may in future enable selection of biotechnological strains lacking this pathogenic marker.
Our preliminary mutagenesis data on the cciI, amiI, and opcI demonstrate that these genes have functional roles during infection assisting in the definition of the cci as a pathogenicity island. The role cciI in virulence and inflammation corroborates previous data on quorum sensing in B. cenocepacia (17, 38). Mutation of the B. cenocepacia CepIR system, the first LuxIR homologs to be identified in this species, did not completely prevent quorum sensing activity (17), suggesting that the bacterium may possess additional signaling systems. Bioinformatic analysis of the B. cenocepacia J2315 genome indicates that, other than the CepIR and CciIR genes, no further LuxIR homologs are present. The difference in virulence between the cciI mutant and K56-2 was similar to that reported for a K56-2cepI mutant in the agar bead model (38). These data suggest that the cciI is part of a second cell signaling system in B. cenocepacia and that both quorum-sensing systems are involved in regulation of virulence factors in this species. Further studies are in progress to determine the role of cciI in cell signaling in B. cenocepacia. Complementation of the cciI mutant has been performed and restores quorum-sensing activity to wild-type levels in vitro; in addition, construction of a double cepI cciI mutant results in a lack of detectable AHL production in B. cenocepacia (R. J. Malott, A. Baldwin, E. Mahenthiralingam, and P. A. Sokol, Abstr. Am. Soc. Microbiol. N.W. Branch Meet. 2003, abstr. 77, p. 97, 2003). However, it is not known if the CciIR system works in isolation or in concert with other genetic systems on the island. The adjacent fatty acid biosynthesis genes are oriented in the same direction as the cciI and cciR genes, and could potentially be transcribed together with them. This raises the possibility that these genes may be involved in modifying in some way the lipid moieties on the AHL molecule(s) generated by this system. It is also likely that the IS element present in all ET12 strains (Table 3) would have an effect on this putative modification.
It is not unusual for bacteria to have multiple cell signaling systems. In addition to the CepIR system, B. vietnamiensis has a second set of LuxIR homologs designated BviIR (8, 20). The role of the bviIR genes in regulation of virulence factors has not yet been determined. In some species multiple sets of quorum-sensing genes have been identified, and for at least some functions, hierarchical regulation exists where one system has control over the other. The most well characterized example of this is in Pseudomonas aeruginosa, where the LasRI system has dominant regulatory control over the RhlRI system (10, 31). Multiple luxIR homolog-based quorum-sensing systems have also been described in Yersinia pseudotuberculosis (3) and Rhizobium leguminosarum (47), and one system has been shown to have a regulatory role over the other. Studies are under way to determine if there is any regulatory relationship between the cciIR and the cepIR genes in B. cenocepacia or if these systems operate independently in regulation of potential virulence factors.
The amidase mutant was the only cci mutant to demonstrate a reduced rate of persistence during infection (Table 4). The AmiI protein has some homology with the GatA subunit of the glutamyl-tRNA amidase (9). However, genes encoding the other two subunits of the heterotrimeric GatABC complex are not present in this locus, and B. cenocepacia has a full set of gatCAB genes on the large chromosome. It is therefore likely that AmiI is involved in modification of the substrate of the putative amino acid transport system described below. Mutation of amiI is not lethal and does not alter basic in vitro growth kinetics, indicating that this system is not essential. Downstream of the amiI gene are a cluster of genes encoding putative amino acid transport proteins with significant homology to the highly conserved ABC-ATPase transporters (14) (BCAM0260 to BCAM0265 [Fig. 2]). The presence of these genes corroborates our hypothesis that amiI is involved in amino acid metabolism. If AmiI is functionally a GatA homolog, then it and the transporter-like proteins downstream will require energy in the form of ATP (14, 33), a possibility that adds further intrigue to our observation that amiI is required for in vivo survival, where one would consider pathogens to adopt energy efficient pathways unless they perform an important function. The opcI gene, which is closely related to the OmpC family of outer membrane bacterial porins (COG3203 [Fig. 2]), is upstream of amiI and the amino acid transport proteins, suggesting that it may function as a pore for the small molecules processed by these genes. Mutagenesis of the OpcI porin demonstrated that it plays a role in mediating inflammation at the site of infection (Table 5) suggesting that protein may, like other OmpC homologs, modulate immunological expression in order to evade host defenses (1). Complementation of the amidase and porin mutants has been achieved (E. Mahenthiralingam, unpublished data); however, as with the knockout mutants no observable phenotypic changes were noted after complementation. Further characterization of the role the amidase and porin genes will require the development of simple assays to detect their expression.
Although the exact functional role of the majority of the genes within the cci during infection remains to be determined, the preliminary analysis presented here indicates that the element is a highly novel and functionally unique genomic island. The island shares many of the attributes of a classical pathogenicity island encoding virulence associated genes; however, several of the gene clusters within the island are also linked to metabolism. Its association with bacteria from BCC is not surprising since these bacteria are metabolically, ecologically, and genetically diverse, capable of growing in a number of environments from the soil to mammalian infection. All BCC bacteria possess very large multireplicon genomes (6 to 9 Mb [22, 30]) and hence have the capacity to encode multiple novel genomic loci. The B. cenocepacia island is the first example of a genomic island to be described in the BCC, and its role in opportunistic infection and the natural environment is worthy of further study.
Acknowledgments
This work was funded by a project grant (PJ501) from the UK Cystic Fibrosis Trust and a grant from the Canadian Institutes for Health Research to P.A.S.
We thank D. E. Woods for the quantitative histopathology analysis and C. Kooi for excellent technical assistance. We are grateful to the Wellcome Trust, Beowulf Genomics, and the Sanger Institute Pathogen Sequencing Unit for nucleotide sequence analysis of B. cenocepacia J2315.
Editor: J. T. Barbieri
REFERENCES
- 1.Achouak, W., T. Heulin, and J. M. Pages. 2001. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Aris, R. M., J. C. Routh, J. J. LiPuma, D. G. Heath, and P. H. Gilligan. 2001. Lung transplantation for cystic fibrosis patients with Burkholderia cepacia complex. Survival linked to genomovar type. Am. J. Respir. Crit. Care Med. 164:2102-2106. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson, S., J. P. Throup, G. S. Stewart, and P. Williams. 1999. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol. Microbiol. 33:1267-1277. [DOI] [PubMed] [Google Scholar]
- 4.Botelho, B. A., S. Y. Bando, L. R. Trabulsi, and C. A. Moreira-Filho. 2003. Identification of EPEC and non-EPEC serotypes in the EPEC O serogroups by PCR-RFLP analysis of the fliC gene. J. Microbiol. Methods 54:87-93. [DOI] [PubMed] [Google Scholar]
- 5.Cash, H. A., D. E. Woods, B. McCullough, W. G. Johanson, Jr., and J. A. Bass. 1979. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am. Rev. Respir. Dis. 119:453-459. [DOI] [PubMed] [Google Scholar]
- 6.Coenye, T., P. Vandamme, J. R. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coenye, T., P. Vandamme, J. J. LiPuma, J. R. Govan, and E. Mahenthiralingam. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 41:2797-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conway, B. A., and E. P. Greenberg. 2002. Quorum-sensing signals and quorum-sensing genes in Burkholderia vietnamiensis. J. Bacteriol. 184:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curnow, A. W., K.-w. Hong, R. Yuan, S.-i. Kim, O. Martins, W. Winkler, T. M. Henkin, and D. Soll. 1997. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA 94:11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Kievit, T. R., Y. Kakai, J. K. Register, E. C. Pesci, and B. H. Iglewski. 2002. Role of the Pseudomonas aeruginosa las and rhl quorum-sensing systems in rhlI regulation. FEMS Microbiol. Lett. 212:101-106. [DOI] [PubMed] [Google Scholar]
- 11.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 12.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 13.Henry, D. A., M. E. Campbell, J. J. LiPuma, and D. P. Speert. 1997. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J. Clin. Microbiol. 35:614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293:381-399. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlin, S. 2001. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9:335-343. [DOI] [PubMed] [Google Scholar]
- 17.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum Sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 19.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 20.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 22.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahenthiralingam, E., and P. Sayre. 2003. Burkholderia cepacia complex, p. 41-44. In M. D. Licker (ed.), McGraw-Hill yearbook of science and technology. McGraw-Hill, New York, N.Y.
- 27.Mahenthiralingam, E., D. A. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahenthiralingam, E., P. Vandamme, M. E. Campbell, D. A. Henry, A. M. Gravelle, L. T. Wong, A. G. Davidson, P. G. Wilcox, B. Nakielna, and D. P. Speert. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469-1475. [DOI] [PubMed] [Google Scholar]
- 29.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 31.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 33.RajBhandary, U. L. 1997. Once there were twenty. Proc. Natl. Acad. Sci. USA 94:11761-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 35.Sajjan, U., Y. J. Wu, G. Kent, and J. Forstner. 2000. Preferential adherence of cable-piliated Burkholderia cepacia to respiratory epithelia of CF knockout mice and human cystic fibrosis lung explants. J. Med. Microbiol. 49:875-885. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 37.Sokol, P. A., P. Darling, D. E. Woods, E. Mahenthiralingam, and C. Kooi. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5-oxygenase. Infect. Immun. 67:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 39.Sokol, P. A., and D. E. Woods. 1984. Relationship of iron and extracellular virulence factors to Pseudomonas aeruginosa lung infections. J. Med. Microbiol. 18:125-133. [DOI] [PubMed] [Google Scholar]
- 40.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun, L., R. Z. Jiang, S. Steinbach, A. Holmes, C. Campanelli, J. Forstner, U. Sajjan, Y. Tan, M. Riley, and R. Goldstein. 1995. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat. Med. 1:661-666. [DOI] [PubMed] [Google Scholar]
- 42.Tomich, M., A. Griffith, C. A. Herfst, J. L. Burns, and C. D. Mohr. 2003. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infect. Immun. 71:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyler, K. D., G. Wang, S. D. Tyler, and W. M. Johnson. 1997. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J. Clin. Microbiol. 35:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 45.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.: a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 46.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wisniewski-Dye, F., J. Jones, S. R. Chhabra, and J. A. Downie. 2002. raiIR genes are part of a quorum-sensing network controlled by cinI and cinR in Rhizobium leguminosarum. J. Bacteriol. 184:1597-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]