Abstract
Cytochrome P450 CYP2 family enzymes are important in a variety of physiological and toxicological processes. CYP2 genes are highly diverse and orthologous relationships remain clouded among CYP2s in different taxa. Sequence and expression analyses of CYP2 genes in diapsids including birds and reptiles may improve understanding of this CYP family. We sought CYP2 genes in a liver cDNA library of the common cormorant (Phalacrocorax carbo), and in the genomes of other diapsids, chicken (Gallus gallus), zebra finch (Taeniopygia guttata), and anole lizard (Anolis carolinensis), for phylogenetic and/or syntenic analyses. Screening of the cDNA library yielded four CYP2 cDNA clones that were phylogenetically classified as CYP2C45, CYP2J25, CYP2AC1, and CYP2AF1. There are numerous newly identified diapsid CYP2 genes that include genes related to the human CYP2Cs, CYP2D6, CYP2G2P, CYP2J2, CYP2R1, CYP2U1, CYP2W1, CYP2AB1P, and CYP2AC1P. Syntenic relationships show that avian CYP2Hs are orthologous to CYP2C62P in humans, CYP2C23 in rats, and Cyp2c44 in mice, and suggest that avian CYP2Hs, along with human CYP2C62P and mouse Cyp2c44, could be renamed as CYP2C23, based upon the nomenclature rules. Analysis of sequence and synteny identifies cormorant and finch CYPs that are apparent orthologs of phenobarbital-inducible chicken CYP2C45. Transcripts of all four cormorant CYP2 genes were detected in the liver of birds from Lake Biwa, Japan. The transcript levels bore no significant relationship to levels of chlorinated organic pollutants in the liver, including polychlorinated biphenyls and dichlorodiphenyltrichloroethane and its metabolites. In contrast, concentrations of perfluorooctane sulfonate and perfluorononanoic acid were negatively correlated with levels of CYP2C45 and/or CYP2J25, suggesting down-regulation of expression by these environmental pollutants. This study expands our view of the phylogeny and evolution of CYP2s, and provides evolutionary insight into the chemical regulation of CYP2 gene expression in diapsids including birds.
Keywords: Anole lizard, Chicken, Common (great) cormorant, CYP2, Cytochrome P450, Diapsid, Zebra finch
1. Introduction
Cytochrome P450 (CYP) monooxygenases in vertebrates have broad roles in physiological and toxicological processes, including oxidative metabolism of steroid hormones, fatty acids, drugs, procarcinogens, and environmental pollutants. It is known that metabolism by some CYP2 enzymes can result in detoxification and/or bioactivation of some foreign chemicals (Ioannides and Lewis, 2004; Karlgren et al., 2005). The CYP2 gene family is the largest and most diverse CYP gene family in deuterostomes from echinoderms to mammals (Goldstone et al., 2006; Nelson et al., 2009), but the evolutionary relationships among most CYP2s remain unknown, even among vertebrates.
Comparison of genes in humans and zebrafish illustrates the complexity. Humans have 16 CYP2 genes in 11 subfamilies, and zebrafish have 42 CYP2 genes in 11 subfamilies, but there are only two subfamilies (and two genes) sufficiently conserved to be identified as orthologs based on sequence identity, CYP2U1 and CYP2R1 (Goldstone et al., 2010; Nelson, 2009). However, identical catalytic specificities for some CYP2s in mammals and fish suggest a common evolutionary origin, even though they are classified in different subfamilies based on the sequences. For example, fish-specific CYP2P3 resembles human CYP2J2 in regio- and enantio-selectivity for oxidation of arachidonic acids (Oleksiak et al., 2003). Identification of CYP2s in other vertebrate groups may help to discern evolutionary relationships in this complex family. Knowledge of CYP2 genes in birds is limited to CYP2Hs (Davidson et al., 2001; Dogra et al., 1999; Hahn et al., 1991; Hansen et al., 1989; Nakai et al., 1992; Sinclair et al., 1990) and CYP2C45 (Baader et al., 2002) in the chicken (Gallus gallus); no report has addressed phylogenetic or functional analyses of CYP2s in reptiles.
Regulation of CYP2 genes also is poorly known. Some genes in the CYP2 family, most notably CYP2B in some mammals and CYP2H in the chicken, are inducible by phenobarbital (PB) and other PB-type inducers acting via the nuclear receptor constitutive androstane receptor (CAR) and chicken xenobiotic-sensing orphan nuclear receptor (CXR), respectively (Handschin and Meyer, 2003; Waxman, 1999). At least one CYP2C in birds appears to be induced by PB (Baader et al., 2002), but otherwise, PB responsive CYP2 genes in vertebrates are not known. Knowledge of CYP2 induction is highly relevant to assessing effects of environmental chemicals. CYP2 inducers include persistent organic pollutants (POPs) such as ortho-chlorine substituted polychlorinated biphenyls (PCBs), dichlorodiphenyltrichloroethane (DDT) and its metabolites (DDE), and chlordane compounds (Connor et al., 1995; Craft et al., 2002; Nims and Lubet, 1995; Wyde et al., 2003), some of which are known agonists for CAR and/or pregnane X receptor (Kretschmer and Baldwin, 2005). There are large interspecies differences in responses of CYP2s to PB-type inducers (Moore et al., 2000, 2002; Sakai et al., 2006, 2009). POPs have worldwide distribution, high-level bioaccumulation, and toxic potential in animals at higher trophic levels (Tanabe, 2002). Developing biomarkers that are indicative of contamination by POPs is important for understanding the effects of various kinds of POPs in wild species.
Knowledge of identity and expression levels of CYP2 genes is quite limited for wild species. Immunoblot analyses revealed that some wild species express one or more proteins that can cross-react with antibodies against rat CYP2B1/2 (Wolkers et al., 1998, 1999). However, there is doubt still about which CYP2s may be valid as possible biomarkers of exposure to PB-type inducers in wild species (Nyman et al., 2001). Identifying CYP2 genes and quantifying their expression in wild species in relation to POPs accumulation will aid in assessing the potential impacts of these contaminants.
This study initially focused on a wild avian model, the common cormorant (Phalacrocorax carbo), a top predator in the ecosystem of Lake Biwa, Japan. Lake Biwa cormorants are contaminated with classic and emerging POPs including known CYP2 inducers (Kubota et al., 2004; Nakayama et al., 2006, 2008). Previously we identified CYP1A4 and CYP1A5 orthologs in the cormorant, and found significant positive relationships between hepatic expression levels of each CYP1A and concentrations of dioxin and related compounds in the Lake Biwa population, indicating the induction of both CYP1As by these pollutants (Kubota et al., 2005, 2006). Here we screened our custom-made hepatic cDNA library of the common cormorant for novel CYP2 sequences. To evaluate CYP2 genes as useful biomarkers of PB-type inducers, we measured hepatic expression levels of CYP2 gene transcripts in the wild cormorant population. We also searched for CYP2 clan sequences (i.e., CYP1s, CYP2s, CYP17s, and CYP21s [Nelson, 1998]) from the three available diapsid genomes, chicken, zebra finch (Taeniopygia guttata), and anole lizard (Anolis carolinensis), and performed a phylogenetic analysis of the130 identified genes, along with 21 human clan 2 genes and six other selected amphibian and mammalian CYP2 genes, to better understand the evolutionary history of CYP2 genes.
2. Materials and methods
2.1. Sample collection
Common cormorants (Phalacrocorax carbo) (n = 16) were randomly captured by shooting under the license from Shiga Prefecture in 2001 from the southern part of Lake Biwa, Japan. The cormorants were killed by decapitation and were immediately dissected on board after measurements of biometry. In the present study, there were seven males and nine females, four adult males (body mass ranging 1,890-2,480 g) and two adult females (b.w. 1,710 and 1,920 g). Juveniles included three males (b.w. 1,840-1,970 g) and two females (b.w. 1,660 and 1,810 g). There also were five females of unspecified stages of development (b.w. 1,520-2,040 g). Due to the limited number of specimens in each developmental stage, we only compared male-female differences in gene expression levels, but not growth stage differences. Subsamples of the liver were flash frozen in liquid nitrogen and stored at −80 °C until RNA isolation. Concentrations of these environmental contaminants in the liver of cormorants were reported elsewhere (Kubota et al., 2004; Nakayama et al., 2006, 2008). Chemicals analyzed included dioxin and related compounds (DRCs; polychlorinated dibenzo-p-dioxins, polychlorinated dibenzofurans, and dioxin-like [coplanar] PCBs), ortho-chlorine substituted PCBs (PCBs), DDT and its metabolites (DDTs; p,p’-DDT, -DDE, -DDD, o,p’-DDT, -DDE, and -DDD), hexachlorocyclohexane isomers (HCHs; α-, β-, and 1γ-HCH isomers), chlordane compounds (CHLs; cis-chlordane, trans-chlordane, oxychlordane, and trans-nonachlor), perfluorooctane sulfonate (PFOS), perfluorooctane sulfonamide (PFOSA), and perfluorononanoic acid (PFNA). These selected environmental chemicals cover inducers of genes belonging to CYP1, 2, 3, and 4 families in mammals.
2.2. Cloning the full-length sequences of CYP2 genes
We used a hepatic cDNA library constructed from an adult male cormorant (Kubota et al., 2006). Randomly selected library clones (6930 in total) were subjected to 5′-end single-pass sequencing using a MegaBACE™ 1000 capillary sequencer (Amersham Biosciences, Piscataway, NJ, USA). BLAST homology searches against NCBI nucleotide sequence databases were used to identify cormorant cDNAs with strong similarities to vertebrate CYP2 genes. The 3′-end sequences of these candidate cormorant CYP2s were determined using ABI PRISM™ 310 genetic analyzer (Applied Biosystems, Foster City, CA, USA). The resulting full-length cDNA sequences were named by the CYP nomenclature committee.
2.3. Phylogenetic analyses
Chicken, zebra finch, anole lizard, zebrafish, and human CYP sequences were downloaded from the Ensembl database (release 55). All other diapsid CYP sequences were obtained from GenBank. All sequences were aligned using Muscle (v3.6; [Edgar, 2004]), and regions of ambiguous alignment were masked using a custom Perl script. Phylogenetic analyses were performed with RAxML (v7.2.6; [Stamatakis, 2006]) using the PROTCATWAG model of amino acid substitution. One hundred bootstrap replicates were performed to test the clade support. A subset of the CYP2 sequences was used to examine the relationship of the CYP2C subfamily in more detail. Both Bayesian (MrBayes 3.1.2 [Ronquist and Huelsenbeck, 2003]) and maximum likelihood (RAxML) phylogenetic analyses were performed. MrBayes estimates posterior probabilities using Metropolis-Hastings coupled Monte Carlo Markov chains (MC3). We performed MC3 estimates with uninformative prior probabilities using the WAG model of amino acid substitution and prior uniform gamma distributions approximated with four categories. Four incrementally heated, randomly seeded Markov chains were run for 4×106 generations, and topologies were sampled every 100th generation. Burnin value was set to 5×105 generations. 500 bootstrap replicates were performed using RA×ML.
2.4. Quantification of CYP2 mRNA expression levels
Total RNA in the liver of cormorants was isolated with TRIzol® (Invitrogen, Carlsbad, CA) and was treated with RNeasy® Mini Kit (Qiagen, Tokyo, Japan). Messenger RNA expression levels of each of four cormorant CYP2 genes (i.e., CYP2C45, CYP2J25, CYP2AC1, and CYP2AF1) were measured with 50 ng of total RNA using TaqMan® One-Step RT-PCR kit (Applied Biosystems) in combination with an ABI PRISM 7700 Sequence Detector System (Applied Biosystems). Primers for each CYP2 gene were designed with the aid of Primer Express™ version 1.0 (Applied Biosystems). Primer sequences applied for the quantification of each target gene are listed in Table 1. The thermal cycling program was as follows; 30 min at 48 °C and 10 min at 95 °C, and 45 cycles of 15 s at 95 °C, and 1 min at 53 °C for CYP2C45 and CYP2AF1 or 1 min at 50 °C for CYP2J25 and CYP2AC1. Quantitative values were calculated from the threshold PCR cycle number (Ct) at which the increase in signal associated with an exponential growth for PCR product could be detected. The calibration curves were generated by plotting Ct values against the logarithmic value of template dilution factors. For the calibration curves, the template was prepared from total RNA of one specimen. The mRNA levels were normalized to 18S ribosomal RNA content that was quantified by TaqMan® Ribosomal RNA Control Reagents (Applied Biosystems). All experiments were performed in triplicate. Expression levels of normalized mRNA determined in the present study were not compared between the CYP genes, since the amplification rates of PCR products for each gene were not determined.
Table 1.
List of primers used in real time RT-PCR for common cormorant CYP2 genes
| Gene | Primer sequences (5’ to 3’) | Fragment size (bp) | |
|---|---|---|---|
| CYP2C45 | F | CAGCAAAATGCAGGAGGAGAAAG | 82 |
| R | GAACAAGTCGAAGGTGCTGGTTAT | ||
| CYP2J25 | F | ACGTGACCCTCAAAGGCTATTT | 97 |
| R | TCTGGTTTCTCCCACTGGGATT | ||
| CYP2AC1 | F | CCATCTTGTGGTTCCTGGTCTG | 102 |
| R | CATCCTTTTGCCATAGGTTGCC | ||
| CYP2AF1 | F | TACCACGATGAGGAGTTCCAAAG | 89 |
| R | GAATTGTACAGCTGGCTCATGAT |
2.5. Statistical Analyses
A Mann-Whitney U-test was conducted to assess gender-difference in transcript levels of each CYP2 gene. Spearman’s rank correlation test was performed to evaluate the relationships between concentrations of each contaminant and expression levels of CYP2 transcripts, and among CYP2 expression levels. Statistical analysis was conducted by StatView for Windows (v5.0; SAS Institute, Cary, NC, USA). P value of < 0.05 was regarded as statistically significant.
3. Results
3.1. Cloning of full-length common cormorant CYP2 cDNAs
We isolated four distinct full-length CYP2 cDNA clones from the hepatic cDNA library of the common cormorant. Those full-length clones have open reading frames of 1479 to 1497 base pairs coding for 492 to 498 amino acids. The four CYP2s share 52-54% identities with one another at the level of cDNA coding sequences and 38-48% identities between deduced amino acid sequences. These cormorant CYP2 cDNAs were classified as CYP2C45 (DDBJ accession no. AB588748), CYP2J25 (AB588749), CYP2AC1 (AB588750), and CYP2AF1 (AB588751).
3.2. New avian and reptilian CYP2 genes
The four cormorant CYP2 sequences were used to search for related genes in the chicken, zebra finch, and anole lizard, the three diapsids for which genomes are available in Ensembl. In each of these species, there were one or more sequences identified as possible orthologs or co-orthologs of the cormorant CYP2 genes. Particularly interesting is a finch CYP2 sequence we found that is 87% identical to the cormorant CYP2C45 at the level of deduced amino acids. The cormorant CYP2C45 shared high identity (81%) also to chicken CYP2C45. This CYP2 gene in the cormorant showed relatively low identities to the anole CYP2C genes, ranging from 49% to 63%, although some with lower identities are based upon partial sequences.
CYP2J subfamily genes were identified in all of the diapsids examined. In the chicken we found a cluster of CYP2Js (five genes, CYP2J19 to 2J23 and a pseudogene, 2J24P). Identity between cormorant CYP2J25 and members of the chicken CYP2J cluster ranged from 55% to 78% at the amino acid level. Similarly, in the finch and anole multiple CYP2J genes were identified that showed 55% to 60% identities to the cormorant CYP2J25.
A sequence identified in the cormorant was classified in the subfamily CYP2AC. The cormorant CYP2AC1 showed 77% amino acid identity to the chicken CYP2AC1. The cormorant CYP2AC1 was 69% and 68% identical to CYP2AC2s in the chicken and finch, respectively, and 52% and 54% to the CYP2AC7s in these respective species. Four genes classified in the CYP2AC subfamily also were identified in the anole genome, and the amino acid identities of those genes to the cormorant CYP2AC1 ranged from 63% to 68%.
CYP2AF1 identified in the cormorant represents a new subfamily, with members found also in the genomes of the finch and the anole. An apparent ortholog, CYP2AF1 in finch, was 81% identical to cormorant CYP2AF1 and a gene in the anole, CYP2AF2, was 54% identical to the cormorant sequence. The current assembly of the chicken genome did not appear to have a CYP2AF1, but this assembly still is incomplete.
In addition to homologs of the cormorant CYP2 genes, numerous other diapsid CYP2 genes were identified in the genomes of the chicken, finch, and anole, with some genes occurring in known subfamilies and some apparent orthologs of mammalian genes. Diapsid CYP genes identified in this group included genes related to the human CYP2Cs, CYP2D6, CYP2G2P, CYP2J2, CYP2R1, CYP2U1, CYP2W1, CYP2AB1P, and CYP2AC1P.
3.3. Phylogenetic analyses of diapsid CYP2 genes
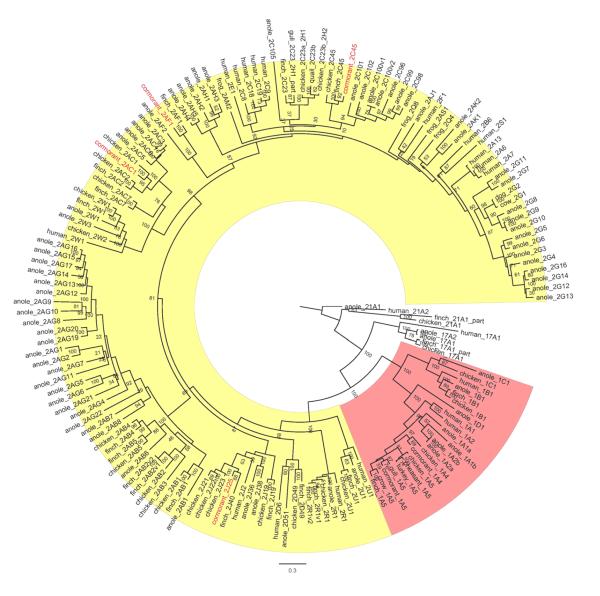
We performed maximum likelihood phylogenetic analyses with all of the available diapsid CYP sequences from the cormorant cDNA library, from GenBank, and from the three available genome projects (chicken, zebra finch, and anole) obtained from Ensembl. Initially we analyzed all of the diapsid CYPs together with selected CYPs from humans and zebrafish, to discern relationships of all genes in a broad context (not shown). Following that phylogenetic analysis, CYP2 clan sequences were chosen, realigned, and reanalyzed to obtain a maximum likelihood phylogeny focused on clan 2 (Figure 1). The tree in Figure 1 contains four cloned cormorant CYP2 genes, as well as 16 human CYP2s, 17 finch CYP2s, 20 chicken CYP2s, and 65 anole CYP2s. The four cloned cormorant genes, CYP2C45, CYP2J25, CYP2AC1, and CYP2AF1 (Figure 1; red), fall either into large clusters of multiple orthologous or co-orthologous genes (CYP2Cs, CYP2Js, and CYP2ACs), or are a part of a smaller, possibly diapsid-specific clade (CYP2AFs).
Figure 1.
Maximum likelihood phylogeny of all available chicken, zebra finch, anole lizard, and human clan 2 CYPs, along with selected other avian, amphibian, and mammalian CYP2 genes. The four cloned common cormorant CYP2s are shown in red. The CYP2 clan includes the CYP1 (highlighted in pink), CYP2 (highlighted in yellow), CYP17, and CYP21 subfamilies. Pseudogenes and some partial gene predictions (part) are excluded from the phylogenetic analysis and do not appear on the tree. The previously named chicken CYP2H1 and CYP2H2 genes are shown as chicken 2C23a_2H1 and chicken 2C23b_2H2, respectively, based upon renaming. Similarly, the previously named gull CYP2H1_part is shown as gull 2C23_2H1_part. Values at the branch points are bootstrap support values of 100 randomly seeded bootstrap replicates using RAxML.
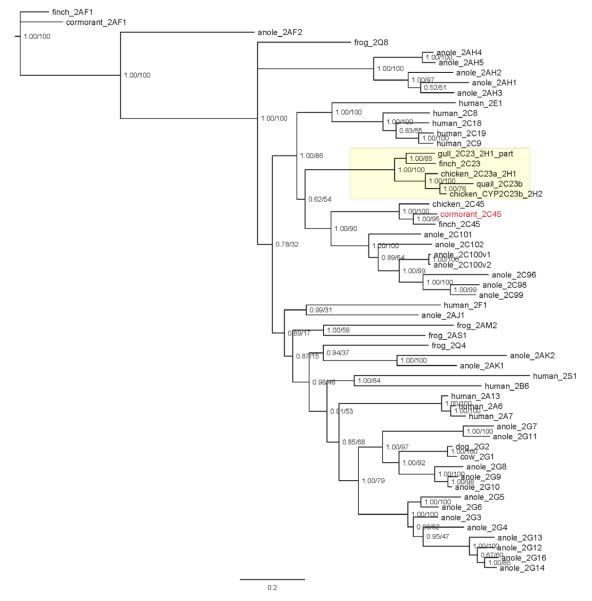
With the addition of diapsid sequences, the CYP2C subfamily at first appears to be more complex, yet the addition of the cormorant sequences clarifies classification of some avian CYPs. Notable is the status of the CYP2H subfamily. The chicken CYP2H subfamily is nested within the CYP2C cluster (Figures 1 and 2). The support for the CYP2C clade in the smaller tree (Figure 2) is reasonably high (1.00 posterior probability; 86% ML bootstrap), although there is some uncertainty in the precise arrangement of the various branches within it, including the CYP2H subfamily (denoted CYP2C23_2H1 and related; see below).
Figure 2.
Phylogenetic tree of CYP2C, CYP2A, CYP2G, and CYP2AH genes, with assorted other CYP2 subfamilies. A Bayesian phylogeny is pictured, with posterior probabilities resulting from 3×106 generation of MC3 and maximum likelihood bootstrap values derived from 500 bootstrap replicates noted at the node points (pp/ML). The cormorant CYP2C45 sequence is in red, while the CYP2C23/CYP2H clade is highlighted in yellow.
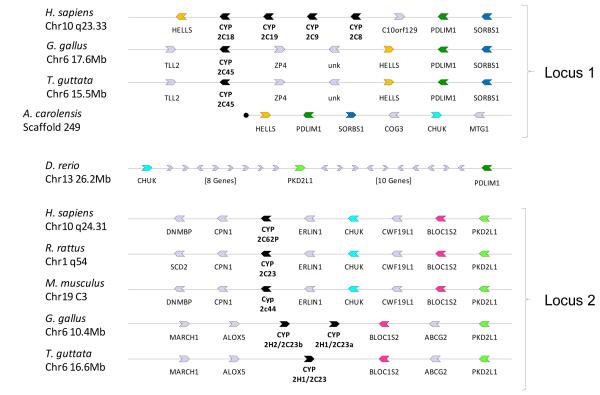
In order to clarify evolutionary relationships of CYP2Hs with CYP2Cs, we searched genomic regions surrounding the CYP2H and/or CYP2C genes in the chicken, finch, anole, zebrafish, human, rat, and mouse (Figure 3), as evolutionary relationships indicating orthology that are not clear from sequence, may be evident in syntenic relationships. Chicken CYP2H1 and CYP2H2 sequences are 92% identical, indicating a very recent duplication probably not found in most birds. Finch only has one gene in this region (Figure 3; locus 2). The chicken and finch CYP2H genes share synteny with rat CYP2C23 and its mouse ortholog Cyp2c44 (Figure 3; locus 2). The human pseudogene CYP2C62P sits between ERLIN1 and CPN1 as in rat and mouse (Figure 3; locus 2) and therefore it also is evolutionarily related to the chicken and finch CYP2H genes, rat CYP2C23, and mouse Cyp2c44.
Figure 3.
Comparative gene synteny for the CYP2C locus in various species. Conserved genes among different taxa are indicated with the same color. Gene length and intergenic distances are not drawn to scale. The end of sequence data for anole scaffold 249 is indicated in circle (•).
A finch sequence was found to share synteny with the chicken CYP2C45 and is therefore an ortholog, labeled as finch CYP2C45 (Figure 3; locus 1). CYP2C genes in the locus 1 in human, chicken, and finch are adjacent to PDLIM1, SORBS1, and HELLS (Figure 3; locus 1). It is apparent that the CYP2C and CYP2H/CYP2C23/Cyp2c44 regions were once united but became separated early in evolution of amniotes (reptiles, birds, and mammals). This can be seen in zebrafish chr13 where CHUK, PKD2L1, and PDLIM1 are in proximity (Figure 3). Frog has SORBS1, PDLIM1, HELLS, BLOC1S2, and PKD2L1 in another small region (not shown) and lizard includes CHUK, SORBS1, PDLIM1, and HELLS on one contig (Figure 3; locus 1). There are no CYP genes identified in this neighborhood until birds. The lizard genome is still too incomplete to judge, though a CYP2C gene may be near HELLS in the lizard.
3.4. Hepatic expression of common cormorant CYP2 transcripts
Comparison of CYP2 mRNA expression levels between male and female cormorant showed a gender specific expression for CYP2AF1 with males having a significantly greater expression level than females (Table 2). The other CYP2 genes showed no significant gender difference in their transcript levels. Therefore, the influence of gender on expression levels was considered only for CYP2AF1, but not for other CYP2s, when further statistical analysis was conducted.
Table 2.
Relative mRNA abundance of individual CYP2 genes in the liver of common cormorants
| Male (n = 7) |
Female (n = 9) |
|
|---|---|---|
| CYP2C45/rRNA | 110 ± 21 | 130 ± 25 |
| CYP2J25/rRNA | 75 ± 18 | 120 ± 32 |
| CYP2AC1/rRNA | 41 ± 7.9 | 46 ± 9.2 |
| CYP2AF1/rRNA | 290 ± 86 * | 87 ± 20 |
Ribosomal RNA (rRNA) was used as a reference gene, and each transcript/rRNA indicates 100 times values of the simple division. Results are expressed as mean ± SEM. The asterisk indicates significant difference in the expression levels between male and female, when Mann Whitney U-test was conducted for the statistical analysis.
The relationships of mRNA expression levels among individual CYP2 genes also were examined (Table 3), since this may give clues concerning the transcriptional regulation of these genes. The results showed that the expression level of CYP2AF1 in female was negatively correlated with that of CYP2C45 (r = −0.97, p = 0.0069), whereas no significant relationship was found for the other CYP genes.
Table 3.
Summary of Spearman’s rank correlations (r values) among individual CYP2 transcript levels in the liver of common cormorants.
| CYP2C45 | CYP2J25 | CYP2AC1 | |
|---|---|---|---|
| CYP2J25 | 0.39 | ||
| CYP2AC1 | 0.46 | 0.26 | |
| CYP2AF1-M | 0.14 | −0.54 | 0.11 |
| CYP2AF1-F | −0.97 * | −0.17 | −0.32 |
M and F represent male and female, respectively. Asterisk indicates significant correlation (p < 0.05).
Environmental pollutants including DRCs, PCBs, DDTs, HCHs, CHLs, and perfluorochemicals (PFCs), all were detected in the same cormorant liver samples as used in the real time PCR analysis for the CYP2 genes we found (Kubota et al., 2004; Nakayama et al., 2006, 2008). None of the chlorinated compounds exhibited any significant relationships between their concentrations (on a wet wt basis) and expression levels of CYP2C45, CYP2J25, CYP2AC1, or CYP2AF1 (Table 4). Combining the concentrations of PB-type inducers (ortho-PCBs, DDTs, and CHLs) provided no significant correlations with expression level of any of the CYP genes (not shown). In contrast, concentrations (on a wet wt basis) of PFCs did show significant relationships with expression levels of some CYP2s. The concentrations of PFOS were negatively correlated with CYP2C45 expression levels (Figure 4A). A similar negative relationship with PFOS concentrations was found for CYP2J25 (Figure 4B). The concentration of another perfluorinated compound, PFNA, also showed a significant negative correlation with CYP2C45 transcript levels (Table 4).
Table 4.
Summary of Spearman’s rank correlations (r values) between residue levels of environmental contaminants (on a wet wt basis) and CYP2 transcript levels in the liver of common cormorants.
| CYP2C45 | CYP2J25 | CYP2AC1 | CYP2AF1-M | CYP2AF1-F | |
|---|---|---|---|---|---|
| DRCs | 0.050 | 0.33 | 0.12 | 0.50 | −0.20 |
| PCBs | 0.10 | 0.29 | 0.085 | 0.50 | −0.33 |
| DDTs | 0.11 | 0.33 | 0.19 | 0.36 | −0.40 |
| HCHs | −0.15 | 0.10 | −0.015 | 0.39 | −0.033 |
| CHLs | 0.13 | 0.20 | 0.34 | 0.43 | −0.47 |
| PFOS | −0.53 * | −0.71 ** | −0.074 | 0.036 | 0.52 |
| PFOSA | 0.074 | 0.038 | 0.31 | 0.00 | 0.63 |
| PFNA | −0.51 * | −0.45 | 0.035 | 0.46 | −0.10 |
M and F represent male and female, respectively. Asterisks indicate significant correlation
p < 0.05
p < 0.01
Abbreviations for chemical name are shown in Materials and Methods.
Figure 4.
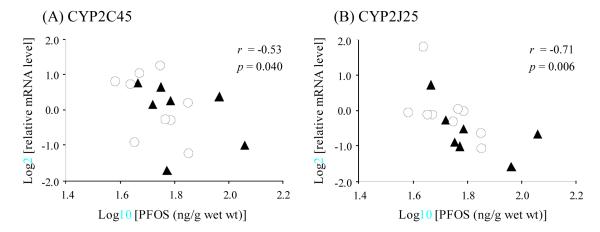
Relationships of perfluorooctane sulfonate (PFOS) concentrations (ng/g wet wt) with expression levels of (A) CYP2C45 and (B) CYP2J25 in the liver of common cormorants collected from Lake Biwa, Japan. Circle and triangle represent the expression levels in female (n = 9) and in male (n = 7), respectively. Values of r and p are indicated in the upper right of each figure. These measurements were logarithmically transformed to make the relationships clearly visible. Concentrations of PFOS were cited from Nakayama et al. (2008), derived from the same specimens used in this study.
4. Discussion
4.1. CYP2 genes in common cormorant and other diapsids
In this study, we identified and cloned novel CYP2 genes from a wild avian species, the common cormorant, which we have used as a model in assessing responses to environmental chemicals. The CYP2 gene family has evolved complex roles in physiology and toxicology, and diverse regulatory mechanisms. This has been determined largely in studies of selected mammalian models and humans. While some CYP2 genes have been examined in the chicken as well (Baader et al., 2002; Davidson et al., 2001; Dogra et al., 1999; Hahn et al., 1991; Hansen et al., 1989; Nakai et al., 1992; Sinclair et al., 1990), the diversity of CYP2 genes and the effects of environmental chemicals on their expression have not been comprehensively investigated in birds. Using the cormorant CYP2 genes, we searched the three available diapsid genomes (i.e., chicken, zebra finch, and anole lizard) for additional clan 2 CYP genes, to construct a vertebrate clan 2 phylogeny that includes these diapsids. Phylogenetic analysis showed that some cormorant CYP2 genes we identified fell within existing CYP2C, CYP2J, or CYP2AC clades, and one in a new subfamily, CYP2AF. While cormorant CYP2C45, CYP2J25, and CYP2AC1 are in large clusters of multiple, possibly orthologous or co-orthologous genes, CYP2AF1 occurs in a smaller, possibly diapsid-specific gene subfamily.
Species represented in the CYP2J cluster have differing numbers of genes at the CYP2J locus, including tandemly duplicated genes, multiple pseudogenes, and fragmentary exons, reflecting the CYP gene birth-death process (Nelson et al., 2004). For example, while humans have one CYP2J (CYP2J2), rats have five and mice have eight. In the phylogenetic tree described in Figure 1, there is evidence for five genes (one gene does not appear on the tree because of a partial sequence available) and one pseudogene (which does not appear on the tree) in the chicken, two finch genes, and two anole genes, as well as twelve zebrafish CYP2J-like genes (Goldstone et al., 2010). All of these genes share synteny with the human CYP2J2. We anticipate that the cormorant CYP2J will occupy the same locus, but whether there are multiple CYP2Js in cormorant is not known. The cormorant CYP2AC1 gene is classified into the CYP2AC clade, where chicken has three, finch has two, and anole has four. The CYP2AC subfamily appears to be relatively conserved throughout tetrapod evolution, since CYP2AC subfamily genes occur also in Xenopus and mammals (Nelson et al., 2009). The finch and anole genomes appear to have orthologs or co-orthologs of the cormorant CYP2AF1 gene. A CYP2AF gene has not been found in the chicken genome, probably due to incomplete assembly of this genome.
Our cloning effort identified one CYP2C gene in the cormorant (i.e., CYP2C45). This cormorant CYP2C gene was previously termed CYP2C84 (Nelson et al., 2009) and based on this study renamed as CYP2C45, for the reasons described below. As the CYP2C gene subsequently uncovered in the finch was 87% identical to the CYP2C in the cormorant, the two genes can be considered as orthologous to one another. The cormorant and finch CYP2C genes bear a high identity (81%) to CYP2C45, previously described in the chicken (Baader et al., 2002), and the finch gene also shares syntenic relationships with the chicken CYP2C45 (Figure 3). Given shared synteny and high identity, the finch gene and the chicken CYP2C45 can be concluded to be orthologs, and therefore the finch CYP2C gene and the cormorant CYP2C should be named CYP2C45. We do not have gene structure information for the cormorant CYP2C, and thus there are some uncertainties about the orthology, yet provisionally, we conclude that the cormorant CYP2C is a CYP2C45.
4.2. Evolutionary relationships of avian CYP2Hs with CYP2Cs
A frequent question concerns the identity of avian CYP genes induced by PB. The CYP2C45 gene found in the cormorant is intriguing in that it may give an answer to the question on the relationships among PB responsive CYP genes, and their evolution. Chicken CYP2C45 has been shown to be inducible by PB type inducers (Baader et al., 2002). Earlier, CYP2H1 and CYP2H2 were identified as PB-inducible CYP genes in the chicken, and were suggested to be avian homologs of the PB-inducible CYP2B genes in mammals (Hansen et al., 1989; Nakai et al., 1992). However, our phylogenetic evidence shows that the CYP2H subfamily in fact is embedded in the CYP2C subfamily, rather than being related to the PB-inducible CYP2B subfamily (Figures 1 and 2). Synteny analysis of CYP2Hs and CYP2Cs (Figure 3) also shows that CYP2H is not an ortholog of CYP2B, but rather is an ortholog of CYP2C62P in humans, CYP2C23 in rats, and Cyp2c44 in mice. Thus far there are no diapsid genes belonging to a CYP2B clade. This means that the induction mechanisms of CYP2H genes may differ from those of CYP2B, while CYP2H and mammalian CYP2B share responsiveness to PB type inducers (Handschin and Meyer, 2003). Interestingly, it is known that the murine CYP2H ortholog, Cyp2c44, is not inducible by PB (DeLozier et al., 2004). This suggests that the PB responsiveness of the CYP2Hs may have arisen separately from Cyp2c44, or that it was lost in Cyp2c44. Thus, it is possible that PB-responsiveness was acquired independently in several CYP2 lines, or that cis-regulatory regions became shared among some CYP2 genes.
CYP2H1 and CYP2H2 in birds were named before the synteny information was available for chicken and finch. Those sequences were different enough not to be named as CYP2C at that time. In the present study, we showed that CYP2Hs in birds share synteny with CYP2Cs in humans, rats, and mice. Based upon the Human Gene Nomenclature Committee, orthologs in vertebrates should be given the same name. Therefore, the mouse Cyp2c44 name should be given the rat CYP2C23 name, since the earlier name has precedence. Similarly, CYP2Hs in birds could be renamed as CYP2C23s, as we have done in Figures 1 and 2. Chicken has a duplication of the gene not seen in the finch or mammals. To distinguish between these co-orthologs, the chicken genes are given the names of CYP2C23a and CYP2C23b. Finch and gull probably have only one gene (i.e., CYP2C23), but the exact number of genes is not clear in quail, as the quail sequence clusters with chicken CYP2C23b (Figures 1 and 2). Thus, there may be two quail CYP2C23 genes.
All the branches from the CYP2C cluster would become CYP2Cs, perhaps with the exception of CYP2E1. The CYP2E subfamily is most likely part of the CYP2Cs (Figure 1), but it is undesirable to rename this subfamily since it is widely known and reported on under the CYP2E1 name. The branch adjacent to the human CYP2Cs exhibits identities to CYP2Cs in the high 40% range. This branch has been named CYP2AH and it does not extend the CYP2Cs up the tree any further (Figure 2).
4.3. Other CYP2 family members
CYP2 genes conserved in diapsids and other vertebrates include CYP2R1 and CYP2U1. These are ancient and may be ancestral to the vertebrate CYP2s, as they are found in all vertebrates examined so far, and are always phylogenetically placed at the base of the CYP2 family (Figure 1). CYP2R1 and CYP2U1 were identified initially in the human genome project. The CYP2R1 enzyme acts as 25-hydroxylase of vitamin D2 and D3 (Cheng et al., 2004), while the CYP2U1 functions in hydroxylation of arachidonic acid and long chain fatty acids (Chuang et al., 2004), suggesting their important and conserved roles in physiological processes.
The anole lizard appears to have a much larger set of CYP2 genes (65 genes) than either chicken (20 genes) or finch (17 genes). Expansions of the CYP2s in the anole are found in subfamilies where avian CYP2s do not occur, CYP2G and CYP2AG (Figure 1). A smaller expansion occurs in the CYP2C subfamily. Similar to the lineage-specific gene subfamily expansion observed in teleost fish (Goldstone et al., 2010) and in rodents (Nelson et al., 2004), these lizard CYP2 families have tandemly duplicated to produce large genomic regions of full and fragmentary CYP genes. Such expansions have been termed blooms by R. Feyereisen (Feyereisen, 2006). The biological significance of these tandemly duplicated CYP gene “blooms” is not known.
4.4. Other CYP2 clan members
In addition to findings on CYP2s, the phylogeny in Figure 1 illustrates a number of interesting features of other clan 2 families, particularly the CYP1s. First, CYP1B1 is conserved in diapsids, and thus the CYP1B subfamily consists of a large cluster of orthologous genes throughout the vertebrates. Secondly, while CYP1D1 in birds has not been found in those genomes, there does appear to be a CYP1D1 in anole lizards. Pseudogenes of CYP1D are found in humans (Nelson, 2009), while this CYP1 subfamily appears to be functional in some other mammals (Kawai et al., 2010). In addition, the relationships between mammalian CYP1A1 and CYP1A2 and the diapsid CYP1As continue to be obscured, probably by gene conversion, as originally described by Goldstone and Stegeman (2006). Nevertheless, synteny would support that CYP1A4 is an ortholog of the CYP1A1 and CYP1A5 is an ortholog of the CYP1A2.
4.5. Hepatic expression of common cormorant CYP2 gene transcripts
The present study revealed that cormorants express all of the CYP2 genes identified, namely, CYP2C45, CYP2J25, CYP2AC1 and CYP2AF1, in the liver at least at the level of transcription. Since members of the CYP2 gene family display a variety of expression profiles, including tissue- and gender-specific regulation (Anderson, 2008; Czerniak, 2001) and induction by drugs and environmental contaminants (Connor et al., 1995; Craft et al., 2002; Nims and Lubet, 1995; Wyde et al., 2003; Waxman, 1999), we investigated whether biological and environmental factors influence the expression levels of these CYP2 genes in cormorant livers. A significant gender difference was observed for CYP2AF, with higher transcript levels in males than in females. Whether the gender difference results in the difference in the gender-related CYP substrate metabolism warrants further investigation. Expression of cormorant CYP2 genes other than CYP2AF1 did not show gender-specific expression profiles.
The correlation analyses between expression levels of CYP2 genes exhibited that there was only a significant negative correlation between CYP2AF1 and CYP2C45 in females. These results suggest that there may be distinct regulatory mechanisms underlying the transcription of these CYPs. On the other hand, it is known that hepatic expression of CYP genes can be influenced by a variety of physiological conditions. Furukawa et al. (2004) showed that there is a substantial difference in the correlation profile of CYP gene expression levels between non-tumorous and tumorous liver tissues; expression levels of CYP2C8 and CYP2C9 were closely associated with CYP2J2 in the non-tumorous tissues, whereas such association was not evident in tumors. Thus, different physiological conditions in individual specimens may affect the hepatic expression levels of these CYP2 genes in an isoform-specific manner.
Besides the physiological conditions, we took into account chemical contamination as a possible environmental factor to alter these CYP gene expressions. Results showed that there were no significant correlations between concentrations of chlorinated compounds (DRCs, PCBs, DDTs, CHLs, or HCHs) and transcript levels of these CYP2 genes, implying that the expression levels were not altered by these contaminants in the liver of wild cormorants. Since the contamination by PB type inducers is widely spread in the environment of Lake Biwa, the lack of association between contaminant levels and CYP2C45 may mean that this CYP2C45 ortholog in the cormorant is not inducible by any of the chlorinated compounds examined, or that the chemical concentrations (e.g., PCBs; 560 ± 250 ng/g wet wt [mean ± SE], DDTs; 270 ± 84 ng/g wet wt) were not sufficient to cause induction.
In contrast to the chlorinated compounds, PFOS and PFNA both showed significant negative correlations between their concentrations and CYP2C45 and/or CYP2J25 transcript levels. It is well known that PFCs can weakly but significantly activate peroxisome proliferator-activated receptor α (PPARα) in mammals (Intrasuksri et al., 1998; Ishibashi et al., 2008; Maloney et al., 1999; Shipley et al., 2004; Takacs et al., 2007; Vanden Heuvel et al., 2006). Several lines of evidence show that treatment of rats with some peroxisome proliferators results in down-regulation of CYP2C genes (Corton et al., 1998; Fan et al., 2004). More direct evidence shows that PPARα is essential in a ligand dependent CYP2C down-regulation (Pozzi et al., 2007; Ripp et al., 2003). Given that PFCs have potential to activate PPARα in cormorants, as in mammals, it is likely that negative relationships of PFC concentrations and CYP2C/2J expression levels observed in this study could involve the PPARα signaling pathway, but further studies focusing on this pathway in diapsids are required.
5. Conclusions
The genes in CYP2C, CYP2J, CYP2AC, and CYP2AF subfamilies cloned from the common cormorant, as well as other CYP2 subfamilies identified in the genomes of the chicken, zebra finch, and anole lizard, show that the CYP2 family in diapsids is highly diverse, as in mammals. Maximum likelihood phylogenetic analyses together with synteny analyses presented here provide new insights into evolutionary relationships among the vertebrate CYP2B/2C/2H subfamilies. Notably, synteny data highlight the necessity to rename certain existing CYP2 genes in vertebrates; CYP2Hs in birds, CYP2C62P in humans, and Cyp2c44 in mice should be renamed as CYP2C23. The present study suggested that PFCs may induce the transcriptional down-regulation of CYP2C/2J genes in the cormorant liver. More information on the regulation and catalytic function of the CYP2 genes will aid in understanding their roles in physiological and toxicological processes.
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research (S) (21221004) to H.I. and (S) (20221003) to S.T., and Grant-in-Aid for JSPS Fellows to A.K. from Japan Society for the Promotion of Science. This work was also supported by NIH grants R01ES015912 and P42ES007381 (Superfund Basic Research Program at Boston University) to J.J.S., and by National Research Foundation of Korea (NRF) to E-Y.K. (2010-0012150). Financial assistance was also provided by 21st Century COE Program and Global COE Program to H.I. and S.T. from the Ministry of Education, Culture, Sports, Science and Technology, and by Basic Research in ExTEND2005 (Enhanced Tack on Endocrine Disruption) to H.I. from the Ministry of the Environment of Japan. The sponsors had no involvement in performing or in the decision to publish this study. The Japanese and U.S. Governments are authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson GD. Gender differences in pharmacological response. Int. Rev. Neurobiol. 2008;83:1–10. doi: 10.1016/S0074-7742(08)00001-9. [DOI] [PubMed] [Google Scholar]
- Baader M, Gnerre C, Stegeman JJ, Meyer UA. Transcriptional activation of cytochrome P450 CYP2C45 by drugs is mediated by the chicken xenobiotic receptor (CXR) interacting with a phenobarbital response enhancer unit. J. Biol. Chem. 2002;277:15647–15653. doi: 10.1074/jbc.M109882200. [DOI] [PubMed] [Google Scholar]
- Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc. Natl. Acad. Sci. USA. 2004;101:7711–7715. doi: 10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SS, Helvig C, Taimi M, Ramshaw HA, Collop AH, Amad M, White JA, Petkovich M, Jones G, Korczak B. CYP2U1, a novel human thymus- and brain-specific cytochrome P450, catalyzes omega- and (omega-1)-hydroxylation of fatty acids. J. Biol. Chem. 2004;279:6305–6314. doi: 10.1074/jbc.M311830200. [DOI] [PubMed] [Google Scholar]
- Connor K, Safe S, Jefcoate CR, Larsen M. Structure-dependent induction of CYP2B by polychlorinated biphenyl congeners in female Sprague-Dawley rats. Biochem. Pharmacol. 1995;50:1913–1920. doi: 10.1016/0006-2952(95)02087-x. [DOI] [PubMed] [Google Scholar]
- Corton JC, Fan LQ, Brown S, Anderson SP, Bocos C, Cattley RC, Mode A, Gustafsson JA. Down-regulation of cytochrome P450 2C family members and positive acute-phase response gene expression by peroxisome proliferator chemicals. Mol. Pharmacol. 1998;54:463–473. doi: 10.1124/mol.54.3.463. [DOI] [PubMed] [Google Scholar]
- Craft ES, DeVito MJ, Crofton KM. Comparative responsiveness of hypothyroxinemia and hepatic enzyme induction in Long-Evans rats versus C57BL/6J mice exposed to TCDD-like and phenobarbital-like polychlorinated biphenyl congeners. Toxicol. Sci. 2002;68:372–380. doi: 10.1093/toxsci/68.2.372. [DOI] [PubMed] [Google Scholar]
- Czerniak R. Gender-based differences in pharmacokinetics in laboratory animal models. Int. J. Toxicol. 2001;20:161–163. doi: 10.1080/109158101317097746. [DOI] [PubMed] [Google Scholar]
- Davidson BP, Dogra SC, May BK. A duplicated HNF-3 binding site in the CYP2H2 promoter underlies the weak phenobarbital induction response. Int. J. Biochem. Cell Biol. 2001;33:1080–1093. doi: 10.1016/s1357-2725(01)00076-0. [DOI] [PubMed] [Google Scholar]
- DeLozier TC, Tsao CC, Coulter SJ, Foley J, Bradbury JA, Zeldin DC, Goldstein JA. CYP2C44, a new murine CYP2C that metabolizes arachidonic acid to unique stereospecific products. J. Pharmacol. Exp. Ther. 2004;310:845–854. doi: 10.1124/jpet.104.067819. [DOI] [PubMed] [Google Scholar]
- Dogra SC, Davidson BP, May BK. Analysis of a phenobarbital-responsive enhancer sequence located in the 5′ flanking region of the chicken CYP2H1 gene: identification and characterization of functional protein-binding sites. Mol. Pharmacol. 1999;55:14–22. doi: 10.1124/mol.55.1.14. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LQ, You L, Brown-Borg H, Brown S, Edwards RJ, Corton JC. Regulation of phase I and phase II steroid metabolism enzymes by PPAR alpha activators. Toxicology. 2004;204:109–121. doi: 10.1016/j.tox.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. Evolution of insect P450. Biochem. Soc. Trans. 2006;34:1252–1255. doi: 10.1042/BST0341252. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Nishimura M, Ogino D, Chiba R, Ikai I, Ueda N, Naito S, Kuribayashi S, Moustafa MA, Uchida T, Sawada H, Kamataki T, Funae Y, Fukumoto M. Cytochrome p450 gene expression levels in peripheral blood mononuclear cells in comparison with the liver. Cancer Sci. 2004;95:520–529. doi: 10.1111/j.1349-7006.2004.tb03243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone HM, Stegeman JJ. A revised evolutionary history of the CYP1A subfamily: gene duplication, gene conversion, and positive selection. J. Mol. Evol. 2006;62:708–717. doi: 10.1007/s00239-005-0134-z. [DOI] [PubMed] [Google Scholar]
- Goldstone JV, Hamdoun A, Cole BJ, Howard-Ashby M, Nebert DW, Scally M, Dean M, Epel D, Hahn ME, Stegeman JJ. The chemical defensome: environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev. Biol. 2006;300:366–384. doi: 10.1016/j.ydbio.2006.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone JV, McArthur AG, Kubota A, Zanette J, Parente T, Jonsson ME, Nelson DR, Stegeman JJ. Identification and developmental expression of the full complement of cytochrome P450 genes in zebrafish. BMC Genomics. 2010;11:643. doi: 10.1186/1471-2164-11-643. (published 18 November 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CN, Hansen AJ, May BK. Transcriptional regulation of the chicken CYP2H1 gene. Localization of a phenobarbital-responsive enhancer domain. J. Biol. Chem. 1991;266:17031–17039. [PubMed] [Google Scholar]
- Handschin C, Meyer UA. Induction of drug metabolism: the role of nuclear receptors. Pharmacol. Rev. 2003;55:649–673. doi: 10.1124/pr.55.4.2. [DOI] [PubMed] [Google Scholar]
- Hansen AJ, May BK, Elferink LA. Sequence of a chicken phenobarbital-inducible cytochrome P450 cDNA: regulation of two P450 mRNAs transcribed from different genes. DNA. 1989;8:179–191. doi: 10.1089/dna.1.1989.8.179. [DOI] [PubMed] [Google Scholar]
- Intrasuksri U, Rangwala SM, O’Brien M, Noonan DJ, Feller DR. Mechanisms of peroxisome proliferation by perfluorooctanoic acid and endogenous fatty acids. Gen. Pharmacol. 1998;31:187–197. doi: 10.1016/s0306-3623(98)00029-9. [DOI] [PubMed] [Google Scholar]
- Ioannides C, Lewis DF. Cytochromes P450 in the bioactivation of chemicals. Curr. Top. Med. Chem. 2004;4:1767–1788. doi: 10.2174/1568026043387188. [DOI] [PubMed] [Google Scholar]
- Ishibashi H, Iwata H, Kim EY, Tao L, Kannan K, Tanabe S, Batoev VB, Petrov EA. Contamination and effects of perfluorochemicals in Baikal seal (Pusa sibirica). 2. Molecular characterization, expression level, and transcriptional activation of peroxisome proliferator-activated receptor alpha. Environ. Sci. Technol. 2008;42:2302–2308. doi: 10.1021/es0720558. [DOI] [PubMed] [Google Scholar]
- Karlgren M, Miura S, Ingelman-Sundberg M. Novel extrahepatic cytochrome P450s. Toxicol. Appl. Pharmacol. 2005;207:57–61. doi: 10.1016/j.taap.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Kawai YK, Ikenaka Y, Fujita S, Ishizuka M. The CYP1D subfamily of genes in mammals and other vertebrates. Mamm. Genome. 2010;21:320–329. doi: 10.1007/s00335-010-9263-9. [DOI] [PubMed] [Google Scholar]
- Kretschmer XC, Baldwin WS. CAR and PXR: xenosensors of endocrine disrupters? Chem. Biol. Interact. 2005;155:111–128. doi: 10.1016/j.cbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Kubota A, Iwata H, Tanabe S, Yoneda K, Tobata S. Levels and toxicokinetic behaviors of PCDD, PCDF, and coplanar PCB congeners in common cormorants from Lake Biwa, Japan. Environ. Sci. Technol. 2004;38:3853–3859. doi: 10.1021/es0494858. [DOI] [PubMed] [Google Scholar]
- Kubota A, Iwata H, Tanabe S, Yoneda K, Tobata S. Hepatic CYP1A induction by dioxin-like compounds, and congener-specific metabolism and sequestration in wild common cormorants from Lake Biwa. Japan. Environ. Sci. Technol. 2005;39:3611–3619. doi: 10.1021/es048771g. [DOI] [PubMed] [Google Scholar]
- Kubota A, Iwata H, Goldstone HM, Kim EY, Stegeman JJ, Tanabe S. Cytochrome P450 1A4 and 1A5 in common cormorant (Phalacrocorax carbo): evolutionary relationships and functional implications associated with dioxin and related compounds. Toxicol. Sci. 2006;92:394–408. doi: 10.1093/toxsci/kfl001. [DOI] [PubMed] [Google Scholar]
- Maloney EK, Waxman DJ. trans-Activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol. Appl. Pharmacol. 1999;161:209–218. doi: 10.1006/taap.1999.8809. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, Lambert MH, Moore JT. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol. 2002;16:977–986. doi: 10.1210/mend.16.5.0828. [DOI] [PubMed] [Google Scholar]
- Nakai K, Ward AM, Gannon M, Rifkind AB. Beta-naphthoflavone induction of a cytochrome P-450 arachidonic acid epoxygenase in chick embryo liver distinct from the aryl hydrocarbon hydroxylase and from phenobarbital-induced arachidonate epoxygenase. J. Biol. Chem. 1992;267:19503–19512. [PubMed] [Google Scholar]
- Nakayama K, Iwata H, Kim EY, Tashiro K, Tanabe S. Gene expression profiling in common cormorant liver with an oligo array: assessing the potential toxic effects of environmental contaminants. Environ. Sci. Technol. 2006;40:1076–1083. doi: 10.1021/es051386m. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Iwata H, Tao L, Kannan K, Imoto M, Kim EY, Tashiro K, Tanabe S. Potential effects of perfluorinated compounds in common cormorants from Lake Biwa, Japan: an implication from the hepatic gene expression profiles by microarray. Environ. Toxicol. Chem. 2008;27:2378–2386. doi: 10.1897/07-614.1. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Metazoan cytochrome P450 evolution. Comp. Biochem. Physiol. C. 1998;121:15–22. doi: 10.1016/s0742-8413(98)10027-0. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Nelson DR. The cytochrome p450 homepage. Hum. Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nims RW, Lubet RA. Induction of cytochrome P-450 in the Norway rat, Rattus norvegicus, following exposure to potential environmental contaminants. J. Toxicol. Environ. Health. 1995;46:271–292. doi: 10.1080/15287399509532035. [DOI] [PubMed] [Google Scholar]
- Nyman M, Raunio H, Taavitsainen P, Pelkonen O. Characterization of xenobiotic-metabolizing cytochrome P450 (CYP) forms in ringed and grey seals from the Baltic Sea and reference sites. Comp. Biochem. Physiol. C. 2001;128:99–112. doi: 10.1016/s1532-0456(00)00177-0. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Wu S, Parker C, Qu W, Cox R, Zeldin DC, Stegeman JJ. Identification and regulation of a new vertebrate cytochrome P450 subfamily, the CYP2Ps, and functional characterization of CYP2P3, a conserved arachidonic acid epoxygenase/19-hydroxylase. Arch. Biochem. Biophys. 2003;411:223–234. doi: 10.1016/s0003-9861(02)00734-8. [DOI] [PubMed] [Google Scholar]
- Pozzi A, Ibanez MR, Gatica AE, Yang S, Wei S, Mei S, Falck JR, Capdevila JH. Peroxisomal proliferator-activated receptor-alpha-dependent inhibition of endothelial cell proliferation and tumorigenesis. J. Biol. Chem. 2007;282:17685–17695. doi: 10.1074/jbc.M701429200. [DOI] [PubMed] [Google Scholar]
- Ripp SL, Falkner KC, Pendleton ML, Tamasi V, Prough RA. Regulation of CYP2C11 by dehydroepiandrosterone and peroxisome proliferators: identification of the negative regulatory region of the gene. Mol. Pharmacol. 2003;64:113–122. doi: 10.1124/mol.64.1.113. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sakai H, Iwata H, Kim EY, Tsydenova O, Miyazaki N, Petrov EA, Batoev VB, Tanabe S. Constitutive androstane receptor (CAR) as a potential sensing biomarker of persistent organic pollutants (POPs) in aquatic mammal: molecular characterization, expression level, and ligand profiling in Baikal seal (Pusa sibirica) Toxicol. Sci. 2006;94:57–70. doi: 10.1093/toxsci/kfl088. [DOI] [PubMed] [Google Scholar]
- Sakai H, Kim EY, Petrov EA, Tanabe S, Iwata H. Transactivation potencies of Baikal seal constitutive active/androstane receptor by persistent organic pollutants and brominated flame retardants. Environ. Sci. Technol. 2009;43:6391–6397. doi: 10.1021/es901120r. [DOI] [PubMed] [Google Scholar]
- Shipley JM, Hurst CH, Tanaka SS, DeRoos FL, Butenhoff JL, Seacat AM, Waxman DJ. trans-Activation of PPARalpha and induction of PPARalpha target genes by perfluorooctane-based chemicals. Toxicol. Sci. 2004;80:151–160. doi: 10.1093/toxsci/kfh130. [DOI] [PubMed] [Google Scholar]
- Sinclair JF, Wood S, Lambrecht L, Gorman N, Mende-Mueller L, Smith L, Hunt J, Sinclair P. Isolation of four forms of acetone-induced cytochrome P-450 in chicken liver by h.p.l.c. and their enzymic characterization. Biochem. J. 1990;269:85–91. doi: 10.1042/bj2690085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Takacs ML, Abbott BD. Activation of mouse and human peroxisome proliferator-activated receptors (alpha, beta/delta, gamma) by perfluorooctanoic acid and perfluorooctane sulfonate. Toxicol. Sci. 2007;95:108–117. doi: 10.1093/toxsci/kfl135. [DOI] [PubMed] [Google Scholar]
- Tanabe S. Contamination and toxic effects of persistent endocrine disrupters in marine mammals and birds. Mar. Pollut. Bull. 2002;45:69–77. doi: 10.1016/s0025-326x(02)00175-3. [DOI] [PubMed] [Google Scholar]
- Vanden Heuvel JP, Thompson JT, Frame SR, Gillies PJ. Differential activation of nuclear receptors by perfluorinated fatty acid analogs and natural fatty acids: a comparison of human, mouse, and rat peroxisome proliferator-activated receptor-alpha, -beta, and -gamma, liver X receptor-beta, and retinoid X receptor-alpha. Toxicol. Sci. 2006;92:476–489. doi: 10.1093/toxsci/kfl014. [DOI] [PubMed] [Google Scholar]
- Waxman DJ. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch. Biochem. Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
- Wolkers J, Witkamp RF, Nijmeijer SM, Burkow IC, de Groene EM, Lydersen C, Dahle S, Monshouwer M. Phase I and phase II enzyme activities in Ringed seals (Phoca hispida): characterization of hepatic cytochrome P450 by activity patterns, inhibition studies, mRNA analyses, and western blotting. Aquat. Toxicol. 1998;44:103–115. [Google Scholar]
- Wolkers J, Burkow IC, Monshouwer M, Lydersen C, Dahle S, Witkamp RF. Cytochrome P450-mediated enzyme activities and polychlorinated biphenyl accumulation in harp seal (Phoca groenlandica) Mar. Environ. Res. 1999;48:59–72. [Google Scholar]
- Wyde ME, Bartolucci E, Ueda A, Zhang H, Yan B, Negishi M, You L. The environmental pollutant 1,1-dichloro-2,2-bis (p-chlorophenyl)ethylene induces rat hepatic cytochrome P450 2B and 3A expression through the constitutive androstane receptor and pregnane X receptor. Mol. Pharmacol. 2003;64:474–481. doi: 10.1124/mol.64.2.474. [DOI] [PubMed] [Google Scholar]