Abstract
Bronchopulmonary dysplasia (BPD) is one of the most common causes of mortality and morbidity in neonatal intensive care units. Persistent inflammation, with an abnormal influx of polymorphonuclear leukocytes (PMNs) followed by monocytes (MONOs), occurs early in the pathogenesis of BPD. Anti-inflammatory therapy with better efficacy and safety than dexamethasone (DEX) is needed. In the present study we determined cell-specific gene expression and cytokine release in response to glucocorticoids versus interleukin-10 (IL-10). Subsequently, we hypothesized that the insensitivity of MONOs to DEX was associated with a failure of the glucocorticoid receptor to translocate to the nucleus. PMNs and MONOs were isolated from umbilical cord blood at birth, and pretreated with PBS vehicle, IL-10 or glucocorticoids prior to endotoxin (LPS)-stimulation for 4 and 18h. Genome-wide gene expressions were determined by microarray and validated by RT-qPCR. Interleukin 8 release in cell culture supernatant was measured by ELISA. To examine the mechanism of monocyte insensitivity to glucocorticoids, nuclear translocation of the glucocorticoid receptor was determined by Western blots. MONOs had 6 times the number of genes changing expression with IL-10 compared to PMNs at 4h. DEX at the therapeutic level for neonates with BPD had no effect on gene expression in MONOs. The order of potency for inhibition of interleukin-8 release from MONOs was IL-10 >betamethasone >dexamethasone and hydrocortisone. Glucocorticoid potency in MONOs was directly related to glucocorticoid receptor translocation to nucleus. Gene expression profiling for IL-10 versus glucocorticoids indicates there may be major differences in therapeutic efficacy for BPD.
Keywords: Bronchopulmonary dysplasia, monocytes, polymorphonuclear leukocytes, gene expression microarray, dexamethasone
Introduction
Bronchopulmonary dysplasia (BPD) is one of the most serious and common disorders which evolves primarily in very low birth weight infants, as well as near term and term infants after treatment for acute hypoxemic respiratory failure [1-3]. While the pathogenesis of this chronic lung disorder involves many factors, inflammation plays a pivotal role and a common pathway leading abnormal lung growth [4]. Neonates who develop BPD have prolonged hospital stays requiring supplemental oxygen and ventilator support. Newborns with BPD are also at increased risk for death and survivors are at increased risk for abnormal pulmonary function and neurologic abnormalities during childhood development [2]. Today, newborns developing bronchopulmonary dysplasia (BPD) are being treated with glucocorticoids by most neonatologists to aid in discontinuance of mechanical ventilation [5]. Dexamethasone (DEX) is the anti-inflammatory therapy for BPD that has been most thoroughly studied and most commonly used so far [1]. However, DEX has only partial efficacy and possible, serious acute and chronic adverse effects [5]. New anti-inflammatory therapy is needed to treat BPD.
Early in the evolution of BPD, abnormal numbers of polymorphonuclear leukocytes (PMNs) and monocytes (MONOs) are recruited sequentially into the airspace of the lung [6-8]. Associated with the influx of these cells is the release of pro-inflammatory and anti-inflammatory cytokines in airway fluid. It is theorized that there is inadequate, innate anti-inflammatory response [9-11]. Several studies have demonstrated absent or extremely low levels of the potent anti-inflammatory cytokine, interleukin-10 (IL-10), in the airway fluid of preterm as well as term infants at high risk for BPD [11-14]. On the other hand exogenous IL-10 has been shown to be equipotent, on a molar basis to DEX, in relation to the inhibition of pro-inflammatory cytokine release from PMNs [15].
Considering that exogenous IL-10 therapy may have potential benefit for the effective and safe treatment of BPD [7,16,17], the present study compared IL-10 versus DEX, in endotoxin-stimulated PMNs and MONOs of the human newborn at the level of genome-wide gene expression. The overall hypothesis was that there would be cell-specific gene expression profiles, in response to exogenous IL-10 versus DEX treatment that would lead to a better understanding of the anti-inflammatory efficacy of these agents. Subsequently, it was hypothesized that the mechanism for the relative insensitivity of the MONO to DEX on gene expression profiling was related to a diminished translocation of the DEX and cytoplasmic glucocorticoid receptor complex from the cytoplasm to the nucleus. As a corollary, we hypothesized that there could be a difference in the insensitivity of MONOs to other glucocorticoids used in the perinatal period such as betamethasone (BETA) and hydrocortisone (HC), on pro-inflammatory cytokine release. The results of this translational study have implications for novel therapy for BPD and possibly other inflammatory diseases of the newborn.
Materials and methods
Subjects and sample collection
Cord blood (approximately 60 ml) was obtained from placentas immediately after elective, term, cesarean section deliveries. One of the investigators attended the delivery to exclude placentas associated with labor, rupture of membranes, clinical chorioamnionitis, antenatal steroids, maternal disorders or maternal medications for underlying diseases. Blood was collected in heparinized preservative-free tubes for immediate transport to the laboratory, followed by immediate cell isolation procedures [15-17]. A total of 15 cord blood samples were used. The study was approved by the Internal Review Board of the North Shore-Long Island Jewish Health System.
Cell isolation
PMNs and MONOs were isolated from cord blood under pyrogen-free conditions at room temperature, as described in previous work [15-17]. PMN isolation involved density centrifugation and red cell lysis with hypotonic saline. PMN purity was >95%, determined by differential staining and viability was >95%, determined by trypan blue exclusion method. MONO isolation used density centrifugation of peripheral blood monocytic cells (lymphocytes and monocytes) followed by magnetic bead negative selection. MONO purity was > 90% as determined by flow cytometry for CD14+ cells and viability was >95% determined as above.
RNA isolation, amplification and labeling
PMNs and MONOs were resuspended in RPMI 1640+10% FCS at 5x106 cells/mL. Cells were pre-incubated with PBS (Gibco-Invitrogen) (vehicle control) or equimolar concentrations (10-8 M) of recombinant human IL-10 (R&D Systems, Minneapolis, MN., USA) or DEX (Abraxis Pharmaceutical Products, Schaumburg, IL, USA) for 1 h at 37°C and 5% CO2, then stimulated with LPS (10 ng/mL) (Sigma-Aldrich Corp. St. Louis, MO, USA) for 4 h. For all experiments in this study, the LPS dose (10ng/ml) was chosen as a submaximal and clinically relevant level based on endotoxin levels measured in amniotic fluid [18]. The 10-8 M concentrations of DEX and IL-10 were chosen based on the plasma levels of DEX that were detected in newborns being treated for BPD [19,20]. Cells were harvested and resuspended in RNAlater (Invitrogen, Grand Island, NY, USA) to preserve RNA. Total RNA was isolated using Qiagen RNeasy mini kit (Qiagen,Valencia, CA, USA) according to manufacturer’s protocol. Total RNA was amplified and labeled (cRNA) using Ambion MessageAmpTM II-Biotin enhanced kit (Invitrogen) according to the manufacturer’s protocol. Both total RNA and cRNA concentration and quality were determined using NanoDrop-1000 (focusing on 260/280 and 260/230 ratios) (Thermo scientific, Wilmington, DE, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Microarray hybridization and scanning
20μg fragmented, biotin-labeled cRNA (n=5) was hybridized to Gene Chip Human U133 plus 2.0 microarray platforms. (Affymetrix, Santa Clara, CA, USA) for 18hrs at 45°C as described in the Affimetrix genechip expression technical manual. After hybridization, washing and staining of the chips were processed by a Gene Chip 450 fluidics station (Affymetrix), and scanned by a Gene Chip 3000 scanner (Affymetrix).
Microarray data analysis
Raw data (n=5 cord blood samples, 30 microarrays) was uploaded onto GeneSifter (Geospiza, Inc. Seattle, WA, USA), log transformed and normalized using GC-RMA. One-way ANOVA was performed on PMNs and MONOs separately, comparing LPS alone to LPS+DEX versus LPS+IL-10. A Benjamin & Hochberg correction for multiple analyses, and an overall p (significance) value of <0.05 indicated at least a 2 fold change in gene expression. In separate analyses we found significantly changing genes for LPS versus LPS + IL-10 and LPS versus LPS + DEX (p<0.05 and fold change ≥ 2) that could be used to determine significant KEGG (Kyoto Encyclopedia of Genes and Genomes) gene pathways (significance= a Z score of -2> Z >2). Gene expression data was submitted to public GEO datasets (GSE35683).
Quantitative reverse transcriptase PCR (RT-qPCR)
In order to validate microarray results, 5 genes with either increased, decreased or no change in expression were chosen for RT-qPCR (IL-6, IL-1 family member 9 (IL-1F9), hepcidin antimicrobial peptide, CCAAT/enhancer binding protein delta and Krupple-like factor 9. Primers and hybridization probes were designed by Roche Universal Probe Library Assay Design Center (Roche, Mannheim, Germany). Gene expression (n=5, same subjects as microarray experiments) were amplified by an Applied Biosystems 7900 HT thermocycler and results were analyzed using the delta-delta crossover threshold method.
Monocyte cytokine release
1x106 MONOs from 5 cord blood samples were plated in each well of 6 well cell culture plates and stimulated with LPS (10 ng/mL) for 18 h after a 1 hr pre-incubation with or without serial doses of dexamethasone sodium phosphate (DEX), betamethasone sodium phosphate and acetate (BETA) (Celestone® Soluspan®, Schering Corporation, Kenilworth, NJ, USA) or hydrocortisone sodium succinate (HC) (Pharmacia & Upjohn, Kalamazoo, MI, USA). PBS was used as the dilutant vehicle for each glucocorticoid and IL-10. Cell culture supernatants were collected after 18 h. In separate experiments (N=5) 1x106 MONOs and 1x106 PMNs were stimulated with LPS (10 ng/mL) for 4 and 18 h after a 1 h pre-incubation with equimolar concentrations (10-8 M) of DEX, BETA or HC. Cell culture supernatants were collected after 4 and 18 h. Interleukin-8 release, (as a representative of other known pro-inflammatory cytokines released under the same experimental conditions) was measured by the Human CXCL8/IL-8 Quantikine ELISA Kit (R&D systems, Minneapolis, MN, USA) [17].
Western blotting for the glucocorticoid receptor
MONOs were stimulated with LPS (10 ng/mL) for 1 hr after a 1 hr pre-incubation with serial doses of DEX, BETA or HC, and IL-10 at specifically at 10-8 M. PBS was used as a control baseline. Nuclear and cytoplasmic extracts were prepared from 1x106 cells as described previously [17]. Cytoplasmic extracts and washed nuclear pellets were boiled with sample buffer (SDS reducing buffer; Bio-Rad Laboratories, Hercules, CA, USA) to denature proteins. Denatured proteins were separated on 10% denaturing polyacrylamide gel and transferred onto a nitrocellulose membrane (Hybond-ECL; Amersham-GE Healthcare, Piscataway, NJ, USA). For the detection of the glucocorticoid receptor, membranes were blocked overnight with 5% (w/v) bovine serum albumin (Sigma-Aldrich Corp. St. Louis, MO, USA) in 10 mM Tris-Cl (pH 7.5), 140 mM NaCl, 1.5 mM MgCl 2 and 0.1% Tween 20 solution (TBST-BSA) before incubating with primary antibody against glucocorticoid receptor (sc-1003 Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 2 h at room temperature, diluted 1: 200 in TBST-BSA. After washing with TBST, membranes were incubated for 1 h with horseradish peroxidase-labeled anti-rabbit secondary antibody (Amersham, Arlington Heights, IL, USA), diluted 1: 20,000 in TBST-BSA. For blocking and incubations with antibodies, TBST-BSA was supplemented with 0.01% (v/v) of each phosphatase inhibitor cocktails A and B (Santa Cruz biotechnology, Santa Cruz, CA, USA). Labeled proteins were detected using enhanced chemiluminescence reagents as described by the manufacturer (ECL PLUS, Amersham-GE Healthcare, Piscataway, NJ, USA). PMNs were used as positive control for ensuring the inhibitory activity of DEX. IL-10 at 10-8 M (AI cytokine) was used as a positive control in MONOs to ensure that cytokine inhibition could occur. Western blot data and ELISA data were analyzed by a one way ANOVA with a Bonferroni correction (overall significance P<0.05).
Results
Genome-wide gene profiling in PMNs and MONOs
Figure 1 shows a microarray heat map for LPS-stimulated PMNs and MONOs exposed to LPS for 4 h, pretreated for 1h with IL-10 or DEX at 10-8 M. Green and red colors indicated significant down regulation and up regulation of gene expression respectively, with at least a 2 fold change from LPS alone, IL-10+LPS and DEX+LPS treated cells. The PMN panel exhibited 4 major clusters of gene expression, grouped along the vertical axis, manifesting significant fold changes. The pattern was striking for up regulation of most genes with little overlap between the two treatments. A very small minority of genes were down regulated by either treatment. The MONO panel showed no visually appreciable changes in gene expression in response to DEX. Yet IL-10 resulted in one large cluster of up regulated genes and slightly larger cluster of down regulated genes.
Figure 1.
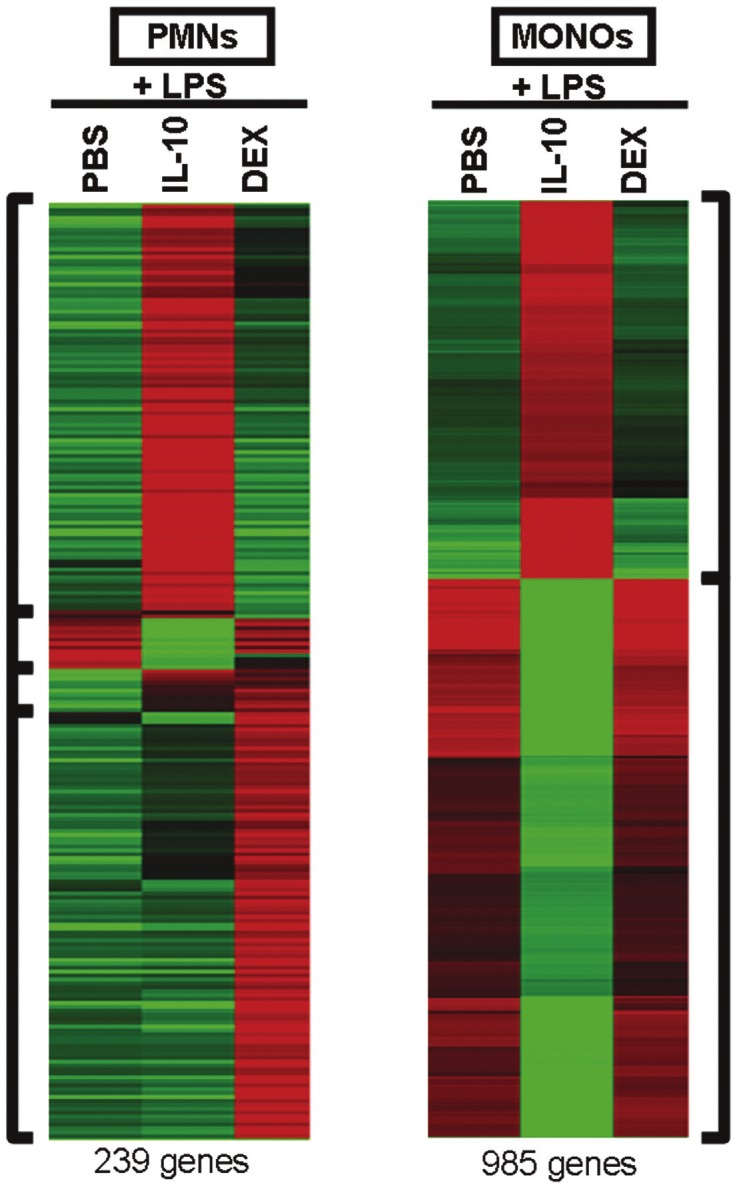
Microarray gene expression heat maps for polymorphonuclear leukocytes (PMNs) and monocytes (MONOs) of the newly born (n=5), pretreated with 10-8 M interleukin-10 (IL-10) or dexamethasone (DEX) for 1 h and then exposed to endotoxin (LPS) for 4 h. Upregulation (green to red color) and downregulation (red to green color) of gene expression changed by IL-10 or DEX were compared to LPS-stimulated cells in phosphate buffer saline (PBS) vehicle alone. For PMNs and MONOs, several clusters of genes (bracketed) changed expression a similar fashion. Only genes were displayed in which a significant changes in gene expression (≥ 2 fold change, adjusted p value < 0.05) were detected.
Differences and similarities in gene expression between PMNs and MONOs, in response to glucocorticoids versus IL-10 is shown in Table 1. This table quantifies the number of individual genes changing expression, by at least 2 fold and statistically significant, for LPS-stimulated PMNs and MONOs separately as well as genes changing in common between treatments and cell type. The major findings were: 1) no changes in MONO gene expression by DEX at 10-8M, 2) MONOs exposed to IL-10 had more than 5 times the number of genes changing compared to PMNs, and 3) only a small number of genes were changing when looking for those in common between cell types or treatments.
Table 1.
Number of genes changing expression caused by interleukin 10 (IL-10) or dexamethasone (DEX) at 4 h in endotoxin-stimulated leukocytes of the newly born
| Cell Type / Treatments | IL10 (10-8 M) | DEX (10-8 M) | Common between treatments |
|---|---|---|---|
| Polymorphonuclear leukocytes | 193 | 202 | 18 |
| Monocytes | 1186 | 0 | 0 |
| Common between cell types | 61 | 0 | -- |
Gene expression was > 2 fold change (up-regulated or down-regulated) using an adjusted p value < 0.05 (N=5) by microarray, compared to endotoxin alone.
Table 2 demonstrates the parallel changes in gene expression between microarray data and RT-qPCR techniques, supporting the validation of microarray results as a screening tool. Five genes from both PMNs and MONOs were selected for RT-qPCR to demonstrate either upregulation, downregulation, or no change in gene expression. A greater than 2 fold change in gene expression was considered significant. In MONOs we observed no effect of DEX on gene expression by microarray or RT-qPCR. In PMNs results by microarray and PCR were similar except for the effect of DEX on IL-6. For PMNs, DEX did not result in a down regulation of IL-6 based on the microarray assay however there was a relatively small (2.8 fold) but significant decrease based on RT-qPCR.
Table 2.
Microarray gene expression data validation by RT-qPCR in endotoxin-stimulated leukocytes of the new born caused by interleukin 10 (IL-10) or dexamethasone (DEX) compared to endotoxin stimulation alone
| GENE DESCRIPTION | Polymorphonuclear leukocytes | Monocytes | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| ARRAY | RT-qPCR | ARRAY | RT-qPCR | |||||
|
| ||||||||
| IL-10 | DEX | IL-10 | DEX | IL-10 | DEX | IL-10 | DEX | |
| Interleukin-6 | ↓24.66 | -- | ↓17.91 | ↓2.77 | ↓8.89 | -- | ↓14.43 | -- |
| Interleukin-1 family member 9 (IL-1F9) | ↓6.20 | ↓4.44 | ↓4.11 | ↓2.30 | ↓26.79 | -- | ↓18.87 | -- |
| Hepcidin anti-microbial peptide | ↑46.92 | -- | ↑178.5 | -- | ↑2.93 | -- | ↑3.60 | -- |
| CCAAT/enhancer binding protein, delta | ↑2.19 | ↑3.29 | ↑2.57 | ↑5.03 | ↑3.11 | -- | ↑3.22 | -- |
| Krupple-like factor 9 | -- | ↑3.69 | -- | ↑4.58 | -- | -- | -- | -- |
↑ = Up regulation; ↓ = Down regulation of gene; -- represents a non-significant change. Value = fold changes from gene expression in endotoxin-stimulated leukocytes; all values are statistically significant, p<0.05.
Genes changing expression (Table 1) were further classified into pathways using the KEGG system. The cytokine-cytokine receptor pathway was chosen for our primary aim because it reflected the greatest number of genes changing significantly for both PMNs and MONOs as shown in Table 3 (Panels A, B, and C). Panel A shows genes that changed expression caused by IL-10 that were unique to PMNs; 3 were up regulated and one was down regulated. Panel B shows genes that changed expression in response to IL-10 which were unique to MONOs; 26 genes were down regulated and 4 genes up regulated. Panel C shows genes changing expression in response to IL-10 which were common to both cell types. One gene TNF ligand superfamily, member 10 was up regulated in PMNs and down regulated in MONOs. IL-6 had a marked down regulation in both cell types in response to IL-10.
Table 3.
Genes changing expression from the cytokine-cytokine receptor pathway a by microarray analysis in response to interleukin 10 (IL-10) at 4 hrs in endotoxin-stimulated monocytes (MONOs) and polymorphonuclear cells (PMNs) of the newly born
| A. Gene Expression Changes Unique to Polymorphonuclear Leukocytes | ||
|
| ||
| Gene Name | Fold Change | |
|
| ||
| Up Regulated | ||
| TNF receptor superfamily, member 10d | 5.5 | |
| Inhibin, beta B | 5.0 | |
| Interleukin 18 receptor accessory protein | 3.6 | |
|
| ||
| Down Regulated | ||
| Tumor necrosis factor (TNF superfamily, member 2) | 2.1 | |
|
| ||
| B. Gene Expression Changes Unique to Monocytes | ||
|
| ||
| Gene Name | Fold Change | |
|
| ||
| Up Regulated | ||
| Platelet-derived growth factor alpha | 5.3 | |
| interleukin 1 receptor, type II | 4.7 | |
| Colony stimulating factor 1 receptor | 3.7 | |
| Interleukin 21 receptor | 3.4 | |
|
| ||
| Down Regulated | ||
| Interleukin 12B | 33.8 | |
| Chemokine (C-X-C motif) ligand 10 | 17.4 | |
| Chemokine (C-C motif) ligand 7 | 17.3 | |
| Chemokine (C-X-C motif) ligand 11 | 13.8 | |
| Interleukin 19 | 11.6 | |
| Chemokine (C-X-C motif) ligand 5 | 7.5 | |
| Chemokine (C-X-C motif) ligand 6 | 7.4 | |
| Chemokine (C-C motif) ligand 20 | 6.7 | |
| Interleukin 15 receptor, alpha | 6.4 | |
| Chemokine (C-X-C motif) ligand 2 | 5.7 | |
| Cytokine receptor-like factor 2 | 5.1 | |
| TNF (ligand) superfamily, member 9 | 4.4 | |
| Chemokine (C-X-C motif) ligand 1 | 4.4 | |
| TNF (ligand) superfamily, member 13b | 4.3 | |
| Met proto-oncogene (MET) | 4.0 | |
| Interleukin 1 receptor, type I | 3.9 | |
| Interleukin 15 | 3.0 | |
| Chemokine (C-C motif) receptor 7 | 2.8 | |
| Chemokine (C-C motif) ligand 22 | 2.8 | |
| Chemokine (C-C motif) ligand 5 | 2.6 | |
| TNF receptor superfamily, member 18 | 2.6 | |
| TNF receptor superfamily, member 9 | 2.6 | |
| Chemokine (C-X-C motif) receptor 7 | 2.6 | |
| Fas (TNF receptor family, member 6) | 2.3 | |
| Chemokine (C-C motif) ligand 23 | 2.2 | |
| Chemokine (C-C motif) ligand 2 | 2.2 | |
|
| ||
| C. Genes Common between Both Cell Types | ||
|
| ||
| Gene Name | Fold Change | |
|
| ||
| Up Regulated | PMNs | MONOs |
| Chemokine (C-X3-C motif) receptor 1 | 3.1 | 11.0 |
| TNF receptor superfamily, member 10c | 3.4 | 3.2 |
|
| ||
| Differentially Regulated | ||
| TNF (ligand) superfamily, member 10 | ↑5.2 | ↓6.5 |
|
| ||
| Down regulated | ||
| Interleukin 6 (interferon, beta 2) | 24.7 | 8.9 |
| Chemokine (C-X-C motif) ligand 3 | 6.3 | 10.9 |
| Colony stimulating factor 3 (granulocyte) | 2.1 | 4.6 |
| Inhibin, beta A | 4.0 | 3.8 |
| Interleukin 1, alpha | 3.4 | 3.8 |
| Interleukin 23, alpha subunit p19 | 4.1 | 8.7 |
Genes were listed from the cytokine-cytokine receptor pathway of the Kyoto Encyclopedia of Genes and Genomes.
All genes were significant by adjusted p<0.05 and fold change ≥ 2.
Table 4 shows PMN genes changing expression in response to DEX belonging to the cytokine-cytokine receptor pathway based from the KEGG system. This pathway also had the highest Z of 7, compared to other pathways in PMNs affected by DEX. The greatest fold changes were seen in the up regulation of IL-18 receptor 1 and inhibin beta B as well as the down regulation of TNF (ligand) superfamily 15. DEX had no effect on genes in this pathway for MONOs.
Table 4.
Genes changing expression from the cytokine-cytokine receptor pathway a by microarray analysis in response to dexamethasone at 4 hrs in endotoxin-stimulated polymorphonuclear cells of the newly born
| Gene Name (Gene ID) | Fold Change |
|---|---|
| Up Regulated | |
| Interleukin 18 receptor 1 | 8.4 |
| Inhibin, beta B | 3.7 |
| Chemokine (C-X-C motif) receptor 7 | 2.6 |
| Interleukin 13 receptor, alpha 1 | 2.6 |
| V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 2.4 |
| Interleukin 18 receptor accessory protein | 2.3 |
| Interleukin 8 receptor, beta | 2.2 |
| Interleukin 7 receptor | 2.0 |
|
| |
| Down Regulated | |
| TNF (ligand) superfamily, member 15 | 3.4 |
| Leukemia inhibitory factor | 2.4 |
| Oncostatin M | 2.2 |
| Interleukin 23, alpha subunit p19 | 2.0 |
| colony stimulating factor 1 (macrophage) | 2.0 |
| Interleukin 2 receptor, beta | 2.0 |
Genes were listed from the cytokine-cytokine receptor pathways of the Koyoto Encyclopedia of Genes and Genomes (KEGG).
All fold changes are significant: p<0.05.
Monocyte cytokine release
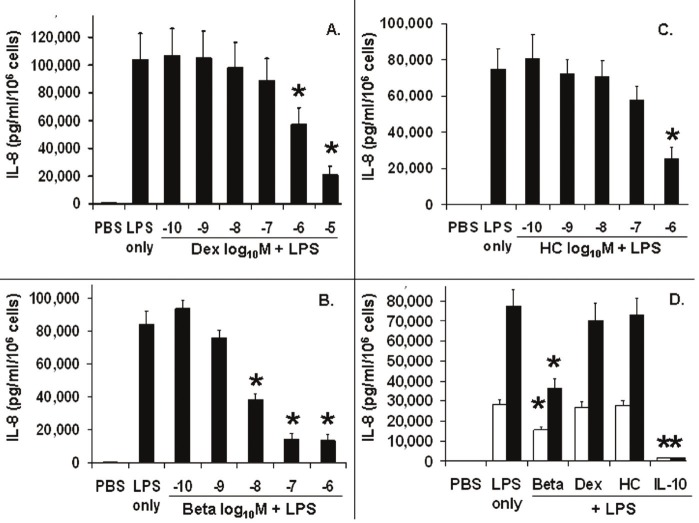
Subsequent experiments focused on the insensitivity of the MONOs to DEX and determined if BETA and HC gave similar results. Figure 2 (panel A) shows the release of IL-8 from LPS-stimulated monocytes pretreated for one hour with serial increasing doses of DEX. IL-8 (pg/ml) release was measured from MONOs after 18 h of incubation. DEX did not cause a significant reduction in IL-8 release, until a dose of 10-6 M. Panel B shows that BETA resulted in a significant inhibition of IL-8 release at 10-8 M. Panel C shows that HC did not cause a significant reduction in IL-8 release until 10-6 M. Panel D shows the comparison of the temporal effect of the different glucocorticoid pretreatment, all at 10-8 M, on IL-8 release from MONOs at 4 and 18h hours of LPS stimulation. Only BETA showed a significant reduction in IL-8 release by 4 hours (44%) and by 18 h (56%). As a positive controI, IL-10 at 10-8 M reduced IL-8 release by 95% and 98% at 4 and 18 hours, respectively.
Figure 2.
Interleukin-8 (IL-8) release into cell culture media from LPS-stimulated monocytes of the newly born (n=5), pretreated with serially increasing doses (log10 M) of either dexamethasone (DEX, panel A), betamethasone (BETA, panel B) or hydrocortisone (HC, panel C) for 1 h. This was followed by stimulation with endotoxin (LPS) in phosphate buffered saline (PBS) for 18 h. Panel D shows IL-8 release for each glucocorticoid at 4□ and 18 ■h at 10-8 M with a comparison to interleukin-10 at 10-8 M. Values are mean±SEM, statistically different (*) from LPS alone (adjusted p value < 0.05).
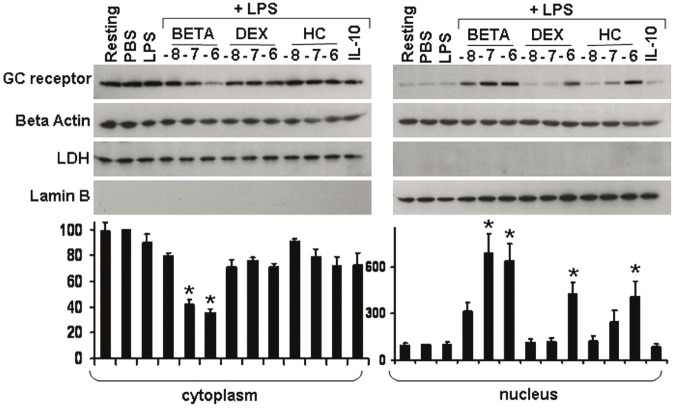
Glucocorticoid receptor nuclear translocation in MONOs
Figure 3 shows the glucocorticoid receptor (GR) protein levels in cytoplasmic and nuclear extracts prepared from LPS-stimulated MONOs at 1 hour. The figure is divided into GR protein levels as detected by Western blot in the cytoplasm versus the nucleus panel (panel B). Actin was used as a control for the amount of protein. Lactose dehydrogenase was used as a cytoplasmic extract marker and is appropriately absent in the nuclear extract. Lamin B was used as a nuclear extract marker and is appropriately absent in the cytoplasmic extract. The upper area of the cytoplasm panel shows dense bands representing the GR in the cytoplasm under resting, PBS, and LPS (alone) conditions; these bands are virtually undetectable in the panel for the nuclear extract under the same conditions. With increasing levels of BETA, prior to LPS-stimulation, there was a decrease in the GR protein levels in the cytoplasm and a concurrent increase in the nucleus. No increase in GR protein levels in the nucleus was seen with DEX or HC until a 10-6 M pretreatment was employed. The bar graph at the base of the figure represents the densitometry analysis of GR/actin densities from 5 cord blood samples.
Figure 3.
Glucocorticoid receptor (GR) protein levels (Western blot) in cytoplasmic (panel A) and nuclear extracts (panel B) prepared from monocytes (MONOs) of the newly born, pretreated for 1 h with serially increasing doses of either dexamethasone (DEX), betamethasone (BETA) or hydrocortisone (HC) in phosphate buffered saline (PBS) for 1 h. The glucocorticoid concentrations are shown as log10 M. This was followed by endotoxin (LPS) stimulation for 1 h. Interleukin-10 at 10-8 M, which produces maximal pro-inflammatory cytokine inhibition, was used as a negative control. The two Western blots show GR protein levels in the cytoplasm and the nucleus from the same subject. Below each panel are combined densitometer analysis of GR/actin densities (percent change from PBS) from 5 different subjects. Values are mean ± SEM, significantly different (*) from LPS alone p<0.05.
Discussion
Because the efficacy and safety of DEX for the treatment of BPD are major concerns for neonatologists, new therapeutic insights from translational research studies have been sought [21]. In addition, in a pilot historical case-controlled study comparing BETA to DEX for the treatment of BPD, BETA showed less short term adverse effects [22].
Persistent inflammation plays a central role in the development of BPD following the recruitment of an abnormal number and prolonged survival of PMNs, followed by MONOs recruitment into the lung [6-8,23]. IL-10 is a homodimer with a molecular mass of 37kDa. IL-10 is one of the most potent anti-inflammatory cytokines known, and has been studied extensively in adults [24]. However, there is little information benchmarking the potency of IL-10 versus DEX as anti-inflammatory agents [15]. Besides its function in terminating the innate immune response, IL-10 regulates differentiation and proliferation of several types of immune cells involved the adaptive immune response [24]. IL-10 has been used in the treatment of various adult inflammatory disorders such as psoriasis and inflammatory bowel disease. Importantly, endogenous airway levels of IL-10 in the newborn may be deficient, creating an imbalance of pro-inflammatory versus anti-inflammatory cytokines in the airway during the development of BPD [12-14,25,26]. Up regulated endogenous IL-10 or exogenous IL-10 replacement therapy has been suggested as a potential treatment for BPD pending more investigation [7,16,17].
The present study provides new information regarding genome-wide gene expression of the effect of IL-10 versus glucocorticoids in leukocytes of newly born. Since this translational research study could have clinical implications for new BPD therapy, microarray methodology provided a screening tool for gene expression that could be important for effective and safe therapy. Fortunately, there has been previous work measuring plasma DEX levels in preterm infants being treated for BPD [19,20] as well as measurements of endotoxin levels in amniotic fluid [18]. Therefore the present microarray studies used a clinically relevant stimulus dose of LPS (10ng/ml) with and without DEX (10-8 M) or an equimolar level of IL-10. The microarray results demonstrated the great difference in the number of genes significantly changing expression between treatments and cell types and the relatively low number of genes changing expression that are common between cell types and treatment. The striking finding was that there was no significant effect on gene expression by DEX at 10-8 M for MONOs as opposed to IL-10 in LPS-stimulated cells; IL-10 had a significant effect on gene expression for a large number of genes in MONOs. For LPS-stimulated PMNs, less than 20% of the affected genes were changing in common between IL-10 and DEX pretreatment. The microarray studies also revealed that IL-10 led to approximately 6 times the number of genes changing expression in MONOs versus PMNs. This difference between cell types may be related to the fact that the monocyte/macrophage cell type is involved in both pro-inflammatory and anti-inflammatory pathways evolving by our sampling time point of 4 h of LPS exposure [16]. KEGG pathways affected by IL-10 versus DEX, were similar however changes in gene expression related to cytokine-cytokine receptor interaction were the most prominent, except for the lack of any gene expression changes for MONOs treated with DEX.
The overall implications from the microarray experiments are that anti-inflammatory therapy with IL-10 will likely show important differences in efficacy and potential side effects compared to DEX. The strength of our microarray statistical analyses was partly due to the subject sample size of 5 microarrays for each condition (cell type and treatment). The limitation of this microarray study is due to the restriction in the number of time points and dose-related responses that were studied. It is likely that gene expressions and their interaction between cell types are occurring before and after the 4 hr time point we chose based on a pilot study. [16] The present study used cord blood from term infants to avoid the confounding effects of antenatal steroids, clinical infection or maternal medications associated with preterm delivery. MONOs from preterm infants versus term human infants may have differences in function as suggested by the lamb model in which monocytes of the preterm lamb showed reduced phagocytosis of apoptotic cells but other functions were similar between preterm and term animals [27]. In addition, toll-like receptor stimulation of PMNs, isolated from pre-term and term infant samples, induce similar level of elevated interleukin 8 release compared to PMNs from adults [28].
To validate microarray results we performed RT-qPCR on selected genes that showed upregulation, no change or downregulation. We chose IL-6 and IL-1 family 9 gene expression because of the marked and highly significant downregulation observed by microarray and the known relationship of these cytokines to the fetal inflammatory response syndrome which is a precursor in some cases for the development of BPD [29,30]. Consistency was seen in the quantitative and qualitative changes in gene expression determined by RT-qPCR except for IL-6; no change in gene expression was associated with DEX in PMNs by microarray compared to a modest down regulation observed by RT-qPCR. Of interest was the marked and highly significant upregulation of hepcidin anti-microbial peptide by microarray for all conditions except for DEX treated MONOs. Hepcidin antimicrobial peptide is involved primarily in cell entry of iron and may have anti-bacterial and anti-fungal properties [31].
In previous work, specifically related to the innate immune response of the newborn, it has been shown that IL-10 is not released by the LPS-stimulated PMNs [16]. Peripheral blood monocytic cells of the newborn do release IL-10 but the level detected in cell culture media is approximately 100 fold less than the level needed to maximally inhibit pro-inflammatory cytokine release [16,17]. The complex mechanism of action of exogenous IL-10 involves the activation of transcription factors STAT-3 and activator protein 1 in MONOs of the newborn [17,32,33]. On the other hand, one of the major anti-inflammatory mechanisms of action of glucocorticoids involves translocation of the glucocorticoid receptor to the nucleus where it inhibits transcription of many pro-inflammatory mediators including cytokines and well as inhibition phospholipase A2 and arachidonic acidderived mediators [34]. In MONOs from adults, a resistance of MONOs to the glucocorticoids such as DEX, resulting in a failure to inhibit pro-inflammatory mediators, has been described [33,35]. The degree of glucocorticoid resistance in macrophages has been linked to specific diseases and the need for high doses of glucocorticoids [33,35].
Based on the microarray and RT-qPCR results showing MONO insensitivity to DEX, the second aim of the present study screened the potency of BETA and HC compared to DEX on an equimolar basis based on IL-8 release. IL-8 is an important and potent chemokine related to the early pathogenesis of BPD [9,11]. Each glucocorticoid was studied in a dose-related fashion over 4 and 18 h, and then glucocorticoid receptor protein translocation from the cytoplasm to nucleus was examined. DEX and HC did not have any effect on IL-8 release from MONOs until 10-6 M. For DEX, a level of 10-6 M would be about a 100 fold increase above the levels found in the plasma newborns treated for BPD (10-8 M) [19,20] and would therefore not be useful clinically owing to expected severe short and long term adverse effects. On the other hand BETA did produce inhibition of IL-8 release at 10-8 M, although not to the same degree as IL-10 at 4 and 18 hours.
The dose-related effect of the three glucocorticoids on GR protein levels correlated with the individual glucocorticoid effect on IL-8 release from MONOs as measured by ELISA on culture media. BETA tended to increase nuclear glucocorticoid receptor protein levels at 10-8 M but this result became statistically significant at 10-7 M. No change in glucocorticoid receptor translocation for DEX and HC was observed until 10-6 M, associated in parallel with a partial inhibition of IL-8 release, compared to the greater inhibition at equimolar levels of BETA, IL-10 served as a strong positive control for the weaker inhibition of IL-8 release by the glucocorticoids.
In conclusion, this translational research study indicates new avenues for future research, aimed at novel potential therapy for BPD, by IL-10 or BETA. BETA may have less central nervous system adverse effects than DEX based on a large retrospective study comparing cystic periventricular leukomalacia in preterm infants who received BETA versus DEX for antenatal lung maturation [36]. The marked difference in cell-specific genome-wide gene expression by IL-10 versus DEX, displayed in the present study, suggests there may be appreciable differences in clinical efficacy and side effects of IL-10 versus glucocorticoids as well. However, since the MONO is the principal precursor for alveolar macrophages, Kupffer cells, Langerhans cells, dendritic cells and microglial cells [37], the effect of IL-10 versus different glucocorticoids on the control of pro-inflammatory cytokine release in this cell line lineage, could have important clinical implications, not only for BPD, but also a variety of serious neonatal inflammatory disorders.
Acknowledgements
Funded: in part by R03-HD048508 (NICHD, PI: Dennis Davidson, MD) by the Lilling Family support of the Neonatal Research Laboratory at The Feinstein Institute for Medical Research.
References
- 1.Stoll BJ, Hansen NI, Bell EF, Shankaran S, Laptook AR, Walsh MC, Hale EC, Newman NS, Schibler K, Carlo WA, Kennedy KA, Poindexter BB, Finer NN, Ehrenkranz RA, Duara S, Sanchez PJ, O’Shea TM, Goldberg RN, Van Meurs KP, Faix RG, Phelps DL, Frantz ID 3rd, Watterberg KL, Saha S, Das A, Higgins RD Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network. Neonatal outcomes of extremely preterm infants from the NICHD Neonatal Research Network. Pediatrics. 2010;126:443–456. doi: 10.1542/peds.2009-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laughon MM, Langer JC, Bose CL, Smith PB, Ambalavanan N, Kennedy KA, Stoll BJ, Buchter S, Laptook AR, Ehrenkranz RA, Cotten CM, Wilson-Costello DE, Shankaran S, Van Meurs KP, Davis AS, Gantz MG, Finer NN, Yoder BA, Faix RG, Carlo WA, Schibler KR, Newman NS, Rich W, Das A, Higgins RD, Walsh MC Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network. Prediction of bronchopulmonary dysplasia by postnatal age in extremely premature infants. Am J Respir Crit Care Med. 2011;183:1715–1722. doi: 10.1164/rccm.201101-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark RH, Kueser TJ, Walker MW, Southgate WM, Huckaby JL, Perez JA, Roy BJ, Keszler M, Kinsella JP. Low-dose nitric oxide therapy for persistent pulmonary hypertension of the newborn. Clinical Inhaled Nitric Oxide Research Group. N Engl J Med. 2000;342:469–474. doi: 10.1056/NEJM200002173420704. [DOI] [PubMed] [Google Scholar]
- 4.Speer CP. Inflammation and bronchopulmonary dysplasia: a continuing story. Semin Fetal Neonatal Med. 2006;11:354–362. doi: 10.1016/j.siny.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Watterberg KL American Academy of Pediatrics; Committee on Fetus and Newborn. Policy statement--postnatal corticosteroids to prevent or treat bronchopulmonary dysplasia. Pediatrics. 2010;126:800–808. doi: 10.1542/peds.2010-1534. [DOI] [PubMed] [Google Scholar]
- 6.Merritt TA, Cochrane CG, Holcomb K, Bohl B, Hallman M, Strayer D, Edwards DK 3rd, Gluck L. Elastase and alpha 1-proteinase inhibitor activity in tracheal aspirates during respiratory distress syndrome. Role of inflammation in the pathogenesis of bronchopulmonary dysplasia. J Clin Invest. 1983;72:656–666. doi: 10.1172/JCI111015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwong KY, Jones CA, Cayabyab R, Lecart C, Khuu N, Rhandhawa I, Hanley JM, Ramanathan R, deLemos RA, Minoo P. The effects of IL-10 on proinflammatory cytokine expression (IL-1beta and IL-8) in hyaline membrane disease (HMD) Clin Immunol Immunopathol. 1998;88:105–113. doi: 10.1006/clin.1997.4510. [DOI] [PubMed] [Google Scholar]
- 8.Jackson JC, Chi EY, Wilson CB, Truog WE, Teh EC, Hodson WA. Sequence of inflammatory cell migration into lung during recovery from hyaline membrane disease in premature newborn monkeys. Am Rev Respir Dis. 1987;135:937–940. doi: 10.1164/arrd.1987.135.4.937. [DOI] [PubMed] [Google Scholar]
- 9.Munshi UK, Niu JO, Siddiq MM, Parton LA. Elevation of interleukin-8 and interleukin-6 precedes the influx of neutrophils in tracheal aspirates from preterm infants who develop bronchopulmonary dysplasia. Pediatr Pulmonol. 1997;24:331–336. doi: 10.1002/(sici)1099-0496(199711)24:5<331::aid-ppul5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 10.Kotecha S, Wilson L, Wangoo A, Silverman M, Shaw RJ. Increase in interleukin (IL)-1 beta and IL-6 in bronchoalveolar lavage fluid obtained from infants with chronic lung disease of prematurity. Pediatr Res. 1996;40:250–256. doi: 10.1203/00006450-199608000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Ambalavanan N, Carlo WA, D’Angio CT, McDonald SA, Das A, Schendel D, Thorsen P, Higgins RD Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network. Cytokines associated with bronchopulmonary dysplasia or death in extremely low birth weight infants. Pediatrics. 2009;123:1132–1141. doi: 10.1542/peds.2008-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oei J, Lui K, Wang H, Henry R. Decreased interleukin-10 in tracheal aspirates from preterm infants developing chronic lung disease. Acta Paediatr. 2002;91:1194–1199. doi: 10.1111/j.1651-2227.2002.tb00128.x. [DOI] [PubMed] [Google Scholar]
- 13.Jones CA, Cayabyab RG, Kwong KY, Stotts C, Wong B, Hamdan H, Minoo P, deLemos RA. Undetectable interleukin (IL)-10 and persistent IL-8 expression early in hyaline membrane disease: a possible developmental basis for the predisposition to chronic lung inflammation in preterm newborns. Pediatr Res. 1996;39:966–975. doi: 10.1203/00006450-199606000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beresford MW, Shaw NJ. Detectable IL-8 and IL-10 in bronchoalveolar lavage fluid from preterm infants ventilated for respiratory distress syndrome. Pediatr Res. 2002;52:973–978. doi: 10.1203/00006450-200212000-00025. [DOI] [PubMed] [Google Scholar]
- 15.Citarella BV, Miskolci V, Vancurova I, Davidson D. Interleukin-10 versus dexamethasone: effects on polymorphonuclear leukocyte functions of the newborn. Pediatr Res. 2009;65:425–429. doi: 10.1203/PDR.0b013e318199384d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson D, Miskolci V, Clark DC, Dolmaian G, Vancurova I. Interleukin-10 production after pro-inflammatory stimulation of neutrophils and monocytic cells of the newborn. Comparison to exogenous interleukin-10 and dexamethasone levels needed to inhibit chemokine release. Neonatology. 2007;92:127–133. doi: 10.1159/000101432. [DOI] [PubMed] [Google Scholar]
- 17.Chusid LA, Pereira-Argenziano L, Miskolci V, Vancurova I, Davidson D. Transcriptional control of cytokine release from monocytes of the newborn: effects of endogenous and exogenous interleukin-10 versus dexamethasone. Neonatology. 2010;97:108–116. doi: 10.1159/000235807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romero R, Roslansky P, Oyarzun E, Wan M, Emamian M, Novitsky TJ, Gould MJ, Hobbins JC. Labor and infection. II. Bacterial endotoxin in amniotic fluid and its relationship to the onset of preterm labor. Am J Obstet Gynecol. 1988;158:1044–1049. doi: 10.1016/0002-9378(88)90216-5. [DOI] [PubMed] [Google Scholar]
- 19.Schild PN, Charles BG. Determination of dexamethasone in plasma of premature neonates using high-performance liquid chromatography. J Chromatogr B Biomed Appl. 1994;658:189–192. doi: 10.1016/0378-4347(94)00192-8. [DOI] [PubMed] [Google Scholar]
- 20.Lugo RA, Nahata MC, Menke JA, McClead RE Jr. Pharmacokinetics of dexamethasone in premature neonates. Eur J Clin Pharmacol. 1996;49:477–483. doi: 10.1007/BF00195934. [DOI] [PubMed] [Google Scholar]
- 21.Wright CJ, Kirpalani H. Targeting inflammation to prevent bronchopulmonary dysplasia: can new insights be translated into therapies? Pediatrics. 2011;128:111–126. doi: 10.1542/peds.2010-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeCastro M, El-Khoury N, Parton L, Ballabh P, LaGamma EF. Postnatal betamethasone vs dexamethasone in premature infants with bronchopulmonary dysplasia: a pilot study. J Perinatol. 2009;29:297–304. doi: 10.1038/jp.2008.194. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen CN, Schnulle PM, Chegini N, Luo X, Koenig JM. Neonatology. Switzerland: Basel: 2010. Neonatal neutrophils with prolonged survival secrete mediators associated with chronic inflammation; pp. 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asadullah K, Sterry W, Volk HD. Interleukin-10 therapy--review of a new approach. Pharmacol Rev. 2003;55:241–269. doi: 10.1124/pr.55.2.4. [DOI] [PubMed] [Google Scholar]
- 25.Viscardi RM. Perinatal inflammation and lung injury. Semin Fetal Neonatal Med. 2012;17:30–35. doi: 10.1016/j.siny.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz C, Temming P, Bucsky P, Gopel W, Strunk T, Hartel C. Immature anti-inflammatory response in neonates. Clin Exp Immunol. 2004;135:130–136. doi: 10.1111/j.1365-2249.2004.02313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer BW, Jobe AH, Ikegami M. Monocyte function in preterm, term, and adult sheep. Pediatr Res. 2003;54:52–57. doi: 10.1203/01.PDR.0000066621.11877.33. [DOI] [PubMed] [Google Scholar]
- 28.Thornton NL, Cody MJ, Yost CC. Neonatology. Switzerland: Basel: 2012. Toll-like receptor 1/2 stimulation induces elevated interleukin-8 secretion in polymorphonuclear leukocytes isolated from preterm and term newborn infants; pp. 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorokin Y, Romero R, Mele L, Wapner RJ, Iams JD, Dudley DJ, Spong CY, Peaceman AM, Leveno KJ, Harper M, Caritis SN, Miodovnik M, Mercer BM, Thorp JM, O’Sullivan MJ, Ramin SM, Carpenter MW, Rouse DJ, Sibai B. Maternal serum interleukin-6, C-reactive protein, and matrix metalloproteinase-9 concentrations as risk factors for preterm birth <32 weeks and adverse neonatal outcomes. Am J Perinatol. 2010;27:631–640. doi: 10.1055/s-0030-1249366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madsen-Bouterse SA, Romero R, Tarca AL, Kusanovic JP, Espinoza J, Kim CJ, Kim JS, Edwin SS, Gomez R, Draghici S. The transcriptome of the fetal inflammatory response syndrome. Am J Reprod Immunol. 2010;63:73–92. doi: 10.1111/j.1600-0897.2009.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroot JJ, Tjalsma H, Fleming RE, Swinkels DW. Hepcidin in human iron disorders: diagnostic implications. Clin Chem. 2011;57:1650–1669. doi: 10.1373/clinchem.2009.140053. [DOI] [PubMed] [Google Scholar]
- 32.Murray PJ. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr Opin Pharmacol. 2006;6:379–386. doi: 10.1016/j.coph.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Barnes PJ, Adcock IM. Glucocorticoid resistance in inflammatory diseases. Lancet. 2009;373:1905–1917. doi: 10.1016/S0140-6736(09)60326-3. [DOI] [PubMed] [Google Scholar]
- 34.de Benedictis FM, Bush A. Corticosteroids in respiratory diseases in children. Am J Respir Crit Care Med. 2012;185:12–23. doi: 10.1164/rccm.201107-1174CI. [DOI] [PubMed] [Google Scholar]
- 35.Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2006;117:522–543. doi: 10.1016/j.jaci.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 36.Baud O, Foix-L’Helias L, Kaminski M, Audibert F, Jarreau PH, Papiernik E, Huon C, Lepercq J, Dehan M, Lacaze-Masmonteil T. Antenatal glucocorticoid treatment and cystic periventricular leukomalacia in very premature infants. N Engl J Med. 1999;341:1190–1196. doi: 10.1056/NEJM199910143411604. [DOI] [PubMed] [Google Scholar]
- 37.Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJ, Liu YJ, MacPherson G, Randolph GJ, Scherberich J, Schmitz J, Shortman K, Sozzani S, Strobl H, Zembala M, Austyn JM, Lutz MB. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74–80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]