Abstract
Pneumocystis carinii is an opportunistic fungal pathogen that causes pneumonia in the immunocompromised host. A protective monoclonal antibody (MAb) termed 4F11 generated against mouse-derived P. carinii was shown by indirect immunofluorescence assay (IFA) to bind surface antigens of P. carinii derived from multiple host species, including humans. We have identified multiple epitopes recognized by MAb 4F11 in two recombinant mouse P. carinii antigens. The epitopes mapped have similar proline content and positive charge distribution. The consensus 8-mer epitope recognized by MAb 4F11 is K/RPA/RPK/QPA/TP. Immune sera raised against intact mouse P. carinii recognized native antigens affinity purified with MAb 4F11 and a recombinant antigen reactive with MAb 4F11. Database searches for short, nearly exact matches to the mapped MAb 4F11 epitopes identified a bacterial surface antigen, Streptococcus pneumoniae PspA, with a similar proline-rich region. In an IFA, MAb 4F11 detected antigens on the S. pneumoniae surface, and Western blotting identified a protein in S. pneumoniae lysates consistent with the Mr of PspA. A fragment of the S. pneumoniae PspA gene was cloned and sequenced, and the deduced amino acid sequence contained a region with strong similarity to the MAb 4F11 epitopes identified in P. carinii. The PspA recombinant polypeptide was recognized by MAb 4F11 in a Western blot. The ability of MAb 4F11 to recognize similar proline-rich epitopes may explain its ability to recognize P. carinii derived from multiple hosts and will permit testing of the epitopes recognized by this antibody in immunization against P. carinii.
Pneumocystis carinii is an opportunistic fungal pathogen that causes pneumonia (P. carinii pneumonia [PCP]) in the immunocompromised host. PCP, as well as other opportunistic infections, underwent a dramatic rise in prevalence with the onset of the AIDS epidemic (28). With the development of highly effective antiretroviral therapy, the prevalence of PCP in AIDS patients has declined, though it remains the most commonly diagnosed serious opportunistic infection in AIDS patients (33). PCP is also prevalent in persons undergoing chemotherapy or other immunosuppressive therapy for cancer and organ transplantations (28). The most common drug treatments for P. carinii infections are trimethoprim-sulfamethoxazole and aerosolized pentamidine. Because adverse side effects, recurrent infections, and poor compliance are problems with these drugs, alternative treatments or preventative measures against PCP are needed to eradicate this serious opportunistic infection.
P. carinii cannot be continuously cultured outside of its host. P. carinii also has a host species-dependent specificity which complicates the ability to use animal-derived organisms to immunize humans. P. carinii organisms derived from different hosts have isoform variants of common antigens, resulting in different (i.e., noncrossreactive) antigenic determinants (11, 13). Attempts to infect laboratory animals with P. carinii isolated from heterologous mammalian species have met with little to no success (1, 3, 14). However, immunocompetent mice immunized with whole mouse-derived P. carinii are protected from developing PCP after T-cell depletion and subsequent challenge, whereas unimmunized cohorts are not protected (21).
The surface glycoprotein gpA is an abundant and immunodominant antigen of P. carinii (18), although immunization with this antigen does not adequately protect against infection in a mouse model of PCP (17). The majority of monoclonal antibodies (MAbs) against P. carinii surface antigens react only with isoforms showing host species specificity identical to that of the immunogen (14). MAb 4F11 was obtained by selective screening of anti-mouse P. carinii hybridomas for recognition of P. carinii antigens other than gpA (25). MAb 4F11 confers passive prophylaxis against development of PCP when administered intranasally to SCID mice (12). Furthermore, MAb 4F11 recognizes surface antigens of P. carinii derived from different hosts, including humans.
A screen of a P. carinii cDNA expression library with MAb 4F11 revealed a number of positive clones, including mouse P. carinii Kex1 (25). Based on sequence homology to its ortholog in Saccharomyces cerevisiae, Kex1 is a member of the kexin family of subtilisin-like proteases (25). Here, we describe another mouse P. carinii antigen, encoded by cDNA clone A12, which is recognized by MAb 4F11. We mapped the epitopes recognized by MAb 4F11 within mouse P. carinii Kex1 and cDNA clone A12. We also demonstrate that a Streptococcus pneumoniae cell surface antigen, PspA, shares epitopes with mouse P. carinii epitopes recognized by MAb 4F11.
MATERIALS AND METHODS
Oligonucleotide annealing and PCR.
The oligonucleotides used in this study are listed in Table 1. Complementary oligonucleotides were purchased from Sigma-Genosys (Woodlands, Tex.). Lyophilized oligonucleotides were resuspended in annealing buffer (10 mM Tris [pH 8.0], 50 mM NaCl, 1 mM EDTA) at 100 pmol/ml. Twenty-five microliters of each were mixed and heated to 95°C for 2 min and then cooled to 25°C at a rate of 0.02°C/s in a PCR Sprint Thermocycler (Hybaid Ashford). Annealed oligonucleotides were electrophoresed on a 2% agarose gel and purified with the Qiaquick gel extraction kit (Qiagen, Valencia, Calif.), then used immediately in DNA ligations. PCR conditions were as follows: 90 s at 95°C; 90 s at (TM lower −4°C), where TM lower signifies the lower melting temperature of each primer pair; and 2 min at 72°C for 30 cycles, with a 10-min 72°C final extension step for addition of 3′ overhangs by Taq polymerase.
TABLE 1.
Oligonucleotides used in this study
| Construct | Oligonucleotide name | Sequence |
|---|---|---|
| Kexin856-872THIO | Kexin Epitope S | AAACCGGCACCTAAACCAACACCACCTAAACCAGCGCCTAAACCAGCACCAA |
| Kexin856-872THIO | Kexin Epitope AS | TGGTGCTGGTTTAGGCGCTGGTTTAGGTGGTGTTGGTTTAGGTGCCGGTTTA |
| Kexin777-787THIO | A39 Epitope2 S | AGACCAGCACCACCTAAACCAACACCTCAACCAA |
| Kexin777-787THIO | A39 Epitope2 AS | TGGTTGAGGTGTTGGTTTAGGTGGTGCTGGTCTA |
| Kexin856-863THIO | A32.1 Epitope S | AAACCGGCACCTAAACCAACACCAA |
| Kexin856-863THIO | A32.1 Epitope AS | TGGTGTTGGTTTAGGTGCCGGTTTA |
| Kexin865-872THIO | A32.2 Epitope S | AAACCAGCGCCTAAACCAGCACCAA |
| Kexin865-872THIO | A32.2 Epitope AS | TGGTGCTGGTTTAGGCGCTGGTTTA |
| Kexin860-868THIO | A32.3 Epitope S | AAACCAACACCACCTAAACCAGCGCCTA |
| Kexin860-868THIO | A32.3 Epitope AS | AGGCGCTGGTTTAGGTGGTGTTGGTTTA |
| Kex Epi 5′ | pSCREEN T7 10 S | CTGGGTAAGGAGATTATTGCG |
| Kex Epi 5′ | A32 Epitope AS2 | TGGTGCTGGTTTAGGCGCTGG |
| Kex Epi 3′ | A32 Epitope S3 | TCTAAATCATCATCTAAACCAACATC |
| Kex Epi 3′ | pSCREEN T7 10 AS | CCCAAGCTTGTCGACGGAG |
| A1262-77THIO | A12 Epitope S | AAACCTCGACCTCAGCCAACGTCAAAACCTCGACCTCAGCCGACGCCAA |
| A1262-77THIO | A12 Epitope AS | TGGCGTCGGCTGAGGTCGAGGTTTTGACGTTGGCTGAGGTCGAGGTTTA |
| A1270-77THIO | A12.2 Epitope S | AAACCTCGACCTCAGCCGACGCCAA |
| A1270-77THIO | A12.2 Epitope AS | TGGCGTCCCCTCACCTCCACCTTTA |
| A1246-53THIO | A12′2 Epitope S | GAACCTCGACCTCAGCCGACGTCAA |
| A1246-53THIO | A12′2 Epitope AS | TGACGTCGGCTGAGGTCGAGGTTCA |
| A1254-61THIO | A12′3 Epitope S | GAACCTCAGCCTCAGCCGGCGCCAA |
| A1254-61THIO | A12′3 Epitope AS | TGGCGCCGGCTGAGGCTGAGGTTCA |
| A121-82/1-142THIO | A12 S | ACCAATATATCCGAACCAGC |
| A121-142THIO | A12 Mid AS | TTCTGATGTTGACTGAGATGG |
| A121-82THIO | A12 Mid AS2 | CCGACGCCAGAACCTCG |
| A12 Sequence | Lambda forward | TGGCGACGACTCCTGGAGCCCG |
| A12 Sequence | Lambda reverse | TGACACCAGACCAACTGGTAATGG |
| URSP2 PspA seq | PspA S2 (22) | GCAAGCTTATGATATAGAAATTTGTAAC |
| URSP2 PspA seq | PspA AS (22) | CCACATACCGTTTTCTTGTTTCCAGCC |
| URSP2 PspA: THIO | PspA S3 | ACAAGTCTAGCCAGCTCGC |
| URSP2 PspA: THIO | PspA AS (22) | CCACATACCGTTTTCTTGTTTCCAGCC |
Bacterial strains, growth conditions, plasmid isolation, and nucleotide sequencing.
PCR-amplified Kex1 or annealed complementary Kex1 or A12 oligonucleotide inserts (Table 1) were cloned into TOPO TA cloning vectors (Invitrogen Co., Carlsbad, Calif.) following the manufacturer's instructions. Escherichia coli transformants were grown at 37°C in Luria-Bertani (LB) medium with ampicillin (100 μg/ml). For colony immunoscreens, E. coli transformants were grown on LB agar plates with ampicillin (100 μg/ml) and tetracycline (50 μg/ml). S. pneumoniae strains were obtained from the Strong Memorial Hospital Clinical Microbiology Laboratory at the University of Rochester. S. pneumoniae was grown at 37°C on trypticase soy agar II-5% sheep blood agar plates or in Todd-Hewitt broth-5% yeast extract (THY). Plasmid DNA was isolated from E. coli with a Qiagen miniprep kit; both strands of each cloned insert were sequenced by the University of Rochester Core Nucleic Acid Sequencing Facility. S. pneumoniae chromosomal DNA was isolated as described (6).
Immunodetection assays.
MAb 4F11 was produced as described (25). For colony immunoscreens and P. carinii indirect immunofluorescence assays (IFAs), MAb 4F11 immunoglobulin M (IgM) ascites fluid was used at a 1:5,000 dilution. For enzyme-linked immunosorbent assays (ELISAs) and Western blots of purified recombinant proteins, an IgG1 switch-variant of MAb 4F11(G1) (12) was prepared by saturated ammonium sulfate precipitation of tissue culture supernatant and used at the indicated dilutions. MAb 2B5 is an IgG1 that recognizes a mouse P. carinii gpA (10) and was used as a negative control in S. pneumoniae immunoblots and IFAs. MAb 1C7 is an anti-mouse P. carinii IgM used as an isotype control in IFAs against P. carinii derived from different hosts. Pooled hyperimmune serum against mouse P. carinii was obtained as described previously (16).
Purified recombinant protein at a concentration of 10 μg/ml in 50 mM carbonate-bicarbonate buffer (pH 9.5) with 0.1% sodium dodecyl sulfate (SDS) was used to coat Costar ELISA plates (Corning, Corning, N.Y.) for 16 to 20 h at 37°C. The plates were blocked with 3% bovine serum albumin (BSA)-Tris-buffered saline (TBST) for 1 h at 4°C. Primary antibody was added in twofold serial dilutions in 3% BSA-TBST and incubated at 4°C for 1 h. Goat anti-mouse IgG-IgM-alkaline phosphatase conjugate secondary antibody was added at a 1:5,000 dilution in BSA-TBST for 1 h at 4°C. Blue Phos substrate (KPL, Gaithersburg, Md.) was added for 30 min, color development was stopped with 2.5% EDTA, and absorbance was read at 655 nm in a Benchmark microplate reader (Bio-Rad).
Mouse P. carinii Kexin 777-787 (RPAPPKPTPQP) and Kexin 131-142 (SGDTGNVNSGEK) peptides were purchased from Alpha Diagnostics (San Antonio, Tex.). ELISA experiments with the synthetic peptides were performed as described above, using 10 μg of peptide/ml in carbonate-bicarbonate buffer to coat the plates. For competitive ELISA studies, peptides were incubated at threefold dilutions with a starting concentration of 100 μg/ml for 2 h with a 1:3,200 dilution of MAb 4F11 at 4°C to reach equilibrium. Mouse P. carinii sonicates in carbonate-bicarbonate buffer were used to coat ELISA plates at the equivalence of 4 × 104 cysts per well overnight at 4°C. The peptide-antibody mixtures, or antibody alone at 1:3,200 dilution, were used as the primary antibody in the ELISA following the above protocol.
P. carinii IFAs were performed as described (16). S. pneumoniae was swabbed from blood agar plates after overnight growth at 37°C and resuspended in 1.5 ml of PBS-3% fetal calf serum (PBS-FCS). Cells were pelleted by centrifugation at 1,000 × g, resuspended in 1.5 ml of PBS-FCS, and 0.5-ml aliquots were placed into three microcentrifuge tubes to which either 0.5 ml of MAb 4F11 IgG1 tissue culture supernatant, MAb 2B5, or PBS-FCS alone was added. After 2 h of incubation at room temperature with rotation, cells were washed twice with PBS-FCS and incubated with goat anti-mouse IgG fluorescein isothiocyanate (FITC)-conjugated antibody (Molecular Probes Inc., Eugene, Oreg.) for 30 min in the dark with rotation. The cells were washed three times with PBS-FCS and resuspended in 100 μl of PBS-FCS; 5 μl of the suspension was allowed to dry on slides overnight in the dark. Immunofluorescence was visualized with an Olympus BX41 microscope (Olympus America, Melville, N.Y.) with a fluorescein filter cube. Images were captured with a Retiga digital camera and Q capture 2.0 software (QImaging, Burnaby, Canada) and processed with Adobe Photoshop version 6.0.1 (Adobe Systems, San Jose, Calif.).
To confirm the reactivity of E. coli pSCREEN transformants with MAb 4F11, 5-ml overnight cultures were pelleted and resuspended in 200 μl of 1x Laemmli-SDS running/sample buffer with 5% β-mercaptoethanol, and 20-μl samples were separated on NuPAGE Bis-Tris 4 to 12% gels (Invitrogen) by SDS-PAGE. S. pneumoniae from blood agar plates was grown in 5 ml of THY overnight, 1-ml aliquots were pelleted, resuspended in 200 μl of 1 × sample buffer, and 20 μl was resolved by SDS-PAGE. Purified recombinant protein was suspended in sample buffer in equal concentrations and separated as described for cell lysates. Western blots were performed as previously described (25).
Epitope mapping.
The epitope recognized by MAb 4F11 within mouse P. carinii Kex1 was mapped with the Novatope system (Novagen, Madison, Wis.) following the manufacturer's instructions with some alterations. Briefly, 30 μg of plasmid A32:pTrcHIS, which contains the C-terminal 620 residues of the mouse P. carinii Kex1 cDNA (25), was partially digested with DNase I, and fragments were separated by agarose gel electrophoresis. Fragments between 50 and 150 bp were gel purified, the ends of the fragments were filled in with T4 polymerase, and single-stranded deoxyriboadenylate tails were added to the 3′ ends with Tth polymerase. Fragments were then ligated into the linearized pSCREEN T-vector, which contains 5′ thymidine overhangs, and used to transform E. coli Nova Blue cells. Following an overnight incubation at 37°C, transformants were lifted onto nitrocellulose filters, lysed in a chloroform vapor chamber, and denatured with 6 M urea. Filters were then processed analogous to a Western blot (see below). Positive reactivity to MAb 4F11 of selected clones was confirmed by Western blot of boiled lysates.
Recombinant protein expression and purification.
Production of 6 × His-tagged thioredoxin fusion proteins with the pBADTHIO expression system in E. coli was performed following the manufacturer's instructions. Briefly, overnight cultures were diluted 1:40 in fresh LB-ampicillin (100 μg/ml) and grown at 37°C with shaking to an absorbance at 600 nm of approximately 0.5. For fusion protein induction, arabinose was added to a final concentration of 0.02%, and cultures were grown an additional 5 h at 37°C with shaking. Cells were pelleted by centrifugation at 1,000 × g and then resuspended in 1/5 the original culture volume with lysis buffer (50 mM sodium phosphate, 6 M guanidine-HCl, 300 mM NaCl) and vortexed vigorously. Debris was pelleted by centrifugation at 10,000 × g at 4°C. The supernatants were passed over a Talon affinity resin column (BD Biosciences, Palo Alto, Calif.) that had been preequilibrated with lysis buffer. The columns were then washed twice with 10 ml of 50 mM sodium phosphate-300 mM NaCl. The fusion proteins were eluted with 2 ml of buffer containing 50 mM sodium phosphate, 300 mM NaCl, and 150 mM imidazole. The eluted protein was dialyzed overnight against PBS at 4°C and concentrated by vacuum centrifugation. Protein concentrations were determined with a BCA microwell assay kit (Pierce, Rockford, Ill.).
Purification of native P. carinii antigen recognized by MAb 4F11.
The MAb 4F11 affinity column was prepared with 4F11(G1) and Reactigel resin (Pierce) following the manufacturer's instructions. Briefly, 10 ml of 1.7-mg/ml 4F11(G1) was coupled to 3 ml of 6× Reactigel overnight at 4°C. The supernatant was removed, and the resin was blocked with pH 9.0 1.0 M ethanolamine and washed with PBS prior to use. P. carinii antigens recognized by MAb 4F11 were purified from 1 ml of sonicated P. carinii-infected SCID mouse lung homogenates (8 × 106 organisms) by passage over the MAb 4F11 affinity column five times, followed by two washes with 10 ml of PBS and elution with pH 2.5 100 mM glycine buffer.
Statistical analysis.
Experimental ELISA results were statistically compared to the control values with a two-tailed Student's t test. Results were considered significant if P was ≤0.05.
GenBank accession numbers, BLAST searches, and protein sequence alignment.
The GenBank accession numbers for sequences presented in the text are as follows: mouse P. carinii cDNA clone A12, AY371664; and URSP PspA clade defining region and proline-rich repeat, AY371665. Searches for short, nearly exact matches to the kexin 17-mer MAb 4F11 epitope in GenBank were conducted with the BLASTp database search algorithm (2). The Kexin856-872 and Kexin777-787 MAb 4F11 epitopes were aligned with the deduced amino acid sequence of the PspA proline-rich repeat region with the Genetics Computer Group Genesys software (GCG, University of Rochester) (30).
Cloning and expression of an S. pneumoniae pspA fragment.
Conserved primers (Table 1) (22) were used to PCR amplify and sequence a portion of the S. pneumoniae strain URSP2 pspA gene. With primers derived from the PspA sequence obtained, an in-frame fragment encoding the N-terminal alpha-helical domain through the proline-rich repeat region of the molecule was cloned into thioredoxin fusion vector pBADTHIO (Invitrogen) for recombinant polypeptide expression.
RESULTS
Recognition of P. carinii from different hosts by MAb 4F11.
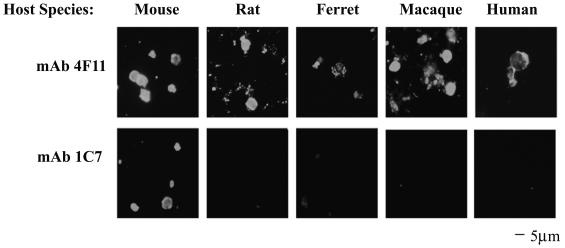
The mAbs 4F11 and 1C7, which were derived from mice immunized with mouse P. carinii, were used as probes against P. carinii isolates derived from mice, rats, ferrets, rhesus macaques, and humans (also termed P. jiroveci). As shown in Fig. 1, MAb 4F11 recognized antigens on the surface of P. carinii derived from all five host species. However, the isotype-matched MAb 1C7 was only capable of recognizing mouse P. carinii, the organism used as the immunogen, a characteristic of most anti-P. carinii antibodies (14). The pattern and intensity of fluorescent staining suggest that the epitope recognized by MAb 4F11 is abundant on the surface of P. carinii cysts and possibly on the trophic form of the organism, which may be represented by the smaller, highly fluorescent particles seen in Fig. 1.
FIG. 1.
MAb 4F11 recognizes P. carinii derived from multiple different host species. MAbs 4F11 and 1C7, both IgM isotypes derived against P. carinii in mice, were used as probes in an IFA of P. carinii isolates from mice, rats, ferrets, macaques, and humans. Each MAb was used at a concentration of approximately 50 ng/ml.
Identification of the epitope within mouse P. carinii Kex1 recognized by MAb 4F11.
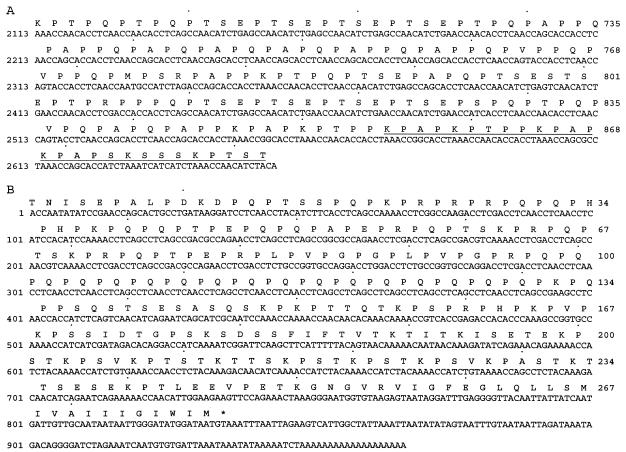
A cDNA clone (A32) encoding the C-terminal 200 amino acid residues of P. carinii Kex1 (25) was used to construct a shotgun cleavage library in E. coli; colony immunoscreening was performed with MAb 4F11. Positive clones were confirmed by Western blotting. Inserts from the plasmids of three positive clones were sequenced and shown to contain the 27-amino-acid region underlined in Fig. 2A. The epitope mapped to the junction of the proline-rich domain and serine/threonine-rich domain of the kexin molecule. To further delineate the epitope, we analyzed two fusion proteins, one containing the first 17 amino acids (Pro-rich domain residues 856 to 872) and the other the last 10 amino acids (Ser/Thr-rich domain residues 873 to 882) of the 27-mer epitope region, in Western blots with MAb 4F11 and an expression control antibody. It was determined that the epitope recognized by MAb 4F11 resides within the first 17 amino acids of the 27-mer (data not shown).
FIG. 2.
Nucleotide and deduced amino acid sequences of the proline-rich domains of P. carinii antigens. (A) Nucleotide and deduced amino acid sequences (GenBank accession no. AF093132) of the proline-rich domain of mouse P. carinii kexin, showing the MAb 4F11 epitope (underlined) mapped in this study. (B) Nucleotide and deduced amino acid sequences of P. carinii cDNA clone A12 (GenBank accession no. AY371664).
Identification of an additional mouse P. carinii protein recognized by MAb 4F11.
In Western blots of P. carinii-infected mouse lung homogenates, several bands are detected by MAb 4F11 (12). A previous immunoscreen of a mouse P. carinii cDNA expression library in λgt11 identified multiple clones containing inserts that encode proteins recognized by MAb 4F11. The primary structure of clone A12 is distinct from mouse P. carinii Kex1 and encodes a 278-amino-acid polypeptide that is rich in proline residues. Clone A12 appears to encode the C-terminal portion of its respective protein based on the presence of a stop codon and poly(A) tail in the nucleotide sequence of the cDNA (Fig. 2B). Outside the proline-rich region, there is no significant homology between the A12 polypeptide and either P. carinii Kex1 or any other protein available in the databases. A hydrophobic C terminus in the A12 polypeptide suggests that the mature protein may be membrane anchored, and the high proline content suggests that the molecule may be cell wall associated (4, 21, 38).
Identification of MAb 4F11 epitope-containing region of mouse P. carinii cDNA clone A12.
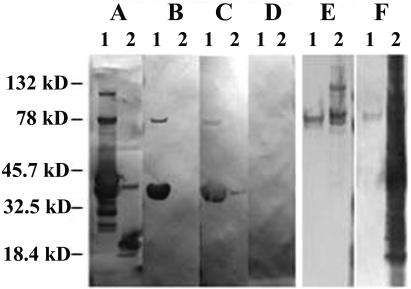
Alignment of the P. carinii Kex1 MAb 4F11 epitope with the deduced amino acid sequence of clone A12 revealed no obvious matches, though some areas of similarity in charge distribution and proline content were observed (not shown). To identify the region of the molecule recognized by MAb 4F11, truncated forms of the A12 molecule were expressed as thioredoxin fusion proteins. Constructs containing amino acid residues 1 to 142 (not shown) and 1 to 82 (Fig. 3B) both reacted with MAb 4F11, while the fusion partner alone was detected by the epitope tag MAb but did not react with MAb 4F11 (Fig. 3A and B). These results narrowed the MAb 4F11 epitope to the first 82 amino acids encoded by clone A12.
FIG. 3.
Western blotting of P. carinii antigens. (A to D) Western blots of A121-82 (lanes 1) and the thioredoxin fusion partner alone (lanes 2) with (A) anti-V5 epitope tag MAb, (B) 4F11(G1), (C) anti-P. carinii hyperimmune mouse serum, 1:250 dilution, and (D) pooled normal mouse sera, 1:250 dilution. (E and F) Western blots of MAb 4F11 affinity-purified P. carinii antigen (lane 1) and P. carinii-infected mouse lung homogenate (lane 2) with (E) MAb 4F11 and (F) anti-P. carinii hyperimmune serum, 1:250 dilution. Pooled normal mouse sera did not react with either affinity-purified antigens or total P. carinii-infected mouse lung homogenates (data not shown).
Recognition of recombinant A12 fusion protein and MAb 4F11 immunopurified native P. carinii antigens by hyperimmune sera from P. carinii-immunized mice.
The fusion protein containing the first 82 amino acids encoded by cDNA clone A12 also reacted with hyperimmune sera from mice immunized with whole P. carinii (Fig. 3C), but not with sera from unimmunized mice (Fig. 3D). This demonstrates that antibodies are made against the MAb 4F11 epitope-containing region of the A12 protein during a protective response to P. carinii. The anti-P. carinii antisera also recognized a P. carinii antigen(s) from the lungs of SCID mice purified with an MAb 4F11 affinity column (Fig. 3F). The faintness of the band recognized in the MAb 4F11 affinity column compared to P. carinii-infected mouse lung homogenates is likely due to the low concentration of antigen recovered in the purification process, as determined by the inability to detect similarly loaded antigen in silver-stained SDS-PAGE gels (not shown).
Fine-structure analysis of the MAb 4F11 epitope constructs in mouse P. carinii Kex1 and clone A12.
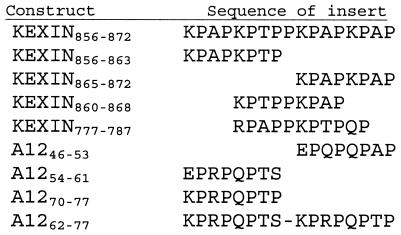
Further examination of both the mouse P. carinii Kex1 and A12 sequences revealed a number of near matches to the Kex1 17-mer epitope identified in the immunoscreen. To determine the sequence constraints of the different possible MAb 4F11 epitopes in Kex1 and A12, fusion proteins containing several of the putative epitopes were made. An alignment of the fusion protein inserts based on conservation of the positions of their proline residues is shown in Fig. 4.
FIG. 4.
Alignment of amino acid sequences of inserts used in epitope analysis based on the positions of conserved proline residues.
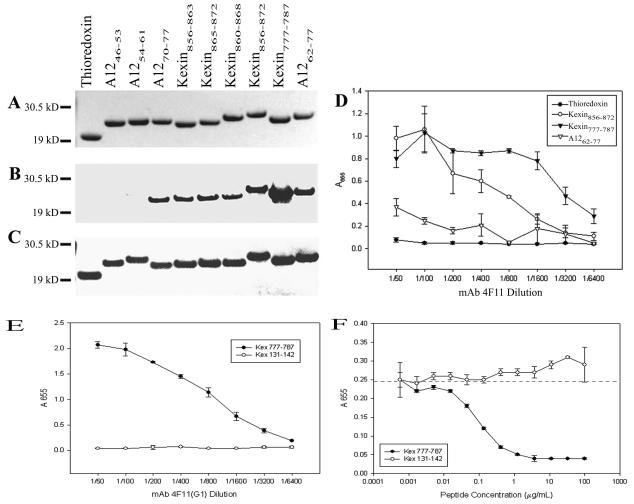
Kexin856-872 contains the 17-mer identified in the original immunoscreen (K856PAPKPTPPKPAPKPAP872). The A1262-77construct (K62PRPQPTSKPRPQPTP77) was chosen because it was the nearest match to the Kex1 17-mer within the Nterminal 82 residues shown to contain an MAb 4F11 epitope (Fig. 3B). The homologous Kexin777-787 construct (R777PAPPKPTPQP787) is located upstream of the region encoded by cDNA clone A32. Because Kexin856-872 and the sequence upstream of this region consist of several nearly exact repeats of eight or nine amino acids, three overlapping constructs, Kexin856-863 (K856PAPKPTP863), Kexin865-872 (K865PAPKPAP872) and Kexin860-868 (K860PTPPKPAP868), were made in an attempt to narrow the epitope further. Three 8-mer fusions were also designed from the A12 sequence, one of which had high similarity to the kexin 8-mers (A1270-77; K70PRPQPTP77). The other two A12 8-mers had charge substitutions at the first position to determine whether a basic residue was required at this position (A1246-53 [E46PRPQPTS53] and A1254-61 [E54PQPQPAP61]).
When 1 μg of each of the purified proteins was separated by SDS-PAGE, the recombinant protein was the only band visible by Coomassie staining (Fig. 5A). Western blots of gels run in parallel showed that A1246-53 and A1254-61, which contain negatively charged glutamic acid residues in the first position, do not react with MAb 4F11, whereas all other constructs that contain positively charged amino acid residues at the first position react with MAb 4F11. These blots also show that the thioredoxin fusion partner alone does not react with MAb 4F11, yet all constructs and the thioredoxin control show equal reactivity to the anti-V5 antibody, which recognizes an epitope within the fusion partner (Fig. 5B and C).
FIG. 5.
Analysis of purified recombinant epitope-thioredoxin fusion proteins. (A) Gel stained with Coomassie blue. (B) Western blot with MAb 4F11(G1), (C) Western blot with anti-V5 epitope tag MAb. (D) ELISA of epitope-thioredoxin fusion constructs with MAb 4F11(G1). Results are plotted as the mean ± standard error of triplicate experiments. (E) ELISA of synthetic P. carinii peptides with MAb 4F11(G1). Results are plotted as the mean ± standard deviation of triplicate experiments. (F) Competitive ELISA with three fold dilutions of synthetic P. carinii peptides as soluble competitors for 4F11(G1) (diluted 1:3,200) binding against plate-bound sonicated mouse P. carinii. Results are plotted as the mean ± standard deviation of triplicate experiments. The dashed line indicates the mean absorbance at 655 nm with no inhibitor and a 4F11(G1) dilution of 1:3,200.
The positive reactivity of A1270-77, Kexin856-863, and Kexin865-872 with MAb 4F11 suggests that MAb 4F11 could recognize an 8-amino-acid peptide. An unanticipated observation was that the Kexin777-787 showed greater binding with MAb 4F11 than the original epitope identified, Kexin856-872, based on the size and intensity of the band detected by Western blotting. Since this sequence of mouse P. carinii kexin falls upstream of the A32 cDNA clone-encoded region used in the mapping experiments, it was not identified by the original epitope mapping strategy. The consensus 8-mer epitope recognized by MAb 4F11 is K/RPA/RPK/QPA/TP, though additional substitutions in the longer epitopes still allow recognition by MAb 4F11. For example, Kexin777-787 and Kexin860- 868 contain an additional proline residue at the fifth position and Kexin777-787 has a charge-conserved arginine at the first position instead of a lysine.
To further evaluate the ability of MAb 4F11 to recognize the larger fusion constructs, we performed ELISA experiments with an MAb 4F11(G1) switch variant or anti-V5 epitope tag control antibody. As shown in Fig. 5D, Kexin856-872 and Kexin777-787 showed the highest reactivity with MAb 4F11 at lower dilutions, whereas A1262-77 showed lower reactivity but significantly higher than the thioredoxin fusion partner alone (P ≤ 0.05). At higher MAb 4F11 dilutions, Kexin777-787 showed greater reactivity with MAb 4F11 than did the original 17-mer epitope, which is in agreement with the Western blot data. At 1:800 or greater dilution, the A12 16-mer showed only background reactivity to MAb 4F11, suggesting that this is the weakest binder of the three epitopes tested.
To confirm the specificity of MAb 4F11 to the epitope of highest apparent affinity (Kex777-787) in the absence of a fusion partner, a synthetic peptide was used in ELISA experiments. The control peptide, Kex131-142 (SGDTGNVNSGEK), did not react with MAb 4F11, whereas Kex777-787 showed high reactivity to MAb 4F11 (Fig. 5E). In an inhibition ELISA, Kexin777-787 was able to completely block binding of MAb 4F11 to native P. carinii antigens at concentrations as low as 1 μg/ml and showed 50% inhibition of binding at 150 ng/ml, whereas Kex131-142 showed no inhibition at the highest concentration tested (Fig. 5F).
Identification of an S. pneumoniae PspA surface protein isoform that is recognized by MAb 4F11.
BLAST searches (2) of GenBank for short, nearly exact matches to the Kex1 17-mer corresponding to the MAb 4F11 epitope revealed a number of proline-rich protein sequences in microbes and plants, but none in mammals. One of these sequences, S. pneumoniae PspA (Gene Bank accession number AAF70097), also contained a large number of lysine residues with similar periodicity to the lysines in Kexin856-872. This proline-lysine repeat motif appears in the majority of isoforms of PspA from different S. pneumoniae strains (22).
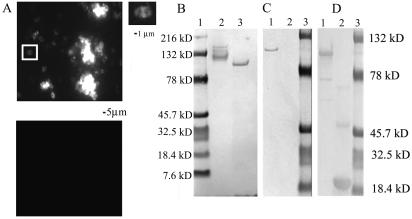
To determine whether MAb 4F11 was capable of recognizing epitopes on the pneumococcal surface, IFAs were performed on four clinical reference strains of S. pneumoniae. All four isolates showed positive staining with MAb 4F11 by IFA, but one strain, URSP2, demonstrated intense reactivity (Fig. 6A). This isolate was chosen for further analysis. To confirm the reactivity between S. pneumoniae and MAb 4F11, extracts of URSP2 cells were analyzed by SDS-PAGE and Western blotting with MAb 4F11. As shown in Fig. 6B, lane 3, a single band of approximately 90 kDa in the S. pneumoniae lysate lane was recognized by MAb 4F11. This band is within the reported size range of S. pneumoniae PspA isoforms (34).
FIG. 6.
MAb 4F11 recognizes surface antigen PspA of S. pneumoniae strain URSP2. (A) (Upper panel) S. pneumoniae was probed with MAb 4F11 and FITC-conjugated secondary antibody; right, enlargement of boxed area showing MAb 4F11 staining of S. pneumoniae diplococcus. (Lower panel) S. pneumoniae probed with isotype-matched MAb 2B5 and FITC-conjugated secondary antibody. (B) Western blot of P. carinii-infected mouse lung homogenates and S. pneumoniae culture lysates probed with MAb 4F11. Lanes: 1, molecular size markers; 2, P. carinii-infected mouse lung homogenate; 3, S. pneumoniae URSP2 lysate. (C and D) Western blots probed with (C) MAb 4F11(G1) and (D) anti-V5 epitope-tagged MAb. Lanes: 1, purified recombinant URSP2 PspA-thioredoxin fusion protein; 2, thioredoxin only; 3, molecular size markers.
A portion of URSP2 PspA containing the nucleotide sequence encoding the proline-rich domain was cloned and sequenced. The deduced amino acid sequence of the PspA fragment was shown to contain a repetitive 88-amino-acid stretch with two regions of 80% or greater similarity to Kexin856-872 and three regions with 80% or greater similarity to Kexin777-787 (Fig. 7). These two Kex1 epitopes showed the strongest reactivity to MAb 4F11 by Western blotting and ELISA (Fig. 5B and 5D). Purified recombinant thioredoxin fusion protein containing the α-helical domain, clade defining region, and proline-rich repeat region of URSP2 PspA was shown to react with MAb 4F11 (Fig. 6C). The thioredoxin fusion partner alone did not react with MAb 4F11 but did react with the anti-V5 epitope tag MAb (Fig. 6C and D). This strongly suggests that the band recognized by MAb 4F11 in Western blots of whole-cell extracts of URSP2 is PspA.
FIG. 7.
Comparison of P. carinii Kex1 epitopes with S. pneumoniae PspA. Computer-assisted alignment of Kexin856-872 and Kexin777-787 to S. pneumoniae URSP2 PspA partial deduced amino acid sequence (GenBank accession no. AY371665). Alignments with greater than 80% similarity and no more than one gap are shown (I, identity, :, similarity, ., gap). The clade-defining region of URSP2 PspA is underlined. The S. pneumoniae sequence is numbered based on the full-length PspA sequence from strain BG8743 (GenBank accession no. AF071803) (22).
DISCUSSION
The ability of MAb 4F11 to recognize P. carinii derived from multiple hosts (Fig. 1) and its ability to recognize antigens other than the immunodominant surface antigen gpA (15, 25) separate MAb 4F11 from most anti-P. carinii antibodies (9, 11). MAb 4F11 takes on added importance because of its ability to confer passive prophylaxis against PCP in a mouse model (12). We describe the mapping of a number of similar peptide epitopes recognized by MAb 4F11 within two different P. carinii antigens. BLAST searches identified epitopes within S. pneumoniae PspA with a high degree of identity to those recognized by MAb 4F11 in P. carinii. The epitopes recognized by MAb 4F11 are not identical but are highly similar in their proline and positively charged amino acid content. Analysis of the deduced amino acid sequences of the two P. carinii antigens containing epitopes recognized by MAb 4F11 suggests that at least one of the two antigens is surface localized. This is in agreement with the IFA data for nonpermeabilized P. carinii organisms (Fig. 1). A rat P. carinii kexin-like molecule has been reported as being localized to the cyst surface (26). If P. carinii Kex1 and the A12 antigen are on the surface of the organism, the proline-rich domains of these molecules would likely be cyst wall associated, based on the proximity of these domains to the hydrophobic C termini of each protein. The potential for MAb 4F11 to recognize multiple similar epitopes may explain its ability to recognize P. carinii isolated from a number of different hosts. This may also explain the apparent abundance of P. carinii surface antigens recognized by 4F11 in IFA.
We report the initial characterization of mouse P. carinii cDNA clone A12. Southern blotting shows a single band recognized by an A12 probe in restriction endonuclease digests of P. carinii-infected mouse lung homogenates and no bands in digests of DNA from uninfected mouse lung homogenates, providing further confirmation that A12 is a P. carinii antigen (L. Fletcher and F. Gigliotti, unpublished observations). Future studies will include cloning and more detailed molecular and antigenic characterization of the full-length molecule. With the identification of the polypeptide fragment encoded by cDNA A12, a total of three mouse P. carinii antigens with proline-rich regions have been identified, including P. carinii Kex1 and gpA (20, 25). However, MAb 4F11 does not bind to gpA (25). The proline-rich domains of these molecules may represent a conserved motif in P. carinii surface antigens. Proline-rich surface proteins have also been identified in the fungi Candida albicans (32) and Saccharomyces cerevisiae (8) and in a number of gram-positive cocci (4, 7, 19, 23, 29). Some of these proline-rich regions are speculated to be cell wall associated (19, 23, 26, 29).
The recognition of recombinant A12 and MAb 4F11 immunopurified P. carinii antigens by hyperimmune sera demonstrates that antibodies are generated against these molecules in an anti-P. carinii response that is protective (21). These data also suggest that in obtaining the B-cell hybridoma that produces MAb 4F11, we did not simply capture a rare immunological event. The culmination of these two points and the ability of MAb 4F11 to confer passive protection against PCP make the P. carinii antigens recognized by MAb 4F11 attractive vaccine candidates.
In silico analysis identified S. pneumoniae PspA as having a region of similarity to the P. carinii MAb 4F11 epitope. This study confirms the presence of shared surface antigen epitopes between these two highly divergent pathogens. The importance of PspA as an S. pneumoniae antigen has been demonstrated by the ability of PspA to induce cross-protection in mice against multiple S. pneumoniae strains (27, 31, 34). Comparison of the clade-defining region of URSP2 PspA to an alignment of a number of different PspA isoforms (22) placed URSP2 PspA in family 1, clade 1. Approximately 50% of S. pneumoniae isolates carry PspA from family 1 (22), suggesting that S. pneumoniae URSP2 PspA represents a potentially prevalent isoform of the molecule.
The proline-rich region of PspA contains at least one protective epitope, since antibodies that react with the proline-rich region of PspA confer cross-protection against multiple S. pneumoniae strains in a mouse model (5). A pilot experiment demonstrated that mice administered MAb 4F11(G1) intraperitoneally prior to intranasal challenge with S. pneumoniae showed a 99.9% reduction in bacteremia compared to control animals, as determined by blood CFU counts 1 day postchallenge (F. Gigliotti and A. G. Harmsen, unpublished observations). In addition, truncated PspA fragments containing the α-helical domain of the molecule, the clade-defining region, and the proline-rich repeat region show increased cross-protective capabilities over fragments containing the α-helical domain and clade-defining region alone (31). Human sera often contain natural, polyreactive antibodies that recognize proline-rich epitopes; these antibodies may represent early defense mechanisms against pathogens (35). Together, these data suggest that exposed proline-rich regions of surface antigens may represent pathogen-associated molecular patterns that are recognized by the immune system (24).
MAb recognition of multiple, proline-rich epitopes, coupled with the prevalence of proline-rich surface antigens on pathogens as different as P. carinii and S. pneumoniae, suggests that it is possible to generate cross-protection by immunization with one or more of these antigens. Such an approach may be of particular interest in prevention of PCP, since P. carinii cannot be grown continuously in culture and organisms derived from one animal host do not provide protection against P. carinii in a different host (14). It will also be of interest to determine whether sera generated against polypeptides containing the MAb 4F11 epitopes are capable of recognizing P. carinii derived from multiple hosts, including humans, as shown for MAb 4F11.
Acknowledgments
We thank Sue Rob of the Clinical Microbiology Laboratory at Strong Memorial Hospital, University of Rochester Medical Center, for providing S. pneumoniae strains. We also appreciate the excellent technical assistance provided by Margaret Chovaniec and Lani Sherrill.
This work was supported by NIH grants AI23302 (F.G.), AI45479 (C.G.H.), and NIAID 5T32AI07362 (Department of Microbiology and Immunology, University of Rochester).
Editor: T. R. Kozel
REFERENCES
- 1.Aliouat, E. M., E. Mazars, E. Dei-Cas, P. Delcourt, P. Billaut, and D. Camus. 1994. Pneumocystis cross infection experiments using SCID mice and nude rats as recipient host, showed strong host-species specificity. J. Eukaryot. Microbiol. 41:71S. [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atzori, C., F. Agostoni, E. Angeli, A. Mainini, V. Micheli, and A. Cargnel. 1999. P. carinii host specificity: attempt of cross infections with human derived strains in rats. J. Eukaryot. Microbiol. 46:112S. [PubMed] [Google Scholar]
- 4.Briles, D. E., J. Yother, and L. S. McDaniel. 1988. Role of pneumococcal surface protein A in the virulence of Streptococcus pneumoniae. Rev Infect Dis 10(Suppl. 2):S372-S374. [DOI] [PubMed] [Google Scholar]
- 5.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y. Y., K. A. Clancy, and R. A. Burne. 1996. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect. Immun. 64:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahnestock, S. R., P. Alexander, J. Nagle, and D. Filpula. 1986. Gene for an immunoglobulin-binding protein from a group G streptococcus. J. Bacteriol. 167:870-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frevert, J., and C. E. Ballou. 1985. Saccharomyces cerevisiae structural cell wall mannoprotein. Biochemistry 24:753-759. [DOI] [PubMed] [Google Scholar]
- 9.Gigliotti, F., B. A. Garvy, C. G. Haidaris, and A. G. Harmsen. 1998. Recognition of Pneumocystis carinii antigens by local antibody-secreting cells following resolution of P. carinii pneumonia in mice. J. Infect. Dis. 178:235-242. [DOI] [PubMed] [Google Scholar]
- 10.Gigliotti, F., B. A. Garvy, and A. G. Harmsen. 1996. Antibody-mediated shift in the profile of glycoprotein A phenotypes observed in a mouse model of Pneumocystis carinii pneumonia. Infect. Immun. 64:1892-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gigliotti, F., and C. G. Haidaris. 1998. Antigenic characterization of Pneumocystis carinii. Semin. Respir. Infect. 13:313-322. [PubMed] [Google Scholar]
- 12.Gigliotti, F., C. G. Haidaris, T. W. Wright, and A. G. Harmsen. 2002. Passive intranasal monoclonal antibody prophylaxis against murine Pneumocystis carinii pneumonia. Infect. Immun. 70:1069-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gigliotti, F., P. J. Haidaris, C. G. Haidaris, T. W. Wright, and K. R. Van der Meid. 1993. Further evidence of host species-specific variation in antigens of Pneumocystis carinii using the polymerase chain reaction. J. Infect. Dis. 168:191-194. [DOI] [PubMed] [Google Scholar]
- 14.Gigliotti, F., and A. G. Harmsen. 1997. Pneumocystis carinii host origin defines the antibody specificity and protective response induced by immunization. J. Infect. Dis. 176:1322-1326. [DOI] [PubMed] [Google Scholar]
- 15.Gigliotti, F., and T. McCool. 1996. Glycoprotein A is the immunodominant antigen of Pneumocystis carinii in mice following immunization. Parasitol. Res. 82:90-91. [DOI] [PubMed] [Google Scholar]
- 16.Gigliotti, F., D. C. Stokes, A. B. Cheatham, D. S. Davis, and W. T. Hughes. 1986. Development of murine monoclonal antibodies to Pneumocystis carinii. J. Infect. Dis. 154:315-322. [DOI] [PubMed] [Google Scholar]
- 17.Gigliotti, F., J. A. Wiley, and A. G. Harmsen. 1998. Immunization with Pneumocystis carinii gpA is immunogenic but not protective in a mouse model of P. carinii pneumonia. Infect. Immun. 66:3179-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graves, D. C., S. J. McNabb, M. H. Ivey, and M. A. Worley. 1986. Development and characterization of monoclonal antibodies to Pneumocystis carinii. Infect. Immun. 51:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guss, B., M. Uhlen, B. Nilsson, M. Lindberg, J. Sjoquist, and J. Sjodahl. 1984. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur. J. Biochem. 138:413-420. [DOI] [PubMed] [Google Scholar]
- 20.Haidaris, C. G., O. F. Medzihradsky, F. Gigliotti, and P. J. Simpson-Haidaris. 1998. Molecular characterization of mouse Pneumocystis carinii surface glycoprotein A. DNA Res. 5:77-85. [DOI] [PubMed] [Google Scholar]
- 21.Harmsen, A. G., W. Chen, and F. Gigliotti. 1995. Active immunity to Pneumocystis carinii reinfection in T-cell-depleted mice. Infect. Immun. 63:2391-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1986. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus. Repetitive structure and membrane anchor. J. Biol. Chem. 261:1677-1686. [PubMed] [Google Scholar]
- 24.Janeway, C. A., Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54:1-13. [DOI] [PubMed] [Google Scholar]
- 25.Lee, L. H., F. Gigliotti, T. W. Wright, P. J. Simpson-Haidaris, G. A. Weinberg, and C. G. Haidaris. 2000. Molecular characterization of KEX1, a kexin-like protease in mouse Pneumocystis carinii. Gene 242:141-150. [DOI] [PubMed] [Google Scholar]
- 26.Lugli, E. B., E. T. Bampton, D. J. Ferguson, and A. E. Wakefield. 1999. Cell surface protease PRT1 identified in the fungal pathogen Pneumocystis carinii. Mol. Microbiol. 31:1723-1733. [DOI] [PubMed] [Google Scholar]
- 27.McDaniel, L. S., J. S. Sheffield, P. Delucchi, and D. E. Briles. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun. 59:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris, A., C. B. Beard, and L. Huang. 2002. Update on the epidemiology and transmission of Pneumocystis carinii. Microbes Infect. 4:95-103. [DOI] [PubMed] [Google Scholar]
- 29.Pancholi, V., and V. A. Fischetti. 1988. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. J. Bacteriol. 170:2618-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ralph, B. A., D. E. Briles, and L. S. McDaniel. 1994. Cross-reactive protection eliciting epitopes of pneumococcal surface protein A. Ann. N. Y. Acad. Sci. 730:361-363. [DOI] [PubMed] [Google Scholar]
- 32.Staab, J. F., C. A. Ferrer, and P. Sundstrom. 1996. Developmental expression of a tandemly repeated, proline-and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans. J. Biol. Chem. 271:6298-6305. [DOI] [PubMed] [Google Scholar]
- 33.Stringer, J. R., and P. D. Walzer. 1996. Molecular biology and epidemiology of Pneumocystis carinii infection in AIDS. AIDS 10:561-571. [DOI] [PubMed] [Google Scholar]
- 34.Tart, R. C., L. S. McDaniel, B. A. Ralph, and D. E. Briles. 1996. Truncated Streptococcus pneumoniae PspA molecules elicit cross-protective immunity against pneumococcal challenge in mice. J. Infect. Dis. 173:380-386. [DOI] [PubMed] [Google Scholar]
- 35.Tchernychev, B., S. Cabilly, and M. Wilchek. 1997. The epitopes for natural polyreactive antibodies are rich in proline. Proc. Natl. Acad. Sci. USA 94:6335-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waltman, W. D., L. S. McDaniel, B. M. Gray, and D. E. Briles. 1990. Variation in the molecular weight of PspA (pneumococcal surface protein A) among Streptococcus pneumoniae. Microb. Pathog. 8:61-9. [DOI] [PubMed] [Google Scholar]