Summary
Background
Fluoroquinolones are broad-spectrum antibiotics widely used in the treatment of bacterial infections such as Staphylococcus aureus isolates. Resistance to these antibiotics is increasing.
Material/Methods
The occurrence of mutations in the grlA and gyrA loci were evaluated in 69 fluoroquinolone-resistant S. aureus isolates from 2 teaching hospitals of Tehran University of Medical Sciences.
Results
Out of the 165 S. aureus isolates, 87 (52.7%) were resistant to methicillin and 69 (41.8%) were resistant to fluoroquinolone. Fluoroquinolone-resistant S. aureus isolates had a mutation at codon 80 in the grlA gene and different mutational combinations in the gyrA gene. These mutational combinations included 45 isolates at codons 84 and 86, 23 isolates at codons 84, 86 and 106 and 1 isolate at codons 84, 86 and 90. Fluoroquinolone-resistant S. aureus isolates were clustered into 33 PFGE types.
Conclusions
The findings of this study show that the fluoroquinolone-resistant S. aureus strains isolated in the teaching hospitals in Tehran had multiple mutations in the QRDRs region of both grlA and gyrA genes.
Keywords: fluoroquinolone resistance, Staphylococcus aureus, QRDR
Background
Staphylococcus aureus, particularly methicillin-resistant S. aureus (MRSA) is of great global concern as it can cause serious infections in both hospitals and the community [1–3]. Increasing resistance to antibiotics among staphylococcal isolates limits the choices of antibiotics available to treat infections caused by these bacteria [4]. Fluoroquinolones are broad-spectrum antibiotics widely used in the treatment of bacterial infections such as gram-positive cocci; however, resistance to these antibiotics has significantly increased world-wide [5,6]. Three different mechanisms of fluoroquinolones resistance have been described in staphylococci. The first is the mutation in the grlA and grlB genes that encodes the subunits of DNA topoisomerase IV, the second is the mutation in the gyrA and gyrB genes that encode the subunits of DNA gyrase, and the third is an active efflux pump mediated by mutations in the norA gene [5–9]. In most cases, mutations occur in the highly conserved quinolone resistance-determining regions (QRDRs) of the grlA and gyrA genes [10].
Despite the high incidence of fluoroquinolones resistance among staphylococci, especially among MRSA, there is currently little information available on the incidence and the types of these resistances in many countries. This lack of information is of particular concern in the Persian Gulf region, where the prevalence of MRSA is high [9,11]. The aim of the present study was to provide information regarding the prevalence of fluoroquinolones resistance among S. aureus isolates in Tehran, Iran.
Material and Methods
Bacterial strains
A total of 165 S. aureus clinical isolates were cultured from patients attending 2 teaching hospitals of Tehran University of Medical Sciences between April 2009 and September 2010 (155 isolates from Imam Khomeini hospital and 10 isolates from Amir Alam hospital). They were cultured from wounds, blood, CSF, body fluid, and urine. Only 1 isolate per patient was included. Isolates were identified to species level using standard biochemical methods including Gram stain, catalase test, tube coagulase, DNase, and fermentation of mannitol [12,13].
Antimicrobial susceptibility test
Antibiotic-containing disks (Mast, UK) were used to determine the susceptibility of S. aureus isolates to several antibiotics according to the criteria established by the Clinical and Laboratory Standards Institute (CLSI) [14]. The antibiotics tested were ciprofloxacin, gatifloxacin, levofloxacin, norfloxacin, ofloxacin, and oxacillin. The minimum inhibitory concentrations (MIC) of ciprofloxacin, ofloxacin and oxacillin were determined using the microbroth dilution method as recommended by the CLSI guidelines. S. aureus (ATCC 29213) was used as control.
Amplification of mecA, grlA, and gyrA genes
Chromosomal DNA was extracted from the clinical S. aureus isolate as previously described [2]. The mecA, grlA, and gyrA genes were amplified by PCR-based methods, using specific primers [10,15,16]. PCR reactions were performed in a 50 μL volume consisting of 1X PCR buffer, 3 mM MgCl2, 0.4 μg/mL of each primer, 1.5 U Taq DNA polymerase, 0.2 mM dNTP Mix and 5 μL of DNA template. The PCR conditions consisted of a pre-denaturation step at 94°C for 5 min, followed by 30 cycles of at 94°C for 40 sec, 51°C for 40 sec and 72°C for 45 sec. A final extension step was performed at 72°C for 5 min. To determine the QRDRs sequences, the PCR products of grlA and gyrA genes were sequenced with both forward and reverse strands at Macrogen (Seoul, South Korea). Sequences were compared with wild-type sequences of grlA and gyrA genes with no mutations [17].
Pulsed Field Gel Electrophoresis (PFGE)
All fluoroquinolone-resistant S. aureus isolates were analyzed by PFGE. The entire genomic DNA was prepared as described previously [15]. After digestion with Sma I endonuclease, DNAs were separated by PFGE (APZoha, Tehran, Iran) for 24 h at 15°C, with an electric field of 6 V/cm3 in 0.5× TBE buffer. The pulse time increased from 1 to 30 sec for 11 h and 1 to 3 sec for 13 h. The gels were stained with ethidium bromide (1 μg/ml) and visualized by UV illumination. DNA from S. aureus NCTC8325 was prepared in the same way and run as molecular size standard.
Results
S. aureus strains were isolated from wounds (76), blood (42), respiratory tract (20), joint fluid (15), urine (11) and CSF (1) in this study.
Out of the 165 S. aureus isolates, 87 (52.7%) and 69 (41.8%) were resistant to methicillin and tested fluoroquinolone antibiotics, respectively. All MRSA isolates contained mecA gene and 67 (77%) isolates were resistant to fluoroquinolones. Overall, the oxacillin MIC50 and MIC90 levels among isolates of MRSA were 128 μg/ml and 512 μg/ml, respectively.
The results of mutations in the QRDRs of the gyrA and grlA genes and the MIC of ciprofloxacin and ofloxacin among fluoroquinolone-resistant S. aureus isolates examined in this study are shown in Table 1. In all the assessed isolates, a mutation in the grlA gene was observed at codon 80. Fluoroquinolone-resistant S. aureus isolates had different combinations of mutations in the gyrA gene. These mutational combinations included 45 isolates at codons 84 and 86, 23 isolates at codons 84, 86 and 106, and 1 isolate at codons 84, 86 and 90.
Table 1.
Mutations in grlA and gyrA genes and susceptibilities in fluoroquinolone resistant Staphylococcus aureus isolates.
| Gene | codon | Mutation | No of isolates | MIC of CIP* (μg/ml) | MIC of OFX (μg/ml) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Range | 50% | 90% | Range | 50% | 90% | ||||
| grlA | 80 | TCC→TTC | 69 | 16–128 | 16 | 128 | 8–32 | 8 | 32 |
| gyrA | 84 + 86 | TCA→TTA + ATT→ATC | 45 | 16–64 | 16 | 32 | 8–32 | 8 | 16 |
| 84 + 86 + 90 | TCA→TTA + ATT→ATC + ATG→AGG | 1 | 16 | 16 | 16 | 8 | 8 | 8 | |
| 84 + 86 + 106 | TCA→TTA + ATT→ATC + GGC→AC | 23 | 16–128 | 32 | 128 | 8–32 | 16 | 32 | |
MIC – Minimum Inhibitory Concentration; CIP – ciprofloxacin; OFX – ofloxacin.
Among the 69 fluoroquinolone-resistant S. aureus isolates, 42 PFGE patterns were generated by digestion with SmaI, which clustered into 33 PFGE types. The majority of the isolates (52) were clustered in 16 pulsotypes (Figure 1), and 4 PFGE types displayed subtypes. Seventeen PFGE types contained a single isolate.
Figure 1.
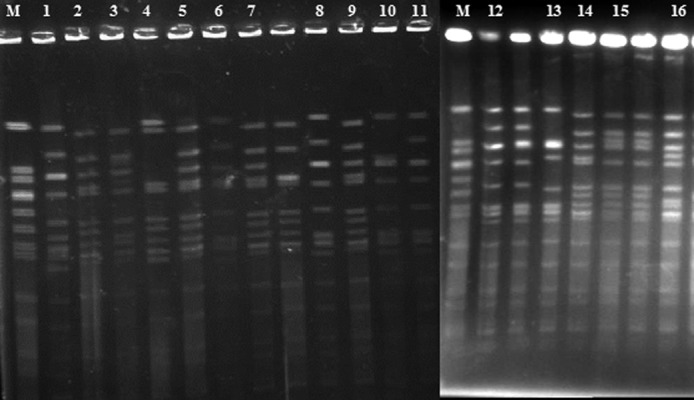
Pulsed-field gel electrophoresis (PFGE) of SmaI macro-restriction fragments of fluoroquinolone-resistant Staphylococcus aureus isolates. Lanes 1–16: Common PFGE types among tested fluoroquinolone-resistant S. aureus. Lane M. S. aureus NCTC 8325.
Discussion
The spread of antimicrobial resistance among S. aureus isolates is a worldwide problem. Since fluoroquinolones, particularly ciprofloxacin and gatifloxacin, are widely used in Iran to treat a variety of infections such as staphylococcal infections [18], fluoroquinolone resistance has posed a serious challenge to the Iranian medical community. This study aimed to characterize the phenotypic and genotypic resistance to fluoroquinolone among S. aureus isolates in Tehran, Iran.
The high prevalence rate of MRSA (52.7%) in the present study is alarming and indicates an increase in the prevalence of MRSA isolates in Tehran. The previous prevalence rate reported in Tehran teaching hospitals in 2008 was 36% [15]. This finding highlights the importance of modified empiric therapy and infection control policies among hospitals in Iran.
The results of this study show a fluoroquinolone resistance rate of 41.8% among S. aureus isolates, which was higher than the 13% to 20.7% reported by similar investigations [19, 20]. Many studies have shown that 1 or 2 point mutations in the QRDRs region of both grlA and gyrA genes are the main mechanisms of fluoroquinolone resistance, in particular ciprofloxacin resistance, among S. aureus isolates [6,16,21,22]. In agreement with many other studies, the results showed that all the fluoroquinolone-resistant isolates had a single mutation at codon 80 in the grlA gene and a mutation at codons 84 in the gyrA gene [16,17,23]. Among the 69 tested isolates, 23 had an additional mutation at codon 106 and 1 had a mutation at codon 90 in the gyrA gene. The mutation at codon 106 has also been reported in a few studies, but its effect on resistance was reported to be unknown [17,21]. According to our literature review, the point mutation at codon 90 (Tyr to Ser) in the gyrA gene has not been reported previously and further studies are needed to determine its effect on fluoroquinolone resistance. In the current study, all the fluoroquinolone-resistant isolates had a mutation at codon 86 in the gyrA gene; this point mutation is a silent mutation and has already been reported by others [8,24].
Our results are consistent with those of others who found that the same mutations in the QRDRs were detected in different PFGE types, and different combinations of mutations were also found in the isolates of the same PFGE type [10,16,25].
Conclusions
In conclusion, our findings show that fluoroquinolone-resistant S. aureus strains isolated from teaching hospitals in Tehran have multiple mutations in the QRDRs region of both grlA and gyrA genes.
Footnotes
Source of support: This research was supported by Tehran University of Medical Sciences & Health Services grant 9678/30-4-88
References
- 1.Brumfitt W, Hamilton-Miller JM. The worldwide problem of methicillin-resistant Staphylococcus aureus. Drugs Exp Clin Res. 1990;16:205–14. [PubMed] [Google Scholar]
- 2.Emaneini M, Taherikalani M, Eslampour MA, et al. Phenotypic and genotypic evaluation of aminoglycoside resistance in clinical isolates of staphylococci in Tehran, Iran. Microb Drug Resist. 2009;15:129–32. doi: 10.1089/mdr.2009.0869. [DOI] [PubMed] [Google Scholar]
- 3.Nicholas BD, Bhargave G, Hatipoglu A, et al. Preoperative prevalence of methicillin-resistant Staphylococcus aureus (MRSA) colonization in patients undergoing intranasal surgery. Med Sci Monit. 2010;16(8):CR365–68. [PubMed] [Google Scholar]
- 4.Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–91. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper DC. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect Dis. 2002;2:530–38. doi: 10.1016/s1473-3099(02)00369-9. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz FJ, Higgins PG, Mayer S, et al. Activity of quinolones against gram-positive cocci: mechanisms of drug action and bacterial resistance. Eur J Clin Microbiol Infect Dis. 2002;21:647–59. doi: 10.1007/s10096-002-0788-z. [DOI] [PubMed] [Google Scholar]
- 7.Horii T, Suzuki Y, Takeshita A, Maekawa M. Molecular characterization of 8-methoxyfluoroquinolone resistance in a clinical isolate of methicillin-resistant Staphylococcus aureus. Chemotherapy. 2007;53:104–9. doi: 10.1159/000098427. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka M, Wang T, Onodera Y, et al. Mechanism of quinolone resistance in Staphylococcus aureus. J Infect Chemother. 2000;6:131–39. doi: 10.1007/s101560070010. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka M, Onodera Y, Uchida Y, Sato K. Quinolone resistance mutations in the GrlB protein of Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:3044–46. doi: 10.1128/aac.42.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iihara H, Suzuki T, Kawamura Y, et al. Emerging multiple mutations and high-level fluoroquinolone resistance in methicillin-resistant Staphylococcus aureus isolated from ocular infections. Diagn Microbiol Infect Dis. 2006;56:297–303. doi: 10.1016/j.diagmicrobio.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 11.Fatholahzadeh B, Emaneini M, Gilbert G, et al. Staphylococcal cassette chromosome mec (SCCmec) analysis and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolates in Tehran, Iran. Microb Drug Resist. 2008;14:217–20. doi: 10.1089/mdr.2008.0822. [DOI] [PubMed] [Google Scholar]
- 12.Mahon CR, Lehman DC, Manuselis G. Text book of diagnostic microbiology. 3rd ed. Philadelphia, PA, USA: 2007. [Google Scholar]
- 13.Reisner SB, Woods GL, Thomson RP, et al. Specimen collection. In: Murray PR, Baron EJ, Pfaller MA, et al., editors. Manual of clinical microbiology. 7th ed. American Society for Microbiology; Washington, DC: 1999. pp. 64–76. [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-First Informational Supplement. 2011;31:M100–S21. [Google Scholar]
- 15.Fatholahzadeh B, Emaneini M, Aligholi M, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus clones from a teaching hospital in Tehran. Jpn J Infect Dis. 2009;62:309–11. [PubMed] [Google Scholar]
- 16.Horii T, Suzuki Y, Monji A, et al. Detection of mutations in quinolone resistance-determining regions in levofloxacin- and methicillin-resistant Staphylococcus aureus: effects of the mutations on fluoroquinolone MICs. Diagn Microbiol Infect Dis. 2003;46:139–45. doi: 10.1016/s0732-8893(03)00037-3. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz FJ, Jones ME, Hofmann B, et al. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob Agents Chemother. 1998;42:1249–52. doi: 10.1128/aac.42.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emaneini M, Aligholi M, Hashemi FB, et al. Isolation of vancomycin-resistant Staphylococcus aureus in a teaching hospital in Tehran. J Hosp Infect. 2007;66:92–93. doi: 10.1016/j.jhin.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Blumberg HM, Rimland D, Carroll DJ, et al. Rapid development of ciprofloxacin resistance in methicillin-susceptible and -resistant Staphylococcus aureus. J Infect Dis. 1991;163:1279–85. doi: 10.1093/infdis/163.6.1279. [DOI] [PubMed] [Google Scholar]
- 20.Marangon FB, Miller D, Muallem MS, et al. Ciprofloxacin and levofloxacin resistance among methicillin-sensitive Staphylococcus aureus isolates from keratitis and conjunctivitis. Am J Ophthalmol. 2004;137:453–58. doi: 10.1016/j.ajo.2003.10.026. [DOI] [PubMed] [Google Scholar]
- 21.Coskun-Ari FF, Bosgelmez-Tinaz G. grlA and gyrA mutations and antimicrobial susceptibility in clinical isolates of ciprofloxacin- methicillin-resistant Staphylococcus aureus. Eur J Med Res. 2008;13:366–70. [PubMed] [Google Scholar]
- 22.Takahata M, Yonezawa M, Kurose S, et al. Mutations in the gyrA and grlA genes of quinolone-resistant clinical isolates of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1996;38:543–46. doi: 10.1093/jac/38.3.543. [DOI] [PubMed] [Google Scholar]
- 23.Yoon EJ, Lee CY, Shim MJ, et al. Extended spectrum of quinolone resistance, even to a potential latter third-generation agent, as a result of a minimum of two GrlA and two GyrA alterations in quinolone-resistant Staphylococcus aureus. Chemotherapy. 2010;56:153–57. doi: 10.1159/000313529. [DOI] [PubMed] [Google Scholar]
- 24.Wang T, Tanaka M, Sato K. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob Agents Chemother. 1998;42:236–40. doi: 10.1128/aac.42.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noguchi N, Okihara T, Namiki Y, et al. Susceptibility and resistance genes to fluoroquinolones in methicillin-resistant Staphylococcus aureus isolated in 2002. Int J Antimicrob Agents. 2005;25:374–79. doi: 10.1016/j.ijantimicag.2004.11.016. [DOI] [PubMed] [Google Scholar]


