ABSTRACT
HIV infection of CD4+ T cells induces a range of host transcriptional changes in mRNAs as well as microRNAs that may coordinate changes in mRNAs. To survey these dynamic changes, we applied next-generation sequencing, analyzing the small RNA fraction of HIV-infected cells at 5, 12, and 24 h postinfection (RNA-Seq). These time points afforded a view of the transcriptomic changes occurring both before and during viral replication. In the resulting small RNA-Seq data set, we detected a phased pattern of microRNA expression. Largely distinct sets of microRNAs were found to be suppressed at 5 and 12 h postinfection, and both sets of changes rebounded later in infection. A larger set of microRNA changes was observed at 24 h postinfection. When integrated with mRNA expression data, the small RNA-Seq data indicated a role for microRNAs in transcriptional regulation, T cell activation, and cell cycle during HIV infection. As a unique benefit of next-generation sequencing, we also detected candidate novel host microRNAs differentially expressed during infection, including one whose downregulation at 24 h postinfection may allow full replication of HIV to proceed. Collectively, our data provide a uniquely comprehensive view of the changes in host microRNAs induced by HIV during cellular infection.
IMPORTANCE
New sequencing technologies allow unprecedented views into changes occurring in virus-infected cells, including comprehensive and largely unbiased measurements of different types of RNA. In this study, we used next-generation sequencing to profile dynamic changes in cellular microRNAs occurring in HIV-infected cells. The sensitivity afforded by sequencing allowed us to detect changes in microRNA expression early in infection, before the onset of viral replication. A phased pattern of expression was evident among these microRNAs, and many that were initially suppressed were later overexpressed at the height of infection, providing unique signatures of infection. By integrating additional mRNA data with the microRNA data, we identified a role for microRNAs in transcriptional regulation during infection and specifically a network of microRNAs involved in the expression of a known HIV cofactor. Finally, as a distinct benefit of sequencing, we identified candidate nonannotated microRNAs, including one whose downregulation may allow HIV-1 replication to proceed fully.
Introduction
HIV infection results in a multitude of changes in infected cells. During periods of active viral replication, these changes occur in multiple classes of molecules, including mRNAs and proteins (1, 2). Infected cells exhibit decreased basic functionality, e.g., in cell signaling (3), culminating in the eventual death of the cell. Using next-generation sequencing (NGS), we recently profiled the transcriptome of HIV-infected lymphoblastoid cells and found that over half of the detectable polyadenylated RNA species were differentially expressed (DE) during infection (1). These changes corroborated clinical results. For example, in CD4+ T cells isolated from HIV+ individuals, a distinct expression profile was observed comprising 260 genes whose expression correlated with viral load; these genes were involved in a variety of functions, including inflammatory signaling, cell cycle, and antigen presentation (4).
Small RNAs constitute an additional class of molecules whose expression is likely to be altered by HIV infection. Among the best-characterized types of small RNAs are microRNAs, single-stranded RNA molecules of approximately 22 nucleotides in length that bind mRNAs through complementary base pairing, resulting in suppressed translation from the mRNAs or degradation of the mRNAs.
HIV infection has been shown to alter microRNA expression, both in cultured cells and in peripheral blood mononuclear cells (PBMCs) isolated ex vivo, e.g., in references 5 and 6. However, technologies such as microarrays that were used in previous studies limited identifications to known, annotated microRNAs. NGS applied to small RNA (small RNA-Seq) circumvents this limitation, detecting microRNAs without the need for prior annotation as well as extending quantitation over a wider dynamic range. Indeed, sequencing has contributed recently to an expansion in the number of known microRNAs (7).
In previous studies where small RNA-Seq has been applied to HIV-infected cells, the focus has typically been on virally encoded microRNAs or on single time points, providing static snapshots of infection (8–11). In this article, we report the use of NGS to observe dynamic changes in cellular small RNAs from HIV-infected CD4+ cells. To this end, we sampled multiple time points, including those preceding detectable HIV replication, to identify signatures of different stages of infection in vitro. To characterize microRNAs, both known and putatively novel, we then integrated our NGS data set with mRNA-Seq data derived from the same experimental samples (1) and thereby identified microRNAs that may contribute to phenotypic changes in infected cells over time. We also provide a full account of putatively novel microRNAs and other small RNAs that were DE upon infection that may in turn provide new avenues for future research.
RESULTS
Abundance distribution of small RNA in HIV-infected cells.
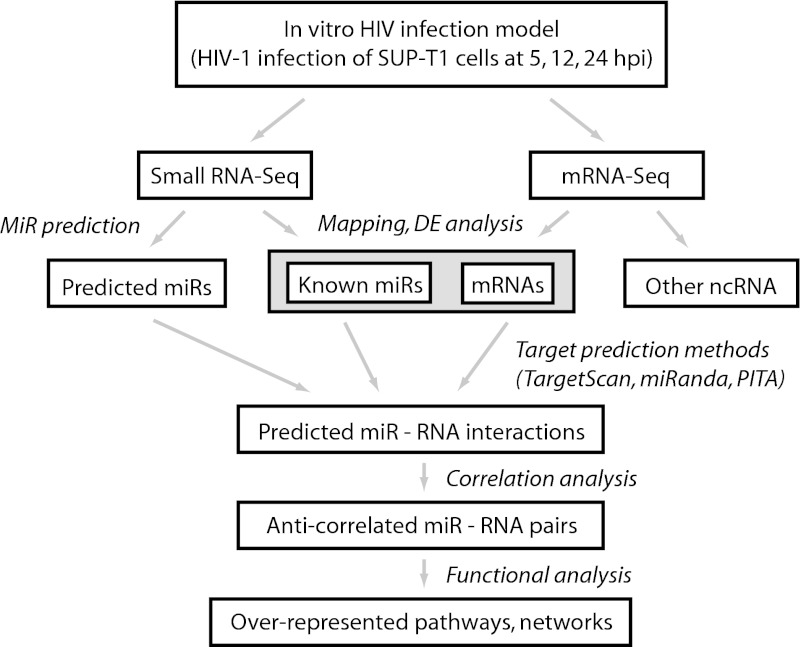
To determine the effect of HIV infection on cellular small-RNA expression, we infected human lymphoblastoid SUP-T1 cells with HIV-1 strain LAI and determined small RNA expression at three different time points: 5, 12, and 24 h postinfection (hpi). In a previous study (1), we found that levels of virus were nearly undetectable at 4 hpi, increasing at 12 hpi and peaking at 24 hpi. Our choice of timing parallels these time points. For controls, we also mock-infected cells using cell culture medium or alternatively infected cells with UV-inactivated HIV-1 (HIVUV). Small RNAs were then size selected, and cDNA libraries were generated and then sequenced by NGS. To analyze the resulting small RNA-Seq data set, we employed a workflow that included mapping sequences to different small RNA annotations, identifying DE microRNAs based on the mapped data, and determining the mRNA targets of the DE microRNAs (Fig. 1).
FIG 1 .
HIV small RNA-Seq analysis workflow. mRNA-Seq data were derived previously from the same set of experimental samples (1).
In the small RNA-Seq data set, we detected several classes of small RNAs, including piwi-interacting RNAs (piRNAs) and microRNAs (see Fig. S1A in the supplemental material). After filtering by a minimum criterion for expression, we found that 155 piRNAs were expressed in the small RNA data set at 12 and 24 hpi; of these, none were DE at 12 hpi, and 21 were DE at 24 hpi, with the majority being upregulated (see Table S1 in the supplemental material). We also found that 531 annotated microRNAs were expressed in the small RNA data set. Levels of expression varied broadly. In particular, a small number of microRNAs were highly expressed. Four microRNAs constituted over half of all microRNA-mapped reads, and 10 microRNAs accounted for over 70% of all microRNA-mapped reads (see Fig. S1B). Among the most highly expressed were several members of the miR-181 family (miR-181a-1, -181b-1, -181d) and let-7 family (let-7a-1, -7f-1) (see Fig. S1B). In contrast, a large number of microRNAs were detected at low levels. Two hundred sixty-three microRNAs had normalized mean read counts of less than 100 in each sample. Overall, the small RNA-Seq data set displayed the broad dynamic range characteristic of NGS data sets with read counts spanning five orders of magnitude (from 1 to over 150,000 mapped reads for the least and most highly expressed species, respectively). These results were consistent with other small RNA-Seq studies, e.g., Peng et al. (12), where over half of the mapped microRNAs from influenza- and severe acute respiratory syndrome (SARS)-infected mouse lungs were attributable to three microRNAs.
Time-specific, phased patterns of microRNA expression in HIV-infected cells.
Because many microRNAs were detected with low read counts, we performed further statistical analysis using DESeq, a negative binomial-based test that treats expression as discrete counts (13). By applying DESeq, we found that small numbers of microRNAs were DE at 5 and 12 hpi (numbers of microRNAs are in Table 1; expression values and microRNA IDs are in Fig. S2A to C in the supplemental material; full information is in Table S1 in the supplemental material). Furthermore, similar numbers of microRNAs were detected as DE in HIV-infected cells and HIVUV-infected cells at these time points (Table 1; see also Fig. S2D to F and Table S1). In contrast, at 24 hpi, a larger number of microRNAs were found DE, approximately 7-fold the number DE at 5 and 12 hpi (Table 1; see also Fig. S2C and Table S1). This expansion was specific to infection with live virus, as cells infected with HIVUV continued to show small numbers of DE genes at 24 hpi (Table 1; see also Fig. S2F and Table S1). This expansion in the number of DE microRNAs was similar to the expansion observed for DE mRNAs in the same samples (1), indicating that microRNA and mRNA expression were similarly affected by HIV-1 infection. However, out of those microRNAs found DE in HIVUV-infected cells at 24 hpi, the majority (13 out of 18) were also found DE in HIV-infected cells (Table 1), suggesting that a small subset of changes occurring later in cells infected with live virus were attributable to viral attachment or internalization.
TABLE 1 .
Changes in cellular microRNA expression during infection with HIV and HIVUV
| Regulation | No. of DE microRNAsa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 hpi |
12 hpi |
24 hpi |
|||||||
| HIV | Inter | UV | HIV | Inter | UV | HIV | Inter | UV | |
| Up | 1 | (1) | 6 | 1 | (1) | 5 | 65 | (11) | 14 |
| Down | 5 | (2) | 3 | 8 | (4) | 5 | 39 | (2) | 4 |
“Inter” and numbers in parentheses indicate overlapping DE microRNAs between HIV and HIVUV infections at each time point. DE microRNAs were determined by comparing HIV- and HIVUV-infected cells to mock-infected cells at each time point using DESeq. Those microRNAs with Benjamini-Hochberg-adjusted P values of <0.05 are shown.
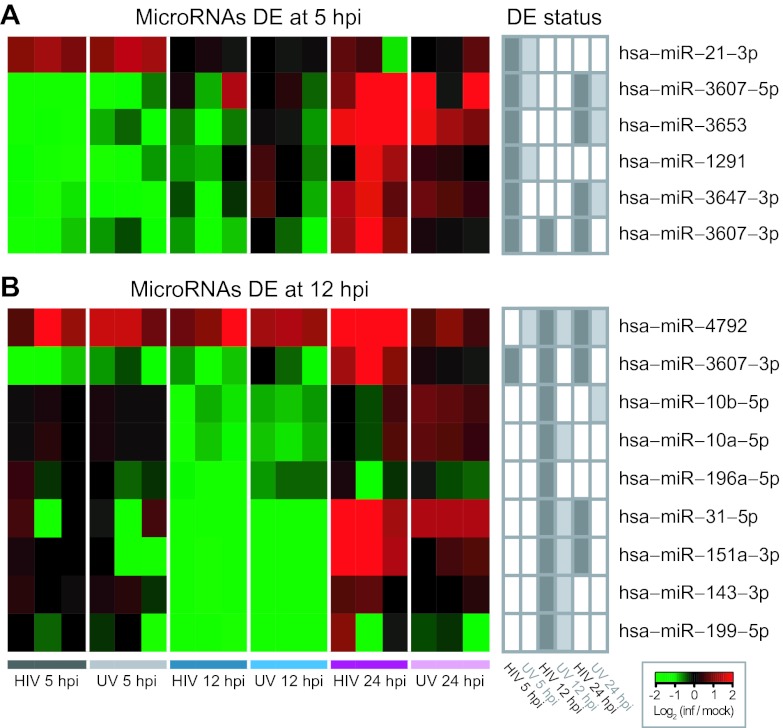
DE microRNAs displayed a strongly phased, time-point-specific pattern of expression. For example, DE microRNAs at earlier time points (5 and 12 hpi) were predominantly downregulated relative to their levels in mock-infected cells (Table 1; Fig. 2A and B). However, by 24 hpi, the majority of these microRNAs were found upregulated relative to mock-infected cells (Fig. 2A and B). This pattern was observed in the majority of DE microRNAs at earlier time points (five out of five downregulated at 5 hpi, and four out of eight downregulated at 12 hpi; Fig. 2A and B). Likewise, many of the microRNAs found DE at 24 hpi but not at earlier time points nonetheless displayed directions of changes in earlier and later time points (see Fig. S2G in the supplemental material). This phased time point specificity suggests that changes in microRNAs tracked specific stages of HIV infection in vitro. Interestingly many of the trends observed in HIV-infected samples were also observed in HIVUV-infected samples, though with reduced magnitudes of change and/or not at statistical significance, indicating that viral attachment or internalization induced a similar but attenuated response, relative to live virus, even later in infection (see Fig. S2G).
FIG 2 .
Heat maps showing magnitude and direction of expression changes in differentially expressed (DE) microRNAs at early time points. (A) DE microRNAs at 5 hpi; (B) DE microRNAs at 12 hpi. Log2 ratios based on normalized read counts in infected replicates versus averaged time-matched mock infections are shown. DE status of the microRNAs at other time points and in other conditions is indicated to the right.
Characterization of DE microRNAs by integration with mRNA-Seq data.
To determine the possible effects of HIV-induced changes in microRNA expression, we performed a correlative analysis with mRNA expression data. We obtained predictions of microRNA complementarity to 3′ untranslated regions (UTRs) in mRNAs from various algorithms and then compared expression profiles from small RNA-Seq and mRNA-Seq from the same samples (1). MicroRNAs and possible target mRNAs having significant, negative correlations (anticorrelations) were identified, and the annotations of the anticorrelated target mRNAs were used to determine possible functional effects of the microRNAs.
Following this protocol, we identified a large number of anticorrelated interactions between DE microRNAs and expressed mRNAs. In total, we identified 5,023 anticorrelated interactions involving 46 DE microRNAs and 1,917 detected mRNAs (see Table S2 in the supplemental material). Out of these 1,917 mRNAs, 1,900 were previously detected as DE by mRNA-Seq (1), indicating that DE mRNAs were overrepresented among anticorrelated target mRNAs (P < 10−15 by Fisher’s exact test). MicroRNAs and mRNAs varied widely in the number of predicted interactions, and individual microRNAs were predicted to target as many as 518 mRNAs, and individual mRNAs were predicted to be targeted by as many as 15 microRNAs (see Table S2).
MicroRNA involvement with transcriptional regulation in HIV-infected cells.
To identify the possible biological effects of DE microRNAs in HIV-infected cells, we examined the annotations (pathways and functions) associated with their predicted, anticorrelated target mRNAs. Specifically, we compared the number of target mRNAs to the number of DE mRNAs associated with each annotation term and determined those annotations with a significant overrepresentation of target mRNAs. We assumed that annotations with an abundance of target mRNAs were more likely to identify pathways regulated by microRNAs. We also examined the set of microRNA-mRNA interactions associated with each annotation to identify possible hub (highly connected) microRNAs and mRNAs. Hub microRNAs may be important regulators of multiple mRNAs, while hub mRNAs may be robustly regulated by multiple microRNAs.
A small number of annotations were enriched with target mRNAs at frequencies greater than expected by chance (by one-sided Fisher’s exact tests with P values of <0.05) (Table 2; see also Table S3 in the supplemental material). These annotations included “transcription regulatory activity,” which was associated with 111 upregulated target mRNAs at 24 hpi (Table 2). These mRNAs were in turn predicted to be targeted by 20 DE microRNAs (see Table S2 and Fig. S3A). Another set of target mRNAs encoding transcriptional regulators (albeit with different specific annotation terms) was found to be downregulated (Table 2; see also Table S2 and Fig. S3B). The presence of up- and downregulated microRNAs associated with multiple annotations related to transcriptional regulation suggest a role for microRNAs in regulating changes in gene expression during HIV infection.
TABLE 2 .
Annotations enriched in target mRNAs
| Annotation | Observed no. of hitsa | Odds ratiob | P valuec |
|---|---|---|---|
| Upregulated mRNA targets at 24 hpi | |||
| GO:0030528, transcription regulator activity | 111 | 1.30 | 0.019 |
| GO:0032868, response to insulin stimulus | 16 | 1.92 | 0.038 |
| GO:0043434, response to peptide hormone stimulus | 18 | 1.76 | 0.049 |
| Downregulated mRNA targets at 24 hpi | |||
| GO:0051252, regulation of RNA metabolic process | 107 | 1.26 | 0.037 |
| GO:0006357, regulation of transcription from RNA polymerase II promoter | 60 | 1.34 | 0.046 |
| GO:0010628, positive regulation of gene expression | 51 | 1.37 | 0.049 |
The number of target mRNAs associated with each annotation term.
Proportion of target mRNAs compared to random expectation.
Frequencies were compared to random selection from up- or downregulated DE mRNAs, and P values were derived from one-sided Fisher’s exact test.
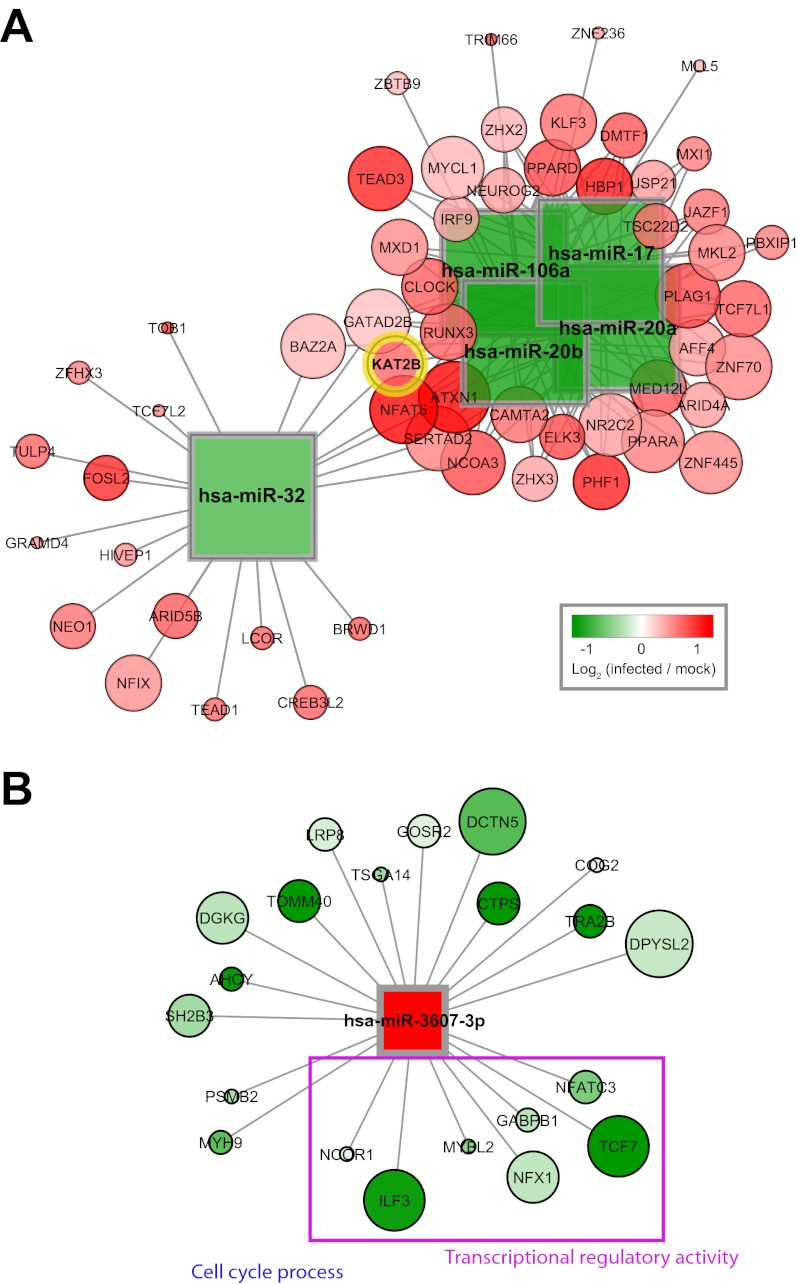
A subnetwork of upregulated mRNAs associated with DE downregulated microRNAs was found to include K(lysine) acetyltransferase 2B (KAT2B), a transcription factor known to contribute to HIV replication and known to be targeted posttranscriptionally by miR-17 and miR-20 (14). This subnetwork showed that these and other microRNAs were predicted to target KAT2B as well as a broader set of mRNAs with functional connections to KAT2B (Fig. 3A). Additional microRNAs in the subnetwork included miR-106a and miR-20b, two paralogs of miR-17 and miR-20, derived from the same host gene cluster (Fig. 3A). Their inclusion in the subnetwork suggests they may also have been downregulated in the earlier experiment, implicating miR-17 and miR-20 (14). In addition, a nonparalogous microRNA, miR-32, was also included in this subnetwork and may have regulated the same target mRNAs (Fig. 3A). Together, these microRNAs may represent an overlapping, robust network of regulators upstream of mRNAs for transcriptional regulators that contribute to HIV replication.
FIG 3 .
Anticorrelated targets of selected DE microRNAs. (A) Subnetwork of DE microRNAs and target mRNAs associated with the annotation “transcription regulatory activity.” The microRNAs targeting the known HIV cofactor KAT2B are shown together with other mRNAs targeted by the same microRNAs. Shape corresponds to molecule type (mRNAs as circles, microRNAs as squares). Size scales with connectivity such that larger mRNAs are targeted by greater numbers of microRNAs. Expression values at 24 hpi (as log2 ratios of average infected to mock-infected measurements) are overlaid. (B) Predicted targets of miR-3607-3p with highly significant, negative correlations to microRNA expression levels. Interactions with adjusted P values of <0.01 are shown, and expression values at 24 hpi are overlaid.
MicroRNA regulators of T cell activation in HIV-infected cells.
We found that several other pathways were enriched with target mRNAs, albeit at a relaxed threshold for significance (P < 0.10 by one-sided Fisher’s exact test; see Table S3 in the supplemental material). By this threshold, some pathways, including “regulation of small GTPase-mediated signal transduction,” “phosphoinositide binding,” and “regulation of Ras protein signal transduction,” were enriched in upregulated targeted mRNAs, suggesting microRNA downregulation may have contributed to increased receptor signaling (see Table S3). By the same criterion, other pathways, including “intracellular transport” and “secretion by cell,” were enriched in downregulated targeted mRNAs, suggesting microRNA upregulation may have contributed to the suppression of intracellular trafficking (see Table S3). T cell activation-related pathways were also enriched with downregulated target mRNAs (as “costimulatory signal during T-cell activation” and “positive regulation of T cell activation” with P values of 0.05 and 0.08, respectively) (see Table S3 and Fig. S3C). We previously found that T cell activation-associated mRNA expression was strongly suppressed at 12 and 24 hpi (1). In this study, we found that microRNAs were predicted to target key molecules involved in T cell activation, such as CD4, CD28, and TP53 (see Fig. S3C). In particular, CD4 mRNA was targeted by five microRNAs (miR-181d, -663, -663b, -1248, and -1303), the most highly targeted hub mRNA in this subnetwork (see Fig. S3C). Possible hub microRNAs and mRNAs for other pathways are shown in Fig. S3.
Targets of microRNAs DE at early time points: cell cycle and transcriptional regulation.
Many of the microRNAs DE early in infection (at 5 or 12 hpi) lacked published target mRNA predictions due to their recent discovery in the literature or limited degree of evolutionary conservation, which is suboptimal for algorithms such as TargetScan. To better characterize the early DE microRNAs, we generated an alternative set of TargetScan predictions using low evolutionary conservation thresholds and performed the correlation analysis between these microRNAs and their alternatively predicted target mRNAs.
Out of the five microRNAs DE at 5 hpi, three (miR-3607-5p, miR-3607-3p, and miR-3653) were significantly, negatively correlated with annotated, target mRNAs from the alternative predictions (see Table S4). One of these, miR-3607-3p, was highly expressed in SUP-T1 cells (see Table S4) and DE at all three time points (downregulated at 5 and 12 hpi and upregulated at 24 hpi), unique among the microRNAs in our study (Fig. 2A and B). In our analysis, miR-3607-3p showed highly significant, anticorrelated expression with 21 target mRNAs at 12 and 24 hpi (Fig. 3B). These were enriched in annotations for “cell cycle process” and “transcriptional regulatory activity,” with interleukin enhancer-binding factor 3 (ILF3) and nuclear receptor corepressor 1 (NCOR1) having both annotations. In addition, four mRNAs in this network were annotated for “transcriptional regulatory activity” and encoded transcription factors: ILF3, GABPB1 (GA-binding protein transcription factor, beta subunit 1), NFATC3 (nuclear factor of activated T cells, calcineurin-dependent 3), and TCF7 (transcription factor 7). The phased expression pattern of miR-3607-3p suggests that these targeted mRNAs may have been subjected to variable degrees of regulation—with reduced suppression early in infection and then increased suppression later in infection—during the course of HIV infection.
Candidate novel microRNAs DE during HIV infection.
The small RNA-Seq data set also included short reads that mapped to the genome such that they suggested the existence of nonannotated microRNAs (i.e., microRNAs not found in miRBase). We designated these sequences as candidate novel microRNAs. We identified these microRNAs using the miRDeep algorithm which detects features (such as read patterns and folding energies) consistent with the presence of microRNAs (15). We then filtered the miRDeep output using a minimum threshold for expression and identified DE candidate novel microRNAs using DESeq.
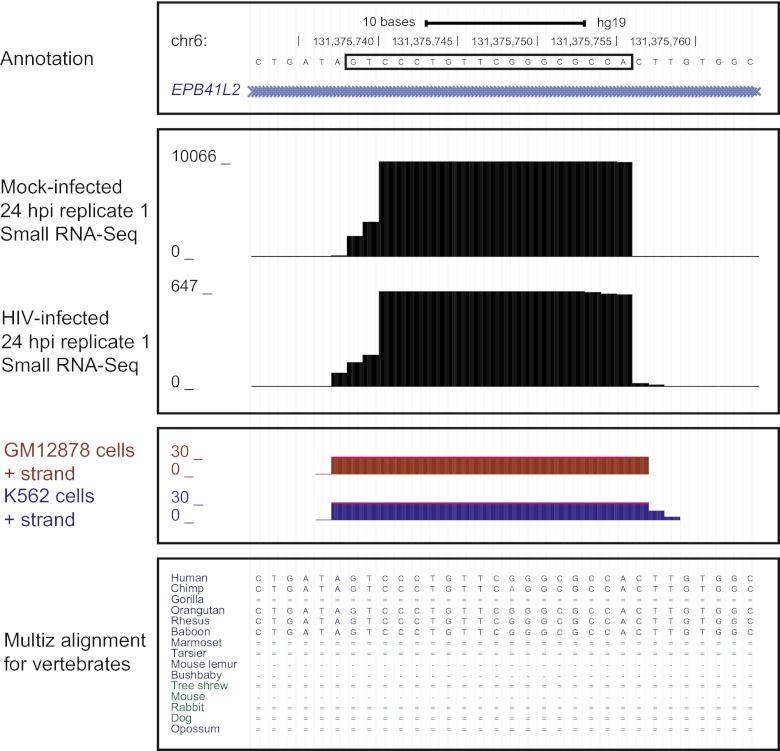
We detected no DE candidate novel microRNAs at 5 or 12 hpi and six DE candidate novel microRNAs at the 24 hpi time point (with nonzero read counts in both mock- and HIV-infected samples; Table 3). The most significantly DE candidate novel microRNA was an 18-mer encoded in the first intron of the EPB41L2 (erythrocyte membrane protein band 4.1-like 2, transcript variant 3) gene (Table 3; Fig. 4). This putative microRNA was found highly expressed in uninfected cells and downregulated by 90% at 24 hpi, a change specific to infection with live virus (Table 3 and data not shown). Other DE candidate novel microRNAs were expressed at lower levels overall or associated with smaller fold changes, leading us to focus on the EPB41L2-encoded candidate microRNA in further analysis.
TABLE 3 .
DE candidate novel microRNAs at 24 hpi
| Sequence | Locationa | Annotationb | Avg read countc |
Padjd | |
|---|---|---|---|---|---|
| Mock24 | HIV24 | ||||
| GUCCCUGUUCGGGCGCCAe | chr6:131375689.131375757: + | EPB41L2 intron | 1795.5 | 171.9 | 5.2 × 10−9 |
| UUAGUGGCUCCCUCUGCCUGCA | chr19:57950479.57950538: + | ZNF749 intron | 138.1 | 27.8 | 2.4 × 10−3 |
| GUCCCUGUUCAGGCGCCA | chr12:69593921.69593965: − | Intergenic | 37.8 | 5.2 | 5.0 × 10−3 |
| UGUAUGUAUGUAGACGUAUAUC | chr17:43642608.43642668: − | Intergenic | 32.3 | 3.7 | 5.6 × 10−3 |
| UGCCCUGAGACUUUUGCUCUAA | chr3:127305953.127306019: − | TPRA1 intron | 36.2 | 6.9 | 2.6 × 10−2 |
| UCACGUCCCUGUUCGGGCGCCA | chr19:58024383.58024432: − | Intergenic | 2138.6 | 854.3 | 3.1 × 10−2 |
Genomic position and strand of predicted pre-microRNA hairpin structure.
RefSeq mRNA annotation overlapping with given locations.
Average, normalized read counts for mock- and HIV-infected replicates at 24 hpi.
P values generated by DESeq and adjusted for total detected candidate novel microRNAs.
Bold indicates candidate microRNA chosen for follow-up analysis.
FIG 4 .
Candidate novel microRNA predicted from the small RNA-Seq data set and found DE during HIV infection at 24 hpi. Boxed nucleotide sequence shows location of mature candidate microRNA. RefSeq annotation shows location within the first EPB41L2 intron. Small RNA-Seq data show number of reads in one mock-infected replicate and one HIV-infected replicate at 24 hpi. ENCODE small RNA-Seq data show reads for other human immune-related cell types. Multiz alignment shows aligned mammalian sequences (with single and double lines indicating zero alignment and one or more unalignable bases, respectively).
Additional data supported the existence of an EPB41L2-encoded microRNA. mRNA-Seq reads mapped to EPB41L2 exons in the same experimental samples, indicating transcriptional activity at this locus (see Fig. S4 in the supplemental material) (1). However, EPB41L2 mRNA expression was found unchanged in HIV-infected cells at 12 and 24 hpi (1), suggesting that while the putative microRNA may have been derived from the EPB41L2 pre-mRNA, regulation occurred at the level of the excised intron, pre-microRNA, or mature microRNA, rather than at the level of the primary transcript. Interestingly, for the other two candidate novel microRNAs encoded within mRNA-encoding transcripts, a similar lack of concordance between microRNA and mRNA levels was observed (Table 3; see Table S4).
External data also supported the existence of a microRNA at this location. Small RNA-Seq data from the ENCODE project showed that other immune cell types—including GM12878, a lymphoblastoid cell line, and K562, a leukemia-derived cell line (16)—expressed a small RNA mapping to the same location and strand detected by miRDeep (Fig. 4). This indicated that the EPB41L2-encoded microRNA was robustly expressed and detected, despite differences in cell type, sample processing, and read mapping. In addition, evolutionary comparisons of aligned sequences from primates, mammals, and other vertebrates showed that the sequence for the EPB41L2-encoded microRNA was conserved in higher primates, particularly Old World monkeys and apes, with the exception of gorillas, but not other vertebrates (Fig. 4).
External data sets provided additional evidence that viral infection results in the downregulation of the EPBL41L2-encoded candidate novel microRNA. We obtained raw small RNA-Seq data from a separate infection model system, human cytomegalovirus (HCMV) infection of human fibroblasts (17), and observed reads mapping to the location of the candidate microRNA in each of four samples (mock- and HCMV-infected at 24 and 72 hpi), suggesting that the microRNA was expressed in human fibroblasts. Moreover, quantification of these reads suggested that HCMV downregulated the expression of this microRNA at 24 hpi (from 13.2 rpm to 5.6 rpm, a change of 58%) and at 72 hpi (from 34.4 rpm to 7.0 rpm, a change of 80%).
To further characterize the EPB41L2-encoded candidate novel microRNA, we generated predictions using the TargetScan algorithm and performed a correlation analysis to identify possible mRNA targets. The novel microRNA was predicted to target a total of 365 mRNAs by TargetScan. Out of these, 53 were significantly, negatively correlated in their expression levels (see Table S4 in the supplemental material). No individual annotation was found to be enriched among these host targets (with adjusted P values of <0.05; data not shown). However, we noted that an identical 18-nucleotide RNA was identified by Yeung et al. in the context of HIV infection, and this RNA was found perfectly complementary to the primer-binding site of HIV RNA (5).
We verified the expression of the EPB41L2-encoded microRNA by performing quantitative PCR (qPCR) using an additional HIV infection experiment sampled at increased frequency, 4-hour intervals through 24 hpi (see Fig. S5A in the supplemental material). The microRNA was detected in all samples by qPCR, but out of the five time points sampled, the largest downregulation was observed at 24 hpi, consistent with the downregulation observed in the small RNA-Seq data (see Fig. S5A). Altered expression of the EPB41L2-encoded microRNA may therefore be specific to late stages of infection with live virus.
Validation and temporal pattern of microRNA expression by qPCR.
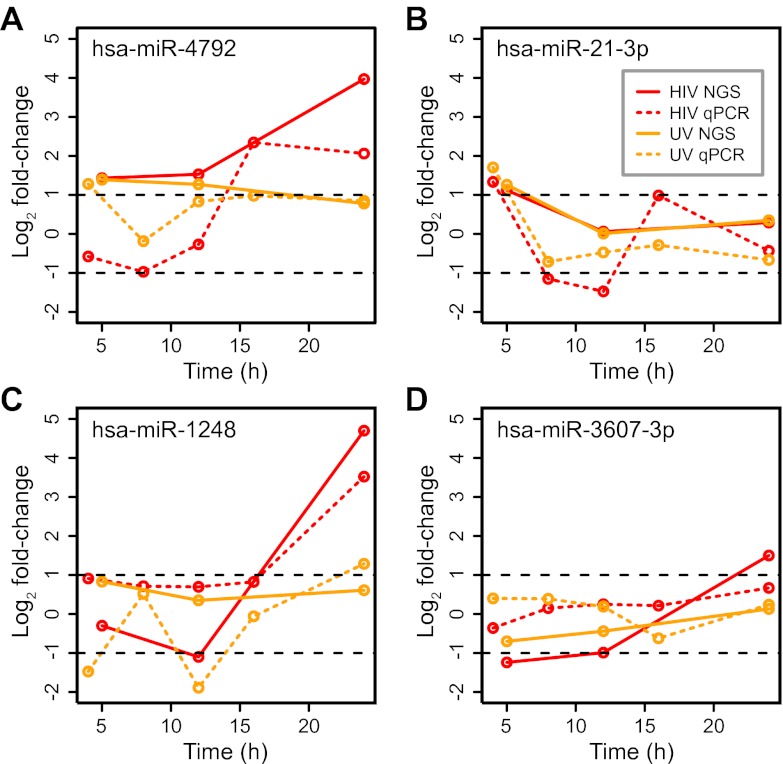
To validate other small RNA-Seq measurements, we performed qPCR on selected known microRNAs. Samples were derived from the same, increased-frequency experiment used to verify expression of the EPB41L2-encoded microRNA. The additional time points allowed a more precise estimation of the timing of expression patterns. Overall, we found that qPCR validated large fold changes in microRNA expression identified by small RNA-Seq, particularly 2-fold or greater upregulation (Fig. 5A to D; see also Fig. S5B to K in the supplemental material). For example, in the small RNA-Seq data set, we observed miR-4792 to be upregulated at 12 hpi and more highly upregulated at 24 hpi (Fig. 2B and 5A). In the qPCR data set, this increase was found to occur more precisely between 12 and 16 hpi (Fig. 5A). Likewise, miR-1248 was expressed at elevated levels at 24 hpi but not at 12 hpi in the small RNA-Seq data set (Fig. 5C). In the qPCR data set, this increase was evident at an intermediate time point, 16 hpi (Fig. 5C). The overall correspondence between qPCR and small RNA-Seq, particularly for upregulated microRNAs, was consistent with previous observations for mRNAs (18).
FIG 5 .
qPCR experiment of microRNAs found upregulated during HIV infection by small RNA-Seq. Samples for qPCR were derived from an independent experiment, and for each time point, log2 ratios of average infected to mock-infected measurements are shown.
DISCUSSION
During the course of infection, HIV is known to affect cellular functions at several levels. In this study, we investigated the effect of HIV infection on small RNA expression in CD4-expressing T lymphoblastoid cells. Other studies have previously used NGS to examine small RNAs in the context of HIV infection, e.g., Yeung et al. and Schopman et al. (8, 10). However, the focus in these studies has largely been on virally encoded microRNAs rather than on cellular microRNAs. These studies typically also examine only a single time point after infection, e.g., two days postinfection in Yeung et al. and Schopman et al. (8, 10). Other studies have profiled microRNA expression in infected patient populations, e.g., Bignami et al. (19), but in these cases, heterogeneous mixtures of cell types served as the source material, complicating data analysis. In contrast, we have focused on the host response at the level of a single infected cell type and profiled this system over time using small RNA-Seq and other NGS-based approaches (1). To our knowledge, this study offers the first unbiased glimpse into the dynamic changes underlying the small RNA population of HIV-infected cells. The specific goals of the study were to (i) identify patterns of DE microRNA expression starting early in infection, (ii) identify anticorrelated targets of DE microRNAs, including those with low evolutionary conservation, and (iii) detect other classes of small RNA altered during infection, including candidate novel microRNAs.
Early, phased microRNA signatures of HIV infection.
In our previous work on this experimental system, we found that viral replication was undetectable at 4 hpi but then proceeded rapidly by 12 hpi (1). In this study, we were particularly interested in detecting microRNA signatures early in infection. Using small RNA-Seq, we identified 14 microRNAs DE at 5 and 12 hpi whose expression appeared to be largely coordinated. Many of these displayed a phased expression pattern with time, suppressed initially and then overexpressed by 24 hpi.
Several of the microRNAs that we observed as DE, particularly at early time points, were either poorly characterized or not previously associated with HIV, e.g., as reviewed by Houzet and Jeang (20) or Swaminathan et al. (21). By identifying target mRNAs with anticorrelated expression patterns, we implicated one of these DE microRNAs, miR-3607-3p, in the regulation of cell cycle- and transcription-associated mRNAs. HIV infection is known to downregulate cell cycle-related genes (1, 22), and differing levels of regulation with time may reflect evolving requirements of the virus following entry. In particular, the upregulation of cell cycle-related transcription factors prior to replication, mediated by the downregulation of microRNAs such as miR-3607-3p, may result in the internal production of raw materials needed for viral replication or convert host chromatin to an open configuration. In particular, the NFATC3 (nuclear factor of activated T cells, cytoplasmic 3) gene, a target of miR-3607-3p in our analysis, encodes a transcription factor that has recently been shown to be involved in T-cell activation (23). Likewise, the TCF7 gene, another predicted target of miR-3607-3p, encodes a T-cell-specific transcription factor (24). Relieving the inhibition of these transcription factors via the suppression of miR-3607-3p and other microRNAs may represent an early event of HIV infection which is reversed later in HIV infection. Consistent with this interpretation, the MYH9 gene (myosin, heavy chain 9, nonmuscle), a target mRNA of miR-3607-3p found downregulated later in infection, was also found to be downregulated in HIV-infected human glomeruli, which may contribute to HIV-associated pathology (25). While we cannot discern the mechanism by which viral entry or internalization might suppress miR-3607-3p and other early DE microRNAs from our data, we speculate that a surface protein such as gp120 initiates a signaling cascade that destabilizes the available pool of miR-3607-3p.
In general, we observed agreement between our results and those from previous studies, particularly for other, highly expressed microRNAs. For example, Hayes et al. measured changes in CEMx174 lymphocytes infected with HIV-1 strain NL4-3 (26). Out of the 42 DE microRNAs observed by Hayes et al., 11 were also found to be DE at 24 hpi in our data set, perhaps indicating cell type specificity (26). Of these, eight displayed concordant changes between the two data sets, with complete concordance observed for the most highly expressed microRNAs (let-7e, miR-92, and miR-106a, all with >10,000 reads per sample in uninfected SUP-T1 cells) (26). A similar concordance among highly expressed microRNAs was observed for clinical samples as well. For example, among the DE microRNAs observed in PBMCs from HIV-infected individuals by Houzet et al., four out of the five mostly highly expressed microRNAs (let-7e, miR-20a, miR-20b, miR-92, and miR-182, again with >10,000 reads per sample in uninfected SUP-T1 cells) displayed directional concordance (6). Likewise, concordance with the clinical data set from Witwer et al. was evident for highly expressed microRNAs, such as miR-155 (upregulated by 1.5-fold in PBMCs from viremic individuals and by 1.8-fold in our study at 24 hpi) and miR-181b (upregulated by 1.8-fold in PBMCs from viremic individuals and in our study at 24 hpi) (11). Highly expressed microRNAs may therefore represent cell type-robust signatures.
Transcriptional regulation impacted by altered microRNA expression.
We identified a small number of additional pathways and functions that were enriched in the predicted mRNA targets of DE microRNAs. These annotations included “transcription regulator activity,” whose associated mRNAs were upregulated at 24 hpi. This dense network of transcriptional regulators includes cofactors that are known to contribute to HIV replication, namely, KAT2B, also known as PCAF (P300/CBP-associated factor) (14). Our analysis showed that microRNAs of the miR-17-92 cluster—known to be relevant to T cell function (27)—and miR-32 may target KAT2B and other transcription factors coordinately regulated with KAT2B. In particular, miR-32 has not previously been associated with HIV cofactor expression. Our results suggest that HIV-1 infection may suppress several microRNAs with overlapping specificities to effect HIV cofactor expression.
We note that our approach to identifying target mRNAs considers the effect of microRNA on target mRNA stability alone. An extension of this approach would be to include other possible effects of microRNA, such as reductions of translational efficiency. Nevertheless, recent data point to destabilization of target mRNAs as the main mode of microRNA activity (28–30); our approach reflects this state of knowledge. In addition, while our approach is based on complementarity of microRNAs to the 3′ UTRs of mRNAs, recent studies suggest that microRNAs may bind other parts of mRNAs, including 5′ UTRs (31, 32). Our approach utilizing predictive algorithms for microRNA-to-3′ UTR complementarity is intended to detect canonical interactions and limit false positives but would necessarily miss these additional possible interactions. Finally, we note that our approach is based ultimately on predictions of sequence complementarity supplemented by expression data. Additional experiments, such as direct cross-linking and immunoprecipitation of the RISC complex, would provide more direct verification of the targets that we have identified (33).
EPB41L2-encoded candidate novel microRNA during HIV infection.
In addition to annotated microRNAs, we also detected changes in other types of small RNAs, including piRNAs at 24 hpi. piRNA expression was previously detected in HIV-infected Jurkat cells (8). In our study, we detected piRNA expression in an additional T lymphoblast cell line, SUP-T1 cells, providing support for the expression of piRNAs in non-germ-line cells. HIV infection resulted in changes in the expression of 21 piRNAs. piRNAs are hypothesized to function in epigenetic regulatory mechanisms; however, due to a lack of consensus about their function in non-germ-line cells, we report these changes for completeness only (34). We also detected changes in several candidate novel microRNAs. In particular, one candidate encoded in the first intron of EPB41L2 was highly expressed in uninfected cells and strongly downregulated in infected cells. Multiple lines of evidence supported the existence of this microRNA, including ENCODE small RNA-Seq data on multiple immune cell types. Interestingly, Yeung et al. detected a sequence identical to this candidate novel microRNA in the context of HIV infection (8). This small RNA was found to have RNA interference activity toward the HIV-1 RNA primer-binding site where tRNA binds to initiate reverse transcription, and antagomirs directed toward the small RNA resulted in increased HIV-1 replication (8). We find a deeper investigation into the connection between this microRNA and HIV to be outside the scope of the present work. However, in the future it would be interesting to determine whether more direct evidence of the association between this microRNA and HIV-1 RNA could be obtained, such as through immunoprecipitation of the two RNA species in complex.
Our finding that this candidate microRNA was downregulated at 24 hpi, despite largely unchanged expression in mRNA from the same locus, suggests that the candidate microRNA may have been selectively inhibited during HIV infection. Our examination of NGS data from another experimental infection system, HCMV in human fibroblasts, both validated the existence of the candidate microRNA and showed it to be downregulated during a different infection, suggesting this microRNA may function more generally during the immune response to viral infection. Finally, we note that this microRNA displays strong specificity to higher primates, and its presence may reflect a response unique to primates and viruses that target them, such as HIV-1 and HCMV. Unlike known microRNAs, this and other candidate novel microRNAs may represent entirely novel targets for intervention or diagnosis during HIV-1 infection and demonstrate a distinct benefit of applying NGS to the profiling of small RNAs.
MATERIALS AND METHODS
In vitro infection of SUP-T1 cells with HIV-1 LAI.
The design of our experiment comprised treating SUP-T1 cells with HIV-1 strain LAI, UV-inactivated HIV-1 strain LAI (HIVUV), or cell-conditioned medium as mock infection. Experimental steps were performed as described in reference 1 but with the following changes. HIVUV was generated by irradiating HIV preparations for 5 min, a dose we found sufficient to abrogate viral replication in MAGI-CXCR4 cells as detected by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. Lack of replication with HIVUV was confirmed in SUP-T1 cells by viral mRNA load by TaqMan qPCR (35). Both live virus and UV-inactivated virus were added to 5 × 106 cells at a multiplicity of infection of 5. This dose of live virus was found to achieve 100% infection at 24 hpi with ~50% cell viability as measured by trypan blue exclusion assay. Mock infections were performed with an identical volume of SUP-T1-conditioned medium. Each treatment was performed in triplicate, and replicates were sampled at 5, 12, and 24 hpi.
RNA preparation and library construction.
Total RNA was extracted using the mirVana kit (Applied Biosystems/Ambion, Austin, TX), and quality and concentration of RNA were determined by an Agilent 2100 Bioanalyzer. Small RNA libraries were prepared with a small RNA version 1.5 sample preparation kit (Illumina, San Diego, CA). Total RNA was ligated with two adaptors: a 3′ RNA adaptor specifically modified to target microRNAs (5′-/5rApp/ATCTCGTATGCCGTCTTCTGCTTG/3ddC/) and a 5′ RNA adaptor that included the sequencing primer (5′-GUUCAGAGUUCUACAGUCCGACGAUC). Reverse transcription (RT)-PCR amplification was performed using the adaptors as primers. The resulting double-stranded cDNA libraries were purified by polyacrylamide gel electrophoresis using 6% Novex Tris-borate-EDTA gels (Invitrogen, Carlsbad, CA) and size selected to eliminate dimerized adaptors. The quality and concentration of libraries were determined by an Agilent 2100 Bioanalyzer and RiboGreen fluorescence on QuBit (Invitrogen, Carlsbad, CA).
Next-generation sequencing and read mapping.
All libraries were sequenced to a 54-nucleotide (nt) read length on individual lanes of a Genome Analyzer IIx (Illumina, San Diego, CA). Small RNA sequences were submitted to Geospiza (PerkinElmer, Seattle, WA) for initial analysis. Raw reads were trimmed to remove adapter sequence. Trimmed reads were aligned using Burrows-Wheeler Aligner (BWA) software against a filter reference containing rRNA, tRNA, snRNA, and mtRNA sequences (36). The remaining reads were aligned to NCBI build 37.2 of the human genome using BWA and subsequently quantified using genomic annotations for microRNAs (miRBase 17) (7), piRNAs, and known genes (RefSeq gene, build 37.2) or classified as unmapped. MicroRNA read count data were obtained from Geospiza (available at http://viromics.washington.edu). By read count, we refer to the number of cDNA fragments that are sequenced and map to a particular genomic feature. Filtering was performed by requiring ≥10 reads in all the samples of at least one condition for a given microRNA. Normalization was applied to the remaining read count data using DESeq, an R software package for testing differential expression, using R version 2.14.1 (13). Briefly, normalization comprised calculating a size factor for each sample (as the median ratio of read counts for each feature and sample to the geometric mean of read counts for each feature across samples) and dividing all of the read counts in a particular sample by the sample size factor (13). DE microRNAs were determined using negative binomial tests implemented in DESeq, where dispersion was estimated using a fitted, parametric curve. For comparison, data from HCMV-infected fibroblasts was obtained through the Short-Read Archive (study identifier [ID] SRP009246) (17).
Integrative analysis of small RNA-Seq and mRNA-Seq data.
To integrate microRNA and mRNA data, we generated target predictions from three prediction algorithms—TargetScan (37), PITA (38), and Miranda (39)—accessed through the MAGIA website (40). We obtained mRNA data from previous mRNA-Seq analysis of the same 12- and 24-hpi HIV- and mock-infected samples used to derive the small RNA-Seq data set 1. mRNA data were represented as log2 fragments per kilobase of exon per million mapped fragments (FPKM) in each replicate and identified by Entrez gene IDs as accessed via Biomart in Bioconductor (41). MicroRNA data were represented as log2 DESeq-normalized counts with miRBase 17 IDs. For each predicted microRNA-mRNA pairing, we calculated a Pearson correlation coefficient as well as a Benjamini-Hochberg-adjusted P value. Significant interactions were identified by negative correlation coefficients associated with adjusted P values of <0.05. These interactions were used to generate networks in Cytoscape (42).
Functional analysis of mRNA targets of DE microRNAs.
To examine annotations associated with target mRNAs, we utilized the NIH David resource (43). We analyzed up- and downregulated target mRNAs separately in David using the Functional Annotation Chart feature with annotations for GO_BP_Fat, GO_MF_Fat, BioCarta, and KEGG_Pathway and human genome as background. An identical analysis was performed on up- and downregulated DE mRNAs from mRNA-Seq data. Overrepresentation of target mRNAs was determined by one-sided Fisher’s exact tests for each annotation term.
Identification of candidate novel microRNAs.
Candidate novel microRNAs were identified by analyzing genome-mapped small RNA-Seq reads from HIV- and mock-infected samples at 5, 12, and 24 hpi using miRDeep with default parameters (15). Candidate novel microRNAs were then filtered to include only those present with ≥10 reads in ≥11 out of 17 samples (i.e., ~two-thirds of samples). DE candidate microRNAs were identified using DESeq. As a final filter, we considered only candidates with nonzero expression in mock- and HIV-infected samples at the DE time point for further analysis. The UCSC Genome Browser (44) was used to access other small RNA-Seq data from the ENCODE project (16) as well as evolutionary conservation data via Multiz (45) and GERP scores (46).
qPCR.
We performed qPCR for each microRNA using a miRCURY LNA universal RT microRNA PCR system (Exiqon, Woburn, MA). For each assay, we utilized 20 ng of total RNA in an ABI 7900 real-time PCR system with SYBR green-based detection (Applied Biosystems, Foster City, CA). Each assay was run in triplicate with SYBR green PCR Master Mix (Exiqon, Woburn, MA) for microRNA detection in a 10-µl reaction volume.
SUPPLEMENTAL MATERIAL
Small RNA composition of experimental samples. (A) Mapping results to all types of small RNAs in each individual sample; (B) mapping results to microRNA only under each condition, with replicates averaged. Download
MA plot visualization of DE microRNAs of HIV- and HIVUV-infected samples at each time point. HIV samples are shown at 5 hpi (A), 12 hpi (B), and 24 hpi (C), and HIVUV samples at 5 hpi (D), 12 hpi (E), and 24 hpi (F). Red dots indicate DE microRNAs at each time point and condition with adjusted P values of <0.05. Blue and purple lines represent 1.5- and 2.0-fold changes, respectively. (G) Heat map of DE microRNAs at 24 hpi. Log2 ratios of normalized read counts in infected and time-matched mock infections were used. Download
Networks comprised of DE microRNAs and target mRNAs associated with “transcription regulator activity” (A) and “positive regulation of gene expression” (B). Expression values at 24 hpi are overlaid with green and red, indicating down- and upregulation, respectively. Below, protein-protein interactions among mRNA targets were queried in ingenuity pathways analysis. (C) DE microRNAs and target mRNAs associated with T cell activation. Download
Data related to EPBL41L2-encoded microRNA. (A) qPCR of novel microRNA. (B) Genome browser view of gene locus. From top to bottom: RefSeq annotation showing exonic and intronic locations; mRNA-Seq and small RNA-Seq wiggle plots of the EPB41L2 locus showing reads in mock- and HIV-infected replicates; ENCODE small RNA-Seq data showing reads in four immune cell types mapping to the same location and strand; Multiz alignment data for vertebrate genomes; and GERP scores showing mammalian conservation. The highlighted yellow box shows the approximate location of the mature candidate novel microRNA. Download
Complete qPCR results of DE microRNAs. Download
DESeq results showing DE microRNAs for each time and treatment (HIV- and HIVUV-infected at 5, 12, and 24 hpi) and detected piRNAs (as log2-normalized counts and DE results) (.xls format).
Complete correlation analysis results showing microRNA, mRNA target, correlation coefficient, and adjusted P value for each predicted microRNA-mRNA interaction (.xls format).
Complete annotation enrichment analysis results showing observed (anticorrelated mRNA targets) and expected (all DE mRNAs) annotations for up- and downregulated mRNAs as well as Fisher’s exact test results comparing observed and expected annotation frequencies (.xls format).
Additional DE microRNA results, including correlation analysis for DE microRNAs at 5 and 12 hpi using TargetScan predictions with low evolutionary conservation thresholds, DE candidate novel microRNAs at 24 hpi, and correlation analysis for EPBL41L2-encoded, candidate novel microRNA.
ACKNOWLEDGMENTS
We thank G. Lynn Law and Marcus Korth for their critical readings of the manuscript.
We gratefully acknowledge funding from P30DA015625 and P51RR000166.
Footnotes
Citation Chang ST, Thomas MJ, Sova P, Green RR, Palermo RE, Katze MG. 2013. Next-generation sequencing of small RNAs from HIV-infected cells identifies phased microRNA expression patterns and candidate novel microRNAs differentially expressed upon infection. mBio 4(1):e00549-12. doi:10.1128/mBio.00549-12.
REFERENCES
- 1. Chang ST, Sova P, Peng X, Weiss J, Law GL, Palermo RE, Katze MG. 2011. Next-generation sequencing reveals HIV-1-mediated suppression of T cell activation and RNA processing and regulation of noncoding RNA expression in a CD4+ T cell line. mBio 2(5):e00134-11 http://dx.doi.org/10.1128/mBio.00134-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Navare AT, Sova P, Purdy DE, Weiss JM, Wolf-Yadlin A, Korth MJ, Chang ST, Proll SC, Jahan TA, Krasnoselsky AL, Palermo RE, Katze MG. 2012. Quantitative proteomic analysis of HIV-1 infected CD4+ T cells reveals an early host response in important biological pathways: protein synthesis, cell proliferation, and T-cell activation. Virology 429:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schweneker M, Favre D, Martin JN, Deeks SG, McCune JM. 2008. HIV-induced changes in T cell signaling pathways. J. Immunol. 180:6490–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rotger M, Dang KK, Fellay J, Heinzen EL, Feng S, Descombes P, Shianna KV, Ge D, Günthard HF, Goldstein DB, Telenti A. 2010. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 6:e1000781 http://dx.doi.org/10.1371/journal.ppat.1000781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang K-T. 2005. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology 2:81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Houzet L, Yeung ML, de Lame V, Desai D, Smith SM, Jeang K-T. 2008. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology 5:118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kozomara A, Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39:D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yeung ML, Bennasser Y, Watashi K, Le S-Y, Houzet L, Jeang K-T. 2009. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37:6575–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Althaus CF, Vongrad V, Niederöst B, Joos B, Di Giallonardo F, Rieder P, Pavlovic J, Trkola A, Günthard HF, Metzner KJ, Fischer M. 2012. Tailored enrichment strategy detects low abundant small noncoding RNAs in HIV-1 infected cells. Retrovirology 9:27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schopman NC, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, Berkhout B, Haasnoot J. 2012. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 40:414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Witwer KW, Watson AK, Blankson JN, Clements JE. 2012. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology 9:5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng X, Gralinski L, Ferris MT, Frieman MB, Thomas MJ, Proll S, Korth MJ, Tisoncik JR, Heise M, Luo S, Schroth GP, Tumpey TM, Li C, Kawaoka Y, Baric RS, Katze MG. 2011. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. mBio 2(6):e00198-11 http://dx.doi.org/10.1128/mBio.00198-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Triboulet R, Mari B, Lin Y-L, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang K-T, Benkirane M. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582 [DOI] [PubMed] [Google Scholar]
- 15. Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 26:407–415 [DOI] [PubMed] [Google Scholar]
- 16. Affymetrix/Cold Spring Harbor Laboratory. ENCODE Transcriptome Project, Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457:1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stark TJ, Arnold JD, Spector DH, Yeo GW. 2012. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J. Virol. 86:226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morey JS, Ryan JC, Van Dolah FM. 2006. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online 8:175–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bignami F, Pilotti E, Bertoncelli L, Ronzi P, Gulli M, Marmiroli N, Magnani G, Pinti M, Lopalco L, Mussini C, Ruotolo R, Galli M, Cossarizza A, Casoli C. 2012. Stable changes in CD4+ T-lymphocyte microRNA expression following exposure to HIV-1. Blood 119:6259–6267 [DOI] [PubMed] [Google Scholar]
- 20. Houzet L, Jeang KT. 2011. MicroRNAs and human retroviruses. Biochim. Biophys. Acta 1809:686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Swaminathan S, Murray DD, Kelleher AD. 2012. The role of microRNAs in HIV-1 pathogenesis and therapy. AIDS 26:1325–1334 [DOI] [PubMed] [Google Scholar]
- 22. Giri MS, Nebozhyn M, Showe L, Montaner LJ. 2006. Microarray data on gene modulation by HIV-1 in immune cells: 2000–2006. J. Leukoc. Biol. 80:1031–1043 [DOI] [PubMed] [Google Scholar]
- 23. Urso K, Alfranca A, Martínez-Martínez S, Escolano A, Ortega I, Rodríguez A, Redondo JM. 2011. NFATc3 regulates the transcription of genes involved in T-cell activation and angiogenesis. Blood 118:795–803 [DOI] [PubMed] [Google Scholar]
- 24. van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. 1991. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 10:123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hays T, D’Agati VD, Garellek JA, Warren T, Trubin ME, Hyink DP, He JC, Klotman PE. 2012. Glomerular MYH9 expression is reduced by HIV-1. AIDS 26:797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hayes AM, Qian S, Yu L, Boris-Lawrie K. 2011. Tat RNA silencing suppressor activity contributes to perturbation of lymphocyte miRNA by HIV-1. Retrovirology 8:36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C. 2007. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 9:775–787 [DOI] [PubMed] [Google Scholar]
- 28. Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. 2009. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7:e1000238 http://dx.doi.org/10.1371/journal.pbio.1000238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lytle JR, Yario TA, Steitz JA. 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. U. S. A. 104:9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ørom UA, Nielsen FC, Lund AH. 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30:460–471 [DOI] [PubMed] [Google Scholar]
- 33. Thomson DW, Bracken CP, Goodall GJ. 2011. Experimental strategies for microRNA target identification. Nucleic Acids Res. 39:6845–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siddiqi S, Matushansky I. 2012. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J. Cell. Biochem. 113:373–380 [DOI] [PubMed] [Google Scholar]
- 35. Li CC, Seidel KD, Coombs RW, Frenkel LM. 2005. Detection and quantification of human immunodeficiency virus type 1 p24 antigen in dried whole blood and plasma on filter paper stored under various conditions. J. Clin. Microbiol. 43:3901–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H, Durbin R. 2009. Fast and accurate short read alignment with burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 [DOI] [PubMed] [Google Scholar]
- 38. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat. Genet. 39:1278–1284 [DOI] [PubMed] [Google Scholar]
- 39. Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol. 5:R1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sales G, Coppe A, Bisognin A, Biasiolo M, Bortoluzzi S, Romualdi C. 2010. Magia, a Web-based tool for miRNA and genes integrated analysis. Nucleic Acids Res. 38:W352–W359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. 2005. Biomart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21:3439–3440 [DOI] [PubMed] [Google Scholar]
- 42. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. David: database for annotation, visualization, and integrated discovery. Genome Biol. 4:3. [PubMed] [Google Scholar]
- 44. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, Haussler D, Miller W. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14:708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15:901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Small RNA composition of experimental samples. (A) Mapping results to all types of small RNAs in each individual sample; (B) mapping results to microRNA only under each condition, with replicates averaged. Download
MA plot visualization of DE microRNAs of HIV- and HIVUV-infected samples at each time point. HIV samples are shown at 5 hpi (A), 12 hpi (B), and 24 hpi (C), and HIVUV samples at 5 hpi (D), 12 hpi (E), and 24 hpi (F). Red dots indicate DE microRNAs at each time point and condition with adjusted P values of <0.05. Blue and purple lines represent 1.5- and 2.0-fold changes, respectively. (G) Heat map of DE microRNAs at 24 hpi. Log2 ratios of normalized read counts in infected and time-matched mock infections were used. Download
Networks comprised of DE microRNAs and target mRNAs associated with “transcription regulator activity” (A) and “positive regulation of gene expression” (B). Expression values at 24 hpi are overlaid with green and red, indicating down- and upregulation, respectively. Below, protein-protein interactions among mRNA targets were queried in ingenuity pathways analysis. (C) DE microRNAs and target mRNAs associated with T cell activation. Download
Data related to EPBL41L2-encoded microRNA. (A) qPCR of novel microRNA. (B) Genome browser view of gene locus. From top to bottom: RefSeq annotation showing exonic and intronic locations; mRNA-Seq and small RNA-Seq wiggle plots of the EPB41L2 locus showing reads in mock- and HIV-infected replicates; ENCODE small RNA-Seq data showing reads in four immune cell types mapping to the same location and strand; Multiz alignment data for vertebrate genomes; and GERP scores showing mammalian conservation. The highlighted yellow box shows the approximate location of the mature candidate novel microRNA. Download
Complete qPCR results of DE microRNAs. Download
DESeq results showing DE microRNAs for each time and treatment (HIV- and HIVUV-infected at 5, 12, and 24 hpi) and detected piRNAs (as log2-normalized counts and DE results) (.xls format).
Complete correlation analysis results showing microRNA, mRNA target, correlation coefficient, and adjusted P value for each predicted microRNA-mRNA interaction (.xls format).
Complete annotation enrichment analysis results showing observed (anticorrelated mRNA targets) and expected (all DE mRNAs) annotations for up- and downregulated mRNAs as well as Fisher’s exact test results comparing observed and expected annotation frequencies (.xls format).
Additional DE microRNA results, including correlation analysis for DE microRNAs at 5 and 12 hpi using TargetScan predictions with low evolutionary conservation thresholds, DE candidate novel microRNAs at 24 hpi, and correlation analysis for EPBL41L2-encoded, candidate novel microRNA.