Abstract
Endonuclease V (EndoV) initiates a major base repair pathway for nitrosative deamination resulting from endogenous processes and increased by oxidative stress from mitochondrial dysfunction or inflammatory responses. We solved the crystal structures of Thermotoga maritima EndoV in complex with a hypoxanthine lesion substrate and with product DNA. The PYIP wedge motif acts as a minor-groove damage sensor for helical distortions and base mismatches and separates DNA strands at the lesion. EndoV incises DNA with an unusual offset nick one nucleotide 3′ of the lesion, as the deaminated adenine is rotated ~90° into a recognition pocket ~8Å from the catalytic site. Tight binding by the lesion recognition pocket in addition to Mg2+ ion and hydrogen-bond interactions to the DNA ends stabilize the product complex, suggesting orderly recruitment of downstream proteins in this base repair pathway.
Nitrate or nitrite metabolism generates mutagenic reactive nitrogen oxides, which can deaminate exocyclic amines of DNA nucleobases. Thus, adenine in DNA is deaminated to hypoxanthine, guanine to xanthine or oxanine, and finally cytosine to uracil (Fig. 1a). Such nitrosative deamination of DNA bases can cause transition mutations and cancer predispositions 1-5. The deaminated bases have strong miscoding properties and produce mutations during subsequent replication, in which hypoxanthine mispairs with cytosine, leading to a A/T to G/C mutation 2. Although until recently missing from otherwise comprehensive analyses of DNA base repair pathways 6, EndoV, encoded by the nfi gene, is the key enzyme for initiating repair of deaminated purine bases 1, 2, 7, 8. EndoV sequence homologues are conserved in all domains of life from bacteria to humans. A multiple-sequence alignment of EndoV reveals residues characteristic of this protein family, including the fully conserved Asp43, Tyr80, Glu89, Asp110, His116 and Lys139 (Supplementary Fig. 1 online).
Figure 1.
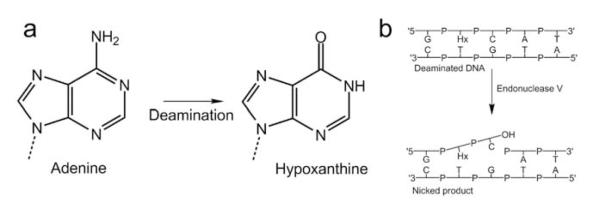
EndoV 3′ incision initiating deaminated adenine repair. (a) Deamination of the exocyclic amino group in adenine yields hypoxanthine. (b) EndoV dependent repair is initiated by cleavage at the second phosphodiester bond 3′ to the lesion (Hx, hypoxanthine) resulting from deamination of adenine.
Under physiological conditions, EndoV hydrolyzes the second phosphodiester bond 3′ of a deaminated base using Mg2+ as a cofactor 9. This unprecedented 3′ offset incision by a DNA repair protein involved in single-base lesion recognition and excision is unique to EndoV (Fig. 1b). In contrast, DNA glycosylases in the Base Excision Repair (BER) pathway remove the damaged base by hydrolyzing the N-glycosylic bond, leaving an abasic site for downstream processing 10. Although downstream processing for the EndoV pathway remains unknown, a 3′-5′ exonuclease activity may generate a DNA repair patch spanning only 2-3 nucleotides to either side of a hypoxanthine base 8. EndoV has high affinity for both the hypoxanthine substrate and the nicked product 11, 12. Further, Endo V has the capability to recognize all possible deaminated DNA bases 13. In vitro, EndoV also shows activity toward the single-base lesions abasic sites, urea 9 and base mismatches 11, 14. Finally, E. coli EndoV is also capable of cleaving insertion/deletion mismatches, flap and pseudo Y structures 15, which are all characterized by a discontinuous/distorted DNA helix.
Although EndoV cleaves a spectrum of DNA lesions, genetic analysis of E. coli nfi insertion mutants and overproducing strains suggest a major role for EndoV in the in vivo repair of deaminated purine bases like hypoxanthine 2, 7, 8, 16. The function of EndoV in eukaryotic cells is less understood, however, the nfi mutant of fission yeast Schizosaccharomyces pombe displays a strong mutator phenotype (I. Alseth, CMBN, Rikshospitalet University Hospital, personal communication), and EndoV from mice comprises DNA repair activities resembling those of the bacterial counterparts 17. Furthermore, nfi−/− mice display a cancer prone phenotype (A. Klungland, CMBN, Rikshospitalet University Hospital, personal communication) supporting a key role for EndoV in genome integrity and for malfunction of the EndoV pathway in cancer pathophysiology.
In order to characterize DNA repair initiation for this prototypic enzyme, we solved the crystal structures of Thermotoga maritima EndoV in complex with a hypoxanthine lesion substrate and product DNA.
RESULTS
Structure determination
To unravel the molecular basis for lesion recognition and the unprecedented 3′ offset nicking of the various substrates of EndoV, we crystallized and solved structures for both inactive EndoV D43A mutant 18 and active wild-type EndoV from Thermotoga maritima (Tma) in complex with DNA harboring a deaminated adenine. The D43A mutant represents the lesion recognition complex (LRC) prior to phosphodiester hydrolysis, whereas the wild-type complex establishes the structure of the product complex (PC). Both complexes crystallized in space group I222 with two copies of EndoV in the asymmetric unit. The structure of the LRC structure was solved by single-wavelength anomalous diffraction (SAD) with 5-bromo-2-deoxyuracil substituted DNA, and refined to 2.1Å resolution (R = 21.6%, Rfree = 25.8%). The wild-type product complex structure was initially refined by rigid-body refinement, using the atomic coordinates of the protein part only from the LRC as a starting model. The LRC was refined to 2.15Å resolution (R = 25.9%, Rfree = 28.8%). We expect these structures to be generally representative of the EndoV enzyme family, as Thermotoga maritima EndoV contains conserved residues characteristic of EndoV enzymes from E. coli to humans (Supplementary Fig. 1 online).
Description of the Structure
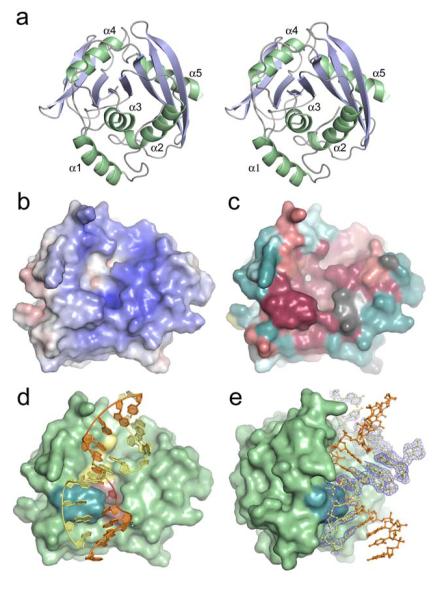
Structure determination of Tma EndoV reveals an αβα protein with a central 8-stranded β-sheet of parallel and anti-parallel strands flanked on either side by α-helices (Fig. 2a). In general, the protein structures of the LRC and PC are nearly indistinguishable, with only minor reorientations of a few side chains surrounding the mutated Asp43. The proteins are superimposable with an r.m.s.d. of 0.30Å for 223 Cα atoms. EndoV contains a ‘ribonuclease H like motif’ like in E. Coli RNase H 19, 20, which is also found in E. coli and yeast Holliday junction resolvase RuvC 21, 22, the catalytic domain of E. coli DNA transposase 23, the PIWI domain of P. furiosus Argonaute 24, and the 5′ endonuclease domain of the nucleotide excision repair protein UvrC from T. maritima25. However, the topology and number of strands in the central β-sheet as well as the number and orientation of the surrounding helices varies between these proteins, and none of these other enzymes with known structure share the substrate affinity and the enzymatic properties of EndoV.
Figure 2.
EndoV overall fold, surface characteristics and protein-DNA complex structure. (a) Stereo pair showing protein fold and ternary structure of T. maritima EndoV. (b) Electrostatic potential of wild-type EndoV mapped onto the solvent accessible protein surface (blue positive regions, red negative regions). Electrostatic potential calculated using APBS. (c) Molecular surface showing conserved residues in the EndoV family [colored from dark burgundy (highly conserved) through neutral grey into dark cyan (highly variable)]. Degree of conservation calculated using ConSeq (http://conseq.tau.ac.il/). (d) Molecular surface with bound DNA (orange and yellow tubes and spheres) showing spatial relationships among key structural elements: the strand-separating PYIP wedge (cyan, left) protrudes out adjacent to residues Asp43, Glu89, Asp110, His214 involved in Mg2+ ion binding and phosphodiester incision (yellow, center), as well as hypoxanthine lesion and surrounding residues (Leu85, Gly111, Gln112, Gly113, Gly136, Leu142) forming the nucleobase pocket (red, center). (e) Molecular surface of wild-type Tma EndoV showing substantial bending of the bound duplex DNA (orange and yellow ball-and-stick representation). PYIP-wedge shown in cyan. Experimental electron density is shown for one of the DNA strands in the duplex (σA-weighted 2Fo-Fc map contoured at 1σ).
The positively charged DNA binding surface of EndoV (Fig. 2b) comprises a distinct central cleft of conserved residues (Fig. 2c) that runs across the β-sheet and includes a base lesion recognition pocket, a strand separating wedge, and a catalytic pocket (Fig. 2d). The liganded DNA forms duplex DNA through homo-dimerization via a 2-fold crystallographic symmetry within the crystal lattice, so that two EndoV molecules bind to one single DNA duplex with two hypoxanthine bases (Supplementary Fig. 2 online). The DNA duplex is sharply bent adjacent to the lesion and bound to EndoV with its minor groove facing the protein (Fig. 2e).
DNA strands are separated by a highly conserved wedge
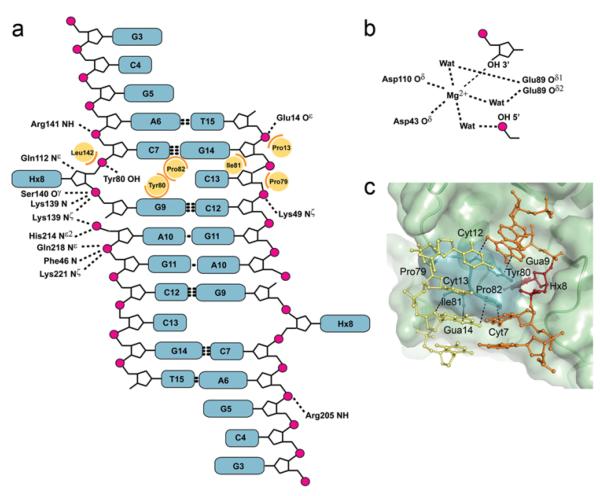
A majority of the interactions between EndoV and the DNA ribose-phosphate backbone are on the 3′ side of the lesion (Fig. 3a and Supplementary Fig. 3 online), including the conserved Lys139 and His214. Several hydrophobic residues are strategically located to play key roles in maintaining the DNA conformation. Particularly, a wedge-like segment on the protein surface, arising from the PYIP motif (Pro79-Tyr80-Ile81-Pro82) divides the duplex DNA strands adjacent to the lesion (Fig. 2d, e and Fig. 3). The Tyr80 aromatic ring stacks face-to-face with the guanine base 3′ to the deaminated base, sterically blocks the vacant hypoxanthine position in the DNA helix, and hydrogen bonds to the DNA phosphate backbone. Pro82 stacks against Tyr80 as well as the base 5′ of the lesion, and together with Ile81, wedge open the duplex (Fig. 3c). This PYIP-wedge identified here thus separates the two DNA strands at the lesion and pushes the base (cytosine) opposite hypoxanthine partly out of the duplex; however, base pairs on either side of the lesion remain paired (Fig. 3c). Tma EndoV Y80A and Y80H mutants are severely compromised in binding to DNA base lesions and the corresponding nicked product, whereas Y80F is similar to the wild type EndoV in this respect, suggesting that the aromatic ring stacking identified here is key to retaining high affinity to the DNA 18.
Figure 3.
Protein-DNA contacts. (a) Protein-DNA contacts in the wild-type product complex. Hydrogen bond/ionic interactions (dashed lines, 3.75Å cut-off), main chain amide nitrogen atoms (N) and steric interactions (orange arcs, 4.25Å cut-off) involving side chains (yellow circles) are shown for one of the two EndoV molecules binding symmetrically to the DNA. (b) Close-up of the coordination around the phosphodiester insicion. (c) Close-up of the strand separating PYIP-wedge with selected distances to DNA bases stacking with residues defining the protein surface. The hypoxanthine (dark red) is partially buried behind the wedge.
The discovery of the strand-separating wedge as part of the DNA binding motif helps explain the broad EndoV substrate range, whereby the PYIP-wedge forms a key element in the recognition of structures with helical distortions such as insertion/deletion mismatches, pseudo Y, flap and hairpin substrates.
The deaminated base is inserted into a specific pocket
The PYIP wedge motif is furthermore well suited to play an important and independent role in search for modified single bases by interrogating duplex DNA and presenting specific bases to the active site (Fig. 3c). In contrast to known DNA base repair mechanisms for damage reversal and BER, which flip the nucleoside ~180° into a lesion recognition pocket via the major groove 26, the hypoxanthine in the EndoV complex is inserted into the predominantly hydrophobic recognition pocket by a ~90° degree rotation in the opposite direction toward the minor groove (Fig. 3c).
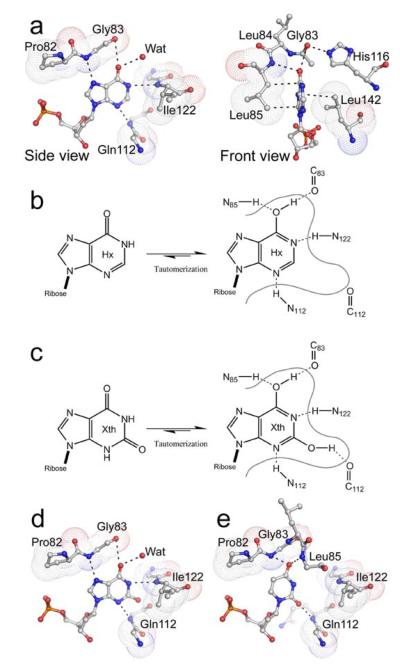
The hypoxanthine base is inserted between the hydrophobic side chains of Leu85 and Leu142, and the high degree of conservation in these positions across the species reflects the importance of hydrophobic stacking with the electron rich, heterocyclic ring (Fig. 4a). The Leu142 side chain appears important for contacting the 5-membered heterocyclic ring of hypoxanthine as well as assisting the associated rotation by close contacts to the DNA backbone. The shape of the pocket is further established in part by key conserved glycine residues (Gly83, Gly111, Gly113 and Gly121) of which Gly11, Gly113 and Gly121 are invariant in the EndoV family (Supplementary Fig. 1 online). Introduction of a valine side chain at either Gly111 or Gly113 in the DGXG-motif reduces endonucleolytic activity for hypoxanthine-containing DNA by 50% 13. Further, the strictly conserved Gly136 has been shown to be important for cleavage of various substrates 13. Gly136 lies in the middle of β-strand 4, and insertion of a larger hydrophobic side chain is likely to shift the neighboring loop containing the DGXG motif, thus displacing both the metal coordinating Asp110 as well as the remaining residues in the DGXG motif. Altogether, these amino acid substitutions validate the importance of the protein fold as a scaffold for the lesion recognition pocket and the metal coordination around the active site Asp110.
Figure 4.
Protein-DNA contacts in the base lesion pocket. (a) Diagram of interactions involved in hypoxanthine recognition, shown in both side-view and front-view. Hydrogen bonds and steric interactions shown with dashed lines; the van der Waals volumes of selected residues involved in hypoxanthine contacts represented by dotted surfaces. (b) Tautomeric forms of hypoxanthine and detailed hydrogen bonding network. (c) Tautomeric forms of xanthine and detailed hydrogen bonding network. (d) Model of uracil binding to EndoV.
Lesion recognition involves protein backbone atoms
The discrimination between adenine and hypoxanthine in particular, as well as between other native DNA bases and their corresponding deaminated analogues in general, arise through several lesion-specific interactions in the recognition pocket. Except for a single solvent water molecule, all polar contacts between EndoV and the hypoxanthine base involve protein backbone atoms only. Consequently, the recognition pocket is fairly rigid. The close proximity of the Ile122 backbone amide NH to the N1 atom in the deaminated purine bases probably leads to hypoxanthine and xanthine binding in their respective tautomeric imidic acid forms (Fig. 4a-c). The resulting hydroxyl group attached to C6 in hypoxanthine/xanthine forms hydrogen bonds with the Gly83 carbonyl oxygen and Leu85 amide nitrogen, which are reinforced cooperatively (Fig. 4b, c). The xanthine model further suggests a hydrogen bond interaction between the Gln112 backbone carbonyl oxygen and the hydroxyl group of C2 in xanthine (Fig. 4d). The invariant His116 side chain also interacts with the backbone carbonyl oxygen of Gly83, securing a stable conformation of the adjacent critical PYIP-wedge loop and closing in the two sides of the pocket. Consistent with our structures, His116 can be replaced by hydrogen bond donating residues like Gln and Thr without notable loss of activity 13.
Modeling of uracil in the EndoV binding pocket suggests that the two carbonyl moieties are recognized by the backbone amide NH groups of Gly83, Leu85 and Gln112, while Gly83 backbone carbonyl and Ile122 amide NH are too distal to form any contacts (Fig. 4e). This model furthermore suggests that rejection of the closely related thymine is governed by steric repulsion between the additional methyl group in thymine and the side chain of Pro82 within the conserved PYIP-wedge. The smaller pyrimidine ring of uracil will share a smaller contact surface with Leu85 and Leu142, which may explain the lower affinity to uracil compared with the larger purines like hypoxanthine27.
EndoV has also been shown to cleave substrates with base-mismatches and helical distortions, like mismatch-loops, hairpins and flap structures, implying that EndoV can possibly also accommodate native bases in the recognition pocket. Interestingly EndoV cleaves mismatched base pairs preferentially at adenine and guanine purines11. However, EndoV binds to cleaved mismatch base pair products with much lower affinity as compared to cleaved deaminated bases27 indicating that the pocket is significantly less favorable for adenine and guanine than for the corresponding deaminated bases. These results, combined with the present structural analysis, suggests that the EndoV nucleobase pocket is optimized for binding deaminated bases, yet capable of accommodating the normal adenine and guanine bases in a context of a base-mismatch or DNA helix distortion only.
Tight binding of DNA ends secure a stable product complex
The crystal structure of wild-type EndoV in complex with DNA reveals the atomic details of the 3′ incised product including the orientation of all catalytic residues and the metal ion cofactor (Fig. 3b and Fig. 5a). The distance between Gua9 free 3′ OH and Ade10 free 5′ phosphate exceeds 5Å at the cleavage site, and the electron density for both terminal groups is distinct, demonstrating complete phosphodiester cleavage (Fig. 5b). The Mg2+ ion is directly coordinated to the 3′ OH group of Gua9 and the two catalytic residues Asp43 and Asp110. Removal of Asp43, Glu89 or Asp110 severely affect catalysis 18. Two water molecules connect the Mg2+ ion to the conserved Glu89 while another water molecule bridges the metal cofactor and the free 5′ phosphate. The terminal 5′ phosphate is additionally held firmly in place by Lys139 and His214 (Fig. 5a). The side-chain of the conserved Lys139 is bridging two DNA phosphate groups on either side of the insicion (Fig. 3a and Fig 5a). Combined, this specific hydrogen bond network appears fine-tuned to secure strong binding of the free DNA ends of the cleavage product after catalysis.
Figure 5.
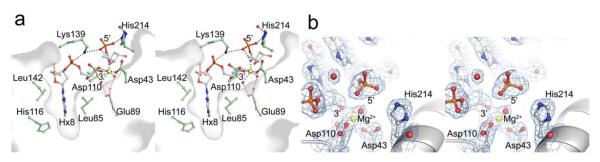
Active site architecture of EndoV-DNA complex. (a) Stereo pair showing active site with the nicked product DNA free 3′ hydroxyl- and 5′ phosphate groups. A thin protein surface slab shows the steric separation of the recognition pocket and the catalytic centre (yellow Mg2+). (b) Stereo pair of the active site region with experimental electron density showing the DNA incision (density gap) and Mg2+ ion coordination (2Fo-Fc map contoured at 1.0σ).
Binding of EndoV to the intermediate cytotoxic single-strand break product is probably crucial for recruitment of and controlled handover to downstream processing factors, as proposed for BER nucleases APE1 and EndoIV 26. The side chain of Leu85 forms a physical barrier between the catalytic site and the lesion recognition pocket, separating the processes of lesion recognition and strand incision by about ~8Å. Thus, the present product complex defines how EndoV binds and protects the single strand break product in the initial step of this pathway. The permanent insertion of the deaminated base in the lesion recognition pocket following catalysis secures tight binding of the nicked product.
The structurally related enzymes Argonaute 24 and UvrC 25 have catalytic triads consisting of two aspartates and one histidine, corresponding to Asp43, Asp110 and His214 (DDH-motif) in Tma EndoV. Both these proteins also bind only one Mn2+ ion, yet these enzymes may bind two metals in the presence of a nucleic acid substrate 25. However, the metal ion in both Argonaute and UvrC is directly coordinated to the histidine side chain (Supplementary Fig. 4 online), while in the product complex of EndoV, His214 is directly involved in DNA binding (Fig. 5a). The second metal ion is not required to bind the product with high affinity. Two metal ions have been observed in the E. coli Tn5 transposase/DNA complex 28 as well as in the B. halodurans RNase HI RNA/DNA hybrid complex 29, binding to a related DDD/DDE motif. In eukaryotic EndoV, His214 is replaced by an aspartic acid (Supplementary Fig. 1 online), so the resultant DDD motif could resemble the 2-metal binding sites in RNase HI and Tn5 transposase. In that case, the second Mg2+ cation could function as a bridge between the negatively charged aspartic acid side chain and the free 5′ phosphate in the incised product, thereby diminishing the phosphate-carboxylate electrostatic repulsion that would otherwise exist in a protein-DNA complex of eukaryotic EndoV.
The D43A mutant seems unable to bind the Mg2+ cofactor – no electron density corresponding to a metal binding to the protein is apparent in the LRC structure. Finally, comparison of the LRC and PC complexes of Tma EndoV reveals no conformational change or substantial shifts in the protein backbone to take place during catalysis (Supplementary Fig. 5 online).
DISCUSSION
Together, the structures of the EndoV-DNA substrate and product complexes presented here provide novel insight into the initial step of what has been a structurally undefined, but biologically critical, DNA base repair pathway 1, 6, 8, 17. The structures reveal a conserved strand-separating PYIP wedge positioned to play an important role as a lesion sensor by presenting deaminated bases to the lesion specific pocket. The wedge may also work as a sensor for detection of base-mismatches, particularly involving purine bases. Moreover, this motif could have an independent role in recognition of DNA structures with helix distortions, by wedging into the more open DNA minor groove in such substrates. The present structural analysis suggests that the EndoV lesion-binding pocket is optimized for recognizing deaminated bases, but normal adenine and guanine bases can possibly also be accommodated if present in a base-mismatch or close to a DNA helix distortion.
Finally, these EndoV structures address the paradox of a base repair pathway that cleaves DNA one nucleotide 3′ of a base lesion: the 3′ downstream nicking is due to a physical barrier between the catalytic site and the lesion recognition pocket that enforces a one-nucleotide offset strand incision. This dual pocket feature secures tight binding of the cytotoxic, nicked repair intermediate to EndoV by permanent insertion of the deaminated base in the lesion recognition pocket in combination with tight hydrogen bonding of both the free 3′ and 5′ ends by conserved residues in the active site.
METHODS
Expression and purification of endonuclease V from Thermotoga maritima
A pET28b plasmid including a full-length Tma endonuclease V (EndoV) mutant D43A or wild-type sequence was transformed into the E. coli expression strain BL21 CodonPlus (DE3) RIL (Stratagen), and overexpressed in LB-broth cultures supplemented with 50 mg l−1 kanamycin. Expression was induced with 0.5 mM IPTG at OD600 ~ 0.75 for 2-3 hours at 37°C. Cell-free extracts were prepared by 3 × 30 s sonication of cell pellets dissolved in 50 mM NaCl, 20mM MES pH 6.5 and 10mM β-mercaptoethanol (buffer A). Cell debris was removed by centrifugation at 20000 – 27000 g for 30 min. The protein extracts were incubated at 70° C for 15 minutes, followed by a second centrifugation step. The cell-free extracts were loaded onto 5 ml HiTrap SP XL columns (GE Healthcare) equilibrated with buffer A. Each protein was eluted using a linear gradient to 1M in NaCl. The proteins were eluted in fractions, analyzed on SDS-PAGE gel, and pure EndoV fractions were pooled and dialysed against buffer A. Aliquots of EndoV (~8mg ml−1) were stored at −20 °C prior to crystallization.
Crystallization and Data Collection of the Lesion Recognition Complex
An 11-mer DNA oligonucleotide (Operon Biotechnologies GmbH) with sequence 5′-GC-5BrU-AC-Hx-GA-5BrU-CG-3′, containing both 5-bromo-deoxyurasil (5BrU) and hypoxanthine (Hx), was annealed with a complementary strand with T opposite Hx. Purified D43A EndoV was thawed on ice, mixed with the DNA in molar ratio 1:1.5 (excess DNA) and equilibrated on ice for > 30 min. Plate shaped crystals of the EndoV-DNA complex were obtained by the vapor diffusion method at room temperature using hanging drops equilibrated against 6 – 12% (w/v) MPEG 2k in 200mM imidazolematate buffer, pH 5.8 – 7.4. Crystals grew to final size of about 0.1mm, and were mounted in cryo loops and flash-frozen in liquid nitrogen following a short soak in mother liquor supplemented with ethylene glycol to a final concentration of 30% (v/v). A complete SAD data set to 2.1 Å resolution (T = 100 K, λ = 0.9117 Å) was collected using beamline BL12.3.1 at the ALS synchrotron, Berkeley Laboratories. Diffraction images were processed and the integrated data were scaled and merged with the HKL2000 suite 30. The crystal belongs to space group I222 with cell dimensions a = 51.98 Å, b = 132.24 Å, c = 192.62 Å (Table 1).
Table 1.
X-ray data collection and refinement statistics
| Wild-type EndoV (PC) | D43A EndoV (LRC, 5-BrU peak) | |
|---|---|---|
| Data collection | ||
| Space group | I222 | I222 |
| Cell dimensions | ||
| a, b, c (Å) | 55.06, 134.29, 194.45 | 51.98, 132.24, 191.62 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50 – 2.15 (2.23 – 2.15) * | 50 – 2.10 (2.19 – 2.10) |
| Rsym (%) | 7.8 (59.1) | 5.3 (34.9) |
| I / σI | 7.8 (2.6) | 69 (5.9) |
| Completeness (%) | 99.2 (98.9) | 87.6 (62.0) |
| Redundancy | 6.0 (5.6) | 13.3 (10.6) |
| Refinement | ||
| Resolution (Å) | 50 – 2.15 | 50 – 2.10 |
| No. reflections | 39299 | 32087 |
| Rwork / Rfree (%) | 25.9 / 28.8 | 21.6 / 25.8 |
| No. atoms | ||
| Protein | 3580 | 3574 |
| Ligand/ion | 435 | 648 |
| Water | 216 | 222 |
| B-factors | ||
| Protein | 44.5 | 54.7 |
| Ligand/ion | 66.0 | 74.1 |
| Water | 47.6 | 58.8 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.006 |
| Bond angles (°) | 1.3 | 1.3 |
Values in parentheses are for highest-resolution shell (2.23 – 2.15 and 2.19 – 2.10 respectively)
Crystallization and data collection of the wild-type EndoV Product Complex
The structure of the LRC revealed that the combination of a low melting point for the 11-mer with the presence of a short self-complementary segment in the DNA, resulted in a dimerization with two EndoV monomers binding to one single duplex DNA with two lesions (Supplementary Fig. 2a online). Hoping to get a new crystal form of EndoV with a single-lesion duplex DNA, purified wild-type EndoV (8mg ml−1) supplemented with 5mM MgCl2 was mixed with a 15-mer DNA-oligo with sequence 5′-ATGCGAC-Hx-GAGCCGT-3′ (complementary strand contain T opposite Hx) in molar ratio 1:1.5 (excess DNA). The protein-DNA mixture was screened at room temperature using an Oryx robot (Douglas Instruments). Plate shaped crystals were obtained with 10% (w/v) polyethylene glycol 4000, 0.2 M ammonium acetate, 0.01 M calcium chloride and 0.05 M sodium cacodylate, pH 6.5. Crystals were soaked in mother liquor supplemented with 20% (w/v) glucose prior to flash freezing in liquid nitrogen. A complete data set to 2.15Å resolution (T = 100 K, λ = 0.9185 Å) was collected using beamline ID14-4 at the European Synchrotron Radiation Facility in Grenoble, France. Diffraction images were processed using Mosflm 31, and the integrated data were scaled and merged with CCP4/Scala. Despite using a different DNA sequence and oligo length (Supplementary Fig. 2 online), wild-type EndoV still crystallized with two EndoV proteins in the asymmetric unit, both bound to a single-stranded, self-priming DNA forming a DNA duplex through crystallographic 2-fold symmetry in space group I222. The unit cell dimensions are a = 55.06 Å, b = 134.29 Å, c = 194.45 Å (Table 1).
Structure Determination
Solve/resolve 32 was used to calculate initial experimental phases from the 2.1Å single-wavelength anomalous dispersion data of the D43A EndoV LRC. Four bromine sites were identified. Phases were improved by density modification prior to automatic building of 295 residues in two polypeptide chains using ARP/wARP 33. The wild-type EndoV PC was initially refined by rigid-body refinement using the atomic coordinates of the protein part from the refined LRC as a starting model.
Model building and refinement
Generally, the two peptide chains in each structure were completed and adjusted by manual inspection and modeling in Coot 34, interspersed with simulated annealing refinement in CNS1.1 35. Improvements of the models were monitored with R-free cross-validation against 8% and 5% of the data for D43A and wild-type EndoV, respectively. The DNA was manually built into each model using both fofc and 2fofc Fourier maps as guidelines.
For the LRC, a 7-mer single-stranded DNA fragment flanking the flipped hypoxanthine base was easily recognized in the initial difference map. A gradual improvement of the model revealed the location of the remaining 4 bases in the lesion strand, as well as 5 nucleotides belonging to the complementary strand (Supplementary Fig. 3 online). Solvent water molecules were located in succeeding difference maps and manually filtered. Refinement of the occupancy factor for the two strands gave an occupancy factor of ~1.0 and ~0.70 for the lesion and complementary strand, respectively. In the final refinement, the occupancy for the complementary strand was fixed at 0.75. The occupancy factor for the first 3 terminal nucleosides at the 5′ end of the lesion strand was also fixed at 0.50/0.75. The final model of the EndoV LRC contains 2 crystallographically independent EndoV-DNA complexes (each with 223 amino acid residues and 16 nucleotides) as well as 222 solvent water molecules.
The initial refinement of the PC structure was carried out by rigid-body refinement and simulated annealing of the protein part of the EndoV LRC. The sidechains of residues Asp43, Glu89 and Asp110 were also initially removed and remodeled in the final steps of the refinement. A short DNA fragment flanking the flipped hypoxanthine base was readily identified in the initial difference maps. Gradual improvement of the model revealed the location of additional nucleotides as well as one Mg2+ ion in the active site. Solvent water molecules were located by difference maps and manual filtering. The final model of the EndoV product complex contains 2 crystallographically independent EndoV-DNA complexes (each with 223 amino acid residues, 8 or 13 nucleotides and 1 Mg2+ ion) as well as 216 solvent water molecules. Refinement statistics for the two models are listed in Table 1. Electrostatic potential of wild-type Tma EndoV was calculated by APBS 36 and the degree of conservation was calculated using ConSeq 37. All structural figures were prepared with PyMol (Delano Scientific, CA, USA; http://www.pymol.org).
Accession codes
Protein Data bank: Coordinates and structure factors have been deposited in the Protein Data Bank with the accession codes 2w35 (PC) and 2w36 (LRC).
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the technical support at the BL12.3.1 beamline at ALS, Berkeley Laboratories and the ID14-4 beamline at ESRF, Grenoble, used to collect X-ray diffraction data. Base repair in the Tainer lab is funded by a grant from the National Institute of Health. This work in the Bjoras lab is funded by EU, Norwegian Research Council (FUGE-CAMST) and the Norwegian Cancer Society.
Footnotes
AUTHOR CONTRIBUTIONS B.D. and M.B. designed and performed experiments, analyzed data and wrote the manuscript. J.A.T., I.A. and W.C. analyzed data and wrote the manuscript. A.S.A designed and performed experiments and analyzed data. I.R., O.E.O, P.H.B. and H.G. designed and performed experiments.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Demple B, Linn S. On the recognition and cleavage mechanism of Escherichia coli endodeoxyribonuclease V, a possible DNA repair enzyme. J. Biol. Chem. 1982;257:2848–2855. [PubMed] [Google Scholar]
- 2.Schouten KA, Weiss B. Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res. 1999;435:245–254. doi: 10.1016/s0921-8777(99)00049-x. [DOI] [PubMed] [Google Scholar]
- 3.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat. Rev. Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen T, et al. DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc. Natl. Acad. Sci. U. S. A. 1992;89:3030–3034. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wink DA, et al. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 6.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 7.Guo G, Weiss B. Endonuclease V (nfi) mutant of Escherichia coli K-12. J. Bacteriol. 1998;180:46–51. doi: 10.1128/jb.180.1.46-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss B. Removal of deoxyinosine from the Escherichia coli chromosome as studied by oligonucleotide transformation. DNA Repair (Amst) 2008;7:205–212. doi: 10.1016/j.dnarep.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao M, Hatahet Z, Melamede RJ, Kow YW. Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem. 1994;269:16260–16268. [PubMed] [Google Scholar]
- 10.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Lu J, Barany F, Cao W. Multiple cleavage activities of endonuclease V from Thermotoga maritima: recognition and strand nicking mechanism. Biochemistry. 2001;40:8738–8748. doi: 10.1021/bi010183h. [DOI] [PubMed] [Google Scholar]
- 12.Yao M, Kow YW. Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem. 1995;270:28609–28616. doi: 10.1074/jbc.270.48.28609. [DOI] [PubMed] [Google Scholar]
- 13.Feng H, Dong L, Klutz AM, Aghaebrahim N, Cao W. Defining amino acid residues involved in DNA-protein interactions and revelation of 3′-exonuclease activity in endonuclease V. Biochemistry. 2005;44:11486–11495. doi: 10.1021/bi050837c. [DOI] [PubMed] [Google Scholar]
- 14.Yao M, Kow YW. Strand-specific cleavage of mismatch-containing DNA by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem. 1994;269:31390–31396. [PubMed] [Google Scholar]
- 15.Yao M, Kow YW. Cleavage of insertion/deletion mismatches, flap and pseudo-Y DNA structures by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem. 1996;271:30672–30676. doi: 10.1074/jbc.271.48.30672. [DOI] [PubMed] [Google Scholar]
- 16.Weiss B. Endonuclease V of Escherichia coli prevents mutations from nitrosative deamination during nitrate/nitrite respiration. Mutat. Res. 2001;461:301–309. doi: 10.1016/s0921-8777(00)00062-8. [DOI] [PubMed] [Google Scholar]
- 17.Moe A, et al. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Lu J, Barany F, Cao W. Mutational analysis of endonuclease V from Thermotoga maritima. Biochemistry. 2002;41:8342–8350. doi: 10.1021/bi015960s. [DOI] [PubMed] [Google Scholar]
- 19.Katayanagi K, et al. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990;347:306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- 20.Yang W, Hendrickson WA, Crouch RJ, Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 21.Ariyoshi M, et al. Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 22.Ceschini S, et al. Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2. EMBO J. 2001;20:6601–6611. doi: 10.1093/emboj/20.23.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 24.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 25.Karakas E, et al. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. EMBO J. 2007;26:613–622. doi: 10.1038/sj.emboj.7601497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Yao M, Kow YW. Further characterization of Escherichia coli endonuclease V. Mechanism of recognition for deoxyinosine, deoxyuridine, and base mismatches in DNA. J. Biol. Chem. 1997;272:30774–30779. doi: 10.1074/jbc.272.49.30774. [DOI] [PubMed] [Google Scholar]
- 28.Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition. Curr. Opin. Struct. Biol. 2004;14:50–57. doi: 10.1016/j.sbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CWJ, Sweet RM, editors. Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 31.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 32.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berezin C, et al. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.