Abstract
We have previously described a 27-kb pathogenicity island of Streptococcus pneumoniae, termed pneumococcal pathogenicity island 1 (PPI1), which contains iron uptake locus piaABCD, required for full virulence in mice, and a further 28 previously uncharacterized genes. We have investigated one of these, Sp1051, which encodes a protein of unknown function. Disruption of Sp1051 does not affect growth in laboratory broth, serum, or blood but impairs virulence in mouse models of infection. When S. pneumoniae capsular serotypes were analyzed by PCR and Southern hybridization, it was found that 33% did not contain Sp1051. Analysis of other genes within PPI1 demonstrated that, compared to the serotype 4 genome published by The Institute for Genome Research (TIGR), the genomes of many strains contain deletions of a variable number of genes between Sp1046 and Sp1064, conforming to one of six different patterns. Amplification by PCR of this PPI1 variable region from a capsular serotype 17 strain and comparison of the sequence to TIGR serotype 4 strain sequence showed that Sp1051 is contained within an 11.3-kb segment of DNA flanked by 7-bp direct repeats within the serotype 4 strain which is not present in the serotype 17 strain. Further comparison of the sequences of this region between the three published S. pneumoniae genomes demonstrated that serotype 19F and strain R6 contain novel complements of genes not present in the serotype 4 strain. These data indicate that there is striking variation in gene content and structure of the 3′ region of PPI1 among strains and that this region includes at least one virulence determinant. Gene variation within horizontally acquired DNA such as that of PPI1 may be one factor modulating differences in virulence among strains.
With many bacterial pathogens, including the gram-positive respiratory pathogen Streptococcus pneumoniae (3, 5), the ability to cause disease varies among strains within a species. One major cause for variations in virulence among strains is differences in gene content, with genes encoding specific virulence functions conferring particular virulence phenotypes. For instance, many strain-specific virulence factors of Escherichia coli are encoded on regions of DNA with atypical nucleotide compositions thought to have been horizontally acquired, termed pathogenicity islands (PAIs) (7). However, with the exception of differences in capsular serotype (3, 5), the reasons for variation in virulence among strains for gram-positive pathogens such as S. pneumoniae are poorly characterized. Recent data have demonstrated that additional regions of probably horizontally acquired DNA are also found within the genomes of gram-positive bacterial pathogens, some of which vary in content among strains and could potentially influence differences in virulence (6, 11, 15). The functions of the majority of proteins encoded within regions of horizontally acquired DNA in S. pneumoniae and other gram-positive pathogens are unknown, but these regions can contain genes which affect virulence and are therefore described as PAIs (9, 14), including a 27-kb region of the S. pneumoniae genome termed pneumococcal pathogenicity island I (PPI1) (1). PPI1 shares many of the features of PAIs from gram-negative bacteria, including variant G+C content, the presence of several genetic mobility genes (including genes encoding a recombinase and a transposase), and lack of similarity of its genes to genes within closely related Streptococcus species. PPI1 contains the operon piaABCD, which encodes an important S. pneumoniae ABC transporter required for iron uptake and full virulence in mouse models of pneumonia or septicemia (1, 2). However, although piaABCD is contained within a PAI, this operon is found in all the S. pneumoniae strains investigated and so does not contribute to differences in virulence among strains.
The annotated sequence of PPI1 in the fully sequenced capsular serotype 4 S. pneumoniae strain KNR.7/87 indicates the presence of another 28 genes in addition to the piaABCD operon (15). As PAIs often contain multiple virulence genes, we have investigated other genes within PPI1 to determine whether they have a role during pathogenesis. One gene, Sp1051, was found to be required for full virulence in mouse models of septicemia and pneumonia. In contrast to piaABCD, Sp1051 was not present in 4 of the 12 capsular serotypes investigated. Detailed analysis of the 3′ region of PPI1 (which contains Sp1051) demonstrated that many strains contained several gene deletions in this region compared to The Institute for Genome Research serotype 4 strain genome sequence. There was striking variation in the pattern of genes deleted, and comparisons of the genome sequences of this region for four strains with different capsular serotypes showed that each strain contained only partially overlapping complements of genes within this region. Hence the 3′ region of PPI1 contains a strain-specific virulence determinant, Sp1051, and is highly variable in structure among strains.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
PCR analysis of the structure of PPI1 was performed on the capsular serotype 3 strain 0100993 (12); 13 strains from M. C. Enright and B. G. Spratt, including 10 capsular serotype 3 strains representing all the main multilocus sequence typing (MLST) lineages (strain names and sequence types [ST] are as follows: IOKOR490, ST180; IOKOR1522, ST180; M110, ST180; M111, ST180; M145, ST180; M146, ST231; M147, ST181; M148, ST182; M149, ST181; M290, ST49) (http://spneumoniae.mlst.net), 2 capsular serotype 4 strains (M313, ST259; M127, ST205), and 1 serotype 6B strain (M158, ST171) (5); and 12 S. pneumoniae clinical isolates (representing capsular serotypes 1, 2, 3, 4, 9N, 12, 14, 16, 17, 19F, and 35F) obtained from J. Paton. S. pneumoniae strains were cultured at 37°C and 5% CO2 on Columbia agar supplemented with 5% horse blood or in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY), supplemented when necessary with chloramphenicol (CM; 4 μg ml−1). Data for growth curves were collected by measuring optical density at 580 nm (OD580) of 1-ml cultures grown in sterilized 1.5-ml cuvettes at 1-h intervals. Strains were stored at −70°C as aliquots of THY broth culture (OD580 of 0.3 to 0.4) containing 10% glycerol. Plasmids were amplified in E. coli strain DH5α and grown at 37°C on Luria-Bertani medium with appropriate selection (13).
Nucleic acid isolation manipulations and analysis.
S. pneumoniae chromosomal DNAs were isolated with Wizard genomic DNA isolation kits (Promega), and plasmid DNA was isolated from E. coli with Qiagen plasmid kits. Reverse transcriptase PCRs (RT-PCRs) were performed as previously described with RNA isolated with the SV total RNA isolation system (Promega) and the Access RT-PCR system (Promega) and target-specific primers (Table 1) (2). Cloning, transformation, restriction digestions, ligations of plasmid DNA, and preparation of membranes for Southern hybridizations were performed according to standard protocols (13). Southern hybridizations were prepared and probed with the Alkphos direct-labeling kit (Amersham Biosciences). Plasmid inserts were sequenced with Applied Biosystems dye terminator chemistry, and cycle sequencing was performed by the Medical Research Council DNA Sequencing Service, Hammersmith Hospital, London, United Kingdom. S. pneumoniae sequence data were obtained from The Institute for Genomic Research website (http://www.tigr.org) or http://bioinformatica.cnio.es/data/Spneumo/ and analyzed with BLAST (available at the National Center for Biotechnology Information website) (http://www.ncbi.nlm.nih.gov/blast/), MacVector (International Biotechnologies, Inc.), and Artemis3 (Genome Research Ltd.). For PCR analysis of PPI1 structure, primer sets were designed from the sequenced genome of strain KNR.7/87 (15), and, to allow primers to anneal to regions with minor sequence variation in different strains, an annealing temperature of 50°C was used. All negative PCRs were repeated twice with positive control reactions.
TABLE 1.
Primers used in this study
| Name | Sequence (5′-3′) |
|---|---|
| 3.1 | GC TCT AGA GTA AAT TAC CAA GTG AGG |
| 3.2 | CGC TCT AGA TCA CCT GTA TAG GGT CG |
| 6.2 | GAA GGA CGG ATC CTC AAA G |
| 6.6 | CAA CTT AAA TCC ATT CCC |
| 6.7 | GCA AGA CTT GGG TAT TAC TG |
| 6.9 | GGC CAC CAT CCG TTT TTC AG |
| 8.1 | GC TCT AGA GCA AGG AAT CTT CGT TCA CTG |
| 8.2 | CGC TCT AGA GGC CAA TTG TAC TTC ATA TC |
| 8.3 | CCA ATT GTA CTT CAT ATC CC |
| 9.4 | GTC TTG TGA AAG CTA CCG |
| 9.6 | AGC CGT CTC ATA TAT AGA CA |
| 9.7 | CTT ATG TTA GCA GAG GCA ATC |
| 9.8 | GCT CTA GAG GGT TTA ATA ACA AG |
| 9.9 | CGC TCT AGA GAC TGC CAT TTG TAA C |
| 10.1 | CGC TCT AGA TCT TGC CTT GGT GTC GGA C |
| 10.2 | CGC TCT AGA GTT GCT TGC CAC GCA CCA C |
| 12.1 | GC TCT AGA CTA TGA AAC TGA AAT AG |
| 12.2 | CGC TCT AGA CGA TAA ATC ACT AGC TTC |
| 14.2 | CGC TCT AGA TTG CGA TAT TTA TCT CAA ACG C |
| 14.3 | GC TCT AGA CTT AGG TTT AAG AGG TCT AG |
| C.1 | CGG AAG AAT CAT ACG TGA C |
| C.2 | CGC CAG TAA ACG GAT CTA C |
| D.1 | GGT CTA TTC GAC CAC CAG |
| D.2 | CTG GTG ACC TGC ATC AGC |
Construction of mutant strains.
Disruption plasmids pPC34 and pPC57 were constructed by ligating into the XbaI site of pID701 (12) an internal fragment of the target gene amplified by PCR from genomic DNA with primers 8.1 and 8.2 (to target Sp1051, pPC34) or primers 9.8 and 9.9 (to target Sp1052, pPC57). Strains PPC34 and PPC57 containing disrupted copies of Sp1051 and Sp1052 were obtained by transformation of strain 0100993 with plasmid pPC34 or pPC57, respectively, using insertional duplication mutagenesis as previously described (1). Mutant identities were confirmed by PCR and shown to be stable through two cycles of growth in THY (approximately 24 cell divisions, with 100% of colonies retaining CM resistance). To avoid the confounding effects of phase variation on survival curve experiments, stocks in which the vast majority of bacteria were in the opaque phase were prepared by passaging the wild-type and mutant strains through mice by intraperitoneal inoculation and recovery from the spleen 20 h later.
In vivo studies using mouse models of S. pneumoniae infection and mixed-infection experiments.
In vivo experiments were performed as previously described using outbred male white mice (strain CD1; Charles River Breeders) weighing from 18 to 22 g and halothane anesthesia for intranasal inoculations (1). Inocula were prepared from thawed glycerol stocks of S. pneumoniae strains appropriately diluted in 0.9% saline stocks. For the pneumonia model, mice were given between 5 × 105 and 2 × 106 bacterial CFU intranasally (i.n.), and for the systemic model mice were given 103 bacterial CFU by intraperitoneal (i.p.) injection. For mixed infections, blood and heat-killed serum (65°C for 20 min; both obtained from human volunteers), THY, or mice inoculated with approximately equivalent CFU of the two strains to be compared were used. Mice were sacrificed after 24 (i.p. inoculations) or 48 h (i.n. inoculations), and target organs were recovered and homogenized in 0.5 ml of 0.9% saline. Dilutions of the homogenized organs or culture medium were plated on nonselective medium and cultured overnight, and the competitive index (CI) was calculated by transferring 100 colonies to Columbia blood agar containing CM to identify mutant colonies. The CI is defined as the ratio of mutant to wild-type strain CFU recovered from the mice divided by the ratio of mutant to wild-type strain CFU in the inoculum (1). A CI close to 1.0 is expected if the mutant strain has no growth or virulence defect under the conditions tested, and the lower the CI the more marked the growth defect for the mutant strain. For survival curves groups of mice were given pure inocula of the test strains, monitored closely, and culled when showing signs of terminal infection as previously described (1).
RESULTS AND DISCUSSION
Descriptions and RT-PCR analysis of Sp1050 to Sp1053.
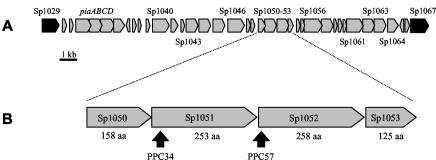
We selected a group of four genes (Sp1050, Sp1051, Sp1052, and Sp1053) within the 3′ portion of PPI1 for further investigation of their possible roles during S. pneumoniae infection (Fig. 1). The genes are transcribed in the same direction and, in common with most genes within PPI1 (except piaABCD), have not been previously investigated apart from their annotation in the sequenced genome (15). Sp1050 encodes a 158-amino-acid putative transcriptional regulator containing a helix-turn-helix motif, Sp1051 encodes a 253-amino-acid conserved hypothetical protein, Sp1052 encodes a 258-amino-acid putative phosphoesterase, and Sp1053 encodes a 125-amino-acid conserved hypothetical protein. The possibility that Sp1050 to Sp1053 are cotranscribed was assessed by RT-PCRs with RNA isolated from S. pneumoniae strain 0100993 (2). Primer pairs were designed by using the sequence of strain KNR.7/87 to amplify products spanning the gene junctions (primers 6.6 and 8.3 for Sp1050 and Sp1051, 8.1 and 9.4 for Sp1051 and Sp1052, and 9.7 and 9.6 for Sp1052 and Sp1053; Table 1). All primer pairs successfully amplified products from genomic DNA. Products were amplified from cDNA across the Sp1051-Sp1052 and Sp1052-Sp1053 junctions, but not across the Sp1050-Sp1051 junction (with three separate sets of primers), indicating that Sp1051, Sp1052, and Sp1053 are transcribed as a single RNA transcript distinct from Sp1050.
FIG. 1.
(A) Map of PPI1, showing the positions of the iron uptake ABC transporter genes piaABCD and of Sp1050 to Sp1053. Genes within PPI1 are in grey, and those flanking the ends of PPI1 are in black. (B) Genetic organization of Sp1050 to Sp1053. The derived numbers of amino acid residues (aa) for each protein are given below the corresponding gene. Arrows, insertion points for the mutant strains.
The in vitro and in vivo phenotype of the Sp1051 mutant.
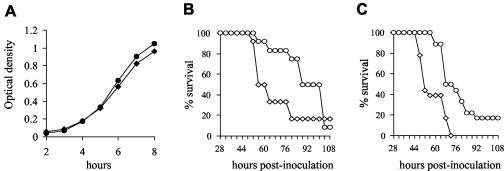
To assess the importance of the Sp1051 Sp1052 Sp1053 operon during infection, a strain containing an insertion in Sp1051 was made by transformation of plasmid pPC34 into strain 0100993 by insertional duplication mutagenesis with selection using CM to make strain PPC34 (Fig. 1B). Strain PPC34 had no growth defect in the rich laboratory growth medium THY broth compared to the wild-type strain, as determined by measuring OD580 (Fig. 2A). The ability of strain PPC34 to grow under various in vitro conditions and its ability to cause disease in animal models of S. pneumoniae infection were compared to those of the wild-type strain by using mixed infections and by calculation of the CI. The CI for strain PPC34 versus the wild-type strain after 6 h growth at 37°C in THY, human blood, or heat-treated serum was close to 1.0 (Table 2), indicating that strain PPC34 has no growth defect under these conditions. In contrast, mixed infections in animal models of systemic (bacteria inoculated via i.p. injection) or respiratory (bacteria inoculated via i.n. instillation of a suspension of bacteria) infection both showed that strain PPC34 had a virulence defect in vivo (Table 2). To help verify that the reduction in virulence was linked to the disruption of Sp1051, mixed infections by i.p. inoculation were performed on two additional PPC34 mutant strains (PPC34 clones 2 and 3) obtained in a separate transformation reaction. Both these strains were attenuated to a degree similar to that to which the original PPC34 strain was attenuated (Table 2). The reduction in the CI of strain PPC34 was detectable 6 h after inoculation and was severe by the time preterminal infection had developed (24 h after i.p. inoculation and 72 h after i.n. inoculation): only 1 of 500 colonies recovered from the lungs of five mice inoculated i.n. was CM resistant. Determination of CIs is a highly sensitive method of detecting differences in virulence among different strains but cannot demonstrate whether a mutant strain is still capable of causing progressive infection. Therefore, to further characterize the degree of attenuation in virulence of strain PPC34, groups of 12 mice were given 5 × 105 CFU of the wild-type or PPC34 strain by i.n. inoculation and monitored for the development of disease. Although mice inoculated with strain PPC34 became ill with a high mortality, the median time to the development of terminal infection was 88 h compared to 56 h for mice inoculated with wild-type S. pneumoniae (Fig. 2B). A second experiment using a higher inoculum of 2 × 106 CFU gave similar results (median time to the development of terminal infection, 68 versus 52 h) (Fig. 2C). These results demonstrate that strain PPC34 is partially attenuated in virulence and is unable to compete during both systemic and respiratory infection with the wild-type strain. Hence the Sp1051 Sp1052 Sp1053 operon affects in vivo growth of S. pneumoniae, and represents a second locus within PPI1 which influences virulence in addition to piaABCD.
FIG. 2.
(A) Growth curves of the PPC34 (circles) and wild-type (diamonds) strains in THY as determined by measuring OD. (B) Survival of groups of 12 mice inoculated i.n. with 5 × 105 CFU of the PPC34 (circles) or wild-type strains (diamonds). (C) Survival of groups of 18 mice inoculated i.n. with 3 × 106 CFU of the PPC34 (circles) or wild-type (diamonds) strains.
TABLE 2.
Comparison of the CIs for mixed inoculations of the Sp1051 mutant strain (PPC34) versus the wild-type strain in vitro and in mouse models of pneumonia or systemic infection
| Strains compared | Mouse modela or growth medium | Time point (h) | CIb (SD) | n |
|---|---|---|---|---|
| Wild type vs PPC34 | SI | 6 | 0.50 (0.11) | 4 |
| Wild type vs PPC34 clone 1 | SI | 24 | 0.04 (0.02) | 4 |
| Wild type vs PPC34 clone 2 | SI | 24 | 0.09 (0.02) | 5 |
| Wild type vs PPC34 clone 3 | SI | 24 | 0.09 (0.04) | 5 |
| Wild type vs PPC34 | PN | 6 | 0.67 (0.19) | 4 |
| Wild type vs PPC34 | PN | 72 | 0.005 (0.008) | 5 |
| Wild type vs PPC34 | THY medium | 6 | 0.97 (0.03) | 4 |
| Wild type vs PPC34 | Whole blood | 6 | 0.90 (0.13) | 4 |
| Wild type vs PPC34 | Heat-treated serum | 6 | 0.84 (0.27) | 4 |
PN and SI, mouse models of pneumonia and systemic infection, respectively.
Similar results were obtained in duplicate experiments.
To determine if the reduced virulence of strain PPC34 is due to disruption of Sp1051 or polar effects on other genes within the operon, a mutant strain containing an insertion terminating transcription of Sp1052 (termed PPC57) was made by insertional duplication mutagenesis using plasmid pPC57 (Fig. 1B). The CI (mean ± standard deviation) for strain PPC57 compared to the wild-type strain in the systemic model of infection 24 h after inoculation was 0.75 ± 0.46, showing that the loss of virulence of strain PPC34 is due mainly to mutation of Sp1051. Sp1051 encodes a protein of unknown function with similarity to a previously undescribed protein found in the Streptococcus agalactiae genome (73% identity over 250 amino acids). The mechanism by which this protein affects virulence requires further investigation, but the absence of a growth defect for the PPC34 strain in whole blood or serum indicates that the reduced virulence of this strain has a more complex basis than simply a reduced growth rate under physiological conditions.
Distribution of Sp1051 among a range of S. pneumoniae strains.
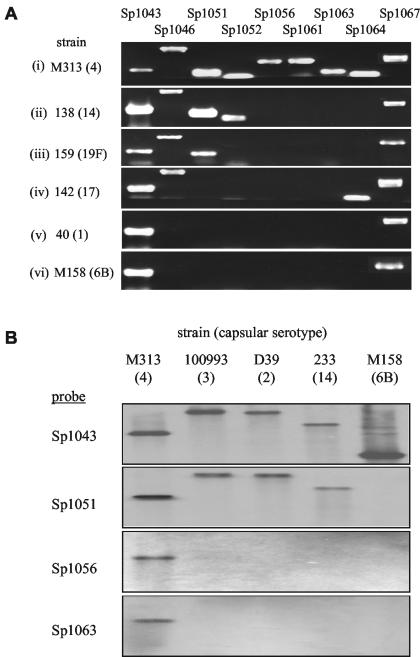
As Sp1051 is contained within PPI1 and hence may have been acquired by horizontal transfer, we investigated the distribution of this gene among 26 different S. pneumoniae strains using primers 8.1 and 8.2 to amplify an internal portion of Sp1051 by PCR. These strains represented 12 different capsular serotypes and included 10 strains representing all the major MLST lineages of capsular serotype 3 strains (http://spneumoniae.mlst.net). Strains 142, 20, 40, and M158 (capsular serotypes 17, 35F, 1, and 6B, respectively) failed to give a product on repeated attempts despite successful positive control reactions (Fig. 3A), suggesting that Sp1051 is absent in these S. pneumoniae strains. A product was obtained for Sp1051 from all the capsular serotype 3 strains despite the wide range of MLST types investigated, indicating that acquisition of Sp1051 is a relatively stable event. As Sp1051 is missing in only a proportion of strains, it is unlikely to be essential for virulence, but rather may modulate the virulence of those strains which carry it. This is in keeping with the in vivo data described above, which demonstrated that, although strain PPC34 has a strong competitive disadvantage in mixed infections, it is still able to cause fatal, albeit delayed, infection when given as a pure inoculum.
FIG. 3.
(A) Results of PCR analysis for genes within PPI1. Each strip is an ethidium bromide-stained agarose gel containing products of PCRs designed to amplify internal fragments of genes within or adjacent to PPI1 from one S. pneumoniae strain. Representative strains are presented for each of the six different patterns of PPI1 identified. The patterns for the 16 strains (capsular serotypes) investigated were as follows: (i) strains 85 (4), M313 (4), and M127 (4); (ii) strains D39 (2), 100993 (3), 154 (3), 152 (9N), 144 (12), 138 (14), 233 (14), and 140 (16); (iii) strain 159 (19F); (iv) strain 142 (17); (v) strains 20 (35F) and 40 (1); (vi) strain M158 (6B). The gene names for each product are listed above the corresponding PCR product. (B) Southern analysis of the distribution of genes within PPI1 from strain KNR.7/87 among other S. pneumoniae strains. Each panel represents a Southern membrane containing HindIII-digested genomic DNA from five different strains representing varied capsular serotypes (listed above) probed with a PCR-amplified internal portion of a gene from within PPI1 (listed to the left).
The gene content of PPI1 has considerable variation among different S. pneumoniae strains.
The observation that Sp1051 is not present in a significant proportion of strains prompted a more detailed analysis of the structure of PPI1 in 16 S. pneumoniae strains (representing 12 different serotypes) using PCR to amplify portions of 12 additional genes within or flanking PPI1 (piaA, Sp1040, Sp1043, Sp1045, Sp1046, Sp1052, Sp1056, Sp1057, Sp1061, Sp1063, Sp1064, and Sp1067). DNA fragments were consistently amplified from all strains for genes within the 5′ end of PPI1 (piaA, Sp1040, Sp1043, and Sp1045) and from Sp1067, which flanks the 3′ end of PPI1 (Fig. 1A and 3A and data not shown). In contrast, PCR products were not obtained for a variable number of adjacent genes between Sp1046 and Sp1067 within the 3′ end of PPI1 for all strains except capsular serotype 4 strains (Fig. 3A). These results suggest that blocks of these genes are not present in strains with other capsular serotypes, and six different patterns of gene deletions were identified (Fig. 3A). Southern analysis using internal fragments of Sp1043, Sp1051, Sp1056, and Sp1063 to probe HindIII-digested DNA from five strains under nonstringent conditions gave a pattern of absent genes for these strains identical to that from the PCR analysis (Fig. 3B). In addition, the pattern of the gene deletions from PPI1 in strain D39 was the same as that obtained by a DNA microarray comparison between the genome sequence of a capsular serotype 4 strain and that of D39 (15). The PCR and Southern analysis data and genome data all demonstrate that genes within the right-hand portion of PPI1 (from Sp1046 to Sp1066), including the virulence gene Sp1051, are present only in a subset of S. pneumoniae strains. We therefore refer to this region as the PPI1 variable region.
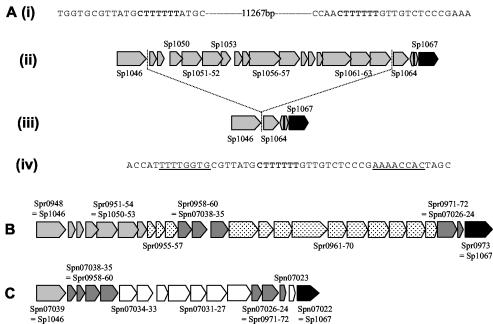
To characterize the PPI1 variable region in more detail, we attempted to amplify the DNA sequence of this region from strains D39, 0100993, 142, and 40 using primers which anneal to the genes flanking this region. However, only a DNA fragment from strain 142 was successfully amplified, possibly because in the other strains too long a segment of DNA was present between these primers for successful amplification. Strain 142 is a capsular serotype 17 strain for which the PCR analysis failed to amplify products from Sp1051, Sp1052, Sp1056, Sp1057, Sp1061, and Sp1063 (Fig. 3A). With primers designed to anneal to Sp1046 and Sp1064 (primers 6.2 and C.2, respectively) a 1.8-kb segment of genomic DNA from strain 142 was amplified, ligated into pCR1-TOPO to make plasmid pPC53, and sequenced. Alignment of this sequence to the available sequence of this region in strain KNR.7/87 (15) confirmed that in strain 142 the genes Sp1047 to Sp1063 are missing and furthermore that Sp1046 is adjacent to Sp1064 with no insertion of extra sequence in this strain compared to the KNR.7/87 strain (Fig. 4A). Hence an 11.3-kb block of genes in the PPI1 variable region, including the virulence gene Sp1051, has either been lost from strain 142 or inserted into the KNR.7/87 strain. PAIs are often flanked by direct repeats of nucleotide sequence (7), and the region of DNA containing Sp1047 to Sp1063 is flanked by a short 7-bp direct repeat in the KNR.7/87 strain, with a single copy of the direct repeat present at the insertion point of this region in strain 142 (Fig. 4A). In addition there are short 8-bp complementary nucleotide sequences flanking the direct repeat (Fig. 4A). These nucleotide features may have resulted from the integration of the region of DNA containing Sp1047 to Sp1063 into the KNR.7/87 genome.
FIG. 4.
(A) Genetic organization of the right-hand part of PPI1 in strains KNR.7/87 (ii) and 142 (iii). Boxes, open reading frames, with the gene name marked for the larger genes. (i and iv) Nucleotide structure at the junction of the insertion of the PPI1 variable region in strain KNR.7/87 (i) and strain 142 (iv). The sequence in boldface is the direct repeat flanking the 11.3 kb of DNA present in the KNR.7/87 strain but not present in strain 142; the underlined sequences are the complementary nucleotide sequences flanking the insertion point in both strains. (B) Genetic organization of the right-hand part of PPI1 in strain R6. (C) Genetic organization of the right-hand part of PPI1 in strain G54. Light grey, genes present within the KNR.7/87 genome and PPI1; black, first gene flanking the 3′ end of PPI1; dark grey, genes present in strains R6 and G54 but not strain KNR.7/87; white, genes unique to strain G54; speckled, genes unique to strain R6.
However, the results of the PCR analysis for genes within PPI1 in different strains indicate that simple integration or excision of the DNA at the site of the direct repeat between Sp1046 and Sp1064 produces only one of the possible structures for the PPI1 variable region. Of the six different structures, the point of divergence from the KNR.7/87 sequence varied from 5′ to Sp1045, Sp1046, Sp1051, or Sp1052 and from 3′ to Sp1063, Sp1064, or Sp1067 (Fig. 3A). Furthermore, comparisons of the genes present in this region in the three different S. pneumoniae strains for which genome sequences are available (strain KNR.7/87, the unencapsulated strain R6, and the capsular serotype 19F strain G54) demonstrated that each strain has a unique complement of genes within the PPI1 variable region (Fig. 4B) (4, 10, 15). Between the homologs of Sp1054 and Sp1067 strain R6 contains 18 genes totaling 20.5 kb of DNA, and between the homologs of Sp1046 and Sp1067 strain G54 contains 15 genes, 7 of which are homologs of genes inserted into this region in strain R6 (Fig. 4B). None of these genes have homologs within the KNR.7/87 genome. They encode a possible lantibiotic system, regulators, and several proteins of unknown function but no known virulence factors. The marked heterogeneity in the structure of this region of PPI1 in the strains investigated to date suggests that further analysis of other strains would probably identify additional novel gene complements for their PPI1 variable regions. In addition, these results suggest that PPI1 is a mosaic PAI with sets of genes such as piaABCD and Sp1051 to Sp1053 having been acquired at different times.
It is estimated that the gene content of S. pneumoniae varies by 10% among strains (8), and genome comparisons of S. pneumoniae strains using a DNA microarray representing the sequenced genome of KNR.7/87 demonstrated that there are 10 clusters of genes which are absent in some S. pneumoniae strains (15). These deletions vary from 9 to 37 kb and include regions with atypical nucleotide structure, such as PPI1, the capsular locus, and another recently identified probable S. pneumoniae PAI involved in adhesion to eukaryotic cells and virulence (9). Using PCR, Southern hybridization, and genome sequences from four strains for this region we have demonstrated that the gene content and structure of the 3′ region of PPI1 show marked variation among different strains. Additional regions of the KNR.7/87 genome may show variation in gene complement between strains, increasing the potential S. pneumoniae population gene pool considerably. Strains of S. pneumoniae are known to vary in their ability to cause invasive disease, and, although this is often related to capsular serotype (3, 5), virulence determinants contained within regions of variable DNA such as Sp1051 and the recently described rrgABC locus (9) may also modulate strain-to-strain variation in virulence. To identify the potential S. pneumoniae genome pool and to fully characterize the basis for differences in virulence among strains, the variable regions from a number of representative strains will need to be sequenced and the genes contained in these regions will need to be analyzed for their role in disease pathogenesis.
Acknowledgments
This work was supported by a Wellcome Trust Advanced Fellowship for Medical and Dental Graduates to J. S. Brown (056586) and by a Medical Research Council Program Grant awarded to D. W. Holden.
Editor: J. N. Weiser
REFERENCES
- 1.Brown, J. S., S. M. Gilliland, and D. W. Holden. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40:572-585. [DOI] [PubMed] [Google Scholar]
- 2.Brown, J. S., S. M. Gilliland, J. Ruiz-Albert, and D. W. Holden. 2002. Characterization of Pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect. Immun. 70:4389-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 4.Dopazo, J., A. Mendoza, J. Herrero, F. Caldara, Y. Humbert, L. Friedli, M. Guerrier, E. Grand-Schenk, C. Gandin, M. de Francesco, A. Polissi, G. Buell, G. Feger, E. Garcia, M. Peitsch, and J. F. Garcia-Bustos. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb. Drug Resist. 7:99-125. [DOI] [PubMed] [Google Scholar]
- 5.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 6.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 7.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 8.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardes, W. Keck, and A. de Saizieu. 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 69:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 10.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 12.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 15.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]