Summary
Background
Overexpression of vascular endothelial growth factor-C (VEGF-C) has been found to play an important role in malignant progression of various cancer cells, in addition to lymphangiogenesis. However, the mechanisms involved are still largely unknown. Our early research has confirmed that the expression of VEGF-C in bladder cancer was markedly higher than that in normal bladder tissues. VEGF-C can also obviously promote proliferation and invasion of bladder cancer T24 cells. In the present work, we attempted to use proteomic analysis to screen out potential VEGF-C-associated proteins involved in malignant progression of the bladder cancer T24 cells.
Material/Methods
Lentivirus vector-based RNA interference (RNAi) was employed to diminish VEGF-C expression of bladder cancer T24 cells. Then we performed comparative proteome analysis to explore differentially expressed proteins in T24 cells with and without VEGF-C siRNA, by two-dimensional difference gel electrophoresis (2D-DIGE).
Results
Twenty-three proteins were identified. Some proteins (matrix metalloproteinase-9, Keratin 8, Serpin B5, Annexin A8) with significant differences were further confirmed by Western blotting.
Conclusions
The 23 potential VEGF-C-associated proteins identified in our study provide us with further insights into the mechanism of VEGF-C promoting malignant progression of bladder cancer cells.
Keywords: vascular endothelial growth factor-C(VEGF-C), bladder cancer, RNAi, proteomics
Background
Bladder cancer is the second-most common genitourinary malignant disease [1], and it has the highest lifetime treatment costs per patient [2]. Bladder cancer is most likely to metastasize through lymphoducts, and once the lymph nodes are involved, the therapy and prognosis are poor [3]. Thus, the metastatic mechanism of bladder cancer is gradually becoming a research focus. VECF-C is widely reported to be associated with lymph node metastasis and a poor prognosis in bladder cancer [4]. It is well established that VEGF-C specifically binds to its receptor, VEGF receptor 3 (VEGFR-3), promoting lymphangiogenesis and increasing the metastatic spread of tumor cells to lymph nodes in bladder cancer [5]. However, it is also indicated that VEGF-C may affect cancer development and metastasis through a direct autocrine mechanism, which has been demonstrated by some recent studies [6–8]. Su et al. [9] demonstrated that VEGF-C/VEGFR-3-mediated invasion and metastasis of human lung adenocarcinoma cells require upregulation of the neural cell adhesion molecule contactin-1 through activation of the Src-p38 MAPK-C/EBP-dependent pathway. He et al. [10] also found that VEGF-C accelerated cervical cancer metastasis by directly driving cancer cell migration and invasion, which were closely related to the effects of VEGF-C on moesin expression and activation through the RhoA/ROCK-2 signaling pathway. In addition, Issa et al. [11] introduced a new mechanism in which VEGF-C secretion and CCR7 expression by tumor cells are positively coupled to synergistically direct and enhance tumor cell invasion toward lymphatic vessels. These findings indicate that the functional mechanisms regarding VEGF-C on tumor cells are complex, and multiple targeting proteins regulated by VEGF-C may be involved. However, these factors are still largely unknown.
Our previous research has confirmed that the expression of VEGF-C in bladder cancer was markedly higher than that in normal bladder tissues [4]. Overexpression of VEGF-C could significantly enhance the proliferation and invasiveness of bladder cancer T24 cells [12]. To identify novel associated proteins of the VEGF-C signaling in bladder cancer, RNAi coupled with 2D-DIGE were introduced in our study. The proteomic approach is a powerful technique that has been widely used in profiling comprehensive protein expressions of diseases [13–15], so it has been commonly used in the identification of potential disease targets or signaling pathways [16,17]. In the present study, we first used lentivirus vector-based RNA interference to knockdown VEGF-C expression of bladder cancer T24 cells. Then, DIGE-based proteomic analysis was employed to reveal the comprehensive protein profiles of T24 cells, and a comparative study was made between normal and VEGF-C-knockdown T24 cells. Potential associated proteins of VEGF-C were screened out and identified by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS). Western blotting was used to validate partial proteins with significant difference in the 2 cell groups. The established functions are also discussed.
Material and Methods
Cell culture
The human bladder cancer cell line T24 was purchased from the Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences. The cell culture medium is RPMI-1640 including 10% calf serum. The normal culture was conducted in a warm box at 37°C with 5% carbon dioxide.
Extraction of total cell proteins
As shown before [12], the lentivirus vector-VEGF-C-siRNA or control vector was added to the cells, so as to establish the siRNA group and the negative control (NC) group. T24 cells without transfection were set up as the control (CON) group. Then those cells in the siRNA group and the CON group were harvested and added to lysis buffer (7 M urea, 2 M thiourea, 4% (W/V) CHAPS, 30 mM Tris). Following lysis of cells, the supernatants were collected by centrifugation at 12000 r/min for 20 min at 4°C. Protein concentrations were determined using Bradford reagent (Sigma) according to the manufacturer’s instructions.
CyDyes labelling
The pH of the protein extract was adjusted to 8.5 using the appropriate volume of NaOH, and protein quantification was performed using the 2D quantity kit (GE), with a Bradford protein assay. Fifty μg of each protein extract were labeled with 400 pmol cyanine dyes on ice for 30 min in the dark. Labeling was terminated by adding 1 μL of 10 mM lysine and incubating for 10 min on ice. Cy3 and Cy5 were used randomly to label samples from the siRNA group or CON group, while a mix sample composed of equal amounts of proteins from each group was labelled with Cy2 as the internal standard.
Two-dimensional electrophoresis
Prior to the first-dimension separation of proteins, three labeled protein samples were combined and added to 450 μL of a rehydration solution (7 M urea, 2 M thiourea, 2% CHAPS, 0.5% IPG 4–7 buffer, 2% dithiothreitol, 0.002% Bromophenol blue), as to rehydrate IPG strips(24 cm, pH 4–7; GE Healthcare) overnight. Then, isoelectric focusing was performed on an Ettan™ IPGphor III isoelectric focusing unit (GE Healthcare). The setting was as follows: 500 V 1 h, 1000 V 1 h, 8000 V 8.5 h. IPG strips were then equilibrated in equilibration buffer (50 mM Tris-HCl, 6 M urea, 30% glycerol, 2% SDS) supplemented with 1% DTT and then 3% iodoacetamide for 15 min, respectively. The strips were overlaid with 1% agarose in 12% SDS-PAGE for the second dimension. It stopped when the blue dye front had run off the bottom of the gels. After the end of electrophoresis, Coomassie Brilliant Blue staining of the gels was carried out [18].
Image analysis and statistics
Images were acquired in a Typhoon 9400 Imager (GE Healthcare). Spot detection, image matching and quantification were performed with DeCyder 6.5 (GE Healthcare). The SPSS 13.0 software package was used to perform statistical analysis. Protein spots that showed a significant change in abundance between the 2 groups were selected for further characterization by mass spectrometry.
MALDI-TOF-MS
Spots of interest were excised from the gel automatically using an Ettan-Picker robot (GE Healthcare) and subjected to tryptic digestion as follows [13]. Firstly, the gels were destained with 50 mM ammonium bicarbonate/50% Acetonitrile (ACN). Then, the gels were dried completely by vacuum-drying. The protein in the gel was digested by treatment with 10 μl tosyl-L-phenylalanine-chloromethyl ketone (TPCK)-treated trypsin (Promega) in 50 mM ammonium bicarbonate at 37°C for 16 h with gentle agitation. After digestion, peptides were extracted with 30 μl extract liquor composed of 50% acetonitrile (1:1) with 0.1% trifluoro-acetic acid (TFA) for 60 min ×2, concentrated and extensively treated with ZipTip (Millipore). Supernatants were collected, and then mixed an equal volume of matrix solution (α-cyano-4-hydroxycinnamic acid, 50% ACN 2:1 with 0.1% TFA). The mixtures were spotted onto a MALDI target plate and subjected to mass spectrometric analysis. Mass spectrometric analysis of tryptic peptides was performed using a Voyager DE-STR Biospectrometry™ Workstation System 4307 MALDI-TOF Mass Spectrometer (ABI) to get a peptide mass fingerprint (PMF). For database search, the PMF was searched with mascot software and the UniProt database. The following search parameters were used in all MASCOT searches: Carbamidomethyl (C) selected for fixed modifications, Oxidation (M) selected for variable modifications, ±50 ppm selected for peptide mass tolerance, and ±0.5 Da selected for fragment mass tolerance.
Western blotting analysis
The cells of the CON group and siRNA group were collected, then added with cell lysis solution (20 mM Tris – HCl pH 7.5,150 mM NaCl, 1%Triton X-100, 30 mM sodium pyrophosphate, 5 μg/ml leupeptin, 10 mM EDTA, 1 mM Na3VO4 and 1 mM PMSF) for lysis on ice for 30 min. Total protein concentration in each lysate was quantified by the Bradford protein assay. Appropriate amounts of protein (20–30 μg) were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were transferred to a polyvinylidene difluoride membrane. The membranes were blocked for 1 h in blocking solution consisting of 5% nonfat milk and Tris-buffered saline containing 0.1%Tween-20 (TBST). Then a dilute solution of primary antibody was incubated overnight with the membrane at 4°C, followed by the corresponding HRP-conjugated secondary antibody. Immunoreactive bands were detected using an ECL-plus kit (Amersham Biosciences, UK). The primary antibodies to Keratin 8, Serpin B5, Annexin A8, matrix metalloproteinase-9 (MMP-9) and secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
T24 cells treated with and without recombinant human VEGF-C (rhVEGF-C)
T24 cells were harvested at their exponential phase of growth, diluted with culture medium to a suspension of 1.25×105/ml, and then seeded into a 96-well culture dish at a concentration of 2×104 T24 cells. The experiment was divided into 4 groups: a treated group with rhVEGF-C (R&D Systems) concentration at 200 ng/ml, the CON, the NC, and the siRNA group without rhVEGF-C. The final volume was 200 ul solution per well and the cells were cultured for 48 h. They were prepared following invasion assay.
Invasion assay
The cell invasive assay was done in Boyden chambers (B-D Company). A polycarbonate filter with pores of 8 μm, which was coated with Matrigel, separates the top of the cell invasion chamber from the bottom. Approximately 1×104 cells were suspended in 100 μL serum-free supplement and were seeded onto the top chamber. Then 1mL of culture medium with 10% FBS was added to the lower chamber. Following incubation for 48 h at 37°C, those cells still on the upper side of the filter were removed with cotton swabs, and the migrating cells on the lower side of the filter were fixed in ethanol and stained with hematoxylin. The number of migrating cells was quantified by manually counting the cells in 5 random fields per well under a microscope at 200× magnification (Olympus). Each assay was repeated 3 times.
Statistical analysis
Statistical analysis was done by using SPSS (version 13.0). The statistical differences between groups were compared using a Student’s t test. Before using the t test, the data were normalized. The t test was applied when normality and homogeneity of variance assumptions were satisfied. The results were considered significant if the P value was less than 0.05.
Results
The 2D-DIGE image analysis
Pooled protein extracts from T24 cells in the siRNA group and the CON group were subjected to DIGE analysis. The images were analyzed using DeCyder software. The 2D-DIGE gel image is seen in Figure 1. The protein profile patterns of the siRNA group and the CON group and the internal standard are displayed by different fluorescence (green, red, blue). All the abundance levels for protein spots in the gels were analyzed to determine the difference. Four biological replicates were used to determine the differential expression between the siRNA group and the CON group. Figure 2 represented the result of No.1516 protein spot. In the end, we found 25 differentially expressed proteins for mass spectrometry.
Figure 1.
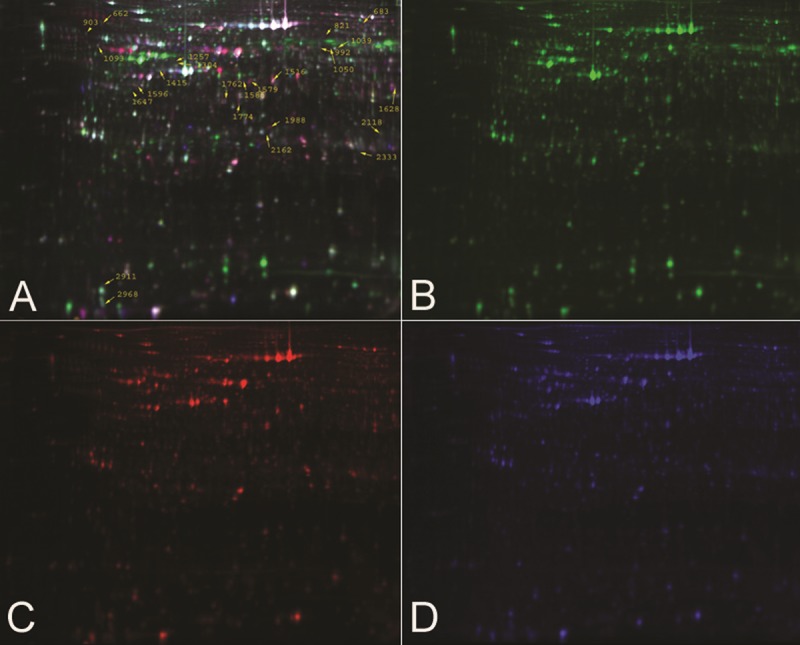
Representative DIGE fluorescence images of T24 cells with and without siRNA. There are twenty-five differential expressed proterins spots we found, as labeled in the image. (A) Overlays of the Cydye-labeled images; (B) Cydye-labeled image of sample of CON group; (C) Cydye-labeled image of sample of siRNA group; (D) Cydye-labeled image of internal standard.
Figure 2.
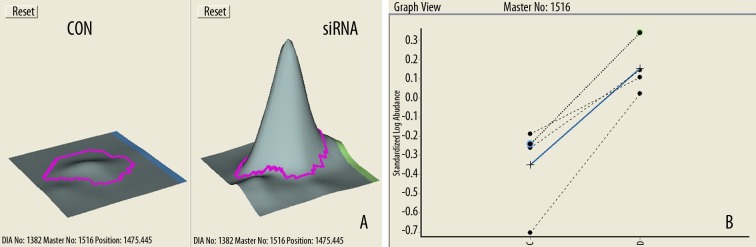
The 2D-DIGE image analysis of spot 1516. (A) The abundance levels for protein spots 1516. (B) The graph view of comparison between CON and siRNA group.
MALDI-TOF-MS
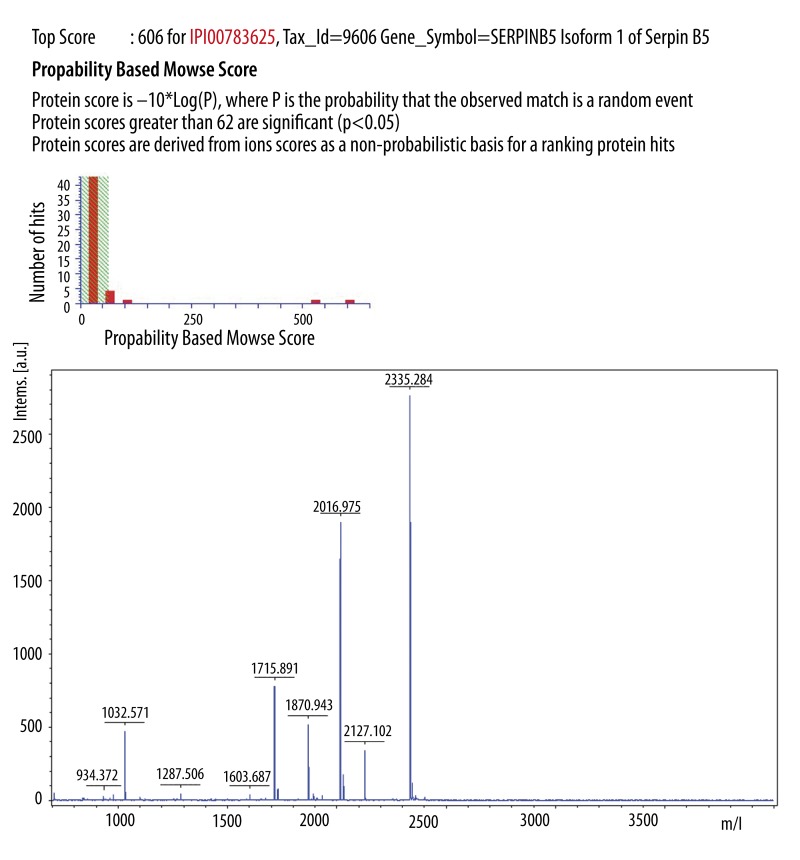
Of the 25 proteins, 23 were finally identified. These proteins were grouped into different functional classes: cytoskeletal proteins, signal transduction proteins, molecular chaperones, cell proliferation and metastasis-related proteins, biosynthesis proteins, energy metabolism proteins, enzymes, and proteins of unknown function. Among them, 10 proteins were overexpressed in the siRNA group compared with the CON group, and 13 proteins were downregulated (Table 1). Four protein spots expressing MMP-9, keratin 8, Serpin B5, and Annexin A8 had the most striking differences. Several spots were identified as containing the same protein. For example, spots 1516, 1579, and 1647 were each identified as containing the Serpin B5 protein (Figure 3). The same protein in several spots may represent different protein modifications that may be of biological relevance [19].
Table 1.
The differentially expressed proteins between siRNA and CON group identified by MS.
| Spot No. | Protein Accession* | Protein | MW(Da) | PI | Sequence coverage (%) | Score | Ratio** | Functions |
|---|---|---|---|---|---|---|---|---|
| 662 | Q96JB5 | Isoform 2 of CDK5 regulatory subunit-associated protein 3 | 46664 | 4.56 | 5 | 71 | −1.62 | Involved in cell proliferation |
| 683 | P02545 | Isoform ADelta 10 of Lamin-A/C | 70903 | 8.55 | 32 | 188 | −1.69 | Structural molecule activity |
| 821 | A6NBZ8 | Putative uncharacterized protein ALB | 73881 | 6.33 | 26 | 139 | −2.3 | Unknown |
| 903 | 075312 | ZPR1 | 51463 | 4.66 | 24 | 394 | −1.4 | Involved in cell proliferation, signal transduction |
| 992 | P14868 | Aspartyl-tRNA synthetase, cytoplasmic | 57499 | 6.11 | 39 | 242 | 1.5 | Aminoacylase activity, aspartate-tRNA ligase activity |
| 1257 | P14780 | matrix metalloproteinase-9 | 78429 | 5.69 | 43 | 139 | −2.1 | Breakdown of extracellular matrix |
| 1304 | P62736 | Actin, aortic smooth muscle | 42381 | 5.23 | 31 | 255 | −1.56 | Regulation of cell motility |
| 1415 | P05787 | Keratin 8 | 26765 | 4.66 | 56 | 264 | −2.2 | Maintaining cellular structural integrity and also functions in signal transduction and cellular differentiation |
| 1516 | P36952 | Serpin B5 | 42530 | 5.72 | 45 | 606 | 3 | As tumor suppressor, blocks the growth, invasion, and metastatic properties of tumors |
| 1579 | P36952 | Serpin B5 | 42530 | 5.72 | 40 | 356 | 2.2 | As tumor suppressor, blocks the growth, invasion, and metastatic properties of tumors |
| 1585 | P02675 | Fibrinogen beta chain | 56577 | 8.54 | 20 | 304 | 1.8 | Component of fibrinogen, acting as a cofactor in platelet aggregation, regulate cell adhesion and spreading |
| 1596 | P07910 | Isoform 4 of Heterogeneous nuclear ribonucleoproteins C1/C2 | 27861 | 4.55 | 34 | 361 | 1.55 | Modulating the stability and the level of translation of bound mRNA molecules, binding nucleic acid |
| 1628 | P09467 | Fructose-1,6-bisphosphatase 1 | 37190 | 6.54 | 29 | 387 | 1.73 | Gluconeogenesis regulatory enzyme |
| 1774 | B4DQE1 | Annexin A8 | 44056 | 6.19 | 46 | 35 | −3.66 | Binding site for calcium and phospholipid |
| 1988 | P78417 | Glutathione S-transferase omega-1 | 27833 | 6.23 | 24 | 261 | 1.57 | Glutathione-dependent thiol transferase and dehydroascorbate reductase activities |
| 2118 | 000233 | Isoform p27-L of 26S proteasome non-ATPase regulatory subunit 9 | 24810 | 6.46 | 46 | 441 | −1.76 | Molecular chaperone |
| 2162 | 095336 | 6-phosphogluconolactonase | 27815 | 5.7 | 68 | 649 | 1.73 | Hydrolysis of 6-phosphogluconolactone to 6-phosphogluconate |
| 1039 | P78371 | T-complex protein 1 subunit beta | 57794 | 6.01 | 25 | 392 | −1.49 | Molecular chaperone |
| 1050 | P78371 | T-complex protein 1 subunit beta | 57794 | 6.01 | 23 | 211 | −1.47 | Molecular chaperone |
| 1093 | Q09028 | Retinoblastoma binding protein 4 | 46415 | 4.9 | 11 | 186 | 1.46 | Regulating chromatin metabolism |
| 1647 | P36952 | Serpin B5 | 42530 | 5.72 | 5 | 54 | 1.44 | As tumor suppressor, blocks the growth, invasion, and metastatic properties of tumors |
| 2333 | Q9H8S9 | Mps one binder kinase activator-like 1B | 16897 | 5.35 | 8 | 99 | −1.63 | Tumor suppression by restricting proliferation and promoting apoptosis |
| 1762 | Q7L5N1 | COP9 subunit 6 | 33896 | 5.65 | 15 | 201 | −1.41 | Component of the COP9 signalosome complex involved in multiple signaling pathways |
Accession number of the UniProt database;
average ratio of siRNA group/CON group.
Figure 3.
Results of MALDI-TOF-MS analysis of the protein spot 1516 (n=4). The protein was identified as Serpin B5.
Validation of the difference proteins by Western blotting analysis
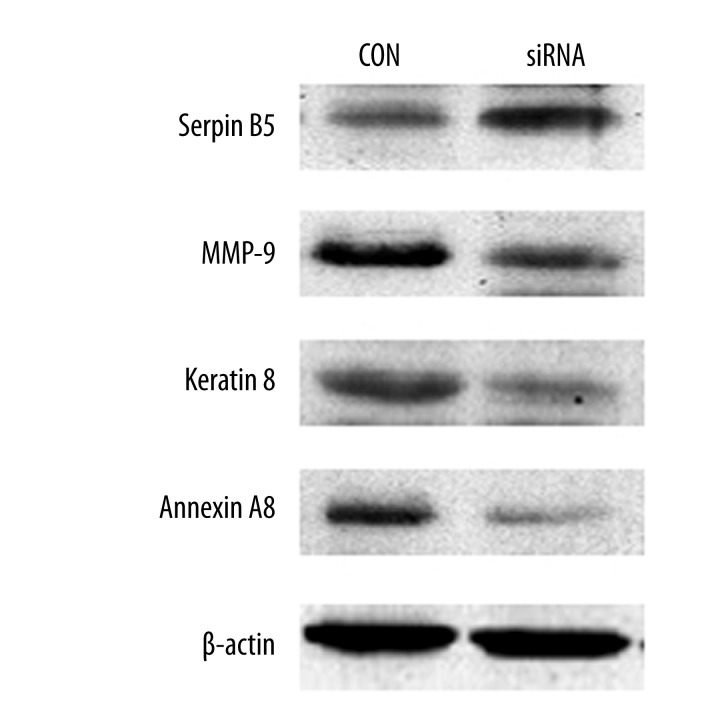
Western blotting analysis was used to determine the expression levels of MMP-9, Keratin 8, Serpin B5 and Annexin A8 in the 2 groups. The results showed that the expression of Serpin B5 in the siRNA group was significantly higher than that of the CON group (Figure 4). But the expression of MMP-9, Keratin 8, and Annexin A8 of the siRNA group showed a remarkable decrease as compared with the CON group. The result was consistent with the results of proteomics research.
Figure 4.
Validation of the different proteins by Western blotting analysis. Compared with CON group, Serpin B5 was significantly up-regulated in CON group, while the expression of MMP-9, Keratin 8 and Annexin A8 showed a remarkable decrease (P<0.05 evaluated by Student’s t test). A representative blot is shown from three independent experiments with identical results. β-actin is as an internal standard.
MMP-9 involved in VEGF-C-mediated cell invasion in T24 cells
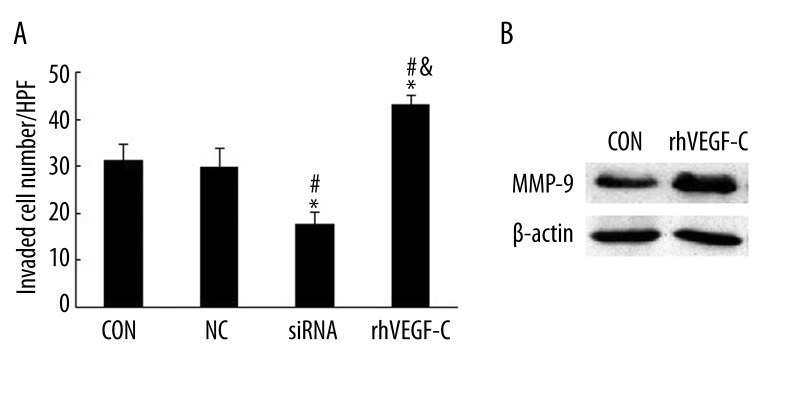
T24 cells treated with VEGF-C-siRNA reduced the invasiveness compared with the CON group and the NC group in the Boyden chamber assay (Figure 5A). There was significant difference between CON and siRNA groups, as well as the NC and the siRNA group (P<0.05).
Figure 5.
The effects on the of T24 cells in the presence or absence of rhVEGF-C. (A) rhVEGF-C reversed the inhibition effect of siRNA on cell invasion. The cell invasive assay was done in Boyden chambers. The total number of cells migrating to the lower chamber per field (magnification ×200) were counted. The results are presented as mean ±SD (n=3). * p<0.05, vs. CON group; # p<0.05, vs. NC group, & p<0.05, vs. siRNA group evaluated by Student’s t test. (B) Expression of MMP-9 detected by Western blotting was greatly enhanced after rhVEGF-C treatment (P<0.05).
MMP-9 was selected for exploring its regulation by VEGF-C. After adding the rhVEGF-C to T24 cells, MMP-9 was found to be significantly up-regulated (Figure 5B). Meanwhile, cell invasion was significantly enhanced (Figure 5A). This further suggests the potential regulatory effect of VEGF-C on MMP-9 expression in invasion of T24 cells.
Discussion
VEGFR-3 (also known as Flt4) [20], a cell surface receptor tyrosine kinase, expresses on the endothelium of lymphatic vessels [21], as well as in some tumor cells [22,23]. VEGF-C binds to and induces autophosphorylation of VEGFR-3, then initiates the downstream cascades. The VEGF-C/VEGFR-3 axis can induce a PKC-dependent activation of the p42/p44 MAPK (ERK1/ERK2) signalling cascade and the PI3-kinase/Akt pathway. These signal transducers are of importance in growth, migration and survival of the lymphatic endothelial cells [24]. Activation of the VEGF-C/VEGFR-3 axis in lymphatic endothelial cells can facilitate metastasis by increasing the formation of lymphatic vessels within and around tumours. In contrast, the pathophysiological role of tumor cell-expressed VEGFR-3 in tumor progression remains to be elucidated. Recently, it was demonstrated that the VEGF-C/VEGFR-3 autocrine loop in tumor cells was a potential enhancer system to promote cancer progression [7], which suggests the VEGF-C/VEGFR-3 axis can directly enhance cancer cell proliferation and invasiveness and contributes to the promotion of cancer cell metastasis. However, because its downstream signaling remains largely unclear, the mechanism involved is still incompletely understood.
Our early research has confirmed that the expression of VEGF-C in bladder cancer was markedly higher than that in normal bladder tissues [4]. Overexpression of VEGF-C could significantly enhance the proliferation and invasiveness ability of bladder cancer T24 cells [12]; therefore, we postulate that some proteins associated with VEGF-C must contribute to the process, which can be screened out of cancer cells with high and low VEGF-C expression.
In the present study, signaling alterations after VEGF-C inhibition in the T24 cells was carried out via a proteomic approach, with further confirmation by Western blotting. We first used siRNA technique to specifically knock down VEGF-C expression in bladder cancer T24 cells. As shown before, the transient transfection of siRNA resulted in the efficient and specific downregulation of VEGF-C mRNA and protein. Thus, we considered that the T24 cells in the CON and siRNA groups are of high and low VEGF-C expression.
Proteomics has the potential to support RNAi experiments by providing a global and subproteomics search platform to robustly resolve and provide relative quantification of proteins within a complex mixture [25]. DIGE-based proteomics has the advantages of adequate sensitivity, high reproducibility, wide linear dynamic range, and minimized experimental variation over conventional proteomics because of internal standards and fluorescence labeling [26]. In the present study, we took advantage of DIGE-based proteomics to explore differences in protein expression levels between the CON group and the siRNA group. Our proteomic study identified a set of proteins that might function associated with VEGF-C in progression of bladder cancer T24 cells. Of the 25 differential protein spots we found, 23 spots were finally identified. Ten proteins were up-regulated and 13 proteins were down-regulated in the siRNA group, compared with the CON group. Among them, 5 proteins were identified with more than 2-fold change in intensity between 2 groups. They are MMP-9, keratin 8, Serpin B5, Annexin A8, and an uncharacterized protein. To some extent, this result is consistent with the data reported by another proteomic analysis of muscle-invasive bladder cancer [27].
In this study, the inhibition of VEGF-C gene in the T24 cells revealed a decrease in the protein level of MMP-9. Matrix metalloproteinases (MMPs) are a family of 23 zinc-dependent endopeptidases. They are involved in tumour development and metastatic spread by degrading basement membrane and different components of the extracellular matrix, including collagen, fibronectin and proteoglycans [28]. Among them, MMP-9 is a critical one involved in tumour invasion. The role of MMP-9 is very complex. It affects not only invasion and metastasis, but also signal pathways involved both in normal physiology and in disease, growth signalling, angiogenesis and lymphangiogenesis [28]. High levels of MMP-9 measured in urine from bladder cancer patients have been linked to poor prognosis [29]. This may reflect that MMP-9 plays a role in bladder cancer invasion and metastasis. Zheng et al. [30] found MMP-9 in conjunction with VEGF-C, promote lymphangiogenesis and lymph node metastasis of breast cancer. Our findings indicated that knockdown of VEGF-C by RNAi significantly suppressed the invasion of T24 cells with MMP-9 down-regulation. When rhVEGF-C was presented, the suppression of invasion and MMP-9 expression in T24 cells was reversed. Taken together, we suggest MMP-9 is a downstream protein of VEGF-C, leading to cell invasion. Recently, Kumar et al. [31] implicated p38 MAPK and MAPKAPK2 in mediating bladder cancer invasion via regulation of MMP-9 at the level of mRNA stability. Thus, we also postulate a potential regulation effect of VEGF-C on MMP-9 expression by the MARK pathway in invasion of T24 cells, which deserves further study.
Keratin 8 (also called CK8 or K8), is a protein member of the type II keratins. It typically dimerizes with keratin 18 to form an intermediate filament (IF) in simple epithelium [32]. This protein plays a role in maintaining cellular structural integrity and also functions in signal transduction and cellular differentiation. It’s noted that keratins are involved in dynamic cell remodeling during cancer progression and, particularly, CK expression patterns have been associated with invasion and metastasis [33]. CK8 presents on the plasma membrane of the tumor cells. The ectoplasmic domain of CK8 can bind to urokinase-type plasminogen activator, activating plasminogen to plasmin. Consequently, plasmin activates growth factors and protease cascades that lead to the disruption of cell-cell and cell-extracellular matrix adhesion via pericellular proteolysis of glycoproteins [34]. Actually, the urinary concentrations of Ck8/CK18 in invasive bladder cancer were significantly higher than those in the superficial bladder cancer [35]. A recent study found that over-expression of Akt1 increased K8/18 proteins and Akt2 up-regulated K18 expression by increased mRNA stability [36]. These results represented the first indication that Akt isoforms regulate IF expression, and support the hypothesis that IFs are involved in PI3K/Akt pathway. Thus, we postulate that keratin 8 may be a downstream target in the VEGF-C/VEGFR-3 axis and play a role in the progress of bladder cancer.
Serpin B5 (also known as Maspin) is a member of the serpin super-family, first identified in a screen for potential tumor suppressors lost in human breast [37]. Although its molecular functions are not well known, maspin is considered to inhibit primary tumor growth, as well as invasion and metastasis [38]. Odero-Marah et al [39] showed that the inhibitory effect of maspin on cell motility correlated with a decrease in Rac 1 effector PAK1, and an increase of PI3K and ERK1/2 activities. Maspin expression was found in high quantities in normal urothelium and was preserved in superficial bladder cancers, but was significantly diminished in invasive carcinomas [40]. These data are in agreement with our result that Serpin B5 was significantly up-regulated in the siRNA group. However, conflicting information exists that maspin and VEGFC are expressed in ovarian tumors with poor prognostic parameters, and seem to play a role in ovarian cancer progression and lymph node metastases. [41]. Thus, maspin might have important roles in the development of different tumor entities, including bladder cancer, and might act through crosstalk with VEGF-C.
Annexins are a super-family of closely related calcium and membrane-binding proteins. More than 50 different annexin isoforms have been identified, which have a variety of roles in tumour development and progression, including involvement in cell signaling pathways, cell motility, tumour invasion and metastasis, angiogenesis, apoptosis, and drug resistance [42]. Annexin A8 has been associated with breast cancers and a recent tissue microarray study has shown a link between annexin A8 expression and a basal-cell like subtype of breast cancers with poor prognosis [43]. Our results revealed that Annexin A8 was down-regulated in the siRNA group as compared with the CON group. Although the mechanisms of Annexins’ involvement in bladder cancer are not yet understood, our finding will attract more interest and efforts to further investigate the roles of annexins in the prognosis of bladder cancer.
Conclusions
In summary, using RNAi coupled with 2D-DIGE, we identified some differentially expressed proteins from bladder cancer T24 cells with high and low VEGF-C expression. The observation was validated by Western blotting analysis. To our knowledge, the present study is the first comprehensive proteome profiling to identify VEGF-C-associated proteins in bladder cancer T24 cells. Our findings are an initial step toward uncovering the molecular mechanism by which VEGF-C promotes malignant development of cancer cells, and provide some markers for predicting metastasis. However, this is only a preliminary study, and further studies on animal models and tissue samples are still needed to elaborate the real roles of these differentially expressed proteins.
Footnotes
Conflict of interest
No potential conflicts of interest were disclosed.
Source of support: This research was supported by the National Natural Science Foundation of China (No.30700832)
Reference
- 1.Gurtowska N, Kloskowski T, Drewa T. Ciprofloxacin criteria in antimicrobial prophylaxis and bladder cancer recurrence. Med Sci Monit. 2010;16(10):RA218–23. [PubMed] [Google Scholar]
- 2.Xu R, Zhang L, Zhao X, et al. Benzalkonium bromide as a new potential instillation drug for bladder cancer: hypothesis and pilot study. Med Sci Monit. 2011;17(12):HY36–39. doi: 10.12659/MSM.882110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vieweg J, Gschwend JE, Herr HW, et al. Pelvic lymph node dissection can be curative in patients with node positive bladder cancer. J Urol. 1999;161:449–54. [PubMed] [Google Scholar]
- 4.Zu XB, Tang ZY, Li Y, et al. Vascular endothelial growth factor-C expression in bladder transitional cell cancer and its relationship to lymph node metastasis. BJU International. 2006;98:1090–93. doi: 10.1111/j.1464-410X.2006.06446.x. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki K, Morita T, Tokue A. Vascular endothelial growth factor-C (VEGF-C) expression predicts lymph node metastasis of transitional cell carcinoma of the bladder. Int J Urol. 2005;12:152–58. doi: 10.1111/j.1442-2042.2005.01010.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Jiang L, She F, et al. Vascular endothelial growth factor-C promotes the growth and invasion of gallbladder cancer via an autocrine mechanism. Mol Cell Biochem. 2010;345:77–89. doi: 10.1007/s11010-010-0562-y. [DOI] [PubMed] [Google Scholar]
- 7.Matsuura M, Onimaru M, Yonemitsu Y, et al. Autocrine Loop between Vascular Endothelial Growth Factor (VEGF)-C and VEGF Receptor-3 Positively Regulates Tumor-Associated Lymphangiogenesis in Oral Squamoid Cancer Cells. Am J Pathol. 2009;175:1709–21. doi: 10.2353/ajpath.2009.081139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kodama M, Kitadai Y, Tanaka M, et al. Vascular Endothelial Growth Factor C Stimulates Progression of Human Gastric Cancer via Both Autocrine and Paracrine Mechanisms. Cl Cancer Res. 2008;14:7205–14. doi: 10.1158/1078-0432.CCR-08-0818. [DOI] [PubMed] [Google Scholar]
- 9.Su JL, Yang PC, Shih JY, et al. The VEGF-C/Flt-4 axis promotes invasion and metastasis of cancer cells. Cancer Cell. 2006;9:209–23. doi: 10.1016/j.ccr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 10.He M, Cheng Y, Li W, et al. Vascular endothelial growth factor C promotes cervical cancer metastasis via up-regulation and activation of RhoA/ROCK-2/moesin cascade. BMC Cancer. 2010;10:170. doi: 10.1186/1471-2407-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issa A, Le TX, Shoushtari AN, et al. Vascular Endothelial Growth Factor-C and C-C Chemokine Receptor 7 in Tumor Cell-Lymphatic Cross-talk Promote Invasive Phenotype. Cancer Res. 2009;69:349–57. doi: 10.1158/0008-5472.CAN-08-1875. [DOI] [PubMed] [Google Scholar]
- 12.Zhang HH, Qi F, Shi YR, et al. RNA Interference-Mediated Vascular Endothelial Growth Factor-C Reduction Suppresses Malignant Progression and Enhances Mitomycin C Sensitivity of Bladder Cancer T24 Cells. Cancer Biother Radiopharm. 2012;27(5):291–98. doi: 10.1089/cbr.2010.0919. [DOI] [PubMed] [Google Scholar]
- 13.Yao HX, Zhang ZQ, Xiao ZQ, et al. Identification of metastasis associated proteins in human lung squamous carcinoma using two-dimensional difference gel electrophoresis and laser capture microdissection. Lung Cancer. 2009;65:41–48. doi: 10.1016/j.lungcan.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 14.Cui YZ, Wu JM, Zong MJ, et al. Proteomic profiling in pancreatic cancer with and without lymph node metastasis. Int J Cancer. 2009;124:1614–21. doi: 10.1002/ijc.24163. [DOI] [PubMed] [Google Scholar]
- 15.Qiu F, Gao YH, Jiang CG, et al. Serum proteomic profile analysis for endometrial carcinoma detection with MALDI-TOF MS. Arch Med Sci. 2010;6(2):245–52. doi: 10.5114/aoms.2010.13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali NA, McKay MJ, Molloy MP. Proteomics of Smad4 regulated transforming growth factor-beta signalling in colon cancer cells. Molecular Biosystems. 2010;6:2332–38. doi: 10.1039/c0mb00016g. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Pineiro AM, Blanco-Prieto S, Sanchez-Otero N, et al. On the identification of biomarkers for non-small cell lung cancer in serum and pleural effusion. J Proteomics. 2010;73:1511–22. doi: 10.1016/j.jprot.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Candiano G, Bruschi M, Musante L, et al. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–33. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 19.Pang J, Liu WP, Liu XP, et al. Profiling Protein Markers Associated with Lymph Node Metastasis in Prostate Cancer by DIGE-based Proteomics Analysis. J Proteome Res. 2010;9:216–26. doi: 10.1021/pr900953s. [DOI] [PubMed] [Google Scholar]
- 20.Joukov V, Pajusola K, Kaipainen A, et al. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. Embo J. 1996;15:290–98. [PMC free article] [PubMed] [Google Scholar]
- 21.Kaipainen A, Korhonen J, Mustonen T, et al. Expression of the fms-like tyrosine kinase 4 gene becomes restricted to lymphatic endothelium during development. Proc Natl Acad Sci USA. 1995;92:3566–70. doi: 10.1073/pnas.92.8.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuchrist C, Erovic BM, Handisurya A, et al. Vascular endothelial growth factor C and vascular endothelial growth factor receptor 3 expression in squamous cell carcinomas of the head and neck. Head Neck. 2003;25:464–74. doi: 10.1002/hed.10235. [DOI] [PubMed] [Google Scholar]
- 23.Witte D, Thomas A, Ali N, et al. Expression of the vascular endothelial growth factor receptor-3 (VEGFR-3) and its ligand VEGF-C in human colorectal adenocarcinoma. Anticancer Research. 2002;22:1463–66. [PubMed] [Google Scholar]
- 24.Bahram F, Claesson-Welsh L. VEGF-mediated signal transduction in lymphatic endothelial cells. Pathophysiology. 2009;17:253–61. doi: 10.1016/j.pathophys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 25.LaCourse EJ, Perally S, Hernandez-Viadel M, et al. A proteomics approach to quantify protein levels following RNA interference: Case study with glutathione transferase superfamily from the model metazoan Caenorhabditis elegans. J Proteome Res. 2008;7:3314–18. doi: 10.1021/pr8001035. [DOI] [PubMed] [Google Scholar]
- 26.Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665–85. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 27.Barboro P, Rubagotti A, Orecchia P, et al. Differential proteomic analysis of nuclear matrix in muscle-invasive bladder cancer: Potential to improve diagnosis and prognosis. Cellular Oncology. 2008;30:13–26. doi: 10.1155/2008/686940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Offersen BV, Knap MM, Horsman MR, et al. Matrix metalloproteinase-9 measured in urine from bladder cancer patients is an independent prognostic marker of poor survival. Acta Oncologica. 2010;49:1283–87. doi: 10.3109/0284186X.2010.509109. [DOI] [PubMed] [Google Scholar]
- 30.Zheng SQ, Huang RQ, Zhang YJ. Role of matrix metalloproteinase (MMP)-2 and -9 and vascular endothelial growth factor C in lymph node metastasis of breast cancer. Zhonghua Bing Li Xue Za Zhi. 2010;39:240–44. [PubMed] [Google Scholar]
- 31.Kumar B, Koul S, Petersen J, et al. p38 Mitogen-Activated Protein Kinase-Driven MAPKAPK2 Regulates Invasion of Bladder Cancer by Modulation of MMP-2 and MMP-9 Activity. Cancer Res. 2010;70:832–41. doi: 10.1158/0008-5472.CAN-09-2918. [DOI] [PubMed] [Google Scholar]
- 32.He T, Stepulak A, Holmstrom TH, et al. The intermediate filament protein keratin 8 is a novel cytoplasmic substrate for c-Jun N-terminal kinase. J Biol Chem. 2002;277:10767–74. doi: 10.1074/jbc.M111436200. [DOI] [PubMed] [Google Scholar]
- 33.Cintorino M, Tripod SA, Santopietro R, et al. Cytokeratin expression patterns as an indicator of tumour progression in oesophageal squamous cell carcinoma. Anticancer Res. 2001;21:4195–201. [PubMed] [Google Scholar]
- 34.Obermajer N, Doljak B, Kos J. Cytokeratin 8 ectoplasmic domain binds urokinase-type plasminogen activator to breast tumor cells and modulates their adhesion, growth and invasiveness. Mol Cancer. 2009;8:88. doi: 10.1186/1476-4598-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hakenberg OW, Fuessel S, Richter K, et al. Qualitative and quantitative assessment of urinary cytokeratin 8 and 18 fragments compared with voided urine cytology in diagnosis of bladder carcinoma. Urology. 2004;64:1121–26. doi: 10.1016/j.urology.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 36.Fortier AM, Van Themsche C, Asselin E, et al. Akt isoforms regulate intermediate filament protein levels in epithelial carcinoma cells. Febs Letters. 2010;584:984–88. doi: 10.1016/j.febslet.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 37.Zou Z, Anisowicz A, Hendrix MJ, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–29. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 38.Sheng SJ. The promise and challenge toward the clinical application of maspin in cancer. Front Biosci. 2004;9:2733–45. doi: 10.2741/1432. [DOI] [PubMed] [Google Scholar]
- 39.Odero-Marah VA, Khalkhali-Ellis Z, Chunthapong J, et al. Maspin regulates different signaling pathways for motility and adhesion in aggressive breast cancer cells. Cancer Biol Ther. 2003;2:398–403. doi: 10.4161/cbt.2.4.471. [DOI] [PubMed] [Google Scholar]
- 40.Beecken WD, Engl T, Engels K, et al. Clinical relevance of maspin expression in bladder cancer. World J Urol. 2006;24:338–44. doi: 10.1007/s00345-006-0085-z. [DOI] [PubMed] [Google Scholar]
- 41.Bolat F, Gumurdulu D, Erkanli S, et al. Maspin overexpression correlates with increased expression of vascular endothelial growth factors A, C, and D in human ovarian carcinoma. Pathol Res Pract. 2008;204:379–87. doi: 10.1016/j.prp.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Mussunoor S, Murray GI. The role of annexins in tumour development and progression. J Pathol. 2008;216:131–40. doi: 10.1002/path.2400. [DOI] [PubMed] [Google Scholar]
- 43.Stein T, Price KN, Morris JS, et al. Annexin A8 is up-regulated during mouse mammary gland involution and predicts poor survival in breast cancer. Clin Cancer Res. 2005;11:6872–79. doi: 10.1158/1078-0432.CCR-05-0547. [DOI] [PubMed] [Google Scholar]