Abstract
Fungal pathogens cause disease in plant and animal hosts. The extent to which infection mechanisms are conserved between both classes of hosts is unknown. We present a dual plant-animal infection system based on a single strain of Fusarium oxysporum, the causal agent of vascular wilt disease in plants and an emerging opportunistic human pathogen. Injection of microconidia of a well-characterized tomato pathogenic isolate (isolate 4287) into the lateral tail vein of immunodepressed mice resulted in disseminated infection of multiple organs and death of the animals. Knockout mutants in genes encoding a mitogen-activated protein kinase, a pH response transcription factor, or a class V chitin synthase previously shown to be implicated in virulence on tomato plants were tested in the mouse model. The results indicate that some of these virulence factors play functionally distinct roles during the infection of tomato plants and mice. Thus, a single F. oxysporum strain can be used to study fungal virulence mechanisms in plant and mammalian pathogenesis.
Fungi are an extremely versatile class of organisms comprised mostly of saprophytes thriving on dead organic material. A relatively small number of fungal species have developed a parasitic lifestyle, associated with the ability to recognize and penetrate a specific host, exploit its nutrient reserves, overcome its innate defense responses, and cause disease. The list of organisms attacked by fungi encompasses evolutionary distinct groups from lower to higher eukaryotes, most prominently plants, insects, and mammals, including humans. To cause disease, fungal pathogens rely on an arsenal of pathogenicity and virulence factors, whose spatially and temporally correct deployment determines the basic pathogenic potential and the extent of infection, respectively. The advent of protocols for targeted gene knockout or random insertional mutagenesis has led to the identification of an increasing number of genes encoding pathogenicity or virulence factors from plant and animal pathogens (12, 34). In spite of these advances, many crucial aspects underlying fungal pathogenesis remain poorly understood.
One of these questions concerns host specificity. Certain fungi cause disease on a single host species, whereas others have extremely broad host ranges. The molecular mechanisms that determine fungal host range specificity are not fully understood. Thus, although a number of virulence determinants are clearly host specific (36), there is also evidence for the existence of universal virulence mechanisms shared by fungal pathogens with highly diverse host ranges (17, 20). Interestingly, common patterns of host defense also are found in evolutionary diverse groups such as plants, insects and mammals (23). Thus, evolutionary ancient mechanisms of fungal virulence and host defense might coexist with highly specific virulence and resistance traits that have arisen during later stages of pathogen-host coevolution.
The availability of the complete genome sequences of fungal plant and human pathogens and their model hosts, as well as the development of high-throughput protocols for gene function analysis, hold a huge potential for advancing our understanding on fungal pathogenesis and host defense (33). However, pathogen host range imposes an important limitation on the comparative analysis of virulence mechanisms in evolutionary distant hosts because it requires gene identification and knockout in different pathogenic organisms. This constraint has been overcome in bacterial systems by developing multihost pathogenesis models, as exemplified by the Pseudomonas aeruginosa strain PA14 (29). Application of these models to phylogenetically diverse host species such as Arabidopsis spp., Caenorhabditis elegans, or mice has led to the identification of both common and contrasting virulence mechanisms in bacterial plant and animal pathogenesis (28, 32). In fungi, no such model system allowing simultaneous testing of virulence factors on plant and mammalian hosts is currently available.
The soil-borne fungus Fusarium oxysporum is the causal agent of vascular wilt, a disease that affects a large variety of economically important crops worldwide (1). Besides its well-studied activity as a plant pathogen, F. oxysporum is known as a serious emerging pathogen of humans due to the increasing number of severe cases reported and to its broad resistance to the available antifungal drugs (2, 24). Fusarium now represents the second most frequent mold causing invasive fungal infections in immunocompromised patients, frequently with lethal outcomes (22, 27, 35). F. oxysporum, together with F. solani and F. verticillioides, are responsible for practically all of the cases of invasive fusariosis in humans (14). Given the dual ability to cause disease both on plants and on humans, we reasoned that F. oxysporum could serve as a universal model for studying fungal virulence mechanisms. In the present study, we tested the ability of F. oxysporum f.sp. lycopersici strain 4287, a well-characterized model pathogen of tomato plants, to cause disease in a mammalian pathogenesis model. We found that strain 4287 is able to produce systemic infections in immunodepressed mice, resulting in a high death rate. By applying the mouse model to a number of knockout mutants previously shown to exhibit altered virulence on tomato plants, we show that specific virulence factors in a single fungal strain play distinct functional roles in plant and animal pathogenesis.
MATERIALS AND METHODS
Fungal isolates and culture conditions.
F. oxysporum f.sp. lycopersici strain 4287 (race 2) was originally obtained from J. Tello, Universidad de Almería, Almería, Spain, and stored at −80°C with glycerol as a microconidial suspension (11). The pathotype of the isolate on tomato was routinely confirmed by plant infection assays. The generation and characterization of the following mutant strains, all derived from wild-type strain 4287, were described previously: mitogen-activated protein kinase (MAPK) mutant Δfmk1 (9), pacC loss-of-function mutant pacC+/−12 and dominant activating mutant pacCc9 (5), and class V chitin synthase mutant D1 and complemented strain C2 (21).
For preparation of inocula, cultures were grown in potato dextrose broth (Difco, Detroit, Mich.) at 28°C and 150 rpm for 4 days, and microconidia were obtained by filtration as described previously (11). The microconidial suspension was centrifuged and the pellet was resuspended in sterile saline. The concentration of conidia was adjusted to 108 CFU/ml with a hemocytometer and checked by plating serial dilutions on potato dextrose agar (PDA; Pronadisa, Madrid, Spain) plates that were incubated at 28°C for 3 days. To prepare heat-killed conidia for testing their activity in the murine model, conidial suspensions were incubated in a thermostatic bath at 60°C for 60 min. Complete heat-killing of conidia was confirmed by plating serial dilutions on PDA plates.
Animal infection.
OF-1 male mice (Charles River, Criffa S.A., Barcelona, Spain) weighing ca. 30 g were used. Five mice were housed per cage in standard conditions with free access to food and water. Conditions were approved by the Animal Welfare Committee of the Faculty of Medicine of Universitat Rovira i Virgili. Mice were immunosuppressed with a single intraperitoneal 200-mg/kg dose of cyclophosphamide (Laboratorios Funk S.A., Barcelona, Spain) and with an intravenous 150-mg/kg dose of 5-fluorouracil (Fluoro-uracil; Roche S.A., Madrid, Spain) on day 0.
Groups of 10 animals were infected by injecting 0.2 ml of an inoculum of 108 conidia/ml into a lateral vein of the tail on day 0. Survival was recorded each day for 13 days. Infection experiments with each individual strain were performed between 3 and 6 times. Survival was estimated by the Kaplan-Meier method and compared among groups by using the log-rank test.
Tissue burden and histopathology.
Randomly chosen surviving mice were sacrificed 13 days after inoculation with an overdose of halotane (Fluothane; Zeneca Farma, S.A., Pontevedra, Spain). Livers, spleens, kidneys, lungs, and brains were aseptically removed, and one-half of each organ was weighed and homogenized in 1 ml of sterile saline. Tenfold serial dilutions of this homogenate were made with sterile saline and spread onto PDA. Plates were incubated at 28°C, colonies were counted after 3 days, and the numbers of CFU per gram of organ were calculated. Fungal colony counts were converted to log10 values and compared by using the analysis of variance test. Calculations were performed by using SPSS for Windows, version 10.0.
The remaining halves of the organs were fixed for 10 days in 10% neutral buffered formaldehyde, embedded in paraffin wax, and automatically processed. Sections of the embedded tissues (3 μm in thickness) were stained with hematoxylin-eosin, periodic acid-Schiff, and methenamine silver (Grocott) for light microscopy observations.
RESULTS
F. oxysporum f.sp. lycopersici causes systemic infection and death in immunodepressed mice.
F. oxysporum strains have been reported either as plant or human pathogens. We tested the hypothesis that a single strain of F. oxysporum can produce disease both on plant and mammalian hosts. As a candidate we chose strain 4287, a tomato pathogenic isolate belonging to F. oxysporum f.sp. lycopersici race 2, because the genetic interaction between this pathogen race and its host plant has been well characterized (1, 31). Isolate 4287 has been previously used in numerous molecular studies, resulting in the availability of genomic and cDNA libraries and optimized plant infection assays (10). Studies in our laboratory with targeted gene knockout mutants (5, 9, 21) have led to the identification of several genes involved in the virulence of F. oxysporum on tomato plants (Fig. 1).
FIG. 1.
Invasive growth of F. oxysporum strains on tomato fruits. Fruits were inoculated by injecting 5 × 105 microconidia of the following strains: wild-type strain 4287, MAPK mutant Δfmk1, pacC loss-of-function mutant pacC+/−, dominant-activating mutant pacCc, chitin synthase mutant ΔchsV complemented with the wild-type chsV allele, and chitin synthase mutant ΔchsV. The photograph was taken after 5 days of incubation at 100% relative humidity.
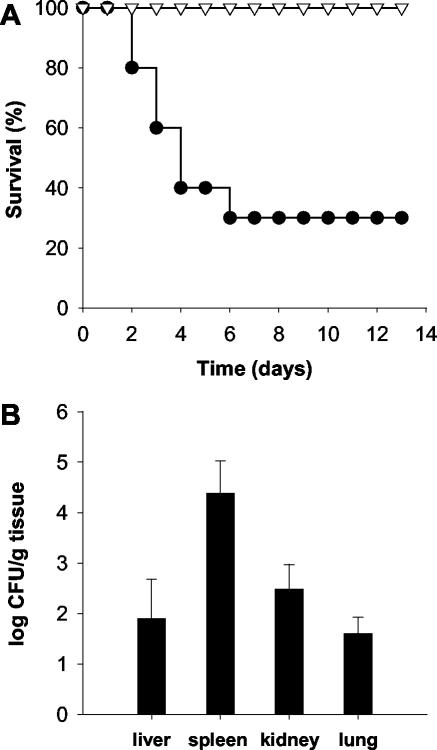
In preliminary experiments, we found that strain 4287 grew and sporulated well in submerged culture on porcine blood plasma at 37°C (data not shown). We next tested the capacity of the strain to cause infections in mice. The murine model was chosen because of its high relevance to infectious diseases in humans (3). Inoculation of immunocompetent mice with microconidia of strain 4287 did not produce any detectable symptoms (data not shown). In contrast, inoculation of immunodepressed mice with 2 × 107 microconidia reproducibly resulted in the death of ca. 70% of the animals (Fig. 2A and Table 1). Importantly, injection of heat-killed conidia caused no mortality, suggesting that death was not simply a consequence of physical presence of the microconidia but required living fungal propagules. To study the ability of F. oxysporum strain 4287 to produce disseminated infections in different organs, fungal tissue burden was determined in the livers, spleens, kidneys, and lungs of surviving mice sacrificed 13 days after challenge. The presence of fungal propagules was detected in multiple organs, with the highest concentration found in the spleen, followed by kidney, liver, and lung (Fig. 2B).
FIG. 2.
F. oxysporum strain 4287 causes systemic infection and death in immunodepressed mice. (A) Groups of 10 immunodepressed mice were infected with 2 × 107 living (•) or heat-killed (▿) microconidia by lateral tail vein injection. The percent survival was plotted for 13 days. The data shown are from one representative experiment. (B) Four surviving mice were sacrificed on day 13 postinfection, and homogenates from the indicated organs were quantitatively cultured on PDA medium.
TABLE 1.
MST of mice infected with the F. oxysporum wild-type strain 4287 and different mutants derived therefroma
| Strain | MST (days) ± SDb | 95% Confidence interval | Significance (P) |
|---|---|---|---|
| 4287 (wild type) | 7.9 ± 0.5 | 6.8-9.0 | |
| Δfmk1 | 5.6 ± 0.8 | 4.2-6.9 | 0.01 |
| pacC+/− | 12.3 ± 0.2 | 11.9-12.8 | <0.01 |
| pacCc | 8.7 ± 0.7 | 7.4-10.0 | 0.57 |
| ΔchsV | 1.7 ± 0.3 | 1.0-2.3 | <0.01 |
| ΔchsV + chsV | 9.2 ± 1.0 | 7.1-11.3 | 0.49 |
Groups of 10 immunodepressed mice were infected with 2 × 107 microconidia by lateral tail vein injection, and survival was recorded daily for 13 days. Data represent the means and standard deviations from at least four separate experiments.
Groups marked with an asterisk were significantly different from the wild type (P < 0.05).
Role of fungal virulence factors in plant and animal infection.
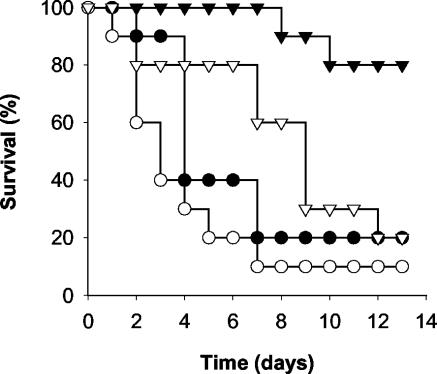
The ability of F. oxysporum 4287 to survive and reproduce in immunodepressed mice, colonize multiple organs, and kill the host suggested that this strain contains the basic pathogenicity determinants required to cause disease on mammalian hosts. To further test the usefulness of the F. oxysporum model for comparing mechanisms of plant and animal pathogenesis, we examined a number of previously obtained knockout mutants in genes involved in virulence on tomato. Among these mutants, Δfmk1 strains that lack an MAPK structurally related to the yeast MAPKs Fus3 and Kss1 exhibit an extreme nonpathogenic phenotype on plants (9). Δfmk1 mutants not only fail to produce vascular wilt symptoms when inoculated onto tomato roots but also are unable to grow invasively on living tomato fruit tissue (Fig. 1). We found that the Δfmk1 mutant was at least as virulent as the wild-type strain in the murine model (Fig. 3 and Table 1), indicating that Fmk1 is dispensable for virulence on immunodepressed mice.
FIG. 3.
Virulence of gene knockout mutants of F. oxysporum on immunodepressed mice. Groups of 10 immunodepressed mice were infected with 2 × 107 microconidia of the wild-type strain 4287 (•), MAPK mutant Δfmk1 (○), loss-of-function mutant pacC+/− (▾), or dominant-activating mutant pacCc (▿). The percent survival was plotted for 13 days. The data shown are from one representative experiment.
We next examined two different classes of mutants in pacC, encoding a zinc finger transcription factor that mediates ambient pH response in fungi (26) and negatively regulates virulence of F. oxysporum on tomato (5). pacC+/− loss-of-function mutants mimic growth at acidic ambient pH and exhibit increased virulence in a root inoculation assay. In contrast, merodiploid pacCc strains expressing a dominant-activating pacC allele mimic growth at alkaline pH and show significantly reduced virulence in the root inoculation assay (5), although invasive growth on tomato fruits is not affected (Fig. 1). Inoculation of immunosuppressed mice with a pacC+/− loss-of-function mutant resulted in a significantly higher survival rate and mean survival time (MST) than in mice inoculated with the wild-type strain (Fig. 3; Table 1). In contrast, survival and MST of mice infected with a dominant-activating pacCc mutant were not statistically different from those infected with the wild-type strain. Thus, PacC is required for full virulence of F. oxysporum on mice but not on tomato plants.
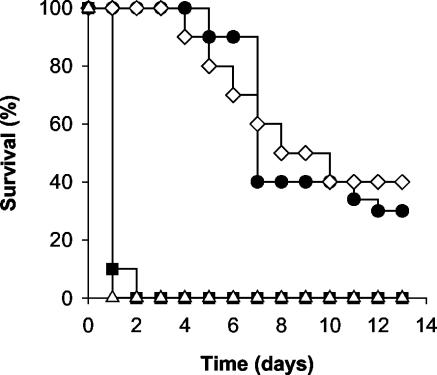
A third class of knockout mutants tested were those lacking a functional copy of the chsV gene (21), encoding a class V chitin synthase required for virulence on tomato (Fig. 1). ΔchsV mutants have defects in cell wall integrity and show morphological alterations such as abnormally enlarged, lemon-like microconidia. When mice were inoculated with 2 × 107 microconidia of the ΔchsV mutant, most of the animals died within 24 h (Fig. 4 and Table 1). We termed this response “fast killing” as opposed to “slow killing” caused by the wild-type strain 4287. A ΔchsV mutant that had been transformed with the wild-type chsV allele reverted to the slow-killing phenotype, indicating that fast killing was determined by lack of a functional ΔchsV allele. Importantly, fast killing was provoked both by living and heat-killed conidia of the chsV mutant (Fig. 4) and occurred both in immunocompetent and in immunosuppressed mice (results not shown). Taken together, these data indicate that the slow- and the fast-killing responses have distinct underlying mechanisms.
FIG. 4.
F. oxysporum mutants lacking the chitin synthase ChsV cause “fast killing” on mice. (A) Groups of 10 immunodepressed mice were infected with 2 × 107 microconidia of the wild-type strain 4287 (•), chitin synthase mutant ΔchsV (▪), heat-killed conidia of the ΔchsV mutant (▵), or a ΔchsV mutant complemented with a wild-type chsV allele (⋄). The percent survival was plotted for 13 days. The data shown are from one representative experiment.
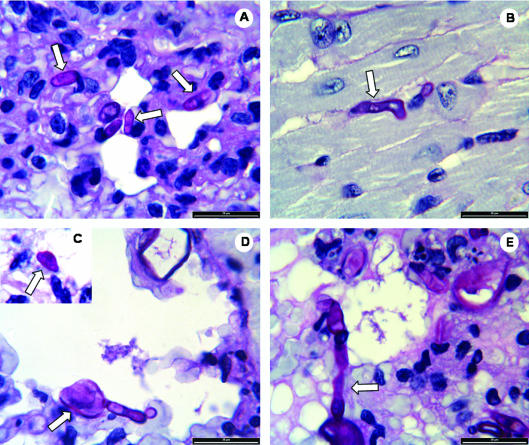
Postmortem studies in mice that had died 24 h after injection with conidia of the ΔchsV mutant revealed an important amount of serum in the upper respiratory tract, suggesting respiratory insufficiency as a possible cause of death. In support of this hypothesis, histolopathological analysis of lung tissue of these animals showed an enlargement of the interstitial walls at the alveolar level. We noted the presence of numerous large (30 by 25 μm), lemon-shaped or irregularly swollen conidia (Fig. 5C to E) which obstructed the blood flow in the interstitial capillaries, leading to congestion, edema, and haemorrhagic foci (Fig. 5E). Some of the conidia had germinated and, ocasionally, branching of the germ tubes was observed. Lung sections of mice infected with heat-killed conidia of the chsV strain revealed a highly similar pattern except that these conidia failed to germinate (data not shown). In contrast, the lungs of animals infected either with the wild-type strain or with the complemented strain were considerably less damaged (Fig. 5A) than those of the group infected with the ΔchsV mutant. The presence of germinated and ungerminated microconidia of normal shape and size (10 by 3 μm) was detected in multiple organs (Fig. 5A and B). Taken together, these findings indicate that the fast-killing response might be caused by respiratory insufficiency due to early lung damage provoked by the conidia of the chsV strain.
FIG. 5.
Histological analysis of mice infected with F. oxysporum strains. Sections of embedded tissues from different organs (A and C to E, lung; B, heart) of mice inoculated with microconidia of F. oxysporum wild-type strain (A and B) or chitin synthase mutant ΔchsV (C to E) and sacrificed 24 h after inoculation were stained with hematoxylin-eosin, periodic acid-Schiff, and methenamine silver and then examined by light microscopy. (A) Characteristic ellipsoidal microconidia of wild-type strain 4287 in the interstitial space; (B) germinated microconidium of wild-type strain 4287; (C) lemon-shaped microconidium of the ΔchsV mutant; (D) swollen ungerminated and germinated microconidia of the ΔchsV mutant; (E) germinated conidia of the ΔchsV mutant growing in the interstitial spaces and provoking an edema. Bar, 20 μm.
DISCUSSION
Within the fungal kingdom, two highly successful groups of pathogens have evolved the ability to infect either plant or animal hosts. Whether these two classes of pathogens use similar mechanisms to cause disease in phylogenetically divergent hosts is currently an open question. Bacterial multihost pathogen models have been highly successful in revealing both conserved and contrasting virulence mechanisms in plant and animal pathogenesis (28, 32). Therefore, the availability of such multihost models for fungal pathogenesis might prove similarly useful. In the present study we have developed a dual plant-animal pathogen system based on a single strain of the vascular wilt fungus F. oxysporum, which allows the simultaneous testing of fungal virulence factors in plant and mammal host models.
In search for a fungal multihost pathogen model, F. oxysporum was an obvious candidate because of its well-documented ability to infect both plants and immunocompromised humans. Due to the economical and historical importance as a causal agent of vascular wilt, a much larger body of information is currently available on the phytopathogenic aspects of F. oxysporum than on its facet as an emerging human pathogen. Plant pathogenic strains of F. oxysporum exhibit a complex pattern of host specificity, prompting their classification into formae speciales and physiological races (1). Whereas race-cultivar specificity within a given forma specialis was recently shown to be controlled by a classical gene-for-gene system (16, 31), the genetic basis underlying plant host specificity in F. oxysporum remains unclear. The finding that isolates from a particular forma specialis which infect the same host plant have independent evolutionary origins suggests that the ability to infect a given plant species may have arisen convergently (25).
We decided to examine the ability of a genetically well-characterized plant pathogenic strain of F. oxysporum to cause disease in a standard animal infection model. An implicit concern with this approach was whether the behavior of a plant pathogenic strain would reflect that of a “true” human pathogen in the murine model. Our results suggest that strain 4287 is not a highly virulent animal pathogen, as shown by the inability to cause disease in immunocompetent mice. However, they also clearly demonstrate that this strain has the basic capacity to infect and to kill immunosuppressed animals in a highly reproducible manner. These findings are consistent with the fact that systemic infections in humans caused by F. oxysporum are predominantly reported in immunocompromised individuals (22, 27, 35). The presence of germinating microconidia in different organs suggests that strain 4287 can grow actively on mammalian tissue and may also undergo cycles of conidiation in the host, as reported previously for pathogenic Fusaria (19). This notion is further supported by the fact that significant amounts of fungal propagules were detected in multiple organs 13 days after inoculation. Even though quantitative CFU data obtained from filamentous fungal pathogens have to be interpreted with caution since they may vary depending on the spore/mycelium ratio, these results clearly indicate that strain 4287 has a high persistency within the mammalian host.
Once we confirmed the basic ability of F. oxysporum strain 4287 to cause disease in a mammalian model, we further tested its usefulness by examining knockout mutants in three functionally distinct classes of genes that were previously found to be involved in virulence on tomato plants. One of them, fmk1, encodes an MAPK that functions in a highly conserved signaling module that is essential for virulence in all fungal plant pathogens examined so far (18). Interestingly, we found that Fmk1 is dispensable for the virulence of F. oxysporum in the disseminated mouse model. This finding is intriguing, because it was suggested that this highly conserved signaling cascade might play functionally similar roles in fungal plant and animal pathogenesis (18). However, a recent study showed that the orthologous MAPK cascade of the human pathogen C. neoformans is also dispensable for virulence in a disseminated mouse model (7). Moreover, the same pathway in C. albicans was previously reported to contribute only partly to virulence (6). Although the role of this pathway has not been tested thus far in topical fungal infection models such as the skin or the oral cavity, these results, taken together, indicate that the Fus3/Kss1 MAPK cascade plays functionally distinct roles in plant and animal pathogenesis. Our data further suggest that such divergent functions not only apply to different pathogen species but also reside within a single fungal strain.
Similarly contrasting results were obtained with mutants in the zinc finger transcription factor PacC. In this case, we found that PacC is required for full virulence on mice, although it was previously shown to be dispensable or even negatively affected virulence on tomato plants (5). Interestingly, the pacC homologue of C. albicans, RIM101, is also required for virulence in a hematogenously disseminated murine model (8). A likely explanation for the contrasting role of PacC in animal and plant pathogenesis is the diverging pH of the host niche: slightly alkaline during systemic animal infection versus acidic in plant tissue. Depending on the ambient pH, PacC may function either as a positive or a negative virulence determinant by activating or preventing expression of distinct groups of pathogenicity genes.
A third class of knockout mutants tested were those affected in the class V chitin synthase ChsV. Chitin synthases catalyze the biosynthesis of chitin, a β-1,4-linked polysaccharide made of N-acetylglucosamine and an essential component of the fungal cell wall (4). F. oxysporum mutants that lack ChsV are unable to infect tomato and hypersensitive to antifungal plant defense compounds such as α-tomatine or H2O2 (21). Surprisingly, most of the mice injected with microconidia of the ΔchsV strain died within 24 h, a phenomenon that we termed “fast killing.” Postmortem studies and histopathological analysis suggested that in these animals death was caused by respiratory insufficiency as a consequence of severe lung damage. In contrast, death in mice infected by filamentous fungi is usually not provoked by initial lung damage but by more generalized lesions affecting numerous organs (13). We found evidence suggesting that this unusual fast-killing effect could be caused by physical obstruction of interstitial capillaries by the abnormally enlarged ΔchsV mutant conidia. The presence of morphological aberrations in fungal class V chitin synthase mutants such as balloon-like swellings and abnormally shaped, lemon-like microconidia, has been reported previously and is probably caused by defects in cell wall integrity, because it can be reversed by addition of an osmoprotectant to the growth medium (15, 21). Two lines of evidence strongly suggest that fast killing is mechanistically unrelated to slow killing caused by the wild-type F. oxysporum strain. First, unlike slow killing, fast killing was induced both by living and heat-killed conidia of the ΔchsV mutant, suggesting that the sole physical presence of the conidia is sufficient to provoke death. Second, in contrast to slow killing, fast killing occurred both in immunocompetent and in immunosuppressed mice, indicating that this mechanism acts independently of the immunological status of the animals.
F. oxysporum strain 4287 is to our knowledge the first fungal model strain reported to reproducibly infect both plant and mammalian hosts. The availability of such a multihost model strain represents a key step toward improving our understanding of how fungal pathogenesis on evolutionary distant hosts has evolved. The results obtained with the F. oxysporum knockout mutants suggest that certain virulence factors essential in plant pathogenesis are dispensable in animal pathogenesis and vice versa. In both cases where such divergent roles were detected between the tomato and the mouse model, the virulence pattern of the F. oxysporum mutants in mice mimics that previously reported for two well-established animal model pathogens, C. albicans and C. neoformans. This finding is important because it addresses the underlying concern whether the virulence pattern of an opportunistic pathogen such as F. oxysporum will reflect that of a “true” human pathogen. Our results with the fmk1 and the pacC mutants suggest that F. oxysporum strain 4287 indeed behaves like a “true” animal pathogen in the immunosuppressed mouse model. Thus, F. oxysporum strain 4287 could be a useful model for the identification of conserved and divergent fungal virulence determinants in plant and animal pathogenesis, similar to the P. aeruginosa strain PA14 (30). Importantly, 4287 is also amenable to random insertional mutagenesis (21), and rapid and reliable in vivo virulence assays on tomato fruits are already available for this strain (9). The F. oxysporum model may therefore prove highly useful in providing new insights into the molecular basis of host specificity in fungal pathogenesis.
Acknowledgments
The research was supported by grants PI-020114 from the Spanish Ministerio de Sanidad y Consumo and BIO2001-2601 from Ministerio de Ciencia y Tecnología. M.O. has a Ph.D. fellowship from Universitat Rovira i Virgili. M.P.M. and Z.C. have Ph.D. fellowships from the Ministerio de Ciencia y Tecnología and the Ministerio de Educación, Cultura y Deporte, respectively. A.D.P. is recipient of a Ramón y Cajal grant from Ministerio de Ciencia y Tecnología.
Editor: T. R. Kozel
REFERENCES
- 1.Beckman, C. H. 1987. The nature of wilt diseases of plants. American Phytopathological Society, St. Paul, Minn.
- 2.Boutati, E. I., and E. J. Anaissie. 1997. Fusarium, a significant emerging pathogen in patients with hematologic malignancy: ten years' experience at a cancer center and implications for management. Blood 90:999-1008. [PubMed] [Google Scholar]
- 3.Buer, J., and R. Balling. 2003. Mice, microbes and models of infection. Nat. Rev. Genet. 4:195-205. [DOI] [PubMed] [Google Scholar]
- 4.Cabib, E., D. H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19679-19682. [DOI] [PubMed] [Google Scholar]
- 5.Caracuel, Z., M. I. Roncero, E. A. Espeso, C. I. Gonzalez-Verdejo, F. I. Garcia-Maceira, and A. Di Pietro. 2003. The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol. Microbiol. 48:765-779. [DOI] [PubMed] [Google Scholar]
- 6.Csank, C., K. Schroppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson, R. C., C. B. Nichols, G. M. Cox, J. R. Perfect, and J. Heitman. 2003. A MAP kinase cascade composed of cell type specific and nonspecific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49:469-485. [DOI] [PubMed] [Google Scholar]
- 8.Davis, D., J. E. Edwards, Jr., A. P. Mitchell, and A. S. Ibrahim. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Pietro, A., F. I. Garcia-MacEira, E. Meglecz, and M. I. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39:1140-1152. [PubMed] [Google Scholar]
- 10.Di Pietro, A., M. P. Madrid, Z. Caracuel, J. Delgado-Jarana, and M. I. G. Roncero. 2003. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4:315-326. [DOI] [PubMed] [Google Scholar]
- 11.Di Pietro, A., and M. I. Roncero. 1998. Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant-Microbe Interact. 11:91-98. [DOI] [PubMed] [Google Scholar]
- 12.Gold, S. E., M. D. Garcia-Pedrajas, and A. D. Martinez-Espinoza. 2001. New (and used) approaches to the study of fungal pathogenicity. Annu. Rev. Phytopathol. 39:337-365. [DOI] [PubMed] [Google Scholar]
- 13.Gow, N. A., A. J. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 14.Guarro, J., and J. Gene. 1995. Opportunistic fusarial infections in humans. Eur. J. Clin. Microbiol. Infect. Dis. 14:741-754. [DOI] [PubMed] [Google Scholar]
- 15.Horiuchi, H., M. Fujiwara, S. Yamashita, A. Ohta, and M. Takagi. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 181:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huertas-Gonzalez, M. D., M. C. Ruiz-Roldan, A. Di Pietro, and M. I. G. Roncero. 1999. Cross protection provides evidence for race-specific avirulence factors in Fusarium oxysporum. Physiol. Mol. Plant Pathol. 54:63-72. [Google Scholar]
- 17.Idnurm, A., and B. J. Howlett. 2002. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot. Cell 1:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, K., D. N. Howell, J. R. Perfect, and W. A. Schell. 1998. Morphologic criteria for the preliminary identification of Fusarium, Paecilomyces, and Acremonium species by histopathology. Am. J. Clin. Pathol. 109:45-54. [DOI] [PubMed] [Google Scholar]
- 20.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83-86. [DOI] [PubMed] [Google Scholar]
- 21.Madrid, M. P., A. Di Pietro, and M. I. Roncero. 2003. Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defense compounds. Mol. Microbiol. 47:257-266. [DOI] [PubMed] [Google Scholar]
- 22.Nucci, M., and E. Anaissie. 2002. Cutaneous infection by Fusarium species in healthy and immunocompromised hosts: implications for diagnosis and management. Clin. Infect. Dis. 35:909-920. [DOI] [PubMed] [Google Scholar]
- 23.Nürnberger, T., and F. Brunner. 2002. Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5:318-324. [DOI] [PubMed] [Google Scholar]
- 24.Odds, F. C., F. Van Gerven, A. Espinel-Ingroff, M. S. Bartlett, M. A. Ghannoum, M. V. Lancaster, M. A. Pfaller, J. H. Rex, M. G. Rinaldi, and T. J. Walsh. 1998. Evaluation of possible correlations between antifungal susceptibilities of filamentous fungi in vitro and antifungal treatment outcomes in animal infection models. Antimicrob. Agents Chemother. 42:282-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Donnell, K., H. C. Kistler, E. Cigelnik, and R. C. Ploetz. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 95:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penalva, M. A., and H. N. Arst, Jr. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponton, J., R. Ruchel, K. V. Clemons, D. C. Coleman, R. Grillot, J. Guarro, D. Aldebert, P. Ambroise-Thomas, J. Cano, A. J. Carrillo-Munoz, J. Gene, C. Pinel, D. A. Stevens, and D. J. Sullivan. 2000. Emerging pathogens. Med. Mycol. 38(Suppl. 1):225-236. [DOI] [PubMed] [Google Scholar]
- 28.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 30.Rahme, L. G., M. W. Tan, L. Le, S. M. Wong, R. G. Tompkins, S. B. Calderwood, and F. M. Ausubel. 1997. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons, G., J. Groenendijk, J. Wijbrandi, M. Reijans, J. Groenen, P. Diergaarde, T. Van der Lee, M. Bleeker, J. Onstenk, M. de Both, M. Haring, J. Mes, B. Cornelissen, M. Zabeau, and P. Vos. 1998. Dissection of the fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell 10:1055-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staskawicz, B. J., M. B. Mudgett, J. L. Dangl, and J. E. Galan. 2001. Common and contrasting themes of plant and animal diseases. Science 292:2285-2289. [DOI] [PubMed] [Google Scholar]
- 33.Sweigard, J. A., and D. J. Ebbole. 2001. Functional analysis of pathogenicity genes in a genomics world. Curr. Opin. Microbiol. 4:387-392. [DOI] [PubMed] [Google Scholar]
- 34.van Burik, J. A., and P. T. Magee. 2001. Aspects of fungal pathogenesis in humans. Annu. Rev. Microbiol. 55:743-772. [DOI] [PubMed] [Google Scholar]
- 35.Vartivarian, S. E., E. J. Anaissie, and G. P. Bodey. 1993. Emerging fungal pathogens in immunocompromised patients: classification, diagnosis, and management. Clin. Infect. Dis. 17(Suppl. 2):S487-S491. [DOI] [PubMed] [Google Scholar]
- 36.Wolpert, T. J., L. D. Dunkle, and L. M. Ciuffetti. 2002. Host-selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40:251-285. [DOI] [PubMed] [Google Scholar]