Summary
Background
Recent studies have suggested that microRNA-10b (miR-10b) acts as a promoter of metastasis in breast cancer, although the underlying mechanism remains largely unknown. In this study, we provide the first evidence that E-cadherin (E-cad) is a potential target of miR-10b.
Material/Method
By applying gain-of-function and loss-of-function approaches in the metastatic breast cancer cell line MDA-MB-231, we demonstrated that miR-10b is necessary and sufficient to regulate the cellular expression of E-cad and in vitro tumor cell invasion.
Results
Comparative expression analysis of miR-10b in benign breast lesions (N=16), primary breast cancers (N=21), and metastatic breast carcinomas (N=23) revealed that miR-10b transcription was uniquely up-regulated in metastatic cancers. The expression level of miR-10b positively correlated with tumor size, pathological grading, clinical staging, lymph node metastasis, Her2-positivity and tumor proliferation, but was negatively associated with estrogen receptor-positivity, progesterone receptor-positivity and E-cad mRNA and protein levels.
Conclusions
These findings indicate the existence of a novel E-cadherin-related mechanism by which miR-10b modulates breast cancer metastasis. In addition, miR-10b may be a useful biomarker of advanced progression and metastasis of breast cancer.
Keywords: microRNA-10b, E-cadherin, breast cancer, metastasis
Background
Breast cancer is the most common malignancy diagnosed in women. More than 1.3 million new cases are reported each year, accounting for approximately 23% of all new cancer cases in either sex worldwide [1]. Several strong risk factors, ranging from genetic to environmental, have been identified for breast cancer and incorporated into public awareness campaigns and clinical monitoring strategies of the general at-risk population [2]. In addition, convenient and accurate early detection techniques have been developed and are in routine use [3]. While these strategies have been able to reduce the rates of mortality associated with breast cancer, nearly half a million women still die of the disease each year; among those, more than 90% are attributed to cancer metastasis. Therefore, substantial research and clinical efforts have been devoted to understanding the underlying molecular mechanisms that control the metastatic spread of tumor cells. It is expected that molecularly targeting these mechanisms, by pharmaceutical or genetic-based approaches, may slow down or stop metastasis and improve the survival of cancer patients.
MicroRNA (miRNA) is a family of endogenous, highly conserved, small non-coding RNA molecules. The in vivo mature miRNA molecules are between 18 to 22 nucleotides (nt) in length, and are generated from the genome by a 2-step process [4,5]. First, the gene for miRNA is transcribed in the nucleus by RNA polymerase II to produce primary miRNA (pri-miRNA), which is then targeted by the ribonuclease Drosha for processing into a stem-loop structure, known as the precursor miRNA (pre-miRNA). Second, with the help of Exportin 5 and Ran-GTP, pre-miRNA is transported to the cytoplasm, where it is further processed by the ribonuclease Dicer to an approximately 22-nt double-stranded RNA (dsRNA) molecule, which is the mature miRNA. The 2 strands of the mature miRNA are thermodynamically asymmetric, and the strand with less 5′-end base-pairing stability, also called the guide strand, is preferentially selected by Argonaute 2 and other proteins for subsequent assembly into the RNA-induced silencing complex (RISC) [6]. By hybridizing, often imperfectly, to a homologous mRNA sequence, the guide strand of miRNA directs RISC to the target mRNA sequence. The miRNA is then cleaved by the endonuclease component of the complex, leading to mRNA degradation and gene silencing. Most miRNA target sites are located within the 3′-untranslated regions (3′-UTR) of genes.
Through this post-transcriptional regulatory mechanism of gene expression, miRNA actively modulates many biological processes, including cell proliferation, apoptosis, differentiation, and motility; likewise, miRNA actions can influence the onset and progression of a number of human diseases, as has been demonstrated for heart disease, hematopoietic disease, neurological disease, immune disease and cancer [4,5,7]. To date, more than 900 mature miRNAs have been identified in the human genome (miRbase database, http://www.mirbase.org/). Although the number of miRNA-coding genes only accounts for approximately 1% of total genes, they are believed to regulate the expression of more than 30% of the protein-coding genes in humans [8].
miRNAs play an important role in normal development of the mammary gland. For example, miR-25 and miR-17–92 are involved in the proliferation of normal mammary ducts and acini, let-7 regulates terminal differentiation of mammary cells, miR-29 mediates breast tissue remodeling, and miR-205 is considered as a marker for normal mammary stem cells [9]. In addition, miRNAs also play key roles in breast cancer development. Abnormal expression levels of miRNAs have been clinically observed in cancerous breast tissues, where they are believed to function as oncogenes or tumor suppressors [5].
Recently, Ma et al. identified the miR-10b gene as a target of the transcription factor Twist, which is highly expressed in metastatic breast cancer cells and stimulates in vitro and in vivo tumor invasion [10]. Up-regulated expression of miR-10b was shown to result in suppression of the miRNA-10b target homeobox gene D10 (HOXD10), which in turn led to induced expression of the pro-metastatic gene RHOC[10]. In the study presented herein, we identified another target of miR-10b silencing activity, E-cadherin (E-cad), an essential component for maintaining epithelial cell polarity and a potent suppressor of breast cancer invasion/metastasis. Moreover, we showed that miR-10b is necessary and sufficient for driving the expression of E-cad in breast cancer cells in vitro. A high negative correlation was found between the expressions of these 2 molecules in human breast cancer tissues, and a higher level of miR-10b significantly correlated with advanced disease and higher lymph node metastasis.
Material and Methods
Cell culture and treatment
The human metastatic breast adenocarcinoma cell line MDA-MB-231 was obtained from the Center of Molecular Medicine, Tongji Hospital (Tongji Medical College, Wuhan, China) and cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA).
The 2′-O-methyl miR-10b inhibitor (IN-10b, an antisense oligonucleotide specifically targeting mature miR-10b; sequence 5′-CACAAAUUCGGUUCUACAGGGUA-3′), a control scrambled oligonucleotide (IN-Ctrl, 5′-CAGUACUUUUGUGUAGUACAA-3′), the 2′-O-methyl miR-10b mimic (MI-10b, dsRNA molecule mimicking the endogenous mature miR-10b molecules; sense 5′-UACCCUGUAGAACCGAAUUUGUG-3′, antisense 3′-CAAAUUCGGUUCUACAGGGUAUU-5′), and its corresponding control scrambled RNA duplex (MI-Ctrl, sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′) were designed and synthesized by GenePharma (Shanghai, China). To observe the transfection efficiency, an FAM-labeled miR-10b inhibitor or mimic was used (GenePharma). For delivery of the inhibitor or mimic into the cells, MDA-MB-231 were seeded at 1.5×106 cells/well in 6-well plates. Twenty-four hours later, the miR-10 inhibitor or mimic was transfected into the cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) by following the manufacturer’s protocol.
Patient selection and clinicopathological information
This study was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China), and written consent was obtained from all subjects. A cohort of 44 breast cancer tissues were freshly isolated from female patients (age range: 35 to 76 years; average: 50.27 years) in either Tongji Hospital (Huazhong University of Science and Technology, Wuhan, China) or the Affiliated Xuzhou Hospital, Medical College of Southeast University (Xuzhou, China). According to the World Health Organization (WHO) guidelines for Histological Grading of Tumors of the Breast (2003), the breast cancer tissue samples included 21 grade I, 14 grade II, and 9 grade III tumors. Pathologically, these tissues included 35 invasive ductal carcinoma, 4 ductal carcinoma in situ, 2 mucinous carcinoma, 1 medullary carcinoma, 1 Paget’s disease, and 1 neuroendocrine carcinoma. Based on the 2002 TNM Classification of Breast Cancers developed by the International Union Against Cancer (UICC) and the American Joint Committee on Cancer (AJCC), the sample cohort was composed of 21 stage I, 9 stage II, and 14 stage III cases.
Other available clinicopathological data associated with each breast cancer patient were collected, including tumor size, positivity for Her2, estrogen receptor (ER) and progesterone receptor (PR), and mitotic index (as indicated by Ki-67 staining). As controls, 16 samples of benign breast lesions, including 12 fibroadenomas, 2 adenosis, 1 mammary hamartoma, and 1 mammary duct ectasia, were freshly obtained from 16 female patients (age range: 20 to 63 years; average: 34.06 years) in the Second Affiliated Hospital of Henan University of Science and Technology. All breast tissues were immediately transferred to liquid nitrogen and stored at −86°C until future use.
Reverse transcription (RT) followed by quantitative real-time PCR (qPCR)
Total RNA was extracted from breast tissues or breast cancer cells using the Trizol reagent (Invitrogen) and following the manufacturer’s instructions. To quantify the miR-10b amount in each of the total RNA samples, the Hairpin-it™ miR-10b qPCR Quantitation Kit (GenePharma) was used according to the manufacturer’s protocol. This procedure involves exclusive stem-loop RT primer hybridization to mature, miRNA, and not to pri-miRNA or pre-miRNA, followed by amplification of the mature miR-10b using a set of gene-specific primers. As an internal control, the small nuclear RNA (snRNA) U6 was amplified by using the U6 snRNA qPCR Normalization Kit (GenePharma).
To examine the expression of E-cad in various samples, total RNA was reverse transcribed into cDNA using MMLV reverse transcriptase (Promega, Madison, WI) and following the manufacturers’ protocol. qPCR was performed using the following primers provided in the Custom RT-qPCR Gene Expression Kit (GenePharma): human E-cad (amplicon size of 176 base pairs (bps)): forward 5′-ATGCCATCGTTGTTCACTGGA-3′, reverse 5′-CATGAGAAGTATGACAACAGCCT-3′; human GAPDH (internal control; amplicon size of 113 bps): 5′-AGTCCTTCCACGATACCAAAGT-3′, reverse 5′-ATTGGAACGATACAGAGAAGATT-3′. The expression of the target gene was calculated using the 2−ΔΔCt method, as previously described [11].
Western immunoblot
Tissue homogenate was prepared in an ice cold buffer containing 50 mM Tris-HCl (pH 6.7), 2% sodium dodecyl sulfate (SDS), 4% glycerol, and 1 mM phenylmethanesulphonylfluoride (PMSF). Cell lysate was prepared in an ice-cold buffer containing 50 mM Tris-HCl (pH 8.0), 0.5% Triton X-100, 10% glycerol, and 0.1 mM ethylenediaminetetraacetic acid (EDTA). After centrifugation at 12,000×g for 10 min, the supernatant containing the total protein sample was collected and used in subsequent analysis. Following resolution by 10% SDS-polyacrylamide gel electrophoresis (PAGE), the separated proteins were electro-transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was then blocked against non-specific binding by room temperature incubation with TBST buffer (10 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 0.2% Tween-20) containing 5% skim milk for 2 h. After which, overnight incubation with rocking at 4°C was carried out with the following primary antibodies: anti-E-cad antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-β-actin antibody (Sigma, St. Louis, MO, USA). The next day, incubation was carried out with HRP-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA), and the signal was developed with SuperSignal West Pico Chemiluminescent Substrates (Pierce, Rockford, IL, USA). Immunoreactive signal intensities were analyzed by the Gel-Pro Analyzer (Media Cybernetics, Bethesda, MD, USA).
Transwell invasion assay
Transwell inserts in 24-well format (8.0-μm pores; Corning, Lowell, MA, USA) were coated on the upper side with diluted matrigel (50 mg/L matrigel: DMEM with 0.5% FBS) and incubated at 37°C for 4 h. After coating, 5×103 cells in 100 μL DMEM medium containing 5% FBS were seeded on top of the insert. To the lower chamber, 700 μL of DMEM medium containing 20% FBS was added. The invasion assay was allowed to proceed at 37°C for 24 h. Then, the transwell insert was taken out, washed once with phosphate buffered saline (PBS), and fixed in methanol at −20°C for 10 min. After another wash with PBS, the non-invasive cells and matrigel on the top of the insert were cleared with a cotton swab, and the insert was washed 3 times with PBS. After air drying, the insert was immersed in 200 μL of 0.1% crystal violet and incubated at 37°C for 30 min; after a final 3 washes with ddH2O, the inserts were evaluated and digitally imaged under a light microscope. For each sample, 6 random fields were imaged under high magnification (200×) and used to calculate the average number of invaded cells.
Statistical analysis
Statistical analysis was performed using SPSS 13.0 software (Chicago, IL, USA). All quantitative data are presented as mean ± standard deviation (SD) for 3 independent experiments or multiple samples, where indicated. Comparison between 2 groups was first performed by Levene’s test, with those showing equal variance being applied to further testing by one-way analysis of variance (ANOVA). If the ANOVA test was positive (P<0.05), a Student-Newman-Keuls test was performed for pairwise comparisons. The variation of miR-10b expression according to distinct clinicopathological features was analyzed using the Student’s t-test with Bonferroni correction, and the correlation between these parameters was examined by a general linear regression model. The correlation between the expression levels of miR-10b and E-cad was analyzed using Spearman’s rank correlation test, with the coefficient r calculated. A P-value of <0.05 was considered statistically significant.
Results
miR-10b up-regulates E-cad and reduces cell invasion
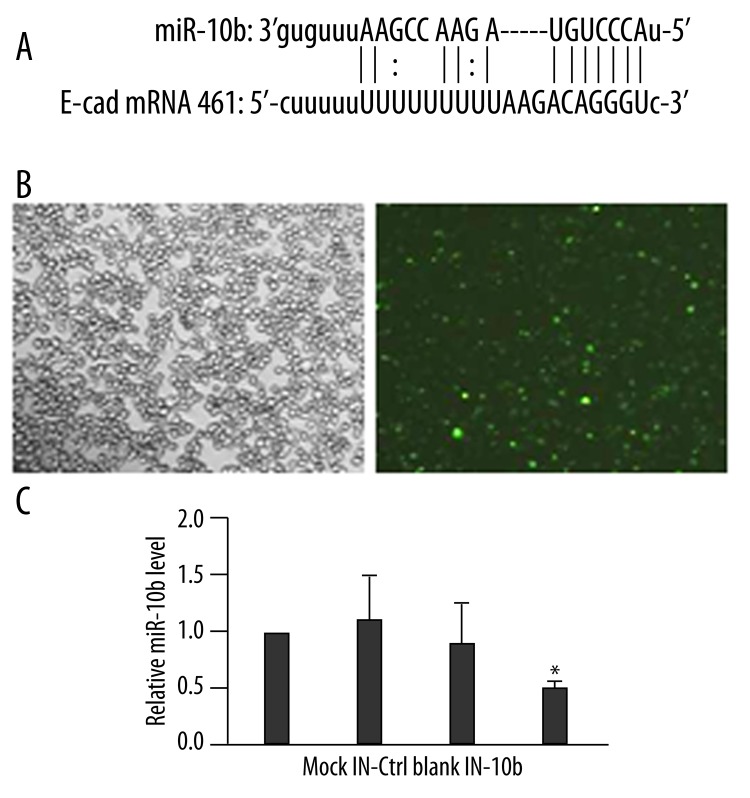
To gain a mechanistic understanding of miR-10b-regulated breast cancer metastasis, we sought potential human miR-10b targets by querying bioinformatics databases using the miRanda (http://www.microrna.org/microrna/home.do) and PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html) algorithms. Both analyses identified the human CDH1 gene that encodes the transmembrane glycoprotein E-cad as a potential target for miR-10b. The potential target site in the E-cad mRNA was located between nucleotides 461 and 481 within the 3′ UTR (Figure 1A).
Figure 1.
miR-10b inhibitor significantly reduces endogenous miR-10b level in MDA-MB-231 cells. (A) Maps of the potential target site of miR-10b in E-cad mRNA. Straight lines indicate canonical Watson-Crick base pairing, and dotted lines indicate non-Watson-Crick base pairing. (B) FAM-labeled miR-10b inhibitor was transiently transfected into MDA-MB-231 cells. Transfection efficiency was estimated 48 h later by imaging under bright light (left) and fluorescence microscopy (right) (200×). (C) The level of miR-10b was examined by RT-qPCR at 48 h after transfection and quantified as the ratio of miR-10b to snRNA U6 (internal control), with the relative level in Mock cells arbitrarily defined as 1.0. Mock, mock-transfected cells; IN-Ctrl, control inhibitor-transfected cells; Blank, non-transfected cells; IN-10b, miR-10b inhibitor-transfected cells. * P<0.01, as compared to all other groups.
To test whether miR-10b is required for the down-regulated E-cad expression commonly observed in breast cancer cells, we transiently transfected MDA-MB-231 cells with the 2′-O-methyl miR-10b inhibitor (IN-10b). Mock-transfected cells (Mock), cells transfected with a scrambled oligonucleotide (IN-Ctrl), or non-transfected cells (Blank) were used as controls to evaluate changes in gene expressions associated with miR-10b silencing. By using FAM-labeled miR-10b, we were able to visually assess transfection efficiency; at 48 h after transfection, the transfection efficiency reached approximately 80% (Figure 1B). Meanwhile, RT-qPCR analysis showed that the level of miR-10b was dramatically reduced in IN-10b cells, by approximately 53% vs. Mock cells, 50% vs. Blank cells, and 58% vs. IN-Ctrl cells (P<0.01; Figure 1C).
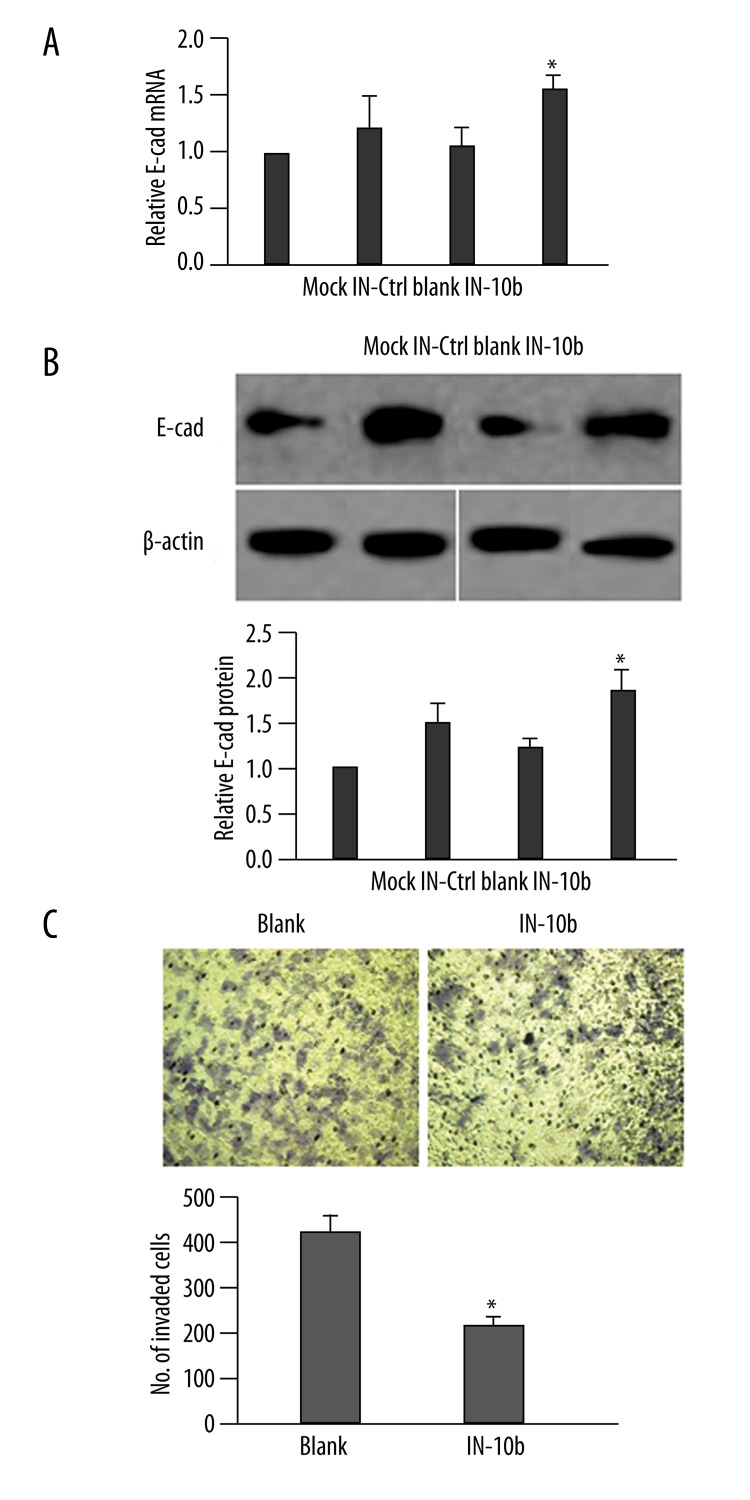
Once reduced levels of miR-10b had been established, we examined the concomitant expression of E-cad at both steady-state mRNA and protein levels. As shown in Figure 2A and 2B, although we detected a slight up-regulation of both E-cad mRNA and protein in IN-Ctrl cells, as compared to Mock and Blank cells, the E-cad expression in IN-10b cells was significantly up-regulated as compared to the other 3 negative control cells (P<0.05). Since E-cad is a crucial regulator of cell migration/invasion, we examined whether inhibition of miR-10b led to any alterations in cell invasive properties, presumably through the dysregulated E-cad. Compared with Blank cells, the invasion capability of IN-10b cells was reduced by approximately 45% (P<0.05; Figure 2C).
Figure 2.
Down-regulation of miR-10b level enhances the expression of E-cad. (A) MDA-MB-231 cells were transfected as indicated. The steady-state mRNA level of E-cad was determined by RT-qPCR 48 h later, and is presented as the ratio to GAPDH (internal control), with the relative level in Mock cells arbitrarily defined as 1.0. (B) MDA-MB-231 cells were treated as in (A), and the protein level of E-cad was examined by a Western immunoblot. A representative gel image is shown (top), along with quantification of the ratio of E-cad to β-actin from three independent experiments (bottom). The level of relative E-cad expression in Mock cells was arbitrarily defined as 1.0. (C) At 48 h after transfection, the invasion capability of MDA-MB-231 cells was examined by Transwell assay. Representative images of invaded cells from each of the groups is presented (top). The number of invaded cells was calculated as the sum of crystal violet-positive cells from six random fields of each sample (bottom). * P<0.05, as compared to all other groups.
Overexpressing miR-10b reduces E-cad and promotes cell invasion
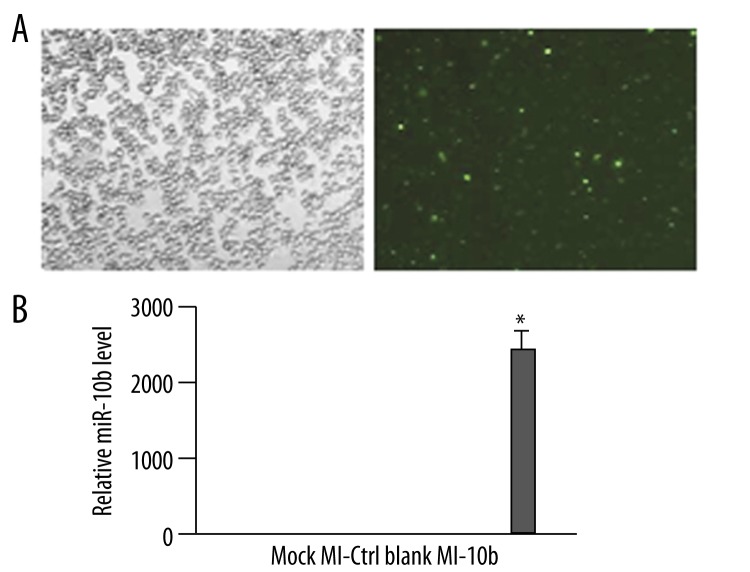
Next, we investigated whether miR-10b is sufficient to down-regulate E-cad in breast cancer cells. To achieve this, we transfected MDA-MB-231 cells with a miR-10b mimic that functions as a mature miR-10b in cells (MI-10b) and would create an environment of miR-10b overexpression. Mock-transfected cells (Mock), cells transfected with a control mimic (MI-Ctrl), or non-transfected cells (Blank) were used as controls to evaluate the change in E-cad expression due to miR-10b overexpression. Again, the transfection efficiency reached approximately 80% at 48 h post-transfection (Figure 3A). As expected, the MI-10b cells showed a dramatically up-regulated miR-10b level, by an approximately 2500-fold increase vs. Mock or Blank cells and 55-fold vs. MI-Ctrl cells (Figure 3B).
Figure 3.
miR-10b mimic-derived overexpression dramatically increases its level in MDA-MB-231 cells. (A) FAM-labeled miR-10b mimic was transiently transfected into MDA-MB-231 cells. The transfection efficiency was estimated 48 h later by imaging the cells under bright light (top) and fluorescence microscopy (bottom) (200×). (B) The level of miR-10b was examined by RT-qPCR at 48 h after transfection and quantified as the ratio of miR-10b to snRNA U6 (internal control), with the relative level in Mock cells arbitrarily defined as 1.0. Mock, mock-transfected cells; MI-Ctrl, control mimic-transfected cells; Blank, non-transfected cells; MI-10b, miR-10b mimic-transfected cells. * P<0.01, as compared to all other groups.
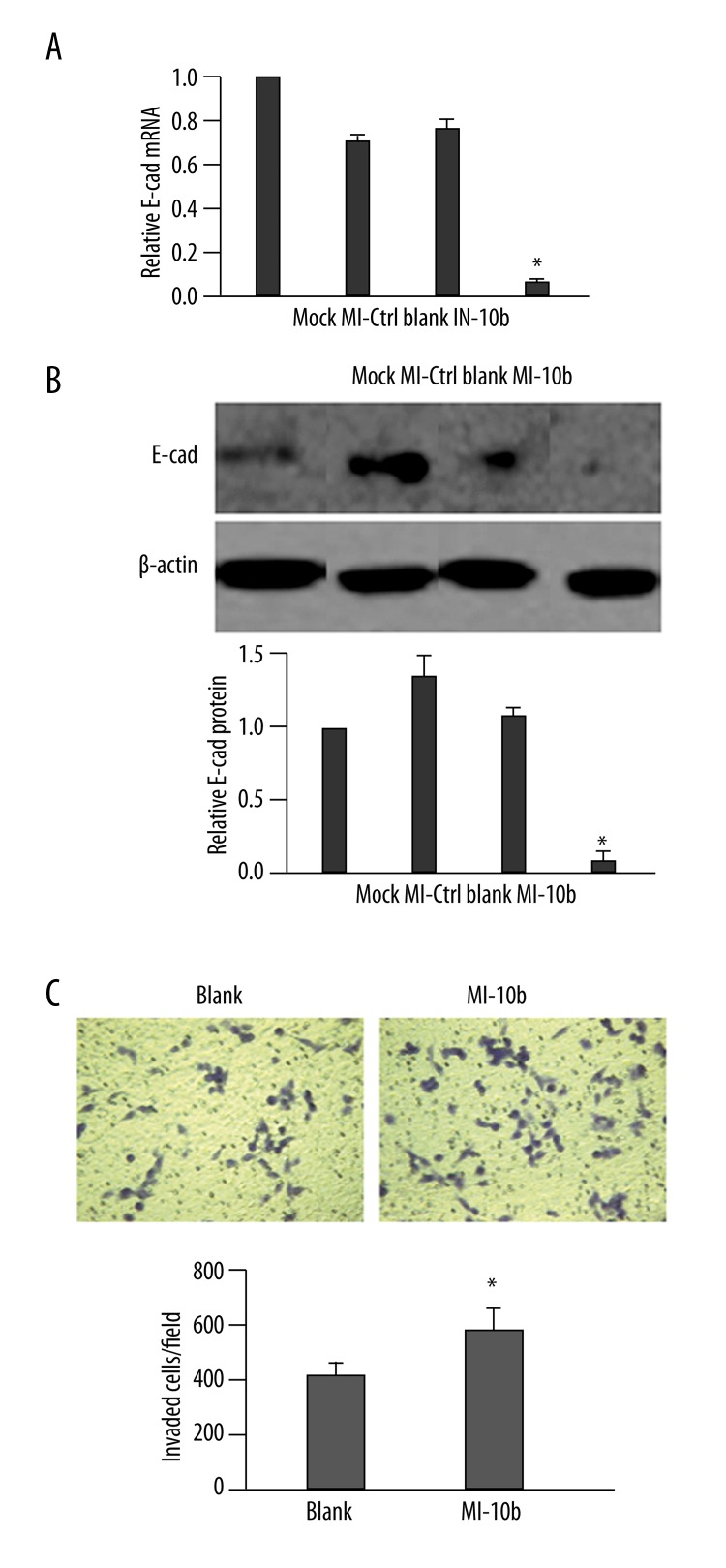
The up-regulation of miR-10b level was accompanied by significant reduction of E-cad mRNA and protein expressions, by more than 90% and 80% vs. Mock cells, respectively (P<0.01; Figure 4A and 4B). In functional analysis, the MI-10b cells also exhibited significantly higher invasion capability than the Blank cells (P<0.05; Figure 4C).
Figure 4.
Up-regulation of miR-10b level reduces the expression of E-cad. (A) MDA-MB-231 cells were transfected as indicated. The steady-state mRNA level of E-cad was determined 48 h later by RT-qPCR and presented as the ratio to GAPDH (internal control), with the relative level in Mock cells arbitrarily defined as 1.0. (B) MDA-MB-231 cells were treated as in (A), and the protein level of E-cad was examined by Western immunoblot. A representative gel image is shown (top), along with quantification of the ratio of E-cad to β-actin from three independent experiments (bottom). The level of relative E-cad expression in Mock cells was arbitrarily defined as 1.0. (C) At 48 h after transfection, the invasion capability of MDA-MB-231 cells was examined by Transwell assay. Representative images of invaded cells from each of the groups is presented (top). The number of invaded cells was calculated as the sum of crystal violet-positive cells from six random fields of each sample (bottom). * P<0.05, as compared to all other groups.
High miR-10b level is detected in metastatic breast cancer and negatively correlates with E-cad expression
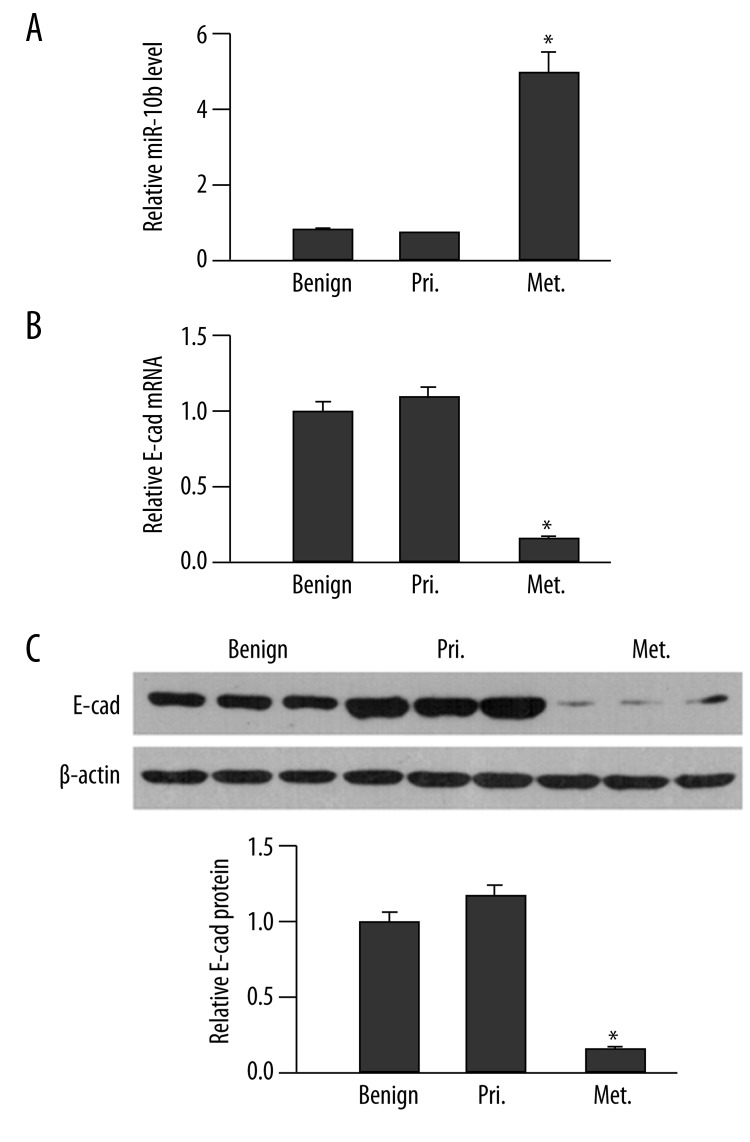
To analyze the clinical significance of the potential cross-talk between miR-10b and E-cad during breast cancer development, we examined the expressions of miR-10b and E-cad in benign breast lesions (Benign, N=16), primary breast cancer that was limited in breast tissue without lymph node metastasis (Pri, N=21), and metastatic breast cancer that was positive for lymph node metastasis (Met, N=23). Compared to cancers from benign breast lesions, primary breast cancers showed no obvious differences in detectable miR-10b level (P>0.05; Figure 5A). Metastatic breast cancers, however, contained significantly higher levels of miR-10b, approximately 5- and 6-fold increases vs. benign tissue and primary breast cancers, respectively (P<0.05).
Figure 5.
miR-10b level negatively correlates with E-cad expression in human breast cancer. (A) The miR-10b level in benign breast tissues (Benign), primary breast cancers (Pri.) and metastatic breast cancers (Met.) was examined by RT-qPCR and quantified as the ratio of miR-10b to snRNA U6 (internal control), with the relative level in normal tissues arbitrarily defined as 1.0. (B) The steady-state E-cad mRNA level in indicated human tissues was determined by RT-qPCR and presented as the ratio to GAPDH (internal control), with the relative level in Mock cells arbitrarily defined as 1.0. (C) The protein level of E-cad was examined by a Western immunoblot. A representative gel image of three samples from each group is shown (top), along with quantification of the ratio of E-cad to β-actin from all tissues in each group (bottom). The level of relative E-cad expression in benign tissues was arbitrarily defined as 1.0. * P<0.05, as compared to the other two groups.
Corresponding to the up-regulated level of miR-10b in metastatic breast cancers, we also found that E-cad was significantly down-regulated at both the mRNA and protein levels in metastatic breast cancers, as compared to that in benign breast lesions and primary breast cancers (P<0.05). The E-cad mRNA and protein levels were not dramatically different among the benign breast lesions and primary breast cancers (P>0.05; Figure 5B and 5C). Spearman’s rank correlation test showed that there was a high negative correlation between miR-10b level and E-cad mRNA or protein level in breast cancer (r<−0.5, P<0.001; Table 1).
Table 1.
Correlation between miR-10b level and E-cad expression in human breast cancer tissues.
| Spearman’s coefficient (r) | miR-10b | E-cad mRNA | E-cad protein |
|---|---|---|---|
| miR-10b | 1.00 | ||
| E-cad mRNA | −0.68 | 1.00 | |
| E-cad protein | −0.52 | 0.87 | 1.00 |
Low negative correlation, 0<r<−0.3; moderate negative correlation, −0.3<r<−0.5; high negative correlation, −0.5<r<−1.0. All P<0.001.
MiR-10b is an indicator of advanced and metastatic breast cancer
The t-test statistical analysis showed that miR-10b level varied significantly in conjunction with the following clinicopathological parameters: tumor diameter (P=0.008), histological grading (P=0.007), clinical staging (P=0.004), status of lymph node metastasis (P=0.003), presence of Her2 (P=0.012), ER and PR expression (P=0.039 and 0.041, respectively), and the level of mitosis (P=0.022; Table 2). Multivariate linear regression analysis revealed that miR-10b level in breast cancer independently and positively correlated with tumor diameter (P=0.0076), histological grading (P=0.0120), clinical staging (P=0.0006), presence of lymph node metastasis (P<0.0001), positivity for Her2 (P=0.0098), and higher mitotic level (P=0.0230), while negatively correlating with positivity for ER and PR (P=0.0184 and 0.0319, respectively; Table 3).
Table 2.
Variation of miR-10b level by distinct clinicopathological features analyzed by the t-test with Bonferroni correction.
| Clincopathological features | Cases, N | miR-10b level | t-statistic | P-value |
|---|---|---|---|---|
| Age, years | 0.375 | 0.341 | ||
| <50 | 25 | 2.31 | ||
| ≥50 | 19 | 2.18 | ||
| Tumor diameter, cm | 7.192 | 0.008 | ||
| ≤5 | 27 | 1.56 | ||
| >5 | 17 | 3.44 | ||
| Histological grading | 7.537 | 0.007 | ||
| I + II | 26 | 1.39 | ||
| III | 18 | 3.62 | ||
| Clinical staging | 8.941 | 0.004 | ||
| I + II | 22 | 1.05 | ||
| III | 22 | 3.70 | ||
| Lymph node metastasis | 9.089 | 0.003 | ||
| No | 21 | 0.63 | ||
| Yes | 23 | 3.76 | ||
| Her2 status | 6.210 | 0.012 | ||
| Negative | 28 | 1.33 | ||
| Positive | 16 | 2.94 | ||
| ER status | 3.830 | 0.039 | ||
| Negative | 24 | 2.86 | ||
| Positive | 20 | 1.15 | ||
| PR status | 3.590 | 0.041 | ||
| Negative | 25 | 3.27 | ||
| Positive | 19 | 2.14 | ||
| Pathological subtypes | 0.045 | 0.780 | ||
| Invasive ductal carcinoma | 29 | 2.75 | ||
| Others | 15 | 2.59 | ||
| Ki-67 mitotic index, (%) | 5.391 | 0.022 | ||
| Low (≤14) | 26 | 1.46 | ||
| High (>14) | 18 | 2.91 |
Table 3.
Correlation between miR-10b level and various clinicopathological features in breast cancer examined by multivariate linear regression analysis.
| Clincopathological features | Expected value | χ2 | F-value | P-value |
|---|---|---|---|---|
| Tumor diameter, cm | ||||
| >5 | 0.8771 | 13.0358 | 5.47 | 0.0076 |
| ≤5 | 0 | |||
| Histological grading | ||||
| III | 0.7952 | 11.1294 | 4.63 | 0.0120 |
| I+II | 0 | |||
| Clinical staging | ||||
| III | 1.3214 | 18.1880 | 7.61 | 0.0006 |
| I+II | 0 | |||
| Lymph node metastasis | ||||
| Yes | 2.3346 | 21.6665 | 9.12 | <0.0001 |
| No | 0 | |||
| Her2 status | ||||
| Positive | 0.5889 | 12.2036 | 5.01 | 0.0098 |
| Negative | 0 | |||
| ER status | ||||
| Positive | −0.6763 | 10.5124 | 4.25 | 0.0184 |
| Negative | 0 | |||
| PR status | ||||
| Positive | −0.4987 | 8.4728 | 3.56 | 0.0319 |
| Negative | 0 | |||
| Ki-67 mitotic index, (%) | ||||
| High (>14) | 0.5328 | 9.4751 | 3.98 | 0.0230 |
| Low (≤14) | 0 | |||
Discussion
E-cad, the prototype member of the classical cadherin family, is of paramount importance in maintaining cell polarity and epithelial integrity. Extensive studies have revealed that a reduction/loss of E-cad expression occurs concomitantly with the progression of breast cancer, from early-stage carcinoma to late-stage invasive carcinoma, and the reduced expression is significantly correlated with higher metastatic risk and worse prognosis [12,13]. Induced re-expression of E-cad, on the other hand, led to reduced motility and reversed the invasive phenotype of breast cancer cells [14,15], illustrating the tumor-suppressor potential of this gene. Several mechanisms at different levels of gene expression have been demonstrated for the loss of E-cad in neoplastic cells [16,17]. On the genetic level, loss of heterozygosity in the chromosome 16q region, where the human E-cad gene is located, was detected in 45–63% of sporadic breast carcinomas [18,19]. In addition, polymorphic mutations in the E-cad gene were associated with development of the lobular subtype of sporadic breast cancer [20,21]. On the epigenetic level, DNA hypermethylation [22,23] and up-regulation of the Snail/Slug zinc-finger transcriptional repressors of the E-cad gene [24] have both been shown to contribute to loss of E-cad expression in breast cancer cells. Other transcriptional repressors for E-cad have been identified, and include the basic helix-loop-helix transcription factors Twist and E12/E47 [25,26] and the zinc-finger E-box binding homeobox 1 and 2 (ZEB1 and ZEB2), the latter of which is controlled by microRNA (miR)-200 family members and miR-205, which have both been characterized as down-regulated in clinical breast cancer specimens lacking E-cad [27,28]. Post-translationally, cell-surface E-cad level is controlled by matrix metalloproteinase (MMP)-mediated proteolytic degradation, as well as by tyrosine phosphorylation-induced E3 ligase-targeted proteosomal degradation. Upon cleavage by MMPs, the extracellular domain of E-cad is released as an 80-kDa soluble fragment. This event not only reduces cell adhesion by depleting intact E-cad available to participate in adhesion complex formation, but also promotes cell proliferation, invasion and migration in a paracrine manner by activating ErbB receptor or up-regulating MMPs [29,30]. Receptor tyrosine kinases (RTKs), such as epidermal growth factor receptor (EGFR), hepatocyte growth factor receptor (HGFR) and fibroblast growth factor receptor (FGFR), are frequently found to be activated in breast cancer cells. These RTKs phosphorylate E-cad, as well as catenins, targeting these molecules for E3 ligase-mediated proteosomal degradation [31,32]. Collectively, tumor cells appear to exploit every possible way to silence E-cad, supporting the theory that loss of E-cad expression is a prerequisite for tumor cell invasion and further metastasis.
In this study, we identified a novel regulator for E-cad expression in tumor cells. The miR-10b controls the level of E-cad at the post-transcriptional level. By using the metastatic breast cancer cell line MDA-MB-231 as a model system, we were able to demonstrate that liposomal-mediated intracellular delivery of the miR-10b inhibitor reduces its endogenous level by approximately 50%. Moreover, this reduction is accompanied by a significant up-regulation of E-cad, at both the mRNA and protein levels. Meanwhile, ectopically expressing miR-10b produced dramatic reductions in E-cad mRNA (by >90%) and protein (by ~80%). In addition, the changes of E-cad expression in response to miR-10b inhibitor or mimics inducing overexpression were accompanied by functional alterations in cell invasion. These data suggest that miR-10b is capable of actively regulating E-cad expression in metastatic breast cancer cells, and that targeted silencing of miR-10b may serve to recover E-cad expression in breast tumors and suppress or eliminate the potential for tumor invasion/metastasis. In addition to the above-mentioned in vitro findings, we found that in clinical samples of breast cancer tissues there is a significant negative correlation between the levels of these 2 molecules. Thus, miR-10b appears to be a biologically important molecule for controlling E-cad expression in human breast cancer and may be amenable to pharmacologic or genetic intervention.
MiR-10b, together with miR-10a, constitutes the entire miR-10 family. MiR-10a and miR-10b genes are localized within the homeobox (Hox) clusters of genes on human chromosomes 17 (17p21) and 2 (2p31.1), respectively. As such, they are co-regulated with the Hox genes, and functionally target many Hox mRNAs [5]. Both genes have been detected as aberrantly expressed in a number of human tumors, including those of the brain, liver, colon and breast [5]. Ma et al. showed that miR-10b is specifically and highly expressed in metastatic breast cancer cells, as compared to its levels in normal mammary epithelial cells or non-metastatic breast cancer cells. Ectopic expression of miR-10b in non-metastatic breast tumors was demonstrated as sufficient to drive potent invasion in vitro and metastasis in vivo[10]. In our study, we compared the expression of miR-10b in 16 samples of benign breast lesions, 21 of primary breast cancers and 23 of metastatic breast cancers. Consistent with the findings from Ma et al., we found that miR-10b was approximately 4-fold higher in metastatic breast cancers than in benign lesions or primary breast cancers (P<0.01), while the levels in the latter 2 types of sample were not dramatically different from each other (P>0.05). More importantly, the higher level of miR-10b independently and significantly correlated with multiple clinicopathological features of advanced progression and metastatic diseases, including larger tumors, grade III tumors, stage III tumors, tumors with lymph node metastasis, tumors with positive Her-2 expression, and tumors showing higher mitotic activity. In contrast, the miR-10b level in breast cancer negatively correlated with ER and PR positivity, 2 key indicators of better prognosis, supporting the notion that miR-10b overexpression can be an effective indicator for breast cancers of advanced and metastatic status.
It has been demonstrated that the basic helix-loop-helix transcription factor Twist controls the expression of miR-10b in breast cancer cells [10]. Twist is also known to directly suppress the expression of E-cad in breast cancer [26]. Therefore, Twist may control E-cad expression, either directly by binding to the promoter region for transcriptional regulation, or indirectly by up-regulating miR-10b for post-transcriptional regulation. It is also possible that breast cancer increases miR-10 expression through genomic amplification, which in turn targets E-cad mRNA for degradation. These potential mechanisms further demonstrate the importance of down-regulated E-cad in maintaining the transformed and metastatic phenotypes of breast cancer.
Conclusions
In summary, we present here the first evidence that E-cad is a potential target for miR-10b and that miR-10b may modulate cancer metastasis through targeting E-cad. MiR-10b is not only necessary but also sufficient for regulating E-cad expression in breast cancer cells, and is biologically significant for suppressing E-cad in metastatic breast cancer. As an independent indicator for advanced and metastatic breast cancer, miR-10b may become a valuable target for cancer therapy.
Acknowledgement
We thank Medjaden Bioscience Limited for assisting in the preparation of this manuscript.
Footnotes
Conflict of interest
There are no any actual or potential conflicts of interest including any financial, personal or other relationships with other people or organizations.
Source of support: Departmental sources
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Rybarova S, Vecanova J, Hodorova I, et al. Association between polymorphisms of XRCC1, p53 and MDR1 genes, the expression of their protein products and prognostic significance in human breast cancer. Med Sci Monit. 2011;17(12):BR354–63. doi: 10.12659/MSM.882121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tvrdik D, Skalova H, Dundr P, et al. Apoptosis – associated genes and their role in predicting responses to neoadjuvant breast cancer treatment. Med Sci Monit. 2012;18(1):BR60–67. doi: 10.12659/MSM.882205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond SM. RNAi, microRNAs, and human disease. Cancer Chemother Pharmacol. 2006;58(Suppl 1):s63–68. doi: 10.1007/s00280-006-0318-2. [DOI] [PubMed] [Google Scholar]
- 5.Le Quesne J, Caldas C. Micro-RNAs and breast cancer. Mol Oncol. 2010;4(3):230–41. doi: 10.1016/j.molonc.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 7.Li M, Marin-Muller C, Bharadwaj U, et al. MicroRNAs: control and loss of control in human physiology and disease. World J Surg. 2009;33(4):667–84. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah AA, Leidinger P, Blin N, Meese E. miRNA: small molecules as potential novel biomarkers in cancer. Curr Med Chem. 2010;17(36):4427–32. doi: 10.2174/092986710794182980. [DOI] [PubMed] [Google Scholar]
- 9.Avril-Sassen S, Goldstein LD, Stingl J, et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genomics. 2009;10:548. doi: 10.1186/1471-2164-10-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–88. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Gould Rothberg BE, Bracken MB. E-cadherin immunohistochemical expression as a prognostic factor in infiltrating ductal carcinoma of the breast: a systematic review and meta-analysis. Breast Cancer Res Treat. 2006;100(2):139–48. doi: 10.1007/s10549-006-9248-2. [DOI] [PubMed] [Google Scholar]
- 13.Jeschke U, Mylonas I, Kuhn C, et al. Expression of E-cadherin in human ductal breast cancer carcinoma in situ, invasive carcinomas, their lymph node metastases, their distant metastases, carcinomas with recurrence and in recurrence. Anticancer Res. 2007;27(4A):1969–74. [PubMed] [Google Scholar]
- 14.Vleminckx K, Vakaet L, Jr, Mareel M, et al. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66(1):107–19. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 15.Sarrio D, Palacios J, Hergueta-Redondo M, et al. Functional characterization of E- and P-cadherin in invasive breast cancer cells. BMC Cancer. 2009;9:74. doi: 10.1186/1471-2407-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranwal S, Alahari SK. Molecular mechanisms controlling E-cadherin expression in breast cancer. Biochem Biophys Res Commun. 2009;384(1):6–11. doi: 10.1016/j.bbrc.2009.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strathdee G. Epigenetic versus genetic alterations in the inactivation of E-cadherin. Semin Cancer Biol. 2002;12(5):373–79. doi: 10.1016/s1044-579x(02)00057-3. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Tanigami A, Yamakawa K, et al. Allelotype of breast cancer: cumulative allele losses promote tumor progression in primary breast cancer. Cancer Res. 1990;50(22):7184–89. [PubMed] [Google Scholar]
- 19.Cleton-Jansen AM, Moerland EW, Kuipers-Dijkshoorn NJ, et al. At least two different regions are involved in allelic imbalance on chromosome arm 16q in breast cancer. Genes Chromosomes Cancer. 1994;9(2):101–7. doi: 10.1002/gcc.2870090205. [DOI] [PubMed] [Google Scholar]
- 20.Kanai Y, Oda T, Tsuda H, et al. Point mutation of the E-cadherin gene in invasive lobular carcinoma of the breast. Jpn J Cancer Res. 1994;85(10):1035–39. doi: 10.1111/j.1349-7006.1994.tb02902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berx G, Cleton-Jansen AM, Strumane K, et al. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996;13(9):1919–25. [PubMed] [Google Scholar]
- 22.Nass SJ, Herman JG, Gabrielson E, et al. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res. 2000;60(16):4346–48. [PubMed] [Google Scholar]
- 23.Cheng CW, Wu PE, Yu JC, et al. Mechanisms of inactivation of E-cadherin in breast carcinoma: modification of the two-hit hypothesis of tumor suppressor gene. Oncogene. 2001;20(29):3814–23. doi: 10.1038/sj.onc.1204505. [DOI] [PubMed] [Google Scholar]
- 24.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62(6):1613–18. [PubMed] [Google Scholar]
- 25.Perez-Moreno MA, Locascio A, Rodrigo I, et al. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276(29):27424–31. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- 26.Vesuna F, van Diest P, Chen JH, Raman V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun. 2008;367(2):235–41. doi: 10.1016/j.bbrc.2007.11.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 28.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–89. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noe V, Fingleton B, Jacobs K, et al. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J Cell Sci. 2001;114(Pt 1):111–18. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- 30.Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J Biol Chem. 2008;283(26):18393–401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita Y, Krause G, Scheffner M, et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol. 2002;4(3):222–31. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- 32.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4(2):118–32. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]