Summary
Background
MicroRNAs (miRNAs) are noncoding RNAs of 18–25 nucleotides that post-transcriptionally regulate gene expression and are involved in a wide range of physiological and pathological conditions. The β-adrenergic signaling pathway plays a fundamental role in regulation of heart function. The present study was designed to investigate the expression profile of miRNAs and functional implications under conditions of β-adrenoceptor activation or inhibition in rat heart.
Material/Methods
Hemodynamic parameters were measured to assess heart function in Wistar rats treated with isoproterenol (ISO) or propranolol (PRO). miRNA expression was analyzed by miRNA Microarray and confirmed by real-time quantitative reverse transcription PCR (real-time qRT-PCR).
Results
Isoproterenol (ISO, a β-adrenoceptor activator) and propranolol (PRO, a β-adrenoceptor inhibitor) induced differential miRNA expression profiles. Out of 349 miRNAs measured, 43 were upregulated and nine downregulated in the ISO group, while five miRNAs were upregulated and 28 downregulated in PRO group. Among these altered miRNAs in both PRO and ISO groups, 11 were cardiac abundant and 11 showed opposite profiles between the PRO and ISO groups. The recognized anti-hypertrophic miRNAs miR-1, miR-21 and miR-27b, and the pro-hypertrophic miRNAs miR-22, miR-24, miR-199a, miR-212 and miR-214, were upregulated in the ISO group. In the PRO group, pro-hypertrophic miRNA miR-30c was upregulated, whereas miR-212 was downregulated.
Conclusions
β-adrenoceptor intervention alters miRNA expression profile, and miRNAs may be involved in the β-adrenoceptor signaling pathway. Cardiomyocyte hypertrophy is a balanced process between pro-hypertrophic and anti-hypertrophic regulation and involves, at the very least, miRNA participation.
Keywords: β-adrenoceptor, miRNAs, antagonistic effect, hypertrophy
Background
β-adrenoceptor (β-AR) stimulation or inhibition is one of the main neurohumoral mechanisms which modulate cardiac function in physiological and pathophysiological states. Three β-AR subtypes – β1-AR, β2-AR, and β3-AR – have been identified in the heart. β1- and β2-ARs are the predominant β-ARs that regulate cardiac contractility, frequency, and rate of relaxation by stimulating the Gs-protein/adenylate cyclase/protein kinase A pathway [1,2]. β3-AR preferentially couples to inhibitory G proteins (Gi) and induces negative inotropic effects [3]. β3-AR stimulation is also related to release of nitric oxide via activation of the endothelial NO-synthase in the heart [3,4]. Stimulation of β-AR in cardiomyocytes activates adenyl cyclase (AC) that catalyzes cyclic adenosine-3′,5′ monophosphate (cAMP) formation, which in turn stimulates cAMP-dependent protein Kinase-A (PKA), resulting in phosphorylation of a series of regulatory proteins including sarcoendoplasmic reticulum Ca2+ transport ATPase (SERCA), transcription factors, and L-type Ca2+ channels. The β-ARs are the main regulator of cardiac hypertrophy, a potential predictor of heart disease [5]. Chronic stimulation or overexpression of β-AR induces cardiomyocyte hypertrophy, which can progress to heart failure [6,7].
MicroRNAs (miRNAs) are small, functional non-coding RNAs 18–25 nucleotides in length that post-transcriptionally regulate mRNA gene expression by acting on its 3′untranslated region (3′UTR). Accumulating evidence has demonstrated that miRNAs are involved in a variety of biological and pathological processes, including development, proliferation, differentiation, apoptosis, cell death, patterning of the nervous system, oncogenesis and drug resistance, cardiac remodeling and heart failure [8–15]. A unique biological property of miRNAs is that one miRNA may repress hundreds of genes, and one mRNA can receive regulation from one to several miRNAs. Thus, miRNAs provide fine-tuning of gene regulation. Altered miRNA expression has been implicated in cellular development [16], oncogenesis [17,18], and heart diseases [19–21]. Eva van Rooij et al reported that more than 12 miRNAs are up- or down-regulated in hypertrophied cardiac tissue in mice and a similar alteration is observed in failing human heart, implying the presence of similar miRNA-controlled hypertrophic mechanisms [20]. Tatsuguchi et al demonstrated that miR-21 and miR-18b are important regulators of cardiac hypertrophy in vivo and in vitro[22]. Notably, miRNAs are potentially involved in cardiac hypertrophy and play dual roles in regulation of cardiac hypertrophy. miR-1, miR-9, and miR-133 [15,23], termed anti-hypertrophic miRNAs, negatively regulate cardiac hypertrophy; in contrast, miR-23a, miR-195, miR-199a-5p, and miR-208a [15,20], termed pro-hypertrophic miRNAs, positively regulate cardiac hypertrophy. miRNAs are emerging as important regulators of cardiac hypertrophy. In view of miRNA involvement in modulating cellular biological functions, we hypothesized that changes of β-AR function may alter miRNA expression profiles. This, in turn, may have implications for the functional performance of the β-AR signaling pathway.
Material and Methods
Hemodynamic measurements
Adult male Wistar rats, weighing between 200–250 g, were studied. The rats were treated with either isoproterenol (ISO; 1.0 mg/kg subcutaneous injection every 12 h for 24 h), or propranolol (PRO; 10 mg/kg/day intragastric administration once a day for 7 consecutive days); the corresponding control group of animals received an equivalent volume of physiological saline. Cardiac function was assessed with hemodynamic parameters measured through a pressure volume control unit (Scisense Inc., London, Ontario, Canada). The rats were anaesthetized with 1% sodium phenobarbital (40–60 mg/kg, i.p.) and the degree of anaesthesia was adjusted to the level where rats just started responding to toe pinch. A pressure-sensing catheter (1.9F, Scisense Inc.) was inserted and advanced in a retrograde direction via the right common carotid artery into the left ventricle. After stabilization for 10 min, the signals were continuously recorded. Measured parameters included heart rate, left ventricular end-systolic pressure (LVESP, mmHg), maximum rate of increase of left ventricular pressure (+dP/dtmax, mmHg/s), and maximum rate of reduction of left ventricular pressure (−dP/dtmax, mmHg/s). After the hemodynamic measurements had been obtained, the rats were sacrificed, their hearts removed promptly, washed in ice-cold 0.9% saline, and were then frozen in liquid nitrogen for later use. Animal experiments involved in this study were approved in advance by the Animal Ethics Committee of Harbin Medical University.
MiRNA microarray
For microRNA array analysis, TRIzol reagent (Invitrogen, CA, USA) was used to extract total RNA from left ventricles of rat hearts from control, ISO, and PRO groups. Hearts were removed from sacrificed rats and RNA samples from three animals in each group were pooled and used for miRNA expression analysis using miRCURY™ Array. Total RNA of 5 μg was labeled by miRCURY™ Array Labelling kit (Exiqon, DK) and hybridized with miRCURY™ Array microarray (Exiqon, DK). The hybridization signal intensities were detected with a GenePix® 4000B microarray scanner (Molecular Devices, CA, USA), and were saved as TIF files. The images were analyzed by Genepix Pro 6.0 software (Axon Instruments, CA, USA), and the results saved in EXCEL files.
Real-time quantitative reverse transcription PCR
Total RNA was isolated from the left ventricle of experimental animals by using the mirVana miRNA Isolation Kit (Ambion Inc., Austin TX). The RNA sample was treated with RNase-free DNase prior to the reverse transcription step. Total RNA 0.5 μl was used for cDNA synthesis with MultiScribe™ Reverse Transcriptase (Applied Biosystem, Foster City, CA, USA) and microRNA-specific primers (Sangon Biotech, Shanghai, China). Real-time quantitative reverse transcription PCR (real-time qRT-PCR) was performed using special miRNA primers and SYBR® Green PCR Master Mix (Ambion Inc.), according to the manufacturer’s protocol. All experiments were conducted in duplicate. DNA was amplified in ABI 7500 fast system (Applied Biosystem, Foster City, CA, USA), using the same cycling parameters as follows: 95°C for 10 min, followed by 40 cycles of a three-stage temperature profile of 95°C for 15 sec and 60°C for 15 sec, then 72°C for 30 sec. All real-time qRT-PCR analyses were carried out using the 2 delta-delta Ct variant (2−ΔΔCT) method with the expression level of U6 as an internal control. A miRNA was considered to be undetectable when cycle threshold values of this miRNA exceeded 35 in the sample tissue.
Statistical analysis
All data are presented as mean ±SD. For relative miRNA expression, the mean value of the control group is defined as 1. Statistical comparisons were performed using the Student’s t-test, with a value of p<0.05 considered significant.
Results
Hemodynamic parameters are presented in Table 1. ISO resulted in significant increase in left ventricular end-systolic pressure (LVESP), maximum rate of increase of left ventricular pressure (+dP/dt), and maximum rate of reduction of left ventricular pressure (−dP/dt), indicating its positive inotropic action on contractility. In contrast, PRO produced negative inotropic effects on cardiac contractility, as indicated by decreases in LVESP and +dP/dt, but changes were not statistically significant. Heart rates (HR) were not significantly altered by ISO or PRO.
Table 1.
Haemodynamic parameters in control, propranolol, and isoproterenol groups.
| Group | HR (beat/min) | LVESP (mmHg) | +dP/dt (mmHg/s) | −dP/dt (mmHg/s) |
|---|---|---|---|---|
| Ctl (8) | 407±23 | 104.7±15.3 | 7127.7±1296.5 | 6420.9±1170.8 |
| PRO (8) | 403±29 | 99.6±15.9 | 6758.0±1610.9 | 6786.8±1642.7 |
| Ctl (9) | 405±24 | 94.6±13.8 | 7220.3±2299.8 | 5998.1±1622.0 |
| ISO (9) | 417±16 | 117.5±15.8* | 10801.9±1316.1* | 7743.1±1671.3* |
Ctl – control; HR – heart rate; LVESP – left ventricular end-systolic pressure; +dP/dt – maximum rate of increase of left ventricular pressure; −dP/dt – maximum rate of reduction of left ventricular pressure. Numbers in brackets indicate number of rats in each group.
P<0.05 vs. Ctl.
We isolated total RNA samples from control, ISO-, and PRO-treated rat hearts and performed miRNA microarray analysis using miRCURY™ Array microarray version 11.0, which included 349 mature rat miRNAs. We found that expression profiles were altered in both ISO- and PRO-treated rat hearts. Compared to the control group, 5 miRNAs were upregulated by >1.3-fold, and 28 downregulated by <30% in the PRO group; 43 miRNAs were upregulated by>1.3-fold and 9 were decreased by <30% in the ISO group (Table 2).
Table 2.
Expression of miRNAs in rat heart treated with PRO or ISO.
| Upregulated miRNAs in PRO group | Upregulated miRNAs in ISO group |
|---|---|
| miR-10a-5p | miR-1, miR-7a, miR-15b, miR-17 |
| miR-30c | miR-19a/b, miR-20a/b, miR-21, miR-22, |
| miR-100 | miR-24, miR-27b, miR-29a/b/c, |
| miR-181c | miR-30b-3p, miR-31, miR-34b/c, miR-92b, |
| miR-192 | miR-99b*, miR-130b, miR-142, miR-146b, |
| miR-184, miR-193, miR-199a-5p, miR-204*, | |
| miR-206, miR-212, miR-214, miR-219-5p, | |
| miR-221, miR-222, miR-223, miR-325-5p, | |
| miR-329, miR-333,miR-338*, miR-340-5p, | |
| miR-363*, miR-382*, miR-409-5p, miR-466c, | |
| miR-471, miR-483, miR-484, miR-532-5p | |
| Downregulated miRNAs in PRO group | Downregulated miRNAs in ISO group |
| miR-30b-3p, miR-34c*, miR-125b-3p | let-7b/c/d/d*/7e, miR-10a-5p, miR-30c-1*, |
| miR-129, miR-183*, miR-184 | miR-30c-2*, miR-185, miR-320, |
| miR-204*, miR-212, miR-290 | miR-331, miR-339-3p, miR-342-3p, |
| miR-291a, miR-300-5p, miR-326 | |
| miR-329, miR-345-3p, miR-378* | |
| miR-330*, miR-338*, miR-340-5p | |
| miR-344-3p, miR-381, miR-382 | |
| miR-382*, miR-434, miR-466b/c | |
| miR-483, miR-551b, miR-743b | |
| miR-877 |
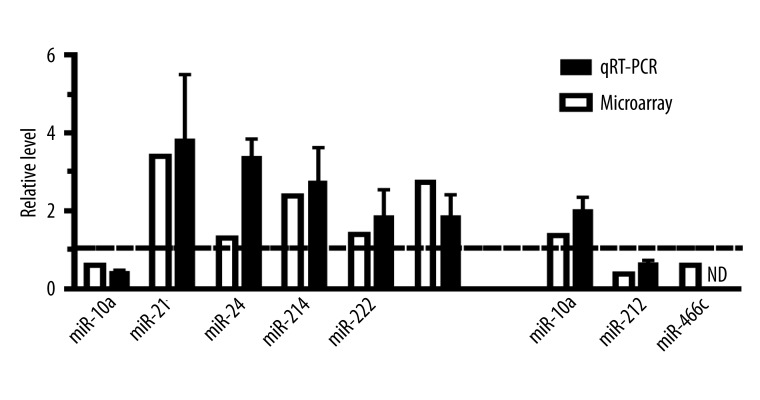
To confirm microarray results, we performed real-time qRT-PCR (qPCR) to quantify the expression of 10 miRMAs randomly chosen from the pool of miRNAs whose expression was significantly altered, as revealed by microarray analysis. As anticipated, the qPCR results were in good agreement with the microarray data; all 10 miRNAs assayed demonstrated the same pattern of changes as with the microarray methods (Figure 1).
Figure 1.
Confirmation of aberrantly expressed miRNAs analysis by quantitative real-time RT-PCR in ISO- or PRO-treated group. ND indicates nondetectable. n=3 independent RNA samples for each group.
Based on a previous report [24], the top 26 abundant miRNAs in rat heart are miR-1, miR-133a/b, miR-26a/b, miR-30a/b/c/d/e/a*/e*, miR-21, miR-24, miR-125a/b, miR-126, miR-126*, miR-23a/b, miR-16, miR-27a/b, miR-34a, miR-100, miR-29a/c, miR-99a, miR-191, miR-195, miR-212, miR-145, miR-22, miR-150, miR-320, miR-378, miR-494, and let-7a/b/c/d/e/f. In the present study, 11 of these cardiac-enriched miRNAs were found to have altered expression in the PRO and ISO groups, including miR-1, miR-21, miR-22, miR-24, miR-27b, miR-29a/c, miR-30c, miR-100, miR-212, miR-320, and let-7b/c/d/e. These were upregulated or downregulated in the PRO or ISO group. Out of these 11 miRNAs, 9 were observed in the ISO group; 2 were observed in the PRO group.
In the ISO group, 43 miRNAs were upregulated, while 9 were downregulated. In contrast, only 5 miRNAs were upregulated in the PRO group, whereas 28 miRNAs were downregulated.
In addition, 11 miRNAs were simultaneously altered in both ISO and PRO groups, out of which miR-10a was upregulated in the PRO group and downregulated in the ISO group. miR-30b-3p, miR-184, miR-204*, miR-212, miR-329, miR-338*, miR-340-5p, miR-382*, miR-466c, and miR-483 were downregulated in the PRO group, but were upregulated in the ISO group.
Discussion
miRNA expression is tissue-specific and plays a significant role in many physiological and pathological processes. In the current study, we demonstrated that either activation or inhibition of the β-AR pathway can alter miRNA expression profiles; β-AR activation primarily upregulates miRNAs, whereas β-AR inhibition downregulates them. Such opposite expression profiles indicate that miRNAs might be involved in opposing effects between β-AR activation and inhibition; for example, the positive inotropic effects of β-AR activation as opposed to the negative inotropic effects of β-AR inhibition.
miRNAs are transcribed from their genes by RNA polymerase II (Pol II) [25,26], which binds to promoter regions for transcription in the presence of transcription factors (TFs). Numerous studies have shown that endogenous miRNAs expression is regulated by TFs [27–30]. β-AR signaling mediates a variety of biological functions in the cardiovascular system, including heart rate [31], cardiac contractility [32], cardiomyocyte apoptosis [33], and hypertrophy [34]. Importantly, TFs are potentially involved in performance of β-AR biological functions; for instance, β-adrenergic stimulation induces cardiac ankyrin repeat protein expression through activation of protein kinase A (PKA) and Ca2+/Calmodulin-Dependent Protein Kinase (CaMK) [35]; cAMP-responsive element modulator (CREM) is important for contractile performance and response to β-AR stimulation in the heart [36]; microphthalmia transcription factor (MITF) is essential for b-AR-induced cardiac hypertrophy [37]; signal transducers and activators of transcription 3 (STAT3) regulates atrial natriuretic factor expression by β-AR stimulation in cardiomyocytes [38]; and serum response factor (SRF) induces miR-1 expression [39].
In the ISO group, 43 miRNAs were upregulated and 9 downregulated; in contrast, only 5 miRNAs were upregulated and 28 downregulated in the PRO group. These results indicate that β-AR stimulation mainly induces miRNAs upregulation, whereas β-AR inhibition results mainly in miRNAs downregulation. It is reasonable to speculate that ISO activates the β-AR signaling pathway, which results in activation of the corresponding TFs and other regulators, in turn inducing the corresponding miRNAs expression. In contrast, PRO induces negative regulation of miRNA expression, likely by inhibiting the β-AR signaling pathway. Interestingly, 11 miRNAs showed opposite expression trends between stimulation and suppression of β-AR signaling pathways in heart tissue. It is speculated that these opposite expressions of miRNAs participate in opposing effects between β-AR activation and inactivation in the heart.
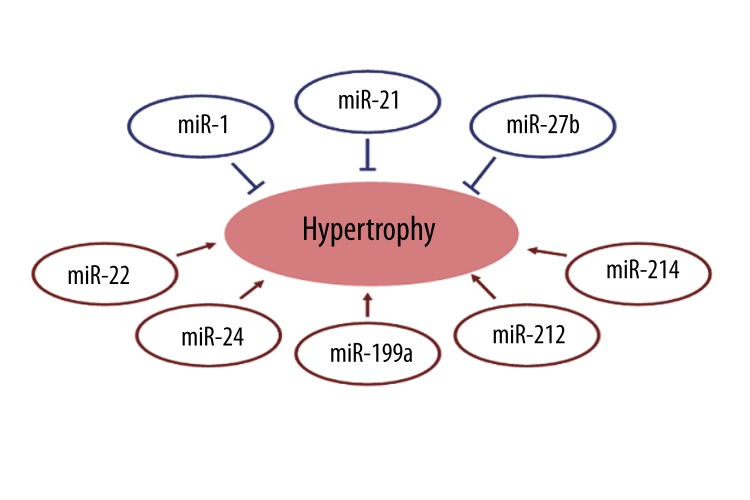
Cardiac hypertrophy is a complex pathological process involving a number of cellular pathways and regulators. Recent studies have demonstrated that miRNA is potentially involved in the process of cardiac hypertrophy and miRNA expressions display very consistent profiles in different models of cardiac hypertrophy. β-AR stimulation is a common pathway for induction of cardiac hypertrophy, which can be a predictor for progression to heart failure [40,41]. Recent studies have documented that the alterations in miRNA expression patterns occur in cardiac hypertrophy [20–24,42]. For example, miR-21 upregulation was observed in hypertrophy-induced aberrant miRNA expression in five studies [20–24,42], miR-214 upregulation in three studies [20,21,42], and miR-27b upregulation in two studies [20,21]. Similar results were also observed in our study. We observed that 18 altered miRNAs in the PRO and ISO groups have been implicated in different models of hypertrophy, including miR-1 [42, 43], miR-10a [42], miR-15b [42], miR-19b [22], 20b [22], miR-21 [20–22, 42], miR-24 [20,42], miR-27b [20,21,42], miR-29a/b/c [20,21,42], miR-30c [42], miR-31 [42], miR-184 [22], miR-185 [21,42], miR-199a-5p [20,42], miR-214 [20, 21, 42], miR-221 [22,42], miR-222 [22,42], and let-7b/c/d* [42]. Among these dysregulated miRNAs, miR-24 [20], miR-214 [20], miR-199a-5p [20], miR-30c [24], miR-22 [24] and miR-212 [24] have been shown to produce pro-hypertrophic effects, while miR-1 [15,42,43], miR-21 [21,22], and miR-27b [24] have anti-hypertrophic effects. Notably, ISO induced upregulation of the recognized anti-hypertrophic miRNAs miR-1, miR-21, and miR-27b, and also of the pro-hypertrophic miRNAs miR-22, miR-24, miR-199a-5p, miR-212, and miR-214; PRO induced upregulation of the pro-hypertrophic miRNA miR-30c and downregulation of the pro-hypertrophic miRNA miR-212. These results indicate that miRNAs may be involved in the process of cardiomyocyte hypertrophy, which results from perturbations in the dynamic and delicate balance between prohypertrophic and antihypertrophic regulators, including miRNAs (Figure 2). Upregulation of miR-199a-5p was also observed in β1AR- and β2AR-overexpressing mice hearts and rat neonatal cardiac myocytes treated with isoproterenol [44]. It plays a role in cardiomyocyte survival in hypoxic states by targeting hypoxia-inducible factor-1 alpha (Hif-1α) and Sirt1. This implies that miR-199a-5p is not only involved in cardiomyocyte survival, but also in cardiac hypertrophy. miRNAs are, at least in part, involved in β-adrenoceptor function.
Figure 2.
Expression profile of anti- and pro-hypertrophic miRNAs in animals treated with ISO. miRNAs in red circles indicate promotion of cardiomyocyte hypertrophy; miRNAs in blue circles indicate suppression hypertrophy.
Conclusions
miRNAs regulate a large number of genes by binding to the 3′UTR of their mRNAs, leading to degradation or translational repression of those targets, which serve a variety of biological functions. It can be speculated that the altered miRNAs induced by β-AR stimulation or inhibition participate in regulation of β-AR-mediated cardiac functions. With increasing numbers of miRNAs being identified, their roles in the β-AR signaling pathway remain to be elucidated. Our results indicate that miRNA may be an important component in the β-AR pathway in the rat heart, thereby providing a new target for modulating β-AR function.
Footnotes
Disclosures
Authors have no conflicts to disclose.
Source of support: This study was supported by the National Nature Science Foundation of China (30971252/C140401 and 81170219)
Reference
- 1.Wallukat G. The beta-adrenergic receptors. Herz. 2002;27:683–90. doi: 10.1007/s00059-002-2434-z. [DOI] [PubMed] [Google Scholar]
- 2.Zhong J, Hume JR, Keef KD. beta-Adrenergic receptor stimulation of L-type Ca2+ channels in rabbit portal vein myocytes involves both alphas and betagamma G protein subunits. J Physiol. 2001;531:105–15. doi: 10.1111/j.1469-7793.2001.0105j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gauthier C, Leblais V, Kobzik L, et al. The negative inotropic effect of beta3-adrenoceptor stimulation is mediated by activation of a nitric oxide synthase pathway in human ventricle. J Clin Invest. 1998;102:1377–84. doi: 10.1172/JCI2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brixius K, Bloch W, Pott C, et al. Mechanisms of beta 3-adrenoceptor-induced eNOS activation in right atrial and left ventricular human myocardium. Br J Pharmacol. 2004;143:1014–22. doi: 10.1038/sj.bjp.0705983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohse MJ, Engelhardt S, Eschenhagen T. What is the role of beta-adrenergic signaling in heart failure? Circ Res. 2003;93(10):896–906. doi: 10.1161/01.RES.0000102042.83024.CA. [DOI] [PubMed] [Google Scholar]
- 6.Osadchii OE. Cardiac hypertrophy induced by sustained beta-adrenoreceptor activation: pathophysiological aspects. Heart Fail Rev. 2007;12:66–86. doi: 10.1007/s10741-007-9007-4. [DOI] [PubMed] [Google Scholar]
- 7.Bisognano JD, Weinberger HD, Bohlmeyer TJ, et al. Myocardial-directed overexpression of the human β1-adrenergic receptor in transgenic mice. J Mol Cell Cardiol. 2000;32:817–30. doi: 10.1006/jmcc.2000.1123. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004:350–55. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Wang JX, Jiao JQ, Li Q, et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nature Medicine. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- 10.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci. 2006;103:7024–29. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, Wang T, Su Y, et al. Recombinant adenoviral microRNA-206 induces myogenesis in C2C12 cells. Med Sci Monit. 2011;17(12):BR364–71. doi: 10.12659/MSM.882122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Li WQ, Li YM, Tao BB, et al. Downregulation of ABCG2 expression in glioblastoma cancer stem cells with miRNA-328 may decrease their chemoresistance. Med Sci Monit. 2010;16(10):HY27–30. [PubMed] [Google Scholar]
- 14.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–76. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 15.Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res. 2008;103:1072–83. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–89. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Xie L, He X, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–22. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda S, Kong SW, Lu J, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–73. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 20.van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Ji R, Yue J, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–40. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatsuguchi M, Seok HY, Callis TE, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–41. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Long B, Zhou J, et al. miR-9 and NFATc3 regulate myocardin in cardiac hypertrophy. J Biol Chem. 2010;285(16):11903–12. doi: 10.1074/jbc.M109.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jentzsch C, Leierseder S, Loyer X, et al. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol. 2012;52(1):13–20. doi: 10.1016/j.yjmcc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 28.Xu H, He JH, Xiao ZD, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–42. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Lu M, Qiu C, Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38:D119–22. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vo N, Klein ME, Varlamova O, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci. 2005;102:16426–31. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohrer DK, Chruscinski A, Schauble EH, et al. Cardiovascular and metabolic alterations in mice lacking both beta1- and beta2-adrenergic receptors. J Biol Chem. 1999;274:16701–8. doi: 10.1074/jbc.274.24.16701. [DOI] [PubMed] [Google Scholar]
- 32.Devic E, Xiang Y, Gould D, Kobilka B. Beta-adrenergic receptor subtype-specific signaling in cardiac myocytes from beta(1) and beta(2) adrenoceptor knockout mice. Mol Pharmacol. 2001;60:577–83. [PubMed] [Google Scholar]
- 33.Communal C, Singh K, Pimentel DR, Colucci WS. Norepinephrine stimulates apoptosis in adult rat ventricular myocytes by activation of the beta-adrenergic pathway. Circulation. 1998;98:1329–34. doi: 10.1161/01.cir.98.13.1329. [DOI] [PubMed] [Google Scholar]
- 34.Adams JW, Brown JH. G-proteins in growth and apoptosis: lessons from the heart. Oncogene. 2001;20:1626–34. doi: 10.1038/sj.onc.1204275. [DOI] [PubMed] [Google Scholar]
- 35.Zolk O, Marx M, Jäckel E, et al. Beta-adrenergic stimulation induces cardiac ankyrin repeat protein expression: involvement of protein kinase A and calmodulin-dependent kinase. Cardiovasc Res. 2003;59:563–72. doi: 10.1016/s0008-6363(03)00476-0. [DOI] [PubMed] [Google Scholar]
- 36.Müller FU, Lewin G, Matus M, et al. Impaired cardiac contraction and relaxation and decreased expression of sarcoplasmic Ca2+-ATPase in mice lacking the CREM gene. FASEB J. 2003;17:103–5. doi: 10.1096/fj.02-0486fje. [DOI] [PubMed] [Google Scholar]
- 37.Tshori S, Gilon D, Beeri R, et al. Transcription factor MITF regulates cardiac growth and hypertrophy. J Clin Invest. 2006;116:2673–81. doi: 10.1172/JCI27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Feng W, Liao W, et al. The gp130/STAT3 signaling pathway mediates beta-adrenergic receptor-induced atrial natriuretic factor expression in cardiomyocytes. FEBS J. 2008;275:3590–97. doi: 10.1111/j.1742-4658.2008.06504.x. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y, Zhang Y, Shan H, et al. MicroRNA-1 downregulation by propranolol in a rat model of myocardial infarction: a new mechanism for ischaemic cardioprotection. Cardiovasc Res. 2009;84:434–41. doi: 10.1093/cvr/cvp232. [DOI] [PubMed] [Google Scholar]
- 40.Ho KK, Anderson KM, Kannel WB, et al. Survival after the onset of congestive heart failure in Framingham Heart Study subjects. Circulation. 1993;88:107–15. doi: 10.1161/01.cir.88.1.107. [DOI] [PubMed] [Google Scholar]
- 41.Levy D, Garrison RJ, Savage DD, et al. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322:1561–66. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 42.Sayed D, Hong C, Chen IY, et al. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–24. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 43.Ikeda S, He A, Kong SW, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193–204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rane S, He M, Sayed D, et al. An antagonism between the AKT and beta-adrenergic signaling pathways mediated through their reciprocal effects on miR-199a-5p. Cell Signal. 2010;22:1054–62. doi: 10.1016/j.cellsig.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]