Summary
Background
Polo-like kinase 1 (PLK1) is an important molecule in proliferation of many human cancers. The aim of study is to clarify the expression patterns and potential function of PLK1 in colorectal cancers.
Material/Methods
Fifty-six colorectal cancers samples were collected and arranged onto a tissue array and the expression of PLK1 were detected by immunohistochemistry and correlated with clinico-pathological characteristics and expression of PCNA. Expression of PLK1 in 9 colorectal cancer cells lines was investigated by RT-PCR and Western blot, then SW1116 cells lines were treated with PLK1 siRNA and the efficiency was examined by Western blot. Transwell test was applied to detect the migration and invasion capability of cancer cells by counting the number of cells passing through the membranes. Cell proliferation and apoptosis were examined by Cell Counting Kit-8 (CCK-8) and Annexin-V Kit.
Results
PLK1 was positively expressed in 73.2% (41/56) of colorectal cancers tissues, but in only 3.6% (2/56) of normal tissues, and was associated with Duke’s stage (P<0.01), tumor size (P<0.01), invasion extent (P<0.05) and lymphatic metastasis (P<0.01). The expression of PLK1 was correlated with expression of PCNA (R=0.553, P<0.01). PLK1 was inhibited in SW1116 cells by treating with PLK1 siRNA oligos, which resulted in a decreased number of cells passing through the membrane as compared with control groups (P<0.01) at 24 hours after transfection. Cell proliferation was inhibited from 48 hours after transfection, while cells apoptosis was induced from 72 hours after transfection.
Conclusions
PLK1 could be a progression marker for colorectal cancer patients and PLK1 depletion can inhibit migration and invasion capability of colorectal cancer cells SW1116, suggesting that PLK1 might be involved in metastasis and invasion of colorectal cancer. Therapeutic strategies targeting PLK1 may be a new approach to colorectal cancer.
Keywords: Polo-like kinase, PCNA, immunohistochemistry, colorectal cancer, migration, invasion
Background
Colorectal cancer is one of the most common gastrointestinal cancers in Western countries and it ranks third in morbidity and mortality of malignant cancer for both sexes [1]. In recent decades its morbidity and mortality have quickly increased in some Asian countries [2]. Although curative surgery is improving and adjuvant/neoadjuvant treatment strategies are now widely used [3], the 5-year survival is still not satisfactory [4]. Our aim in this study was to search for biological markers of colorectal cancer with high sensitivity and specificity, not only for the prediction of prognosis of patients, but also for the direction of individual treatment.
Cancers are widely considered to be genetic diseases. Abnormalities of cell proliferation and apoptosis resulting from chromosomal instability (CIN) are involved in tumorigenesis [5]. Moreover, the centrosome plays a key role in stabilizing the chromosomal [6]. In cancer cells, abnormal centrosomes, such as supernumerary centrosomes, are often observed [7]. We also know that dysregulated cell cycle control contributes to tumorigenesis. If some proteins are involved in both the formation of centrosomes and the cell cycle, the proteins must be very important factors in tumor development and may be potential oncogenes. The Polo-like kinase 1 (PLK1) is one of these proteins.
PLK1, mammalian homologue of Polo (Drosophila) and CDC5 (Saccharomyces cerevisiae), is a highly conservative serine/threonine kinase [8]. Evidence shows that PLK1 plays an important role in G2/M transition [9], bipolar spindle formation and centrosome maturation [10]. Knock-down of the expression of PLK1 in human cancer cells can lead to the failure of spindle assembly [11]. It has been reported that PLK1 is overexpressed in many human cancers, including non-small-cell lung cancer [12], head and neck cancer [13], esophageal carcinoma [14], breast cancer [15], endometrial carcinoma [16], and non-Hodgkin’s lymphomas [17]. It has been suggested that PLK1 might be used as a diagnostic biomarker [12–14]. Moreover, overexpression of Plk1 in NIH3T3 cells could lead to oncogenic focus formation and tumors in nude mice [18] and some studies have found that depletion of PLK1 can induce apoptosis and anti-proliferation effects in some cancers [19,20], suggesting that PLK1 could be a good target for cancer therapy.
Some papers also showed overexpression of PLK1 in colorectal cancer [21,22], but little is known about the other functions of PLK1 in progression of cancer, especially in colorectal cancer. In this study we analyzed the expression of PLK1 in colorectal cancer tissues, comparing it with the adjacent normal tissues; and we postulate that PLK1 could be a cell proliferation marker, just like proliferating cell nuclear antigen (PCNA) [23]. Finally, we knocked down the expression of PLK1 in colorectal cell line SW1116 by small RNA interference method to observe its effect on cell motility, proliferation and apoptosis.
Material and Methods
Patients and tissue samples
Fifty-six colorectal cancer samples and matched adjacent normal tissues from diagnosed patients were obtained by surgery between March 2005 and May 2008 at Ruijin Hospital, Shanghai Jiaotong University School of Medicine, China. The adjacent normal tissues were taken more than 5 centimeters away from the cancer tissues. Clinicopathological information regarding these 56 colorectal cancer cases were as follows: 34 males, 22 females; age from 32 to 89 years old, median age 67 years old; carcinoma locations were 13 in right-hemicolon, 3 in left-hemicolon, 14 in sigmoid colon, 26 in rectum; carcinoma differentiation was 5 well differentiated, 45 moderately differentiated, 6 poorly differentiated; and pathological TNM staging (American Joint Committee on Cancer [24]) was 13 in stage I, 10 in stage II, 29 in stage III, 4 in stage IV. All the samples were formalin-fixed and paraffin-embedded before performing tissue array and immunohistochemical analysis.
Immunohistochemistry
After defining the area of tumors, all the formalin-fixed and paraffin-embedded samples were cored and arranged on a tissue array block by Shanghai Outdo Biotech Co., Ltd; each core was 1.5 millimeter in diameter, and each section was 4 micrometer thick. The slides were deparaffinized in xylene and rehydrated in concentration gradient ethanols. For antigen retrieval, the slides were boiled in 0.01 mol/L sodium citrate buffer, pH 6.0 for 15 minutes, and then the endogenous peroxidase activity was blocked in 3% hydrogen peroxide for 30 minutes, followed by blocking non-specific binding in 1% BSA for 15 minutes. The slides were incubated with primary antibody diluted 1:50 in Tris-buffered saline pH 7.6 (TBS) with 1% BSA of mouse anti-human PLK1 (Santa Cruz, USA) for 2 hours at room temperature. Next, after washing in TBS, the slides were incubated with secondary biotinylated antibody and peroxidase-labeled streptavidin for 10 minutes. Finally, 3, 3′-diaminobenzidine was used as chromogenic agent for 20 minutes. Before being examined with light microscopy, the slides were counterstained with Mayer’s hematoxylin. Negative controls were incubated without primary antibody. Slides were prepared following the same protocol with mouse anti-human PCNA monoclonal antibody (Santa Cruz, USA).
Evaluation of immunohistochemistry
Staining intensity of the slides was detected independently by 2 pathologists who were blinded to our study. The intensity of staining was scored as follows: 0, negative; 1, weak; 2, moderate; 3, strong. The percentage of positive cells was scored as: 0, <5%; 1, 5–30%; 2; 30–75%; 3, >75%. The final score was calculated by multiplication of the 2 parameters (mean of the scores from the pathologists): −, 0–2; +, 3–5; ++, 6–7; +++, 8–9. The cases were grouped as PLK1 & PCNA negative (≤5) and PLK1 & PCNA positive (>5).
Cell culture
Human colorectal cancer cell lines SW1116, SW480, SW620, Colo205, DLD-1, and Lovo were cultured in RPMI 1640 (10% fetal bovine serum) and HCT116, HT29, and Caco-2 were cultured in DMEM (10% fetal bovine serum).
RNA extraction and PCR
The total cellular RNA of the 9 colorectal cancer cell lines were isolated from cells by TRIzol reagent (Invitrogen, USA) one-step method. AMV Reverse Transcriptase (AMV RT) system was obtained from Promega, USA. Primers were: PLK1 sense 5′-TGTTCGCGGGCAAGATTGT-3′ and antisense 5′-GGCTGCGGTGAATGGATATTTC-3′; GAPDH sense 5′-GGACCTGACCTGCCGTCTAG-3′ and antisense 5′-GTAGCCCAGGATGCCCTTGA-3′. The PCR cycling condition was 31 cycles, 95°C for 15s (denaturation), 55°C for 30s (annealing), 72°C for 30s (extension). The PCR products were detected in 2% agarose gel under ultraviolet radiation.
Real-time quantitative PCR
After transcription, 1 μl cDNA was mixed with SYBR Green PCR Master Mix (Applied Biosystems, USA) as the technical manual. PCR cycling condition was 40 cycles, 95°C for 15s (denaturation), 55°C for 30s (annealing), and 72°C for 30s (extension). The reactions were carried out in 96-well plates on ABI Prism 7000 (Applied Biosystems, USA). The results of PCR products were quantitatively confirmed by using melting curves and 2(−ΔCt) method; the expression amount of PLK1 relative to GAPDH was expressed as 100*2(−ΔCt); H2O was defined as negative control.
Silencing of PLK1 by small RNA interference
The siRNA oligos targeted PLK1 (NCBI Reference Sequence: NM_005030.3) and negative controls (NC, scrambled sequence) were designed by Shanghai GenePharma Co., Ltd; the sequences of the siRNA oligos are listed in Table 1. Before transfection, cells were cultured in 6-well plates overnight; each well contained 2×105 cells. When the abundance ratio of cells was up to about 60–70%, cells were transfected with siRNA oligos by using Lipofectamine 2000™ (Invitrogen, USA) according the manufacturer’s instructions. The time of changing medium (6 hours after adding siRNA oligos) is considered as 0 hour after transfection. Cytoplasm total protein was extracted at 48 and 72 hours after transfection, and the efficiency of the siRNA oligos was examined by Western blot and real-time PCR.
Table 1.
Sequences of small synthetic oligonucleotides unique to PLK1.
| Sense strand | Antisense strand | |
|---|---|---|
| siRNA1 | ACGGCAGCGUGCAGAUCAATT | UUGAUCUGCACGCUGCCGUTG |
| siRNA2 | CCAUUAACGAGCUGCUUAATT | UUAAGCAGCUCGUUAAUGGTT |
| siRNA3 | GGGUAUCAGCUCUGUGAUATT | UAUCACAGAGCUGAUACCCAA |
| Negative control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Western blot
Total protein lysates (50 μg) were separated on 10% SDS-PAGE gel and transferred onto PVDF membranes at 15 volt for 1 hour. After being blocked in 5% skim milk for 2 hours at room temperature, the membranes were incubated with primary antibody of PLK1 (diluted 1:200) and GAPDH (diluted 1:2000) for 2 hours at room temperature, followed by incubated with HRP-linked secondary antibody (Santa Cruz, USA) for 1 hour at room temperature. Finally, the bands were detected by using DAB reagent (Dako Corporation, Denmark).
Migration and invasion assay
For migration assay, 6.5mm Transwell® with 8 μm pore Polycarbonate Membrane Insert (Corning, USA) was applied. At 0 hours after transfection, 200 μl of serum-free cell suspension containing 3×104 PLK1-siRNA-transfected cells were added into the upper chamber and 600 μl of medium containing 20% fetal bovine serum was added into the lower chamber. After 24 hours of incubation, the cells were stained by methyl violet and scraped off the cells on the bottom of upper chamber. For invasion assay, the same steps were performed except that the bottom of the upper chamber was covered with 100 μl of diluted growth factor-reduced Matrigel (BD Corporation, USA). The result was determined by counting the stained cells passing through the pores in the membrane; data are expressed as means ±SD of counting 5 random fields of vision.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8, Dojindo, Japan) uses a type of highly water-soluble tetrazolium salt of WST-8. When WST-8 is reduced by dehydrogenases in cells it produces a yellow-colored product (formazan), so the amount of the formazan dye generated by the activity of dehydrogenases in cells is directly proportional to the number of living cells. The detection sensitivity of CCK-8 is higher than other tetrazolium salts such as MTT, XTT, MTS or WST-1, according to the CCK-8technical manual.
In this study, cells were cultured in 96-well plates with 100 μl medium/well, and transfected with PLK1 & NC siRNA oligos according the manufacturer’s instructions of Lipofectamine 2000™. At 24, 48, 60, 72, 96, and 120 hours after transfection, 10 μl CCK-8 was added. After incubation for 2 hours, absorbance was measured at 450nm by a Microplate reader (μQuant, Bio-Tek, USA). The PLK1-siRNA-treated group was compared with NC group and Mock group in the proliferation curves chart.
Apoptosis assay
Cell apoptosis was evaluated by using the Annexin V kit (BD Corporation, USA). At 24, 48, and 72 hours after transfection, cells were incubated with Annexin V antibody and PI according to the manufacturer’s instructions. Apoptosis ratio of the 3 groups (PLK1-siRNA, NC, and Mock) was detected by flow cytometry (FCM).
Statistical analysis
χ2 test or Fisher’s exact test was used to examine the difference in positive expressed rate of PLK1, PCNA between colorectal cancer tissues and normal tissues; correlation of PLK1 expression level and clinicopathological characteristics were examined by χ2 test or Fisher’s exact test and χ2 test for trends; Pearson correlation test was performed to examine the correlation between PLK1 expression and PCNA expression. One-way analysis of variance (ANOVA) was used to test the difference in apoptosis rate between the 3 groups and the different effect on cell proliferation between the 3 groups at every moment. P<0.05 was considered as statistically significant.
Results
Expression of PLK1 and PCNA in colorectal cancer tissues and adjacent normal tissues
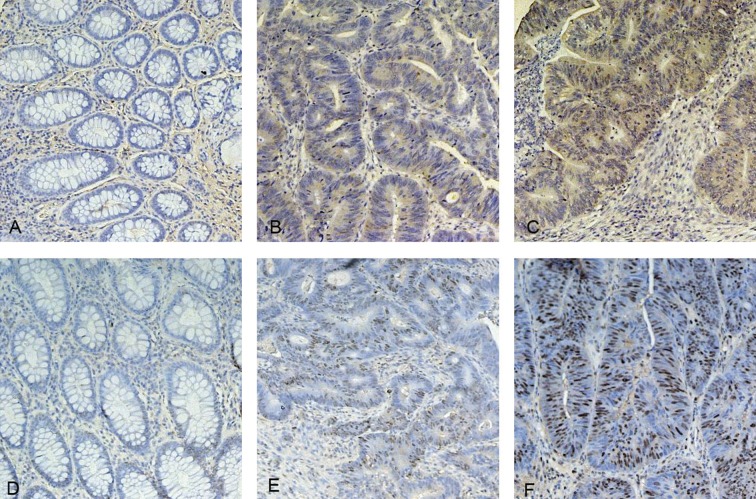
In colorectal cancer tissues, PLK1 was cytoplasmically stained and scored positively in 41 out of 56 cases (Table. 2, Figure 1B and C); PCNA was stained and scored positively in 49 out of 56 cases (Table 2, Figure 1E and F). In adjacent normal tissues, PLK1 and PCNA of almost all cases were stained negatively (Figure 1A and D), but 2 and 6 cases of PLK1 and PCNA group were stained positively. Positive expressions of PLK1 and PCNA in colorectal cancer tissues were significantly greater than in normal tissues – 73.2% (41/56) in cancer tissues vs. 3.6% (2/56) in normal tissues of PLK1 (P<0.01), 87.5% (49/56) vs. 10.7% (6/56) of PCNA (P<0.01, Table 2).
Table 2.
PLK1 and PCNA expression in colorectal tissues, n (%).
| Total | PLK1 positive | PLK1 negative | P-value | PCNA positive | PCNA negative | P-value | |
|---|---|---|---|---|---|---|---|
| Normal tissues | 56 | 2 (3.6) | 54 (96.4) | 0.000 | 6 (10.7) | 50 (89.3) | 0.000 |
| Cancer tissues | 56 | 41 (73.2) | 15 (26.8) | 49 (87.5) | 7 (12.5) |
Figure 1.
Expression of PLK1 and PCNA in colorectal tissues. (A) PLK1 negatively expressed in normal colorectal tissues; (B) PLK1 moderately expressed in colorectal cancer tissues; (C) PLK1 strongly expressed in colorectal cancer tissues; (D) PCNA negatively expressed in normal colorectal tissues; (E) PCNA moderately expressed in colorectal cancer tissues; (F) PCNA strongly expressed in colorectal cancer tissues. Original magnifications ×200.
Association between expression of PLK1 and clinicopathological characteristics of colorectal cancers simples
Statistically significant associations were not observed between PLK1 expression and sex, age, histological differentiation, tumor location and distant metastasis (Table 3). However, there was a statistically significant association with Duke’s stage (P<0.01), tumor size (P<0.01), invasion extent (P<0.05) and lymphatic metastasis (P<0.01).
Table 3.
Association between PLK1 expression and clinical characteristics of colorectal cancers, n (%).
| Characteristics | All cases | PLK1 positive | PLK1 negative | P-value | |
|---|---|---|---|---|---|
| All cases | 56 | 41 (73.2) | 15 (26.8) | ||
| Sex* | 0.122 | ||||
| Male | 34 (60.7) | 22 (64.7) | 12 (35.3) | ||
| Female | 22 (39.3) | 19 (86.4) | 3 (13.6) | ||
| Age | 0.643 | ||||
| ≤67 years old | 29 (51.8) | 22 (75.9) | 7 (24.1) | ||
| >67 years old | 27 (48.2) | 19 (70.4) | 8 (29.6) | ||
| Differentiation | 0.704 | ||||
| Well | 5 (8.9) | 3 (60.0) | 2 (40.0) | ||
| Moderate | 45 (80.4) | 34 (75.6) | 11 (24.4) | ||
| Poor | 6 (10.7) | 4 (66.7) | 2 (33.3) | ||
| Tumor location | 0.683 | ||||
| Right-hemicolon | 13 (23.2) | 11 (84.6) | 2 (15.4) | ||
| Left-hemicolon | 3 (5.4) | 2 (66.7) | 1 (33.3) | ||
| Sigmoid colon | 14 (25.0) | 9 (64.3) | 5 (35.7) | ||
| Rectum | 26 (46.4) | 19 (73.1) | 7 (26.9) | ||
| Duke’s stage2 | 0.001 | ||||
| A | 3 (5.4) | 1 (33.3) | 2 (66.7) | ||
| B | 22 (39.3) | 12 (54.5) | 10 (45.5) | ||
| C | 25 (44.6) | 22 (88.0) | 3 (12.0) | ||
| D | 6 (10.7) | 6 (100.0) | 0 (0.0) | ||
| Tumor size (cm2)** | 0.000 | ||||
| ≤10 | 2 (3.6) | 0 (0.0) | 2 (100.0) | ||
| >10, ≤12 | 27 (48.2) | 14 (51.9) | 13 (48.1) | ||
| >12, ≤25 | 21 (37.5) | 21 (100.0) | 0 (0.0) | ||
| >25 | 6 (10.7) | 6 (100.0) | 0 (0.0) | ||
| Tumor invasion** | 0.021 | ||||
| T1 | 3 (5.4) | 1 (33.3) | 2 (66.7) | ||
| T2 | 16 (28.6) | 10 (62.5) | 6 (37.5) | ||
| T3 | 32 (57.1) | 25 (78.1) | 7 (21.9) | ||
| T4 | 5 (8.9) | 5 (100.0) | 0 (0.0) | ||
| Lymphatic metastasis** | 0.001 | ||||
| N0 | 25 (44.6) | 13 (52.0) | 12 (48.0) | ||
| N1 | 21 (37.5) | 18 (85.7) | 3 (14.3) | ||
| N2 | 10 (17.9) | 10 (100.0) | 0 (0.0) | ||
| Distant metastasis* | 0.565 | ||||
| M0 | 52 (92.9) | 37 (71.2) | 15 (28.8) | ||
| M1 | 4 (7.1) | 4 (100.0) | 0 (0.0) |
Fisher’s exact test;
χ2 tests for trends.
Correlation between expression of PLK1 and PCNA in colorectal cancer
According to the immunohistochemistry results, 15/56 colorectal cancer cases showed +, 32/56 showed ++, and 9/56 showed +++ expression of PLK1. While 11/56 showed +, 31/56 showed ++, and 14/56 showed +++ expression of PCNA. There was a statistically significantly correlation between the expression of PLK1 and PCNA; correlation coefficient is 0.553 (P<0.01, Table. 4).
Table 4.
Correlation between expression of PLK1 and PCNA in colorectal cancer.
| PCNA | PLK1 | R | P-value | ||
|---|---|---|---|---|---|
| + | ++ | +++ | |||
| + | 6 | 5 | 0 | 0.553 | 0.000 |
| ++ | 9 | 20 | 2 | ||
| +++ | 0 | 7 | 7 | ||
Expression and inhibition of PLK1 in colorectal cancer cell lines
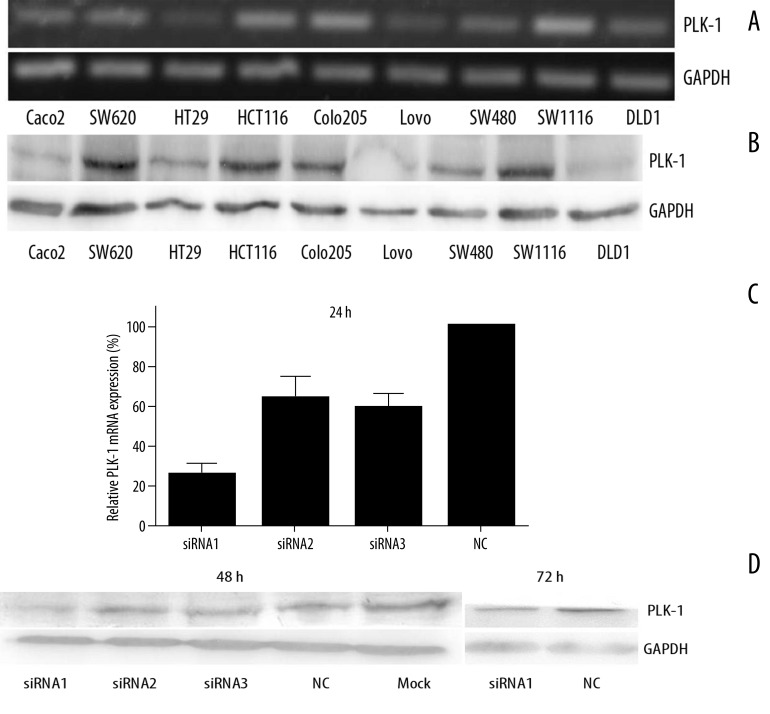
For researching the function of PLK1 in colorectal cancer cells, PLK1 mRNA level and protein level were detected in 9 colorectal cancer cell lines by PCR and Western blotting for 68 kDa (Figure 2A and 2B). As the photographs show, PLK1 mRNA and protein was expressed in all detected cell lines, and was much higher in SW1116. We then treated SW1116 with PLK1 siRNA oligos and the efficiency of the 3 oligonucleotides (siRNA1, siRNA2, siRNA3) were examined by real-time PCR at 24 hours and Western blotting at 48 and 72 hours after transfection (Figure 2C and 2D), compared with the NC group (scrambled siRNA-treated group) and the Mock group (Lipofectamine 2000™-treated group). The results suggest that the siRNA oligos could knock down PLK1 expression on both the mRNA level and the protein level. Besides, the siRNA1 was the most effectual, but the effect at 72 h was weaker than at 48 h.
Figure 2.
Expression and inhibition of PLK1 in colorectal cancer cells lines. (A) PLK1 mRNA expression detected in 9 colorectal cancer cell lines by PCR; (B) PLK1 protein expression detected in 9 colorectal cancer cell lines by Western blotting; (C) PLK1 mRNA expression detected in siRNA-treated SW1116 by real-time PCR at 24 hours after transfection. Values are mean ±SD of the expression amount of PLK1 relative to GAPDH; (D) PLK1 protein expression detected in siRNA-treated SW1116 by Western blotting at 48 hours and 72 hours after transfection.
Inhibition of migration and invasion of colorectal cancer cells by PLK1 siRNA
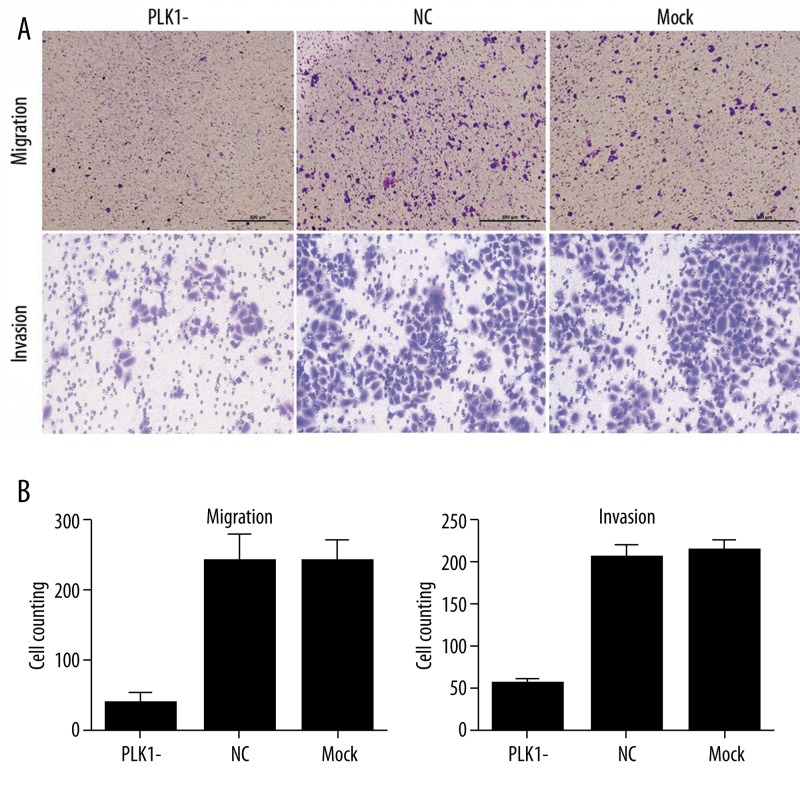
Our study showed that the PLK1 protein expression was correlated with Duke’s stage, tumor size, invasion extent and lymphatic metastasis, which suggests that PLK1 might be involved in advanced stage tumor progression and metastasis. We used the Transwell® assay to determine the effect of PLK1 on colorectal cancer cell migration and invasion (Figure 3A).The results showed: in migration assay, the numbers of cells passing through the membrane were 39±14 (mean ±SD) in the PLK1-siRNA-treated group, 242±41 in the NC group and 236±34 in the Mock group; in invasion assay, the results are 63±5 in the PLK1-siRNA-treated group, 203±11 in the NC group and 209±11 in the Mock group. Cells passing through the membrane in the PLK1-siRNA-treated group were significantly less than in the NC group and the Mock group (P<0.01, Figure 3B). It was demonstrated that down-expression of PLK1 led to inhibition of cell motility of SW1116.
Figure 3.
Effect of PLK1 depletion on migration and invasion of SW1116 cells. (A) 6.5 mm Transwell® with 8 μm pore Polycarbonate Membrane Insert were used for migration and invasion (with Matrigel) assay. The chambers were stained with methyl violet (×4 for migration, ×10 for invasion). (B) The migration and invasion cells were counted in 5 random fields of vision and the number of cells is expressed as mean ±SD for 3 replication experiments.
Influence of proliferation and apoptosis of colorectal cancer cells by PLK1 siRNA
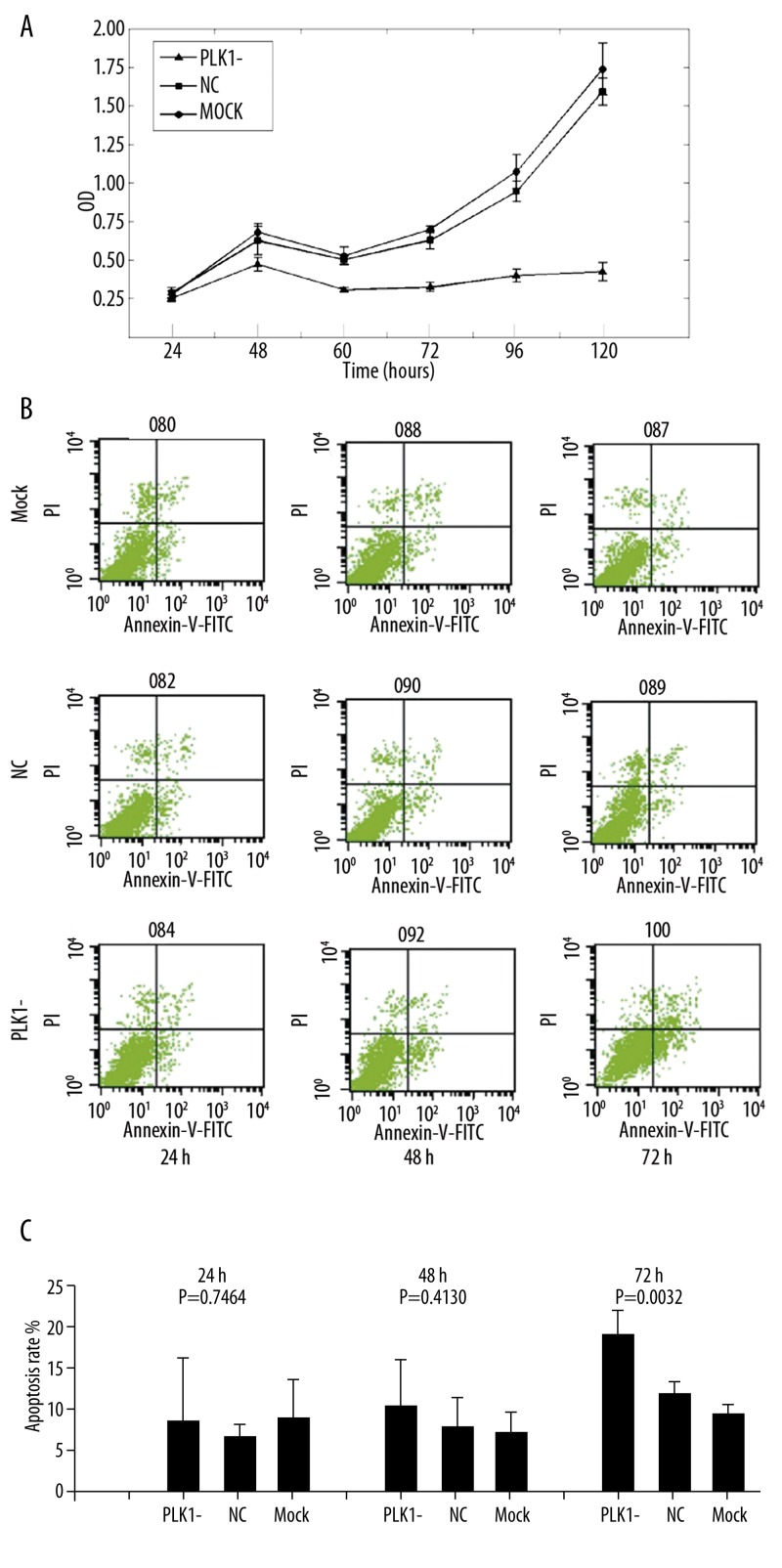
To investigate the relationship between the effect of PLK1 siRNA on cell growth & apoptosis and migration & invasion, CCK-8 was used to determine the proliferation of SW1116 at 24, 48, 60, 72, 96 and 120 hours after transfection (Figure 4A). As shown in the proliferation curves, there was no significant difference between the 3 groups at 24 hours, but PLK1-siRNA-treated cells proliferate more slowly than cells in the NC and Mock groups (P<0.01) at other moments. In the subsequent apoptosis assay, the apoptosis rate in the PLK1-siRNA-treated group, NC group and Mock group at 3 moments were (including early and late apoptosis): 8.84±7.20%, 6.45±1.78%, 9.35±3.86% at 24 hours; 10.39±4.80%, 6.64±4.49%, 6.08±2.76% at 48 hours; and 19.75±3.50%, 11.72±1.07%, 10.08±0.69% at 72 hours. There was no significant difference between the 3 groups at 24 and 48 hours, but increased apoptosis rate was observed in the PLK1-siRNA-treated group at 72 hours after transfection (P<0.01, Figure 4B and C). This demonstrates that down-expression of PLK1 led to suppression of cell proliferation in SW1116 from 48 h after transfection, but resulted to increase of apoptosis rate in SW1116 from 72h after transfection.
Figure 4.
Effect of PLK1 depletion on cells proliferation and apoptosis. (A) CCK-8 was used for determining the role of PLK1 in regulating SW1116 proliferation at different moments. Values are the mean ±SD of absorbance at 450 nm for 6 replication experiments. (B,C) FCM was used to determine the role of PLK1 in regulating apoptosis of SW1116 at different moments. Annexin V and PI stained cells were detected by FCM. Values are the mean ±SD of apoptosis rate (including early and late apoptosis) for the 3 replication experiments.
Discussion
PLK1 was first cloned and reported by Golsteyn in 1994 [25]. Subsequent studies have shown that PLK1 alone was the important protein activating the cdc2-cyclinB complex, which is also known as mitosis-promoting factor (MPF) [9,26]. In the transition of G2 to M phase, PLK1 activate the cdc25, which is a promoter of cdc2-cyclinB complex [27] and also plays a key role in bipolar spindle formation, centrosome maturation, and the separation of the chromosome [10]. Expression of PLK1 is clearly increased in G2/M phase transition [27], and the expression of PLK1 on both the mRNA and protein levels is positively correlated to cell proliferation activity [28].
We confirm that PLK1 is an indispensable protein in cell mitosis and proliferation. Depletion of PLK1 mRNA by small RNA interference can inhibit the mitosis of HeLa and U2OS cells [29]. Cancer is widely considered as a disease resulting from damage to the cell cycle regulation mechanism. Abnormality in initiation, running and termination of the tumor cell cycle can lead to significant increased degree of tumor proliferation; therefore, it is predictable that PLK1 is overexpressed in tumors. Ito et al [30] reported that PLK1 was more strongly expressed in tissues with high cell proliferative activity and many other studies have shown that PLK1 is overexpressed in major cancers on both the mRNA [12–14] and protein levels [15–17,31].
In the present study we detected the expression of PLK1 in colorectal cancer tissues by immunohistochemistry. Our results show that PLK1 was overexpressed in 73.2% (41/56) of cases of colorectal cancers, which is similar to the outcome of 66.7% (102/153) and 73.1% (57/78) reported by Wilko et al. [21] and Takao et al [22], respectively, from colorectal cancers. However, in normal tissues, PLK1 was positively expressed only in 3.6% (2/56) of cases in this study, so it is obvious that overexpressed PLK1 is not necessary for the normal colon tissues. Smith et al [18] reported that overexpression of Plk1 in NIH3T3 cells by plasmid transfection could lead to oncogenic focus formation, and the transfected cells could form tumors in nude mice, suggesting that overexpression of PLK1 might contribute to the progression of cancers and supports the role of PLK1 as an oncogene. Furthermore, chromosomal instability (CIN) is a cause of aneuploidy and loss of heterozygosity [5,32], which is one of the mechanisms of activating oncogenes and inactivating tumor suppressor genes. Dunican et al. [33] found that PLK1 was 1 of the 3 genes that were overexpressed in CIN colorectal cancer cell lines. All these results suggest that CIN results from defects in chromosomal segregation and telomere stability, and that the DNA damage response [32] might cause the mutations of the PLK1 gene and activate the oncogene. Subsequently activated PLK1 gene might lead to the overexpression of PLK1, which might result in inactivation of some tumor suppressor genes, and finally, promote the tumor development. In addition, this consecution explains why PLK1 is overexpressed in major human cancers is but negative expressed in normal adjacent colon mucous tissues, which also have a rapid proliferation rate, according to the findings of the present study.
PCNA is a cycle-associated protein, which combines with cyclinD and cyclin-dependent kinases (CDK), and its expression is highest in late G1 and S phases of the cell cycle. PCNA is a popular mark used to reflect the cell proliferation rate. In the present study, PCNA was overexpressed in colorectal cancer tissues and positively expressed in 87.5% (49/56) of cases. Statistical analysis demonstrated that the expression of PLK1 significantly correlated with the expression of PCNA, proving the correlation between expression of PLK1 and cell proliferation activity, as well as suggesting that PLK1 might be a candidate marker for colorectal cancer because proliferation activity is a crucial marker for cancer progression. Our results also revealed that the expression of PLK1was associated with tumor TNM stage, suggesting positive expression of PLK1 might predict a poor prognosis [34]. Wilko et al. [21] found that colorectal cancer patients with positive expression of PLK1 had a 5-year survival rate of 65%, compared to the 86% rate in PLK1-negative patients (P<0.01); in patients without distant metastasis, PLK1 expression had a significant impact on patient prognosis by multivariate survival analysis (RR=3.3, P=0.019). Zi-Li et al. [35] demonstrated that in hepatocellular carcinoma, 3- and 5-year survival rates of the PLK1-positive expressed group were significantly lower than in the negative group (50.3% and 67.8%, 42.3% and 20.9%), and similar results have been reported in studies of other cancers [12–14].
Expression of PLK1 was also correlated with tumor size, lymph node metastasis and depth of invasion in addition to TNM stage, consistent with the results from Takahashi et al. [22].These clinicopathological characteristics determine the extent of tumor development. Accordingly, we hypothesized that PLK1 might have additional functions and may be involved in the proliferation, migration and invasion of colorectal cancer cells, so our study design was in vitro. The results showed that PLK1 was expressed in all the colorectal cancer cells lines we could obtain, and was expressed much higher in SW1116. Then we effectively knocked down the expression of PLK1 by small RNA interference, the subsequent Transwell test revealed that the number of cells passing through the membranes in the siRNA-treated group was significantly lower than in the NC group and Mock group at 24 hours after transfection, while the cells’ growth and apoptosis were not influenced by the PLK1 siRNA at the moment. From 48 hours and 72 hours after transfection, the cells’ growth and apoptosis began to be inhibited and induced, respectively, by the PLK1 siRNA. The results suggest that the decreased capability in migration and invasion of SW1116 after PLK1 depletion was separate from the effect of PLK1 on growth and apoptosis, and also proved our hypothesis. However, there is little data about the role of PLK1 in migration and invasion of cancer cells. It is widely considered that polymerization of actin coupled with myosin generates the major forces for migration and microtubule also participates in the process [36]. In some studies [37,38] cdc5 (homologue of PLK1) was shown to mediate actin through RhoA to promote cytokinesis; PLK1 also could regular the dynamic change of microtubules [39]. Whether the regulation of actin and microtubules by PLK1 exists in colorectal cancer cells, and the precise mechanism of PLK1-mediated migration and invasion of cancer cells, remain to be elucidated.
Conclusions
We have demonstrated that PLK1 was overexpressed in colorectal cancer tissues and is correlated with Duke’s stage, tumor size, invasion extent and lymphatic metastasis. By using the siRNA method, we showed that knocking down the expression of PLK1 inhibited cell proliferation and motility of colorectal cancer cells. Therefore, PLK1 is likely to play an important role in the progression of colorectal cancer. In addition, the expression of PLK1 in colorectal cancer tissues was correlated with the expression of PCNA, which suggests its potential role as a biomarker of PLK1 for colorectal cancer diagnosis. Further research is needed to prove the value of PLK1 as a molecular target, such as the relationship between PLK1 expression and 5-year survival rate of patients, and interaction between PLK1 and other molecules.
Footnotes
Source of support: Natural Science Foundation of Shanghai, China, grant number: 08ZR1414000
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Sung JJ, Lau JY, Goh KL, Leung WK Asia Pacific Working Group on Colorectal Cancer. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6:871–76. doi: 10.1016/S1470-2045(05)70422-8. [DOI] [PubMed] [Google Scholar]
- 3.Boxberger F, Albrecht H, Konturek PC, et al. Neoadjuvant treatment with weekly high-dose 5-fluorouracil as a 24h-infusion, folinic acid and biweekly oxaliplatin in patients with primary resectable liver metastases of colorectal cancer: long-term results of a phase II trial. Med Sci Monit. 2010;16(2):CR49–55. [PubMed] [Google Scholar]
- 4.Gennari L, Russo A, Rossetti C. Colorectal cancer: what has changed in diagnosis and treatment over the last 50 years? Tumori. 2007;93:235–41. doi: 10.1177/030089160709300301. [DOI] [PubMed] [Google Scholar]
- 5.Michor F, Iwasa Y, Vogelstein B, et al. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol. 2005;15:43–49. doi: 10.1016/j.semcancer.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Dobles M, Liberal V, Scott ML, et al. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–45. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 7.Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- 8.Anger M, Kues WA, Klima J, et al. Cell cycle dependent expression of PLK1 in synchronized porcine fetal fibroblasts. Mol Reprod Dev. 2003;65:245–53. doi: 10.1002/mrd.10289. [DOI] [PubMed] [Google Scholar]
- 9.Roshak AK, Capper EA, Imburgia C, et al. The human polo-like kinase, PLK, regulates Cdc2/cyclin B through phosphorylation and activation of the cdc25C phosphatase. Cell Signal. 2000;12:405–11. doi: 10.1016/s0898-6568(00)00080-2. [DOI] [PubMed] [Google Scholar]
- 10.Dai W, Wang Q, Traganos F. Polo-like kinases and centrosome regulation. Oncogene. 2002;21:6195–200. doi: 10.1038/sj.onc.1205710. [DOI] [PubMed] [Google Scholar]
- 11.Spänkuch-Schmitt B, Bereiter-Hahn J, Kaufmann M, Strebhardt K. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst. 2002;94:1863–77. doi: 10.1093/jnci/94.24.1863. [DOI] [PubMed] [Google Scholar]
- 12.Wolf G, Elez R, Doermer A, et al. Prognostic significance of polo-like kinase (PLK) expression in non-small cell lung cancer. Oncogene. 1997;14:543–49. doi: 10.1038/sj.onc.1200862. [DOI] [PubMed] [Google Scholar]
- 13.Knecht R, Elez R, Oechler M, et al. Prognostic significance of polo-like kinase (PLK) expression in squamous cell carcinomas of the head and neck. Cancer Res. 1999;59:2794–97. [PubMed] [Google Scholar]
- 14.Tokumitsu Y, Mori M, Tanaka S, et al. Prognostic significance of polo-like kinase expression in esophageal carcinoma. Int J Oncol. 1999;15:687–92. doi: 10.3892/ijo.15.4.687. [DOI] [PubMed] [Google Scholar]
- 15.Spänkuch B, Heim S, Kurunci-Csacsko E, et al. Down-regulation of Polo-like kinase 1 elevates drug sensitivity of breast cancer cells in vitro and in vivo. Cancer Res. 2006;66:5836–46. doi: 10.1158/0008-5472.CAN-06-0343. [DOI] [PubMed] [Google Scholar]
- 16.Takai N, Miyazaki T, Fujisawa K, et al. Polo-like kinase (PLK) expression in endometrial carcinoma. Cancer Lett. 2001;169:41–49. doi: 10.1016/s0304-3835(01)00522-5. [DOI] [PubMed] [Google Scholar]
- 17.Mito K, Kashima K, Kikuchi H, Daa T, et al. Expression of Polo-Like Kinase (PLK1) in non-Hodgkin’s lymphomas. Leuk Lymphoma. 2005;46:225–31. doi: 10.1080/10428190400015709. [DOI] [PubMed] [Google Scholar]
- 18.Smith MR, Wilson ML, Hamanaka R, et al. Malignant transformation of mammalian cells initiated by constitutive expression of the polo-like kinase. Biochem Biophys Res Commun. 1997;234:397–405. doi: 10.1006/bbrc.1997.6633. [DOI] [PubMed] [Google Scholar]
- 19.Spänkuch B, Kurunci-Csacsko E, Kaufmann M, Strebhardt K. Rational combinations of siRNAs targeting Plk1 with breast cancer drugs. Oncogene. 2007;26:5793–807. doi: 10.1038/sj.onc.1210355. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Erikson RL. Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci. 2003;100:5789–94. doi: 10.1073/pnas.1031523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weichert W, Kristiansen G, Schmidt M, et al. Polo-like kinase 1 expression is a prognostic factor in human colon cancer. World J Gastroenterol. 2005;11:5644–50. doi: 10.3748/wjg.v11.i36.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi T, Sano B, Nagata T, et al. Polo-like kinase 1 (PLK1) is overexpressed in primary colorectal cancers. Cancer Sci. 2003;94:148–52. doi: 10.1111/j.1349-7006.2003.tb01411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods AL, Hall PA, Shepherd NA, et al. The assessment of proliferating cell nuclear antigen (PCNA) immunostaining in primary gastrointestinal lymphomas and its relationship to histological grade, S1G21M phase fraction (flow cytometric analysis) and prognosis. Histopathology. 1991;19:21–27. doi: 10.1111/j.1365-2559.1991.tb00890.x. [DOI] [PubMed] [Google Scholar]
- 24.Sobin LH, Wittekind C. TNM classification of malignant tumours. 5th ed. New York: Wiley-Liss Inc; 1997. pp. 66–69. [Google Scholar]
- 25.Golsteyn RM, Schultz SJ, Bartek J, et al. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J Cell Sci. 1994;107:1509–17. doi: 10.1242/jcs.107.6.1509. [DOI] [PubMed] [Google Scholar]
- 26.Chen XH, Lan B, Qu Y, et al. Inhibitory effect of Polo-like kinase 1 depletion on mitosis and apoptosis of gastric cancer cells. World J Gastroenterol. 2006;12:29–35. doi: 10.3748/wjg.v12.i1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyoshima-Morimoto F, Taniguchi E, Nishida E. Plk1 promotes nuclear translocation of human Cdc25C during prophase. EMBO Rep. 2002;3:341–48. doi: 10.1093/embo-reports/kvf069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan J, Hörlin A, Hock B, et al. Polo-like kinase, a novel marker for cellular proliferation. Am J Pathol. 1997;150:1165–72. [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Erikson RL. Activation of Cdc2/cyclin B and inhibition of centrosome amplification in cells depleted of PLK1 by siRNA. Proc Natl Acad Sci. 2002;99:8672–76. doi: 10.1073/pnas.132269599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito Y, Yoshida H, Matsuzuka F, et al. Polo-like kinase 1 (PLK1) expression is associated with cell proliferative activity and cdc2 expression in malignant lymphoma of the thyroid. Anticancer Res. 2004;24:259–63. [PubMed] [Google Scholar]
- 31.Jang YJ, Kim YS, Kim WH. Oncogenic effect of Polo-like kinase 1 expression in human gastric carcinomas. Int J Oncol. 2006;29:589–94. [PubMed] [Google Scholar]
- 32.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–72. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunican DS, McWilliam P, Tighe O, et al. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene. 2002;21:3253–57. doi: 10.1038/sj.onc.1205431. [DOI] [PubMed] [Google Scholar]
- 34.Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643–60. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- 35.He ZL, Zheng H, Lin H, et al. Overexpression of polo-like kinase1 predicts a poor prognosis in hepatocellular carcinoma patients. World J Gastroenterol. 2009;15:4177–82. doi: 10.3748/wjg.15.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5–14. doi: 10.1007/s10555-008-9166-3. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida S, Kono K, Lowery DM, et al. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 2006;313:108–11. doi: 10.1126/science.1126747. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Wang J, Jiao H, et al. Cytokinesis and cancer: Polo loves ROCK’n’ Rho(A) J Genet Genomics. 2010;37:159–72. doi: 10.1016/S1673-8527(09)60034-5. [DOI] [PubMed] [Google Scholar]
- 39.Dai W, Wang Q, Traganos F. Polo-like kinases and centrosome regulation. Oncogene. 2002;21:6195–200. doi: 10.1038/sj.onc.1205710. [DOI] [PubMed] [Google Scholar]