Summary
Background
MicroRNAs (miRNAs) are small non-coding nucleotides that regulate mRNA stability and protein expression by imperfect base pairing with the 3′-untranslated region (3′UTR) of target mRNAs. Many miRNAs have been documented to be aberrantly expressed in human cancers, but the role of miRNAs in endometrioid endometrial cancer (EEC) remains poorly understood. The objective of this study was to investigate the effect of miR-125b on EEC development and to explore its molecular mechanism in EEC carcinogenesis.
Material/Methods
Real-time quantitative PCR was applied to evaluate the expression level of miRNA-125b in EEC and normal endometrium (NE) samples. The invasion ability of miR-125b in EEC HEC1B cells was analyzed by Transwell assay after pre-miR-125b or anti-miR-125b transfection. For the invasion mechanism analysis of miR-125b on HEC1B cells, miRBase, TargetScan, miRanda and PicTar were used to predict the possible target gene of miR-125b. Luciferase activities assay, cotransfection and Western blot were used to reveal that the predicted target genes of miR-125b were direct and specific. RNA interference technology was used to confirm that the invasion inhibition of miR-125b was directly induced by ERBB2.
Results
Our study showed that miR-125b was down-regulated in human EEC specimens compared to that in NC specimens. Over-expression of miR-125b in HEC1B cells inhibited EEC invasion and this inhibitory effect on HEC1B cells could be restored by miR-125b knock down. Mechanism analysis revealed that ERBB2 was a direct and specific target of miR-125b. The inhibitory effect on EEC cell invasion was mediated by miR-125b inhibition of the translation of a proto-oncogene, ERBB2.
Conclusions
Aberrantly expressed miR-125b contributes to HEC1B cells invasion partly through directly down-regulating ERBB2 protein expression in EEC. This miRNA signature offers a novel potential therapeutic strategy for EEC.
Keywords: endometrial cancer, miR-125b, ERBB2, invasion
Background
Endometrial cancer is the most frequent malignant tumor of the female genital tract world-wide, and more than 80% of these cases are endometrioid endometrial cancer (EEC) [1]. The morbidity of EEC has been increasing and the main treatment strategy for EEC is surgery, radiation, chemotherapy or hormone therapy [2]. The majority of patients with advanced stage or recurrence have poor prognosis due to limitation of effective treatment [3]. Therefore, understanding the molecular mechanism of EEC carcinogenesis and metastasis may provide insights for developing novel therapeutic strategies.
Recently, microRNAs (miRNAs) have emerged as key regulators of gene expression stability, which give a new dimension to tumor research [4–6]. These small, non-protein-coding RNAs are molecules of −22 nucleotides transcribed from primary transcripts (pri-miRNAs) and precursor miRNA (pre-miRNA) cleaved by Dicer. The mature miRNAs incorporate into the RNA-induced silencing complex (RISC) that regulate gene expression post-transcriptionally through imperfect base pairing with the 3′-untranslated region (3′UTR) of target mRNAs, causing transcript degradation and translational inhibition [5]. Through this mechanism, miRNAs influence various cellular activities, including cell differentiation, proliferation and apoptosis under normal and disease conditions [7]. Collected data indicated that approximately 20–30% of human genes are regulated by miRNAs; around 580 miRNAs have been identified and more than 1000 predicted in humans [8]. Importantly, more than 50% of the human miRNA genes are located in chromosomal fragile sites, leading to DNA copy number abnormalities, epigenetic alterations, mutations, transcriptional deregulation and defective miRNA biogenesis pathways, which further contribute to miRNA dysregulation [9]. Since the 2002 discovery that miRNA dysregulation is associated with tumors, there have been many studies describing the relationship between miRNAs, cancer progression and metastasis [6,10]. Recent findings have provided compelling evidence that certain miRNAs can function as oncogenes or tumor suppressors. Genome-wide miRNA expression profiling showed that miR-125b was aberrantly expressed in human EEC [11,12]. However, the mechanism by which miR-125b might be involved in EEC progression is unknown.
ERBB2 encodes a member of the epidermal growth factor (EGF) receptor family of receptor tyrosine kinases, always binds tightly to ERBB3 and forms ERBB2-ERBB3 heterodimers, stabilizing ligand binding and enhancing kinase-mediated activation of downstream signalling pathways, such as those involving mitogen-activated protein kinase and phosphatidylinositol-3 kinase. For example, overexpressed ERBB2 is thought to be most critical for the aggressive growth and metastatic potential of human breast cancers, since it can recruit up to 6 individual p85 subunits and constitutively activate intracellular phosphatidylinositol 3-kinase, induce serine phosphorylation of AKT, and thereby drive cell survival, proliferation, motility, and invasion [13–15]. Furthermore, amplification and/or overexpression of ERBB2 had been reported in other numerous cancers, including head and neck cancer, gastric cancer and ovarian cancers [16–18]. However, until now, there has been no report about ERBB2’s role and regulation mechanism in EEC.
We examined miRNA-125b expressions in EEC and normal endometrium (NE) via real-time quantitative PCR analysis. The invasion ability of miR-125b and in EEC and mechanism were investigated by in vitro studies. We found that miR-125b were expressed at a lower level in EEC than in NE. Over-expression of miR-125b inhibited invasion of human EEC cells through regulating of ERBB2 expression at the translational level. Our novel findings suggest that aberrant expression of miR-125b is critical for the invasion of human EEC. Targeting miR-125b may aid to the development of novel therapeutic strategies against EEC.
Material and Methods
Patients and specimens
A total of 50 EEC and 30 NE specimens were surgically collected after informed consent from patients ages 46 to 59 years. Specimens from women with the same age group undergoing hysterectomy for nonmalignant conditions were collected for NE controls. Histological identification of EEC was confirmed according to World Health Organization criteria. All protocols were reviewed and approved by the Ethics Committee.
Quantitative real-time PCR
Total RNA was isolated using Trizol reagent according to the manufacturer’s instructions. Then cDNA or small RNA cDNA were reverse-transcribed. Quantitative mRNA or miRNA expression carried out using SYBR green obtained using the ABI 7500 system, and the 18<CT<30 were calculated with 2−ΔΔCT method. miRNA and mRNA expression were normalized by 18s and GAPDH, respectively. MiR-125b, 18s and GAPDH primers were selected from reference [19]. ERBB2 forward primer was 5′-AGCCGCGAGCACCCAAGT-3′, reverse primer was 5′-TTGGTGGGCAGGTAGGTGAGTT-3′ and the PCR product for ERBB2 was 147bp. The qPCR condition were 95°C 15 sec, 40 cycles of 95°C 5 sec, 60°C 34 sec, following by 95°C 15 sec, 60°C 1 min, 95°C 15 sec for getting the melt curve.
Cell culture
The human EEC cell line HEC1B was stored by our laboratory. The cells were cultured in DMEM medium supplemented with 10% fetal calf serum and incubated at 37° with 5% CO2.
Transfection
An appropriate concentration (about 80%) of resuspended HEC1B cells was seeded into a 24-well plate for transfection assay using Lipofectamine™ 2000 reagent according to the manufacturer’s instructions. Total 100 nM microRNA mimics or controls were used for the functional assays. Suitable amounts of ERBB2 dsRNA and the same procedures were adopted for ERBB2 knockdown using siERBB2 with Transmessenger regent following the manufacturer’s instructions.
Invasion assays
After a pretreatment with miRNA mimics or siRNA for 3 days, transwell assay was carried out by using Transwell chamber with pore size of 8.0 μm. Total 2×105 cells were resuspended in 200 μl serum-free medium and seeded in the upper compartment of the chamber. The lower compartment was loaded with 750 μl full culture medium containing 10% FBS. After being incubated at 37° for 12 hr, the chamber was fixed, hematoxylin-stained and counted.
Vector construction and Luciferase reporter assay
ERBB2 3′UTR luciferase reporter constructs were formed according to the reference [20]. In brief, EST clones containing the 3′UTR sequences from ERBB2 cDNA were used as templates for constructing the wild type and mutation type of vector. Sequencing was used to verify the constructs.
For the relative luciferase reporter assay, cells were seeded in a 24-well plate 1 day before. After 24 hr, 100ng ERBB2 3′UTR luciferase construct and 400 ng microRNA mimics were cotransfected using Lipofectamine™ 2000 reagent. Luciferase activity was quantitated 48 hr after cotransfection using Dual-Glo Luciferase assay. The β-galactosidase expression vector was cotransfected with each experiment for normalization of transfection efficiency. The luciferase activity was calculated by relative luciferase activity of firefly to Renilla.
Western blotting
Total protein was extracted using RAPI buffer. Constant 50 μg protein was loaded into SDS-PAGE 8–10% for electrophoresis, transferred onto PVDF membranes and hybridized with a primary antibody followed by a horseradish peroxidase-conjugated secondary antibody. After ECL reaction, gray assay was performed by ImageQuant 5.2 software. β-actin was used for the reference.
Statistical analysis
Statistical analysis was done by SPSS 13.0 software. Data were reported as means ± standard deviation. Differences were assessed by one-way analysis of variance (ANOVA) in nonparametric statistics and Student’s unpaired t-test. P<0.05 means a significant difference.
Results
Decreased expression of miR-125b in human EEC specimens
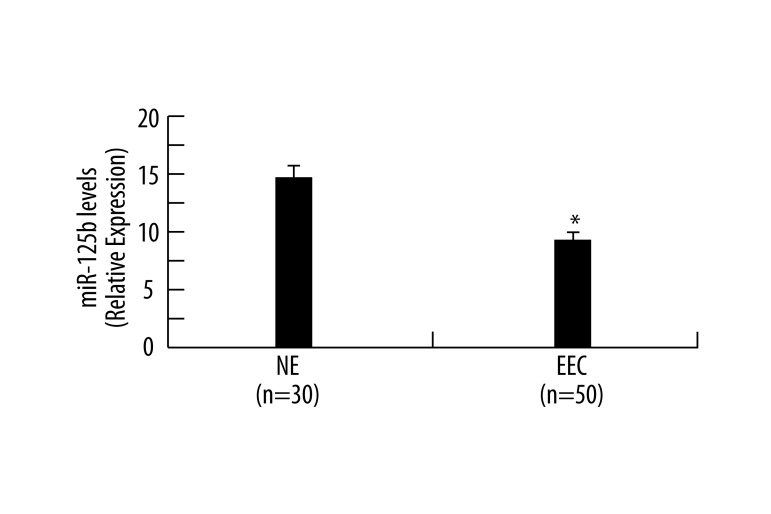
To explore the possible role of miR-125b in EEC development, we first examined the expression of miR-125b in 50 ECC samples and 30 NE samples by SYBR-Green stem-loop qRT-PCR. As shown in Figure 1, the expression level of miR-125b in EEC samples was much lower than that in NE samples, which provided initial evidence that miR-125b may play a role in the development of human EEC.
Figure 1.
The expression of miR-125b was decreased in EEC samples. Quantitative analysis of the expression levels of miR-125b normalized to those of 18s rRNA by qRT-PCR. Data were shown as RQ ±SD in NE (n=30) and EEC (n=50) samples. * p<0.05 vs. NE.
Overexpressing miR-125b suppressed EEC cell invasion
We next examined whether miR-125b affects EEC cell invasion using HEC1B cells. This cell line was chosen because HEC1B cells imitate EEC biology better than other cell lines, especially for cell invasion analysis [9]. As shown in Supporting Information Figure 1, qRT-PCR revealed that pre-miR-125b and anti-miR-125b were efficiently introduced into the cells and altered (up-regulated/down-regulated) miR-125b expression level. Transwell assay showed that the invasion ability of HEC1B cells was markedly reduced by overexpression of miR-125b (Figure 2A–C). To provide further evidence that miR-125b was indeed involved in EEC cell invasion, we studied the effect of inhibitor of miR-125b on the cell invasion. The invasion of the cells transfected with anti-miR-125b was increased compared with that of the cells transfected with control miR; the trans-membrane cells ranged from 18% and 30% (p<0.05) (Figure 2B, D). These results suggest that miR-125b indeed contributes to the invasive properties of EEC cells.
Figure 2.
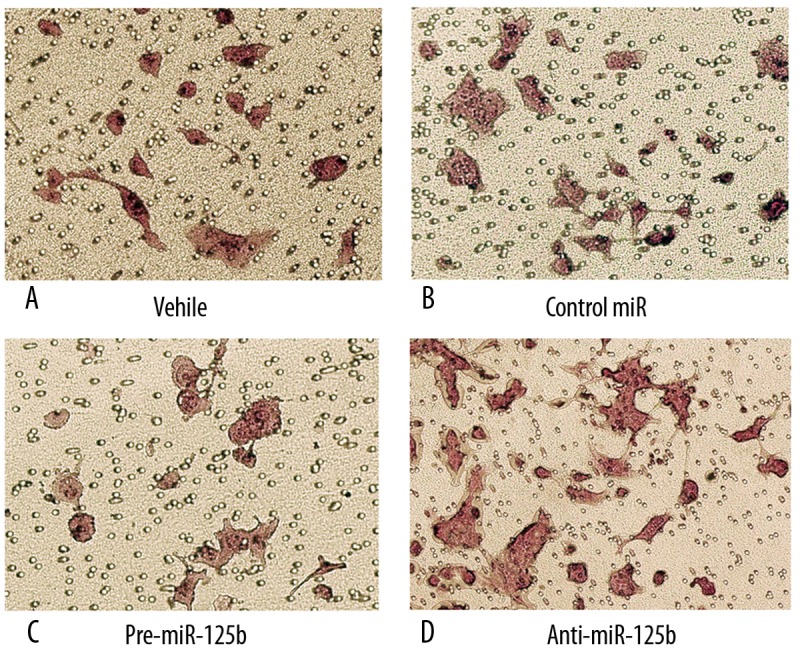
Effects of miR-125b on HEC1B cell invasion. (A–D) represent photographs of invasive cells on the membrane by transfection vehicle, control miR, pre-miR-125b and anti-miR-125b respectively, magnification, 400×. The membrane was formaldehyde fixed and hematoxylin-stained. Randomly 5 fields were selected to calculate the trans-membrane cells, which represented the invasion ability of HEC1B cells.
ERBB2 was a target for miR-125b in EEC
We then investigated the mechanisms by which miR-125b regulates the invasive phenotype. Online search for miR-125b targeting genes by miRBase, TargetScan, miRanda and PicTar revealed that ERBB2, a proto-oncogene associated with increased invasion, could be a potential target of miR-125b (Figure 3A, Table 1).
Figure 3.
ERBB2 is a validated target of miR-125b. (A) 3′UTR of ERBB2 is a target of miR-125b predicted by miRBase, TargetScan, miRanda and PicTar. (B) The reporter assay result with each bar representing values from three independent experiments. The transfection efficiency was normalized by co-transfected renilla luciferase and the light units were calculated by relative luciferase activity of firefly to renilla. * P<0.05. (C) Representative image of the protein level of ERBB2. GAPDH was used as a reference control. (D) quantitative analysis of the relative protein levels of ERBB2 normalized to those of GAPDH was shown. Data were mean ±SD of three independent experiments. * P<0.05.
Table 1.
Predicted microRNAs that targeting ERBB2*.
| microRNAs | miRBase analysis | Target Scan analysis | miR and a analysis | PicTar analysis | ||||
|---|---|---|---|---|---|---|---|---|
| Score | Energy | Context score | Aggregate PCT | Score | Pearson’s correlation | PicTar score | Probabilities | |
| has-miR-23b | 17.985 | −26.99 | ||||||
| has-miR-23a | 17.8792 | −26.64 | ||||||
| has-miR-185 | 17.3502 | −27.89 | ||||||
| has-miR-612 | 17.2231 | −34.49 | ||||||
| has-miR-593 | 17.1937 | −20.28 | ||||||
| has-miR-770-5p | 17.1637 | −20.26 | ||||||
| has-miR-922 | 17.0584 | −20.99 | ||||||
| has-miR-502-5p | 16.7981 | −21.99 | ||||||
| has-let-7b | 16.6096 | −21.77 | ||||||
| has-miR-657 | 16.5319 | −24.35 | −0.11 | – | ||||
| has-miR-337-3p | 16.5038 | −20.58 | ||||||
| has-miR-331-3p | 16.3728 | −21.06 | −0.34, −0.20 | – | ||||
| has-let-7a | 16.3728 | −16.00 | ||||||
| has-miR-129-5p | 16.3728 | −22.03 | ||||||
| has-miR-516a-3p | 16.3214 | −22.76 | ||||||
| has-miR-378 | 16.2665 | −23.91 | ||||||
| has-let-7f-1 | 16.1865 | −22.83 | ||||||
| has-miR-422a | 16.1865 | −23.67 | −21.20 | −0.46 | ||||
| has-miR-125a-5p | 16.1436 | −16.37 | −0.25 | 0.136 | ||||
| has-miR-888 | 16.0807 | −19.08 | ||||||
| has-miR-499-5p | 16.0539 | −18.55 | ||||||
| has-miR-181b | 16.0054 | −23.88 | ||||||
| has-miR-628-5p | 15.9749 | −13.82 | ||||||
| has-miR-516b | 15.9749 | −17.31 | ||||||
| has-miR-498 | 15.9001 | −18.78 | ||||||
| has-miR-485-3p | 15.8691 | −19.8 | ||||||
| has-miR-660 | 15.8691 | −18.00 | −0.11 | N/A | −14.20 | 0.15 | ||
| has-miR-125b | 15.8691 | −16.35 | −0.26 | 0.136 | −16.50 | −0.42 | ||
| has-miR-611 | 15.7948 | −25.66 | ||||||
| has-miR-623 | 15.7948 | −25.32 | ||||||
| has-miR-614 | 15.5842 | −26.73 | ||||||
| has-miR-630 | 15.4459 | −20.01 | ||||||
| has-miR-891a | 15.3401 | −17.95 | ||||||
| has-miR-617 | 15.3401 | −19.32 | ||||||
| has-miR-125a-5p | 15.305 | −20.75 | ||||||
| has-miR-611 | 15.163 | −25.21 | ||||||
| has-miR-410 | 15.097 | −15.24 | ||||||
| has-miR-770-5p | 14.9524 | −16.80 | ||||||
The microRNAs that targeting ERBB2 were predicted using PicTar, miRanda, TargetScan, and miRBase. MicroRNAs that predicted by at least 3 of the 4 methods were indicated with bold font.
To ascertain direct miRNA-target interaction, we set up a luciferase reporter assay. The 3′-UTR of ERBB2, including miR-125b target sites, was cloned into the downstream of pGL3-promotor vector, along with corresponding point mutations within the miRNA seed of these predicted sites. The HEC1B cells were cotransfected with the reporter vector and pre-miR-125b. As shown in Figure 3B, the luciferase activity in HEC1B cells was decreased with WT construct by enhanced miR-125b level, which could be partly restored with mutant constructs. These results suggest that 3′UTR of ERBB2 is a direct target of miR-125b.
We next examined whether miR-125b could regulate endogenous ERBB2 expression. The result showed that compared with controls, endogenous ERBB2 mRNA level was not significantly changed in HEC1B cells (data not shown), whereas the ERBB2 protein level was significantly suppressed with miR-125b enhancement (Fig. 3c and 3d). Considering these findings together, we can conclude that miR-125b represses endogenous ERBB2 expression through a translational pathway.
ERBB2 expression mediates the effect of miR-125b on EEC cell invasion
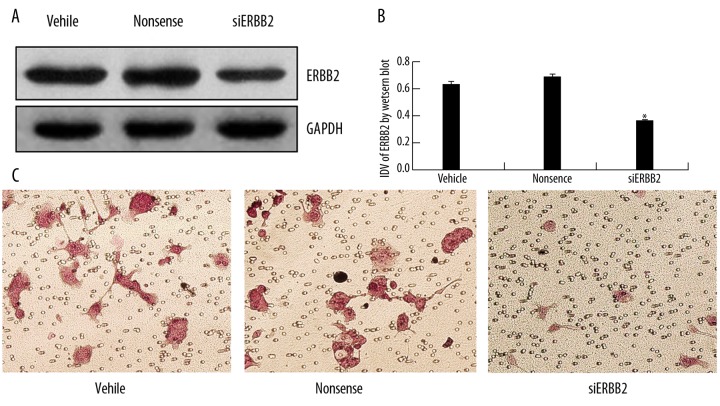
Since miR-125b was involved in invasion of EEC cells and ERBB2 was a target of miR-125b, we next tested the physiological role of miR-125b-target ERBB2 in EEC cells after RNA interference ERBB2. As shown in Figure 4, after siERBB2 treatment, protein expression of ERBB2 was significantly reduced and the number of invasion cells was also sharply reduced (11%, p<0.001). Combined with our substantial evidence that miR-125b inversely regulates ERBB2 expression, we can be certain that miR-125b-controlled EEC cells invasion is mediated to a large extent through ERBB2 down-regulation.
Figure 4.
miR-125b inhibits HEC1B cell invasion by down-regulating ERBB2. (A) Western blot showing ERBB2 knockdown by siERBB2 in HEC1B cells. GAPDH was used as a reference control. (B) quantitative analysis of the relative protein levels of ERBB2 normalized to those of GAPDH was shown. Data were mean ±SD of three independent experiments. * P<0.05. (C) Transwell assay was performed on HEC1B transfected with siERBB2 and controls.
Discussion
MicroRNAs are known to regulate the expression of genes involved in the control of development, proliferation, apoptosis and stress response. Recent studies showed a direct link between miRNAs and human cancers. Several studies have shown that down-regulation of miR-125b gene expression in breast cancer and ovarian cancer is followed by enhanced tumor cell invasion and poor prognosis [19,21]. Also, miR-125b participates in regulating the initiation and progression of osteosarcoma and prostate cancer [22,23]. In this study we found that miR-125b was also down-regulated in EEC; however, its role in EEC tumorigenesis is not entirely clear. Aberrant miRNA expression can regulate critical biological processes, including cell proliferation and invasion, which may promote EEC development and lead to poor prognosis [24,25]. Here, we examined the role of miR-125b in cell invasion in HEC1B cells by knockdown or over-expression of miR-125b, and found that overexpression of miR-125b could suppress the EEC cell invasion ability, while anti-miR-125b could restore this effect. Therefore, our results indicate that miR-125b is critical for the development of human EEC.
Since miRNA exerts its influence by targeting different genes, such as let-7 and Ras, miR-16 and Bcl-2 and miR-17-5p and E2F1 [26–28], we speculate that miR-125b can regulate a single gene to modify the regulation network and trigger cell invasion in EEC. Computational algorithms are effective tools to predict and validate the miRNA gene targets [29]. Through analysis using miRBase, TargetScan, miRanda and PicTar, a number of important candidate targets for miR-125b were predicted. Among these potential targets, ERBB2 is the most intriguing, as ERBB2 has been shown to modulate microRNA activity by binding to the microRNA targeting sequences on the 3′UTR of the targeted mRNAs [30,31]. Also, previous studies demonstrated that ERBB2 is essential for the anchorage-dependent growth potential and even the motility and invasive capabilities in human breast cancer cell lines [20]. ERBB2, encoded as a member of the epidermal growth factor (EGF) receptor family of receptor tyrosine kinases, is associated with increased invasion as a proto-oncogene [32,33]. Our results obtained from gain-of-function and loss-of-function approaches confirmed that ERBB2 is a direct target of miR-125b. First, over-expression of miR-125b significantly reduces the activity of a luciferase reporter containing the 3′UTR sequence of ERBB2. Second, mutation at the miR-125b target site in the 3′UTR of ERBB2 could significantly decrease the miR-125b regulation effect. Third, over-expression of miR-125b down-regulates ERBB2 protein expression post-transcriptionally. Collectively, our results suggest that ERBB2 is a key and direct miR-125b target gene.
In addition, our studies have found that down-regulation of ERBB2 by RNA interference could inhibit metastasis in EEC, which could be restored by over-expression of miR-125b. All these results indicate that miR-125b acts as an endogenous siRNA for ERBB2, and cell invasion induced by ERBB2 can be decreased by the over-expression of miR-125b. Thus, the identification of ERBB2 as a miR-125b target gene provides a possible explanation as to why the over-expression of miR-125b can function as a tumor suppressor in EEC. However, further research is needed to gain a full understanding of the underlying molecular mechanism.
Conclusions
MiR-125b, down-regulated in EEC, functions as a tumor suppressor partly through targeting the ERBB2 gene. This finding not only helps us understand the molecular mechanism of carcinogenesis, but also gives us a strong rationale to further investigate miR-125b as a potential treatment target for EEC.
Footnotes
Source of support: This study was supported by Macao Polytechnic Institute Fund (RP/ESS-03/2009) and National Natural Science Foundation of China (81172408, 30700980)
References
- 1.Farooq MU, Chang HT. Intracranial and scalp metastasis of endometrial carcinoma. Med Sci Monit. 2008;14(9):CS87–88. [PubMed] [Google Scholar]
- 2.Dewdney SB, Mutch DG. Evidence-based review of the utility of radiation therapy in the treatment of endometrial cancer. Womens Health (Lond Engl) 2010;6(5):695–703. doi: 10.2217/whe.10.49. quiz 704. [DOI] [PubMed] [Google Scholar]
- 3.Temkin SM, Fleming G. Current treatment of metastatic endometrial cancer. Cancer Control. 2009;16(1):38–45. doi: 10.1177/107327480901600106. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134(9):1635–41. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–29. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4(9):1179–84. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 8.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 9.Chung TK, Lau TS, Cheung TH, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130(5):1036–45. doi: 10.1002/ijc.26060. [DOI] [PubMed] [Google Scholar]
- 10.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 11.Hsu SD, Chu CH, Tsou AP, et al. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36(Database issue):D165–69. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Q, Luo X, Toloubeydokhti T, et al. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13(11):797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 13.Marone R, Hess D, Dankort D, et al. Memo mediates ErbB2-driven cell motility. Nat Cell Biol. 2004;6(6):515–22. doi: 10.1038/ncb1134. [DOI] [PubMed] [Google Scholar]
- 14.Seton-Rogers SE, Lu Y, Hines LM, et al. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci USA. 2004;101(5):1257–62. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holbro T, Beerli RR, Maurer F, et al. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA. 2003;100(15):8933–38. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao VH, Kandel A, Lynch D, et al. A positive feedback loop between HER2 and ADAM12 in human head and neck cancer cells increases migration and invasion. Oncogene. 2011 doi: 10.1038/onc.2011.460. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao W, Fu HJ, Xie QS, et al. HER2 Interacts With CD44 to Up-regulate CXCR4 via Epigenetic Silencing of microRNA-139 in Gastric Cancer Cells. Gastroenterology. 2011;141(6):2076–87. doi: 10.1053/j.gastro.2011.08.050. [DOI] [PubMed] [Google Scholar]
- 18.Wang KF, Cui H, Feng J, et al. Effect of reduced expression of Her2 by RNA interference on the biological characters of ovarian carcinoma cells. Zhonghua Fu Chan Ke Za Zhi. 2008;43(8):622–25. [PubMed] [Google Scholar]
- 19.Guan Y, Yao H, Zheng Z, et al. MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer. 2011;128(10):2274–83. doi: 10.1002/ijc.25575. [DOI] [PubMed] [Google Scholar]
- 20.Scott GK, Goga A, Bhaumik D, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282(2):1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 21.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219(2):214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 22.Liu LH, Li H, Li JP, et al. miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem Biophys Res Commun. 2011;416(1–2):31–38. doi: 10.1016/j.bbrc.2011.10.117. [DOI] [PubMed] [Google Scholar]
- 23.Shi XB, Xue L, Ma AH, et al. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate. 2011;71(5):538–49. doi: 10.1002/pros.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 25.Pan Q, Chegini N. MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin Reprod Med. 2008;26(6):479–93. doi: 10.1055/s-0028-1096128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–49. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 28.O’Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–44. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adachi R, Horiuchi S, Sakurazawa Y, et al. ErbB2 down-regulates microRNA-205 in breast cancer. Biochem Biophys Res Commun. 2011;411(4):804–8. doi: 10.1016/j.bbrc.2011.07.033. [DOI] [PubMed] [Google Scholar]
- 31.Epis MR, Giles KM, Barker A, et al. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem. 2009;284(37):24696–704. doi: 10.1074/jbc.M109.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu SR, Cheng TS, Chen WC, et al. Matriptase is involved in ErbB-2-induced prostate cancer cell invasion. Am J Pathol. 2010;177(6):3145–58. doi: 10.2353/ajpath.2010.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treekitkarnmongkol W, Suthiphongchai T. High expression of ErbB2 contributes to cholangiocarcinoma cell invasion and proliferation through AKT/p70S6K. World J Gastroenterol. 2010;16(32):4047–54. doi: 10.3748/wjg.v16.i32.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]