Abstract
Hybridoma HB-8696 produces monoclonal antibody (mAb) 520C9 (mouse IgG1), which recognizes breast cancer oncoprotein c-erbB2. The objective of this study was to optimize the medium recipe of HB 8696 cell for production of mAb 520C9. The optimization consisted of two steps: (1) screening of significant nutrients to make subsequent experiments more efficient with less runs and (2) locating their optimal concentrations. 29 variables including essential and non-essential amino acids, glucose, serum and 6 salts, namely NaCl, KCl, CaCl2, NaH2PO4, MgSO4 and Na-pyruvate were chosen in screening phase. The Plackett–Burman method was used to screen the variables influencing mAb production. Seven factors namely glucose, serum, asparagine, threonine, serine, NaCl and NaH2PO4 were identified to have a positive influencing role on mAb production with a confidence level >90 % (p < 0.1). Finally, Response surface methodology revealed the optimal level of the variables. The mAb production and average specific mAb production rate were enhanced by 111.05 and 105 %, respectively, compared to control medium.
Keywords: Monoclonal antibody, 520C9, Hybridoma cell HB 8696, Nutrient screening, Central composite design
Introduction
Monoclonal antibodies (mAb) are extensively used in medical research, therapeutics and diagnostics (Waldmann 2003; Jorgensen et al. 2007). They are also used in the study of cell surface antigens, affinity purification of proteins, histocompatibility testing, and radio immunoassays. The in vitro hybridoma culture method is a suitable option for producing monoclonal antibody in the culture supernatant (Marx and Merz 1995; Falkenberg et al. 1995). Hybridoma HB-8696 produces monoclonal antibody 520C9 (mouse IgG1), which recognizes the breast cancer oncoprotein c-erbB2 that is expressed in 30 % of breast cancer cases (Slamon et al. 1989; Slamon et al. 2001; Shi et al. 1991). Monoclonal antibody 520C9 has been extensively employed in developing bispecific antibody MDX-210 and MDX-H210 to detect c-erbB2 expression and breast cancer treatment where c-erbB2 is expressed at high level (Repp et al. 2003; Repp et al. 1995; Press et al. 2005). Recently, mAb 520C9 has been used in developing trifunctional antibody Ertumaxomab that eliminates tumor cell lines expressing c-erbB2 at low level. Ertumaxomab thus provides therapeutic option for beast cancer patients who are not eligible for Trastuzumab treatment (Jager et al. 2009).
The increased demand for the monoclonal antibody has led to continuous effort in developing and improving hybridoma cell culture that entails optimizing the nutrient medium in order to support cell growth, reduce cell death, and enhance mAb production (Kelley 2009; Butler 2005). The amino acids provided in a medium have been a focus because they are primary source of nitrogen and thus are involved in biosynthesis of structural proteins and enzymes. They can also serve as an energy source. Supplemental feeding of amino acids has proven effective for improving cell viability (Duval et al. 1991) and protecting cells from apoptosis-induced death and nutrient deprivation (Ducommun et al. 2001). Other reports indicate their important role in improving cell growth (Luan et al. 1987) and specific mAb production rate (Ducommun et al. 2001). In general, a limitation of extracellular amino acids affects the intracellular amino acid pool, which consequently influences the whole metabolic network for cell growth and mAb production (Seewoster and Lehmann 1995).
Previously several researchers focused on glucose and glutamine feeding to develop a strategy for acquiring improved cell performance (Luan et al. 1987; Glacken et al. 1986). In several cases, glutamine supplementation did not stimulate cell proliferation (Geaugey et al. 1989) or display a moderate growth supporting effect (Franek 1995). Therefore, several researchers carried out total or partial amino acid supplementation in order to replenish depleted nutrients (Luan et al. 1987; Reuveny et al. 1985; Feder and Tolbert 1985), but that approach could result in the inefficient use of expensive nutrients (Glacken et al. 1986). This indicates that amino acid requirements are cell line specific (McCarty 1962) and in several studies researchers determined the utilization pattern of amino acids and accordingly restored the consumed amino acids (Jo et al. 1990; Duval et al. 1991; Martial-Gros et al. 2001). But sometimes that approach has proven to be unsuccessful for boosting cell growth (Duval et al. 1991) and improving the specific mAb production rate (Hiller et al. 1994) because all amino acids have an individual influence and their coordinated interplay effects cell growth, longevity, and mAb production.
Therefore, in the present study, individual amino acids responsible in enhancing the final secreted mAb were first identified in a rational approach for medium enrichment. Besides glucose is the energy source and supplementation with glucose supports viable cells in extended stationary phase (Ljunggren and Haggstrom 1994; Miller et al. 1989). The specific growth rate of hybridoma cell was found to be controlled by the level of certain serum components (Dalili and Ollis 1989). Serum supports cell growth and phosphate salt is essential in nucleotide formation (deZengotita et al. 2000; Lee et al. 1989; Martens et al. 1992). Besides several researchers have found that an increase of medium osmolarity enhances the specific mAb production rate, but the final mAb titer was not improved because cell growth was suppressed by hyperosmolar stress condition (Oh et al. 1993; Ozturk and Palsson 1991). Considering all the above factors of the nutrients on cell growth and mAb production, twenty-nine variables including all essential and non-essential amino acids, glucose, serum, and six salts (NaCl, KCl, CaCl2, NaH2PO4, MgSO4 and Na-pyruvate) were selected in the initial screening phase to improve the final mAb titer. In the first step, the Plackett–Burman method was used to screen the variables that have a positive influencing role on mAb production. In the next step, concentration range near optimum level of the identified variables were selected by individual batch experiments, beyond that level they have inhibitory effect on cell growth and mAb production. Finally, using the Response surface methodology revealed the optimal levels of the variables that maximize the mAb concentration produced by the hybridoma cell culture.
Materials and methods
Cell line and cell maintenance
The mouse–mouse hybridoma cell line HB 8696 used in this study secretes IgG1 monoclonal antibody which recognizes the human c-erbB-2 oncoprotein, specifically expressed in 30 % of human breast cancer cases. The cell line was procured from ATCC (Designations: 520C9.C3B10T). The basal medium used for cell maintenance was Hybricare-46X (ATCC). This was supplemented with 20 % (v/v) heat inactivated fetal bovine serum (Biological industries), penicillin–streptomycin (100 IU/ml). The hybridoma cell was cultured and maintained at pH 7.2 and temperature 37 °C in a 5 % CO2 atmosphere and 95 % relative humidity of CO2 incubator. The cells were grown in T-flasks and passaged every 48 h using an inoculum size of approximately 1.2 × 105 cells/ml.
Medium
Hybricare-46X medium (ATCC) was used for control experiments and additional nutrients were added in the same medium for other experiments. The composition of the control medium is given in Table 1. All additional nutrients, twenty-one amino acids, glucose and six salts are cell culture grade and procured from Himedia (Mumbai, India).
Table 1.
Medium composition of control experiment
| Variable name | Concentration (g/l) | Variable name | Concentration (g/l) |
|---|---|---|---|
| Amino acids | Vitamins | ||
| Glutamine | 0.615 | Vitamin A Alcohol | 0.00001744 |
| Lysine | 0.102 | Vitamin D-2 | 0.00001744 |
| Arginine | 0.064 | Menadion | 0.00001744 |
| Isoleucine | 0.076 | Para-amino-benzoic acid | 0.00000872 |
| Leucine | 0.076 | Ascorbic acid | 0.00348 |
| Threonine | 0.064 | Vitamin B-12 | 0.000696 |
| Valine | 0.064 | Biotin | 0.000001744 |
| Tyrosine | 0.038 | Choline chloride | 0.00287104 |
| Glycine | 0.027 | Folic acid | 0.00278576 |
| Cysteine | 0.034 | i-Inositol | 0.0050199 |
| Histidine | 0.031 | Nicotinic acid | 0.00000438 |
| Phenylalanine | 0.047 | Niacinamide | 0.00278838 |
| Serine | 0.037 | d-calcium pantothenate | 0.00278576 |
| Alanine | 0.009 | Pyridoxal-HCl | 0.00278838 |
| Asparagine | 0.010 | Pyridoxine-HCl | 0.000004384 |
| Aspartic acid | 0.010 | Riboflavin | 0.00028016 |
| Glutamic acid | 0.010 | Thiamine-HCl | 0.00000174 |
| Methionine | 0.022 | Alpha-tocopherol phosphate | 0.00000174 |
| Proline | 0.013 | Others | |
| Tryptophan | 0.013 | Insulin zinc bovine | 0.005568 |
| Ornithine | 0.001 | d-glucuronolactone | 0.00012528 |
| Sugar | 5-Methylcytosine | 0.00000696 | |
| Glucose | 0.760 | Oxalic acid | 0.091872 |
| Inorganic salts | Phenol red | 0.011832 | |
| NaCl | 4.893 | HEPES | 1.658568 |
| CaCl2 | 0.153 | NAD DPN | 0.0004872 |
| KCl | 0.254 | FAD disodium hydrate | 0.0000696 |
| NaH2PO4 | 0.097 | NADP TPN | 0.0000696 |
| MgSO4 | 0.076 | Uridine 5 triphosphate | 0.0000696 |
| Na-pyruvate | 0.046 | 2-Deoxycytidine-HCl | 0.000696 |
| Na-glucuronate-H2O | 0.00012528 | 2-Deoxyguanosine | 0.000696 |
| Ferric nitrate-9H2O | 0.0000696 | 2-Deoxyadenosine | 0.000696 |
| Na-acetate-3H2O | 0.00348 | Thymidine | 0.000696 |
| Glutathione Reduced | 0.000696 | ||
| Taurine | 0.0002912 | ||
| Cocarboxylase | 0.0000696 | ||
| Tween 80 | 0.00087 | ||
| Coenzyme A | 0.000174 | ||
Cell culture
Batch cultures were carried out in triplicate in 6-well plates under the condition already mentioned in methodology. Each well contained 4 ml medium and was inoculated with an approximate cell density of 1.2x105/ml from the same batch. Samples were collected finally. Cell density was determined and cell free culture supernatants were stored at −20 °C for mAb analysis. In control experiments cells were cultured in Hybricare medium (Table 1) with 20 % fetal bovine serum.
Analysis of samples
Cell density was measured in a Neubauer hemocytometer under phase contrast microscope with Trypan blue dye-exclusion method. The cell free culture supernatant was obtained by centrifugation (12,000 rpm for 10 min). Glucose was measured by Glucose kit (Biolab Diagnostics, Mumbai, India). Lactic acid was measured by HPLC (Agilent, Santa Clara, CA, USA) using HP-Aminex-87H column (Biorad, Hercules, CA, USA) with column temperature at 60 °C and mobile phase of 5 mM sulfuric acid with flow rate of 0.6 ml/min. (Gey et al. 1990). Ammonia was measured by Nessler’s reagent (Morrison 1971). Amino acids were determined by HPLC (Agilent, Santa Clara, CA, USA) with Novapak C18 column (Waters, Eschborn, Germany), using gradient mobile phase A (9 mM sodium dihydrogenphosphate, 4 % dimethylformamide and 0.1–0.2 % triethylamine (TEA), was titrated to pH 6.55 with phosphoric acid) and mobile phase B (80 % (v/v) aqueous acetonitrile) described by Krause et al. (1995). mAb concentration in supernatant was measured with an ELISA. Microtiter plates (Nunc, Roskilde, Denmark) were coated with sheep anti-mouse antibody (Fc specific, Roche, Mumbai, India) at 25 °C for 30 min. The wells were washed with 0.9 % (w/v) NaCl containing 0.1 % (v/v) Tween 20. The plates were incubated at 25 °C for 30 min with a blocking reagent (Roche) consisting of a peptide mixture obtained by proteolysis of gelatin in Tris–HCl buffer and NaCl. After washing, samples were incubated at 25 °C for 30 min. After three washings, plates were incubated with a peroxidase-conjugated anti-mouse κ chain antibody from sheep (Fab-fragments, Roche). The reaction was detected with ABTS subatrate and absorbance was measured using a 405 nm filter. The concentration of each sample was measured from a calibration curve. The standard used was mouse-IgG (Roche).
Experimental design
Previously optimization was done by implementing the variation of one component at a time. This approach was time consuming and assumed that the process response was a function of a single parameter which was varied, not of their interaction. Recently statistical optimization method was carried out where interaction of variables in generating the response was accounted. In this study the optimization consisted of screening of significant nutrients to make subsequent experiments more efficient with less runs using the Plackett–Burman method and locating their optimal concentrations using response surface methodology.
Screening of important nutrients
The Plackett Burman (Plackett and Burman 1946) method was adopted to screen the significant variables for mAb production. Significant factors were screened from a large number of independent factors (N) in a (N + 1) no of experiments. Analysis for the Plackett Burman experiment was carried out as follows:
First, the effect of all variables including dummies was calculated.
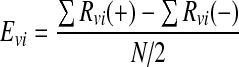 |
1 |
where, Evi is the effect of variable i (g/l); Rvi (+) and Rvi (−) represents the response (mAb production μg/ml) of an assembly in the screening design which contains the high and low levels of variable i, respectively (g/l); N is the number of assemblies (N = 32).
The standard deviation (SD) of dummies which serves as the population standard deviation in the Student’s t test was calculated
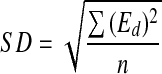 |
2 |
where Ed is the effect of dummy variables; n is the number of dummy variables (n = 2).
Then a student’s t test was performed to identify the significant factors.
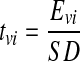 |
3 |
The larger the absolute value of the t value, the more significant is the variable. In the current study of mAb production amino acid requirement by hybridoma cell was for biosynthesis and as energy source. Twenty-nine variables including twenty-one amino acids (essential and non essential), glucose, serum and six salts: NaCl, KCl, CaCl2, NaH2PO4, MgSO4 and Na-pyruvate were included in screening phase.
Locating optimum concentration of significant variables
Response surface methodology (RSM) is a collection of statistical and mathematical approaches and applied to locate the optimal level of the important factors. The interaction of multiple factors is evaluated in minimal experimental trials by RSM. It is a two-level factorial design where contour plots are developed by quadratic effects of the important key factors and a model equation is developed to fit the experimental data for calculating the optimal response of the system (Box et al. 1978). In the current study the Plackett Burman design was followed by RSM (Myers and Montgomery 1995) where the optimum concentration values of the variables were determined assuming the interaction effects of the variables. The dependent variable, mAb production (Y) was assumed to be mainly affected by the seven significant variables. Using least squares the regression model was fit to evaluate the linear interaction and quadratic effects of the variables. The second order polynomial model was described as follows:
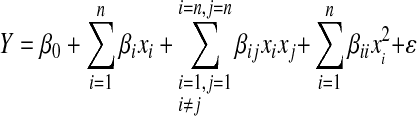 |
where, Y: mAb production, xi: ith variable, βz= regression coefficient (z = i, ii or ij, where i < j) and ε: residual error. The R2 value was used to evaluate model sufficiency. Response surface plots were generated to locate the optimum concentration of the variables. All computations were done using MATLAB.
Results and discussion
Hybricare-46X medium used in the present study, was specially designed by ATCC by combining modified DMEM and NCTC 135 with insuline for fastidious hybridoma cell lines such as HB 8696. ATCC already designed this medium in such a way that it promotes the growth of HB 8696. But it is not designed for optimal mAb production. As Hybricare-46X medium is growth promoting, it was selected as control medium in preference to other commercially available media such as DMEM, RPMI etc. for initial screening and optimization.
Phase 1: Screening of important nutrients by Plackett–Burman design
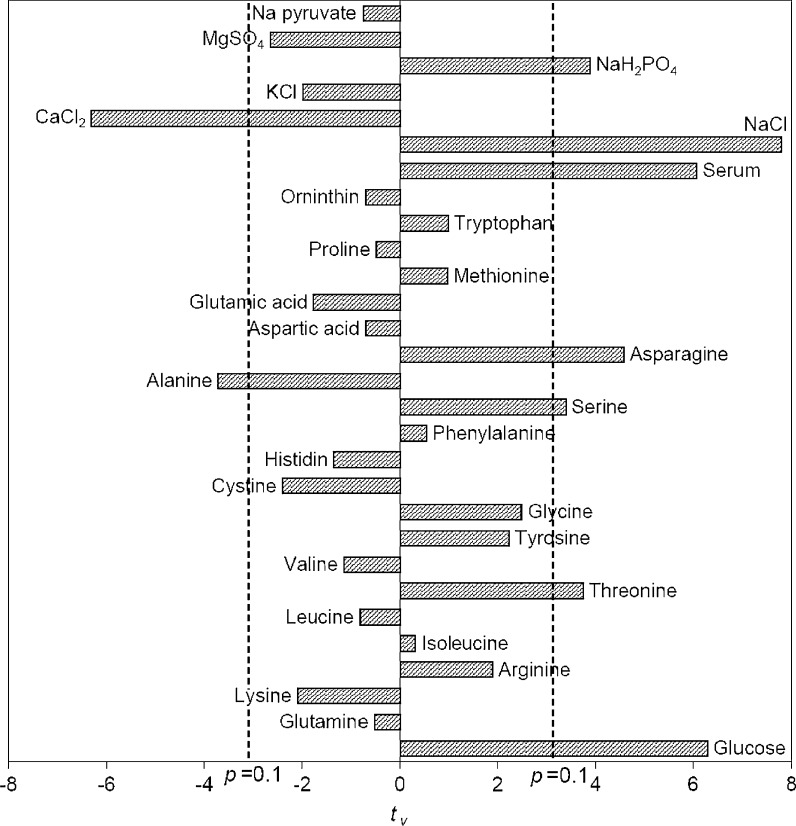
Initial screening of important variables was achieved by minimal trials in Plackett–Burman design. The high and low levels of each variable used in Placket–Burman are shown in Table 2. A design of thirty two experiments was formulated for twenty-nine variables (Table 3) using MATLAB 7.0. Here (+) sign and (−) sign indicated the upper and lower level of each variable in the experimental design. Two dummy variables which were not changing in the design, were introduced to estimate the population standard deviation. Response was measured in terms of mAb production and t values of all the variables were determined and are shown in Fig. 1 where the confidence interval greater than 90 % (pvi < 0.1) was accepted. Nine variables namely glucose, asparagine, threonine, serine, alanine, serum, NaCl, CaCl 2, NaH2PO4 were identified for their significant effect in mAb production. Among them alanine and CaCl2 had a negative effect (negative t value). Therefore the other seven variables namely asparagine, threonine, serine, glucose, serum, NaCl and NaH2PO4 having highly positive t values were significant and considered for further study. Note that glutamine having a negative t value was not influencing mAb production. It is well known that glutamine usually enhances cell growth and the same phenomenon was also found for this cell line (data not shown). Though glutamine was found to be growth stimulatory mAb production was decreased due to lowering of the specific mAb production rate. Parampalli et al. (2007) reported similar preference of other amino acids except glutamine for mAb production. Amino acid requirements are different for different cell lines. Therefore the amino acids identified as important in the current study may or may not be important for other cell lines. Similar phenomena were also found in Zac3, a murine hybridoma where antibody production was increased by addition of serine and threonine (Ducommun et al. 2001). Threonine was also identified as significant for mAb production by hybridoma 130-8F (Alwis et al. 2007). Asparagine was also found as mAb production enhancer for the hybridoma cell line 4A2 (Hiller et al. 1994). On the other hand none of these three amino acids were identified as important for mAb production in the case of hybridoma cell line ATCC-CRL1606 (Selvarasu et al. 2010).
Table 2.
Upper and lower level of variables for Plackett Burman experiments
| SI. no | Variable name | The upper level (g/l) (+) | The lower level (g/l) (−) |
|---|---|---|---|
| 1. | Glucose | 6.0 | 0.598 |
| 2. | Glutamine | 1.2 | 0.484 |
| 3. | Lysine | 0.5 | 0.08 |
| 4. | Arginine | 0.3 | 0.05 |
| 5. | Isoleucine | 0.3 | 0.06 |
| 6. | Leucine | 0.3 | 0.06 |
| 7. | Threonine | 0.3 | 0.05 |
| 8. | Valine | 0.3 | 0.05 |
| 9. | Tyrosine | 0.06 | 0.03 |
| 10. | Glycine | 0.15 | 0.021 |
| 11. | Cysteine | 0.15 | 0.027 |
| 12. | Histidine | 0.15 | 0.024 |
| 13. | Phenylalanine | 0.15 | 0.037 |
| 14. | Serine | 0.15 | 0.029 |
| 15. | Alanine | 0.04 | 0.007 |
| 16. | Asparagine | 0.04 | 0.008 |
| 17. | Aspartic acid | 0.04 | 0.008 |
| 18. | Glutamic acid | 0.04 | 0.008 |
| 19. | Methionine | 0.075 | 0.017 |
| 20. | Proline | 0.04 | 0.01 |
| 21. | Tryptophane | 0.05 | 0.01 |
| 22. | Ornithine | 0.01 | 0.0005 |
| 23. | Serum (%) | 20 | 5 |
| 24. | NaCl | 8 | 3.85 |
| 25. | CaCl2 | 0.75 | 0.12 |
| 26. | KCl | 1.0 | 0.2 |
| 27. | NaH2PO4 | 0.4 | 0.076 |
| 28. | MgSO4 | 0.3 | 0.06 |
| 29. | Na Pyruvate | 0.2 | 0.036 |
Table 3.
Plackett Burman design experiment
| Exp | Variable | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Glutamine | Lysine | Arginine | Isoleucine | Leucine | Threonine | Valine | Tyrosine | Glycine | Cysteine | Histidine | Phenylalanine | Serine | Alanine | |
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − |
| 3 | + | − | − | + | + | − | − | + | + | − | − | + | + | − | − |
| 4 | − | − | + | + | − | − | + | + | − | − | + | + | − | − | + |
| 5 | + | + | + | − | − | − | − | + | + | + | + | − | − | − | − |
| 6 | − | + | − | − | + | − | + | + | − | + | − | − | + | − | + |
| 7 | + | − | − | − | − | + | + | + | + | − | − | − | − | + | + |
| 8 | − | − | + | − | + | + | − | + | − | − | + | − | + | + | − |
| 9 | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| 10 | − | + | − | + | − | + | − | − | + | − | + | − | + | − | + |
| 11 | + | − | − | + | + | − | − | − | − | + | + | − | − | + | + |
| 12 | − | − | + | + | − | − | + | − | + | + | − | − | + | + | − |
| 13 | + | + | + | − | − | − | − | − | − | − | − | + | + | + | + |
| 14 | − | + | − | − | + | − | + | − | + | − | + | + | − | + | − |
| 15 | + | − | − | − | − | + | + | − | − | + | + | + | + | − | − |
| 16 | − | − | + | − | + | + | − | − | + | + | − | + | − | − | + |
| 17 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 18 | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − |
| 19 | + | − | − | + | + | − | − | + | + | − | − | + | + | − | − |
| 20 | − | − | + | + | − | − | + | + | − | − | + | + | − | − | + |
| 21 | + | + | + | − | − | − | − | + | + | + | + | − | − | − | − |
| 22 | − | + | − | − | + | − | + | + | − | + | − | − | + | − | + |
| 23 | + | − | − | − | − | + | + | + | + | − | − | − | − | + | + |
| 24 | − | − | + | − | + | + | − | + | − | − | + | − | + | + | − |
| 25 | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| 26 | − | + | − | + | − | + | − | − | + | − | + | − | + | − | + |
| 27 | + | − | − | + | + | − | − | − | − | + | + | − | − | + | + |
| 28 | − | − | + | + | − | − | + | − | + | + | − | − | + | + | − |
| 29 | + | + | + | − | − | − | − | − | − | − | − | + | + | + | + |
| 30 | − | + | − | − | + | − | + | − | + | − | + | + | − | + | − |
| 31 | + | − | − | − | − | + | + | − | − | + | + | + | + | − | − |
| 32 | − | − | + | − | + | + | − | − | + | + | − | + | − | − | + |
| Exp | Variable | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asparagine | Aspartic acid | Glutamic acid | Methionine | Proline | Tryptophan | Ornithine | Serum | NaCl | CaCl2 | KCl | NaH2PO4 | MgSO4 | Na pyruvate | mAb production (µg/ml) | |
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 5.39 |
| 2 | + | − | + | − | + | − | + | − | + | − | + | − | + | − | 4.29 |
| 3 | + | + | − | − | + | + | − | − | + | + | − | − | + | + | 4.78 |
| 4 | + | − | − | + | + | − | − | + | + | − | − | + | + | − | 5.19 |
| 5 | + | + | + | + | − | − | − | − | + | + | + | + | − | − | 4.79 |
| 6 | + | − | + | − | − | + | − | + | + | − | + | − | − | + | 5.40 |
| 7 | + | + | − | − | − | − | + | + | + | + | − | − | − | − | 5.78 |
| 8 | + | − | − | + | − | + | + | − | + | − | − | + | − | + | 5.06 |
| 9 | + | + | + | + | + | + | + | + | − | − | − | − | − | − | 5.65 |
| 10 | + | − | + | − | + | − | + | − | − | + | − | + | − | + | 2.61 |
| 11 | + | + | − | − | + | + | − | − | − | − | + | + | − | − | 4.96 |
| 12 | + | − | − | + | + | − | − | + | − | + | + | − | − | + | 5.03 |
| 13 | + | + | + | + | − | − | − | − | − | − | − | − | + | + | 3.33 |
| 14 | + | − | + | − | − | + | − | + | − | + | − | + | + | − | 4.49 |
| 15 | + | + | − | − | − | − | + | + | − | − | + | + | + | + | 5.45 |
| 16 | + | − | − | + | − | + | + | − | − | + | + | − | + | − | 2.12 |
| 17 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 4.47 |
| 18 | − | + | − | + | − | + | − | + | − | + | − | + | − | + | 4.26 |
| 19 | − | − | + | + | − | − | + | + | − | − | + | + | − | − | 5.06 |
| 20 | − | + | + | − | − | + | + | − | − | + | + | − | − | + | 0.97 |
| 21 | − | − | − | − | + | + | + | + | − | − | − | − | + | + | 4.00 |
| 22 | − | + | − | + | + | − | + | − | − | + | − | + | + | − | 2.56 |
| 23 | − | − | + | + | + | + | − | − | − | − | + | + | + | + | 4.38 |
| 24 | − | + | + | − | + | − | − | + | − | + | + | − | + | − | 1.86 |
| 25 | − | − | − | − | − | − | − | − | + | + | + | + | + | + | 4.20 |
| 26 | − | + | − | + | − | + | − | + | + | − | + | − | + | − | 4.15 |
| 27 | − | − | + | + | − | − | + | + | + | + | − | − | + | + | 4.74 |
| 28 | − | + | + | − | − | + | + | − | + | − | − | + | + | − | 5.72 |
| 29 | − | − | − | − | + | + | + | + | + | + | + | + | − | − | 4.86 |
| 30 | − | + | − | + | + | − | + | − | + | − | + | − | − | + | 4.46 |
| 31 | − | − | + | + | + | + | − | − | + | + | − | − | − | − | 4.32 |
| 32 | − | + | + | − | + | − | − | + | + | − | − | + | − | + | 4.61 |
Fig. 1.
Identification of significant variables for mAB production by Placket Burman
Phase 2: Locating near the optimum region
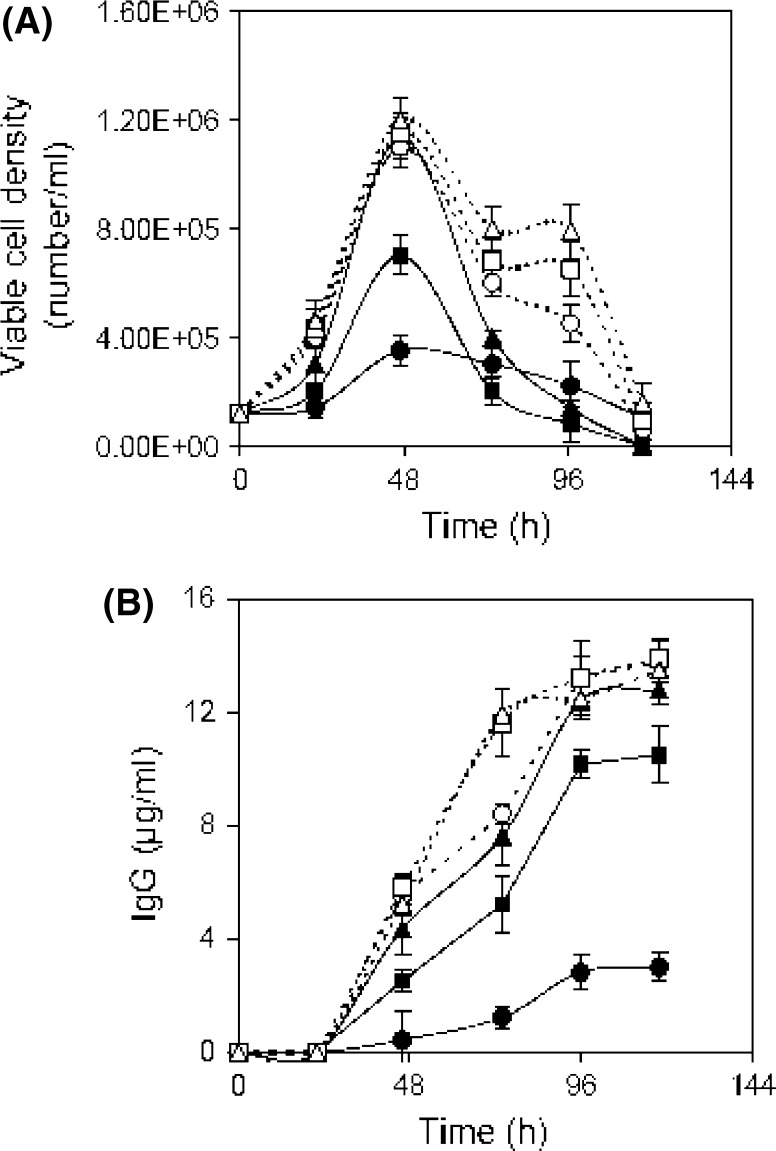
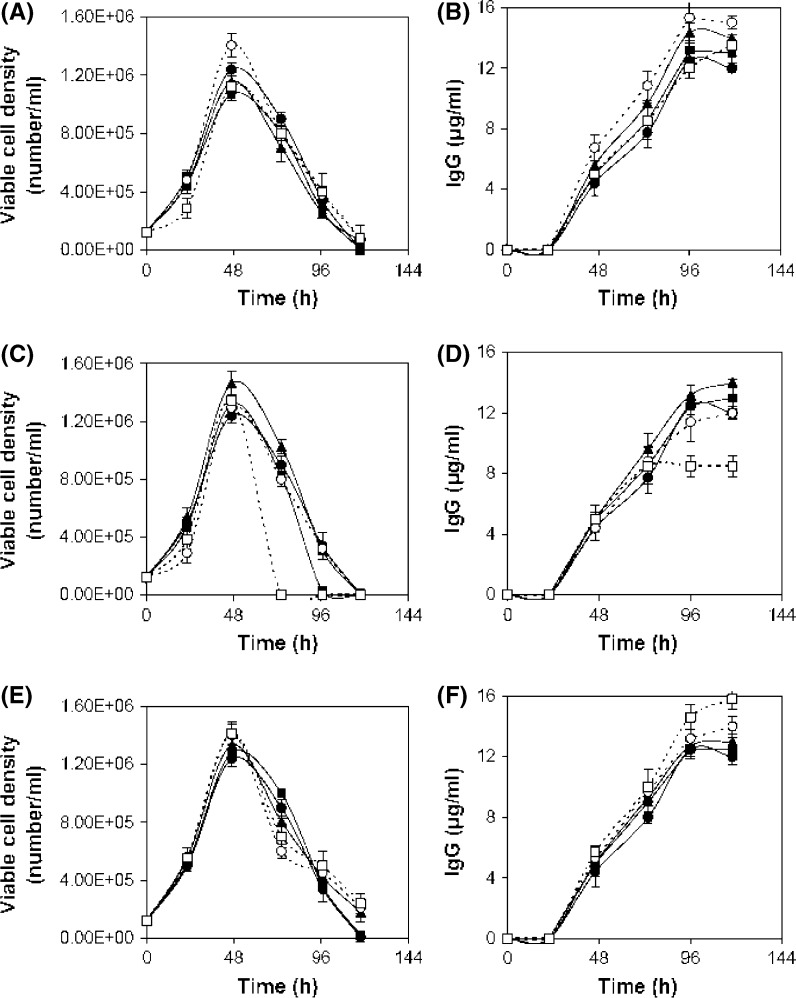
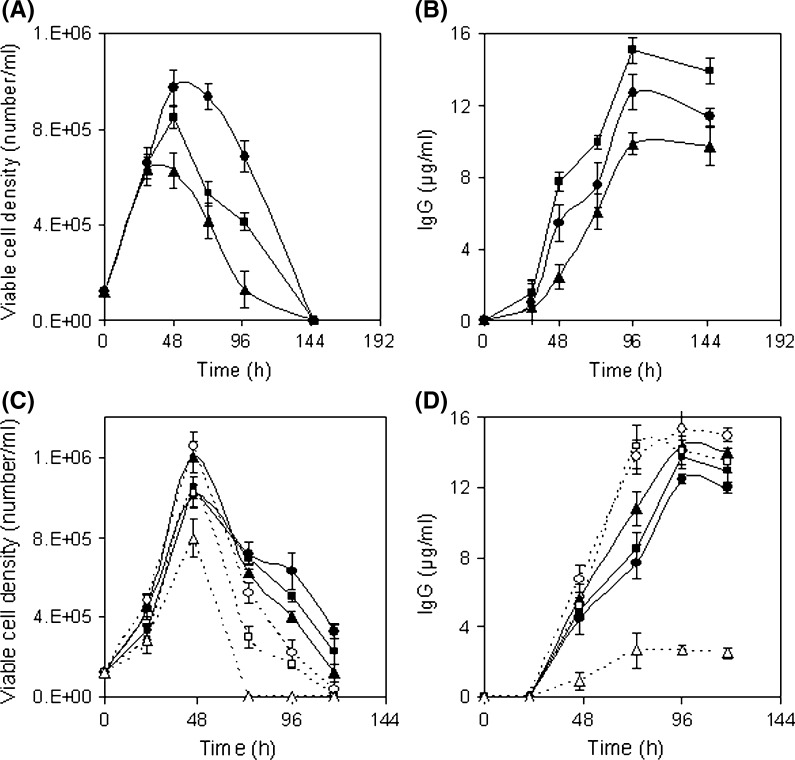
In this step, the region near the optimum value of positive influencing variables was determined. For that, separate batch cultures were carried out varying the concentration of selected components to study their effect on cell growth and mAb production. To understand the sole impact of a single variable, experiments were performed allowing one variable to vary while keeping the other variables constant. Batch cultures were carried out varying glucose concentration from 0.76 to 18 g/l. Growth patterns at higher level of glucose were almost similar to control (0.76 g/l) up to 24 h except 18 g/l (Fig. 2a). After 24 h cell viability was maintained for another 4 days by consuming the remaining glucose (Fig. 2a, b, d). Similar observation was found by Reuveny et al. (1986) where cell viability was maintained over prolonged period of time by glucose feeding because deprivation of glucose can induce apoptosis accelerating steady decline phase (Mercille and Massie 1994). In the case of 18 g/l glucose experiment cell growth was repressed probably due to the inhibition of the high glucose level. The lactic acid was produced marginally higher in the case of higher levels of glucose experiments (lactic acid 15–17 mM) compared to control (lactic acid 13 mM) (Fig. 2e). In the case of high glucose level (2.7–9 g/l) though cell viability was extended, the IgG production was found slightly higher at 4.5 g/l glucose (15.3 μg/ml) as compared to the control (11.5 μg/ml) (Fig. 2c). Similarly a comparative study was carried out by growing hybridoma cell in Hybricare-46X medium supplemented with different concentrations of asparagine from 0.01 to 1.5 g/l along with a control experiment in order to study the effect of asparagine on cell growth and mAb production. Cell growth was improved and a final mAb concentration of 15.27 μg/ml was obtained at 0.75 g/l of asparagine (Fig. 3a, b). To study the effect of threonine, hybridoma cells were cultured varying threonine concentration from 0.064 to 2.4 g/l. Exponential growth phase was stimulated in the presence of threonine but at 2.4 g/l a rapid decline phase was found (Fig. 3c). The final mAb titer was increased with increase of threonine up to 1.2 g/l, compared to control (Fig. 3d). Likewise to study the effect of serine, hybridoma cells were cultured in the presence of additional serine supplemented medium. Cell growth was stimulated in the exponential phase and a slight prolongation of the cell viability was observed at higher concentrations of serine (Fig. 3e). The final mAb titer was improved to 15.8 μg/ml with serine supplementation at 2.1 g/l (Fig. 3f). Serum was screened as another influencing factor by the Plackett Burman method. Separate batch cultures were carried out varying serum percentage from 0 to 30 %. Cell growth was affected at a lower serum level (Fig. 4a). But exponential growth phase was stimulated similarly with serum supplementation from 10 % onwards and a sharp decline phase of viable cells was prevented with the increment of serum percentage (Fig. 4a). The maximum mAb titer was obtained with 20 % serum and no further significant increment of mAb was found with increase of the serum percentage and that may be due to no further increment of specific mAb production rate at higher serum levels (Fig. 4b). Separate batch cultures were run varying NaCl from 4.9 (control) to 9.8 g/l. Cell growth was decreased at hyperosmolar condition and that may be due to stressful condition (Fig. 5a). Cell growth was considerably reduced at 9.8 g/l NaCl may be due to elevated medium osmolarity of 477 mOsm/l where the osmolarity of the control medium was ~300 mOsm/l (Osmolarity of the control medium was taken from the ATCC Hybricare medium data sheet and the osmolarity of other media was calculated based on the added NaCl). But the culture produced slightly enhanced final titer of 13.9 μg/ml at 7.4 g/l NaCl compared to the control (Fig. 5b). This indicated an apparent increase of specific mAb production rate at this level of NaCl. The decrease in cell growth (Oyaas et al. 1989; Ozturk and Palsson 1991; Zhu et al. 2005; Chua et al. 1994) and increase in specific mAb production rate in presence of hypertonic environment were observed by other researchers (Oh et al. 1995; Lin et al. 1999; Ryu and Lee 1997). Phosphorous being an essential part of many cell components like DNA, RNA, phospholipids was selected as significant variable in the initial screening. Previously phosphorous supplementation was found to boost hybridoma cell growth (deZengotita et al. 2000). In the current study cell growth was improved with increase of phosphate up to 1.1 g/l (Fig. 5c) resulting in a correspondingly higher mAb concentration (Fig. 5d). Phosphate addition at 2.7 g/l was found inhibitory to cell growth and mAb production. According to the effect of seven variables on cell growth, mAb production and their respective inhibitory levels, their concentration range near optimum value was selected in RSM experiment to avoid their inhibition.
Fig. 4.
Influence of serum percentage on hybridoma culture. Symbols: 0 % (filled circle), 5 % (filled square), 10 % (filled triangle), 15 % (open circle), 20 % (open square), 30 % (open triangle)
Fig. 2.
Influence of glucose concentration on hybridoma culture a viable cell density, b viability, c IgG, d glucose, e lactic acid and f ammonia. Serum concentration was kept at 20 %. Symbols: Control (0.76 g/l) (filled circle), 1.4 g/l (filled square), 2.7 g/l (filled triangle), 4.5 g/l (open circle), 9 g/l (open square), 18 g/l (open triangle)
Fig. 3.
Influence of amino acid concentration on hybridoma cell culture. a, b asparagine, symbol: different concentrations of asparagine, Control (0.01 g/l) (filled circle), 0.075 g/l (filled square), 0.375 g/l (filled triangle), 0.75 g/l (open circle), 1.5 g/l (open square). c, d threonine, symbol: different concentrations of threonine, Control (0.064 g/l) (filled circle), 0.3 g/l (filled square), 0.6 g/l (filled triangle), 1.2 g/l (open circle), 2.4 g/l (open square). e–f serine, symbol: Different concentration of serine, Control (0.037 g/l) (filled circle), 0.105 g/l (filled square), 0.525 g/l (filled triangle), 1.05 g/l (open circle), 2.1 g/l (open square). The serum concentration was kept at 20 %
Fig. 5.
Influence of NaCl and NaH2PO4 concentration on hybridoma cell culture. a, b NaCl, symbol: different concentrations of NaCl, Control (4.95 g/l) (filled circle), 7.35 g/l (filled square), 9.8 g/l (filled triangle). c, d NaH2PO4, symbol: different concentrations of NaH2PO4, control (0.097 g/l) (filled circle), 0.28 g/l (filled square), 0.55 g/l (filled triangle), 1.1 g/l (open circle), 1.66 g/l (open square), 2.8 g/l (open triangle). The serum concentration was kept at 20 %
Phase 3: Determination of optimum concentration of important variables by response surface methodology
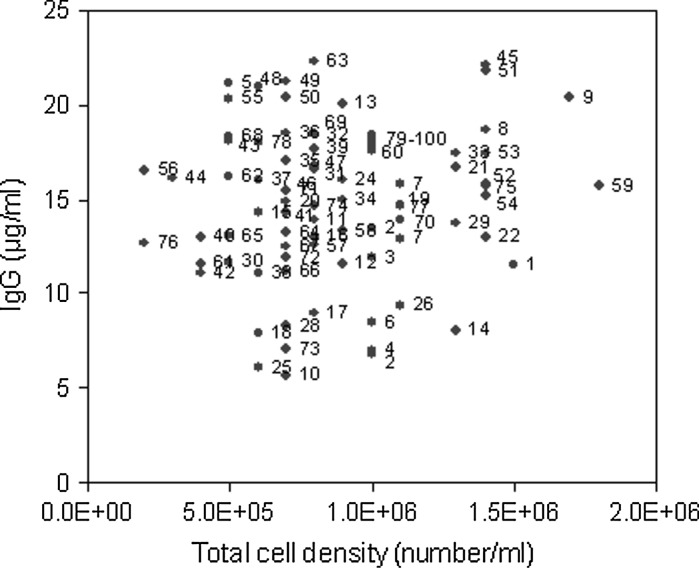
Response surface methodology is a stepwise approach to explore the interactive effect of the significant variables with the obtained response. The seven significant variables with high positive t value were taken into consideration in RSM. Based on Central composite design (CCD), experiments were designed in RSM to optimize the variable concentration. In the current work seventy eight experiments (done in duplicate) along with twenty-two experiments at the central position were carried out. In terms of coded values the RSM experimental design has been shown in Table 4. The corresponding experimental values of each coded variable are given in Table 5. The final mAb titer and respective total cell density are plotted in Fig. 6. The figure elucidated that the IgG production was not only related to cell growth but also dependent on the quality of the cell line for high specific mAb production rate. For example, comparing experiments 14 and 45, both had almost similar cell growth but very different productivity due to high specific production rate in experiment 45. The experiment 63 had a comparatively lower cell growth than that of experiment 45 but had slightly higher mAb production due to a better specific production rate at experiment 63. Using all experimental data a quadratic regression equation was developed by MATLAB 7.0 in terms of mAb production. The second order polynomial regression model is described below:
 |
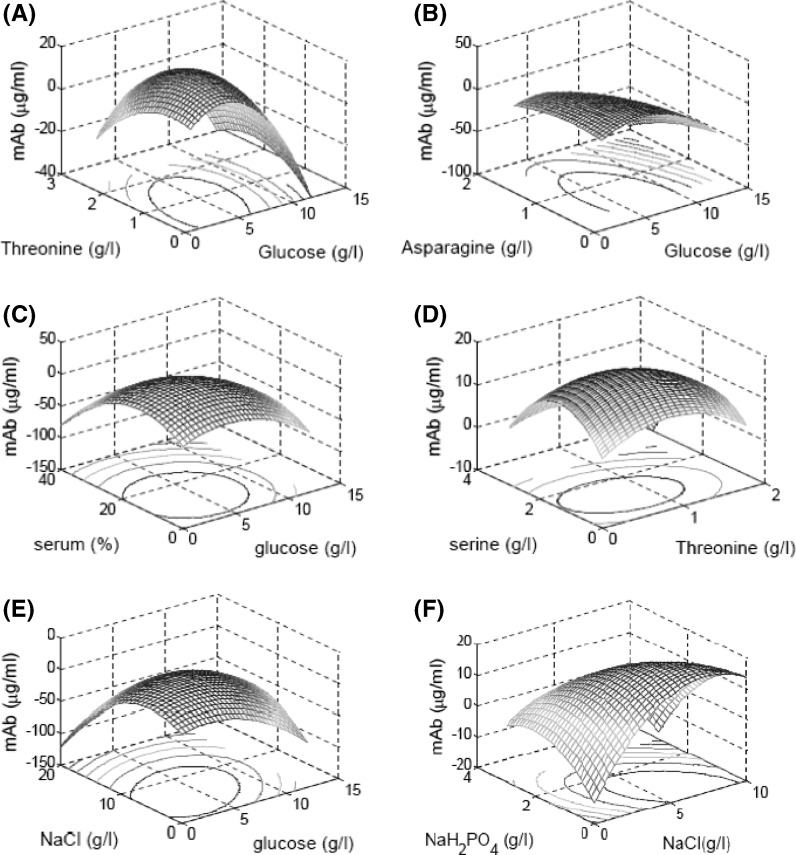
Here 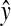 is the mAb production. The R2 value is 0.63 and used to evaluate model sufficiency. The response surfaces were generated for all possible combinations of variables by keeping other parameters constant at a time (Fig. 7). The contour plots were studied and optimized concentrations of the seven variables were located. The optimum values of the variables obtained in this way are given in Table 6.
is the mAb production. The R2 value is 0.63 and used to evaluate model sufficiency. The response surfaces were generated for all possible combinations of variables by keeping other parameters constant at a time (Fig. 7). The contour plots were studied and optimized concentrations of the seven variables were located. The optimum values of the variables obtained in this way are given in Table 6.
Table 4.
Experimental design of RSM in coded values of variables
| Exp no. | Glucose | Threonine | Asparagine | Serine | Serum | NaCl | NaH2PO4 | mAb production (µg/ml) |
|---|---|---|---|---|---|---|---|---|
| 1 | −1 | −1 | −1 | −1 | −1 | −1 | 1 | 11.5 |
| 2 | −1 | −1 | −1 | −1 | −1 | 1 | −1 | 6.8 |
| 3 | −1 | −1 | −1 | −1 | 1 | −1 | −1 | 11.9 |
| 4 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 7.0 |
| 5 | −1 | −1 | −1 | 1 | −1 | −1 | −1 | 21.1 |
| 6 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | 8.5 |
| 7 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 15.8 |
| 8 | −1 | −1 | −1 | 1 | 1 | 1 | −1 | 18.7 |
| 9 | −1 | −1 | 1 | −1 | −1 | −1 | −1 | 20.4 |
| 10 | −1 | −1 | 1 | −1 | −1 | 1 | 1 | 5.7 |
| 11 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 13.9 |
| 12 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 11.6 |
| 13 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | 20.1 |
| 14 | −1 | −1 | 1 | 1 | −1 | 1 | −1 | 8.1 |
| 15 | −1 | −1 | 1 | 1 | 1 | −1 | −1 | 14.3 |
| 16 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | 13.0 |
| 17 | −1 | 1 | −1 | −1 | −1 | −1 | −1 | 9.0 |
| 18 | −1 | 1 | −1 | −1 | −1 | 1 | 1 | 7.9 |
| 19 | −1 | 1 | −1 | −1 | 1 | −1 | 1 | 14.7 |
| 20 | −1 | 1 | −1 | −1 | 1 | 1 | −1 | 14.9 |
| 21 | −1 | 1 | −1 | 1 | −1 | −1 | 1 | 16.7 |
| 22 | −1 | 1 | −1 | 1 | −1 | 1 | −1 | 13.0 |
| 23 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 12.9 |
| 24 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | 16.0 |
| 25 | −1 | 1 | 1 | −1 | −1 | −1 | 1 | 6.1 |
| 26 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 9.4 |
| 27 | −1 | 1 | 1 | −1 | 1 | −1 | −1 | 13.5 |
| 28 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | 8.3 |
| 29 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 13.7 |
| 30 | −1 | 1 | 1 | 1 | −1 | 1 | 1 | 11.7 |
| 31 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | 16.6 |
| 32 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 18.4 |
| 33 | 1 | −1 | −1 | −1 | −1 | −1 | −1 | 17.4 |
| 34 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 15.0 |
| 35 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 17.0 |
| 36 | 1 | −1 | −1 | −1 | 1 | 1 | −1 | 18.5 |
| 37 | 1 | −1 | −1 | 1 | −1 | −1 | 1 | 16.0 |
| 38 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | 11.1 |
| 39 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 17.7 |
| 40 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | 13.0 |
| 41 | 1 | −1 | 1 | −1 | −1 | −1 | 1 | 14.3 |
| 42 | 1 | −1 | 1 | −1 | −1 | 1 | −1 | 11.1 |
| 43 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | 18.1 |
| 44 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 16.1 |
| 45 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | 22.1 |
| 46 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 15.5 |
| 47 | 1 | −1 | 1 | 1 | 1 | −1 | 1 | 16.8 |
| 48 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | 21.0 |
| 49 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | 21.2 |
| 50 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 20.4 |
| 51 | 1 | 1 | −1 | −1 | 1 | −1 | −1 | 21.8 |
| 52 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 15.8 |
| 53 | 1 | 1 | −1 | 1 | −1 | −1 | −1 | 17.4 |
| 54 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | 15.2 |
| 55 | 1 | 1 | −1 | 1 | 1 | −1 | 1 | 20.3 |
| 56 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | 16.5 |
| 57 | 1 | 1 | 1 | −1 | −1 | −1 | −1 | 12.6 |
| 58 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | 13.3 |
| 59 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 15.7 |
| 60 | 1 | 1 | 1 | −1 | 1 | 1 | −1 | 17.7 |
| 61 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 11.6 |
| 62 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 16.2 |
| 63 | 1 | 1 | 1 | 1 | 1 | −1 | −1 | 22.3 |
| 64 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13.2 |
| 65 | −2.8284 | 0 | 0 | 0 | 0 | 0 | 0 | 13.1 |
| 66 | 2.8284 | 0 | 0 | 0 | 0 | 0 | 0 | 11.2 |
| 67 | 0 | −2.8284 | 0 | 0 | 0 | 0 | 0 | 12.5 |
| 68 | 0 | 2.8284 | 0 | 0 | 0 | 0 | 0 | 18.3 |
| 69 | 0 | 0 | −2.8284 | 0 | 0 | 0 | 0 | 18.5 |
| 70 | 0 | 0 | 2.8284 | 0 | 0 | 0 | 0 | 13.9 |
| 71 | 0 | 0 | 0 | – | 0 | 0 | 0 | 15.5 |
| 72 | 0 | 0 | 0 | 2.8284 | 0 | 0 | 0 | 11.9 |
| 73 | 0 | 0 | 0 | 0 | −2.8284 | 0 | 0 | 7.1 |
| 74 | 0 | 0 | 0 | 0 | 2.8284 | 0 | 0 | 14.6 |
| 75 | 0 | 0 | 0 | 0 | 0 | −2.8284 | 0 | 15.7 |
| 76 | 0 | 0 | 0 | 0 | 0 | 2.8284 | 0 | 12.7 |
| 77 | 0 | 0 | 0 | 0 | 0 | 0 | −2.8284 | 14.6 |
| 78 | 0 | 0 | 0 | 0 | 0 | 0 | 2.8284 | 18.0 |
| 79 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17.6 |
| 80 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.1 |
| 81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.2 |
| 82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17.7 |
| 83 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.0 |
| 84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17.6 |
| 85 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.0 |
| 86 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.4 |
| 87 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.0 |
| 88 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.2 |
| 89 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.0 |
| 90 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.2 |
| 91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17.7 |
| 92 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.0 |
| 93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.2 |
| 94 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.1 |
| 95 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17.6 |
| 96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17.8 |
| 97 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.0 |
| 98 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.2 |
| 99 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.0 |
| 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18.1 |
Table 5.
Coded values of six variables in RSM experiments
| Code variable | Glucose (X1) (g/l) | Threonine (X2) (g/l) | Asparagine (X3) (g/l) | Serine (X4) (g/l) | Serum (X5) (%) | NaCl (X6) (g/l) | NaH2PO4 (X7) (g/l) |
|---|---|---|---|---|---|---|---|
| −2.2884 | 0.7656 | 0.0674 | 0.0098 | 0.0373 | 5.000 | 4.9277 | 0.0967 |
| −1 | 2.4575 | 0.4335 | 0.2491 | 0.6717 | 9.483 | 6.5672 | 0.5503 |
| 0 | 3.3828 | 0.6337 | 0.3799 | 1.0186 | 12.50 | 7.4638 | 0.7984 |
| +1 | 4.3081 | 0.8339 | 0.5108 | 1.3656 | 15.1517 | 8.3605 | 1.0464 |
| +2.2884 | 6.0000 | 1.2000 | 0.7500 | 2.0000 | 20.00 | 10.000 | 1.5000 |
Fig. 6.
Relationship of cell growth and mAb production. Each data labeled by the number indicates the corresponding RSM experiments (see Table 4)
Fig. 7.
Response surface and contour plot of different variables a threonine and glucose, b asparagine and glucose, c serum and glucose, d serine and threonine, e NaCl and glucose and f NaH2PO4 and NaCl
Table 6.
Optimum and control medium concentration
| Optimum | Control | |
|---|---|---|
| Variables | ||
| Glucose (g/l) | 4.4 | 0.72 |
| Threonine (g/l) | 0.8 | 0.063 |
| Asparagine (g/l) | 0.16 | 0.009 |
| Serine (g/l) | 0.64 | 0.035 |
| Serum (%) | 14 | 20 |
| NaCl (g/l) | 6.6 | 4.62 |
| NaH2PO4 (g/l) | 0.35 | 0.09 |
| mAb production | ||
| Experimental (μg/ml) | 24.3±1.1 | 11.5±1.5 |
| Prediction (μg/ml) | 22.3 | – |
Validation
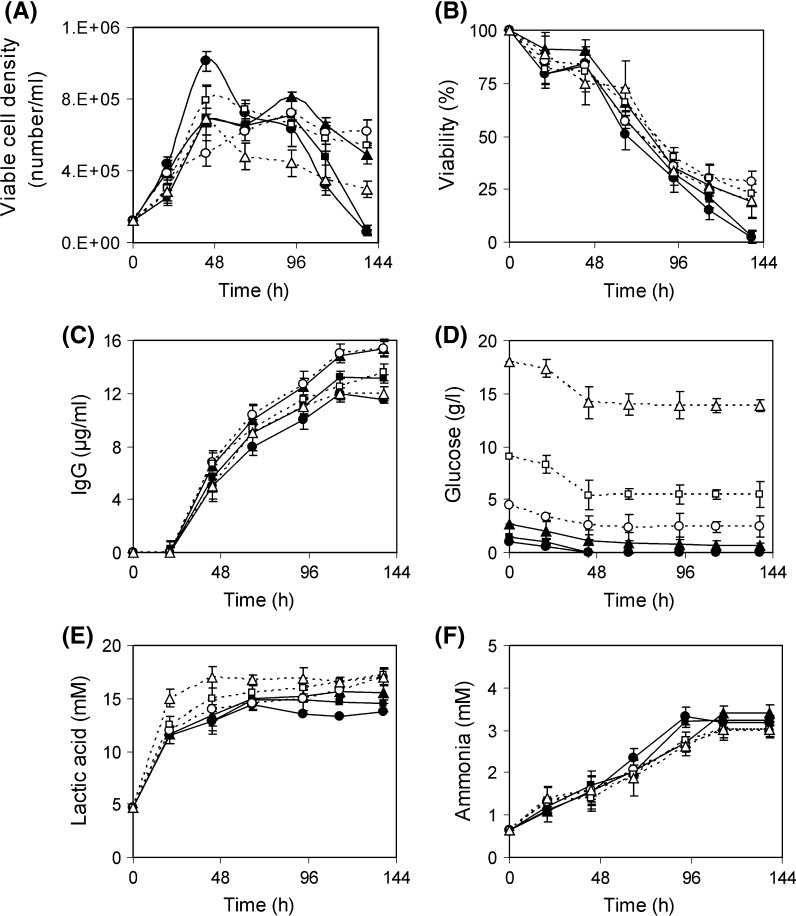
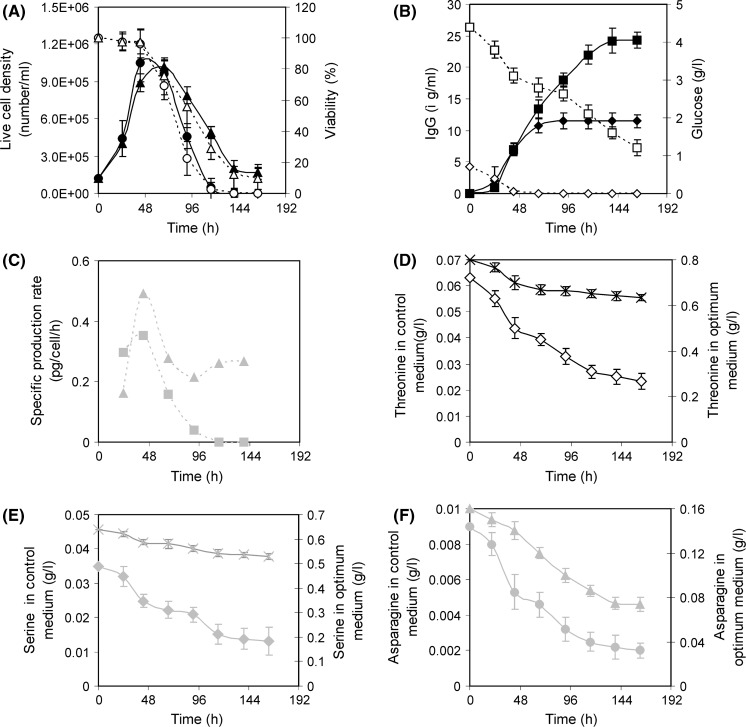
The mathematical model generated during RSM implementation was validated by conducting an experiment at optimal concentrations of the seven variables. The control experiment was carried out simultaneously using the inoculum of the same batch. The cell growth was found almost similar in the exponential phase for both control and optimum experiment but viable cell number did not decline so sharply in the optimal experiment as compared to control. Viability was also better maintained in the optimum experimental condition (Fig. 8a). Under optimal condition the mAb production was increased to 24.3 ± 1.1 μg/ml and it was enhanced by 111.05 % compared to the control medium (11.5±1.5 μg/ml) (Fig. 8b). The mAb produced at optimal condition agreed well with the predicted value (22.3 μg/ml) (Table 6). The average specific mAb production rate was almost double at optimum condition compared to control (Fig. 8c). This increment of mAb was a combined effect of amino acids, optimal levels of sugar, serum and NaH2PO4 and moderate osmolarity stress (Osmolarity of optimum medium was 390 mOsm/l, determined based on added component where as in control it was 300 mOsm/l). Parampalli et al. (2007) statistically optimized medium formulations that improved antibody production by 50 %. As product titer = viability × total cell × specific productivity, product titer can be increased by increasing of viability, total cell and specific productivity. Both the viability and specific production rate of the optimum medium were increased compared to control medium (HB medium), resulting in the increase of the total titer (Fig. 8a, c). Earlier it was indicated that more cells were not responsible for more product or high specific production rate because there are a number of medium compositions where the number of cells was high but product formation was reduced (Fig. 6). So, only viability may not be the driving force for the increase of the specific production rate. Instead, the optimum medium composition itself was responsible for both the increase of the viability and increase of the specific production rate.
Fig. 8.
Hybridoma cell growth and IgG production at control and optimal condition. Symbol: Live cell density at control (filled circle) and optimum condition (filled triangle).Viability at control (open circle) and optimum condition (open triangle). IgG production at control (filled diamond) and optimum condition (filled square). Glucose at control (open diamond) and optimum condition (open square). Specific IgG production rate at control (grey square) and optimum condition (grey triangle). Threonine at control (open diamond) and optimum condition (times). Serine at control (grey diamond) and optimum condition (grey times). Asparagine at control (grey circle) and optimum condition (grey triangle)
It has been reported elsewhere that the amino acids show varying patterns of production or consumption, depending upon oxygen level, culture mode and cell type (Ozturk and Palsson 1991). In our study consumption of all three important amino acids, serine, threonine and asparagine was observed in the batch culture of the hybridoma (Fig. 8d–f). In the control medium (HB medium) serine, threonine and asparagine were consumed to 0.013, 0.023 and 0.002 g/l during batch culture (Fig. 8d–f). So these amino acids were acting as limiting substrate as their levels were very low. This was overcome by adding amino acids to the optimum medium.
In this study some limiting nutrients like glucose and amino acids as well as NaCl and NaH2PO4 have been supplemented initially in the optimum culture. These would be added at a later stage when they appeared to be limiting. But there are already few studies suggesting that early supplementation of hybridoma cultures with rapidly consumed amino acid prolongs their survival, whereas late addition does not modify cell demise (Duval et al. 1991). A profound reduction in the maximal cell density associated with an accelerated drop in cell viability was found in the late addition of amino acid. Also it has been seen in that study that amino acid supply in a later stage does not increase the specific production rates. Therefore in the current study the limiting nutrients and nutrients which maintain osmotic pressure were added early in the culture.
Conclusion
As mAb 520C9 has potential applications, optimization of its production is necessary but optimization strategy is not available yet for this antibody. The purpose of the study was to develop an optimum medium formulation through statistical analysis to maximize mAb 520C9 production by the hybridoma cell HB8696. The methodology by combining the Plackett–Burman design, finding near optimum solution and response surface methodology were effective in screening significant variables and optimizing their concentration. The initial screening procedure by the Plackett Burman method was able to identify the three positively influencing amino acids, namely, asparagine, threonine and serine out of twenty-one amino acids. Four other nutrients, serum, NaCl, glucose and NaH2PO4 were also found important in the initial screening phase. The next step exhibited effect of individual nutrients on cell growth and mAb production along with near optimum level. A significantly higher mAb titer was obtained under optimum medium formulation. The mAb produced at optimal condition is 24.3 ± 1.1 μg/ml whereas mAb was 11.5 ± 1.5 μg/ml for the control condition. The average specific mAb production rate has been enhanced by 105 % compared to control medium. The applied strategy in the current study is systematic, and simple to screen the important nutrient, exhibited positive effect of the key nutrients and determined their optimum concentration.
Acknowledgments
We are thankful to Department of Biotechnology (DBT), Govt. of India, New Delhi for providing financial support during the work done.
References
- Alwis DMD, Dutton RL, Scharer J, Young MM. Statistical methods in media optimization for batch and fed-batch animal cell culture. Bioprocess Biosyst Eng. 2007;30:107–113. doi: 10.1007/s00449-006-0107-7. [DOI] [PubMed] [Google Scholar]
- Box GEP, Hunter WG, Hunter JS. Statistics for experimenters: an introduction to design, data analysis, and model building. NewYork: Wiley; 1978. [Google Scholar]
- Butler M. Animal cell cultures: recent achievements and perspectives in the production of biopharmaceuticals. Appl Microbiol Biotechnol. 2005;68:283–291. doi: 10.1007/s00253-005-1980-8. [DOI] [PubMed] [Google Scholar]
- Chua FKF, Yap MGS, Oh SKW. Hyper-stimulation of monoclonal-antibody production by high osmolarity stress in Erdf medium. J Biotechnol. 1994;37:265–275. doi: 10.1016/0168-1656(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Dalili M, Ollis DF. Transient kinetics of hybridoma growth and monoclonal antibody production in serum-limited cultures. Biotechnol Bioeng. 1989;33:984–990. doi: 10.1002/bit.260330807. [DOI] [PubMed] [Google Scholar]
- deZengotita VM, Miller WM, Aunins JG, Zhou WC. Phosphate feeding improves high-cell-concentration NS0 myeloma culture performance for monoclonal antibody production. Biotechnol Bioeng. 2000;69:566–576. doi: 10.1002/1097-0290(20000905)69:5<566::AID-BIT11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Ducommun P, Ruffieux PA, von Stockar U, Marison I. The role of vitamins and amino acids on hybridoma growth and monoclonal antibody production. Cytotechnology. 2001;37:65–73. doi: 10.1023/A:1019956013627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval D, Demangel C, Munierjolain K, Miossec S, Geahel I. Factors controlling cell-proliferation and antibody-production in mouse hybridoma cells.1. Influence of the amino-acid supply. Biotechnol Bioeng. 1991;38:561–570. doi: 10.1002/bit.260380602. [DOI] [PubMed] [Google Scholar]
- Falkenberg FW, Weichert H, Krane M, Bartels I, Palme M, Nagels HO, Fiebig H. In vitro production of monoclonal-antibodies in high-concentration in a new and easy to handle modular minifermenter. J Immunol Methods. 1995;179:13–29. doi: 10.1016/0022-1759(94)00266-Y. [DOI] [PubMed] [Google Scholar]
- Feder J, Tolbert WR. Mass culture of mammalian cells in perfusion systems. Am Biotech Lab. 1985;3:24–36. doi: 10.1038/nbt0185-24. [DOI] [Google Scholar]
- Franek F. Starvation-induced programmed death of hybridoma cells, prevention by amino-acid mixtures. Biotechnol Bioeng. 1995;45:86–90. doi: 10.1002/bit.260450112. [DOI] [PubMed] [Google Scholar]
- Geaugey V, Duval D, Geahel I, Marc A, Engasser JM. Influence of amino acids on hybridoma cell viability and antibody secretion. Cytotechnology. 1989;2:119–129. doi: 10.1007/BF00386144. [DOI] [PubMed] [Google Scholar]
- Gey M, Klossek P, Becker U. Characterization of biotechnological processes and products using high-performance liquid-chromatography (Hplc).6. Determination of lactic-acid and short-chain carboxylic-acids C1–C5. Acta Biotechnol. 1990;10:459–468. doi: 10.1002/abio.370100516. [DOI] [Google Scholar]
- Glacken MW, Fleischaker RJ, Sinskey AJ. Reduction of waste product excretion via nutrient control—possible strategies for maximizing product and cell yields on serum in cultures of mammalian-cells. Biotechnol Bioeng. 1986;28:1376–1389. doi: 10.1002/bit.260280912. [DOI] [PubMed] [Google Scholar]
- Hiller GW, Clark DS, Blanch HW. Transient responses of hybridoma cells in continuous-culture to step changes in amino-acid and vitamin concentrations. Biotechnol Bioeng. 1994;44:303–321. doi: 10.1002/bit.260440308. [DOI] [PubMed] [Google Scholar]
- Jager M, Schoberth A, Ruf P, Hess J, Lindhofer H. The trifunctional antibody ertumaxomab destroys tumor cells that express low levels of human epidermal growth factor receptor 2. Cancer Res. 2009;69:4270–4276. doi: 10.1158/0008-5472.CAN-08-2861. [DOI] [PubMed] [Google Scholar]
- Jo EC, Park HJ, Park JM, Kim KH. Balanced nutrient fortification enables high-density hybridoma cell-culture in batch culture. Biotechnol Bioeng. 1990;36:717–722. doi: 10.1002/bit.260360709. [DOI] [PubMed] [Google Scholar]
- Jorgensen JT, Nielsen KV, Ejlertsen B. Pharmacodiagnostics and targeted therapies: a rational approach for individualizing medical anticancer therapy in breast cancer. Oncologist. 2007;12:397–405. doi: 10.1634/theoncologist.12-4-397. [DOI] [PubMed] [Google Scholar]
- Kelley B. Industrialization of mAb production technology. The bioprocessing industry at a crossroads. Mabs. 2009;1:443–452. doi: 10.4161/mabs.1.5.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause I, Bockhardt A, Neckermann H, Henle T, Klostermeyer H. Simultaneous determination of amino acids and biogenic amines by reversed-phase high-performance liquid chromatography of the dabsyl derivatives. J Chromatogr A. 1995;715:67–79. doi: 10.1016/0021-9673(95)00578-B. [DOI] [Google Scholar]
- Lee GM, Huard TK, Palsson BO. Effect of serum concentration on hybridoma cell-growth and monoclonal-antibody production at various initial cell densities. Hybridoma. 1989;8:369–375. doi: 10.1089/hyb.1989.8.369. [DOI] [PubMed] [Google Scholar]
- Lin JQ, Takagi M, Qu YB, Gao PJ, Yoshida T. Enhanced monoclonal antibody production by gradual increase of osmotic pressure. Cytotechnology. 1999;29:27–33. doi: 10.1023/A:1008016806599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljunggren J, Haggstrom L. Catabolic control of hybridoma cells by glucose and glutamine limited fed-batch cultures. Biotechnol Bioeng. 1994;44:808–818. doi: 10.1002/bit.260440706. [DOI] [PubMed] [Google Scholar]
- Luan YT, Mutharasan R, Magee WE. Strategies to extend longevity of hybridomas in culture and promote yield of monoclonal-antibodies. Biotechnol Lett. 1987;9:691–696. doi: 10.1007/BF01024599. [DOI] [Google Scholar]
- Martens DE, Degooijer CD, Beuvery EC, Tramper J. Effect of serum concentration on hybridoma viable cell-density and production of monoclonal-antibodies in cstrs and on shear sensitivity in airlift loop reactors. Biotechnol Bioeng. 1992;39:891–897. doi: 10.1002/bit.260390902. [DOI] [PubMed] [Google Scholar]
- Martial-Gros A, Goergen JL, Engasser JM, Marc A. Amino acids metabolism by VO 208 hybridoma cells: some aspects of the culture process and medium composition influence. Cytotechnology. 2001;37:93–105. doi: 10.1023/A:1019908310300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx U, Merz W. In vivo and in vitro production of monoclonal antibodies. Bioreactors versus immune ascites. In: Davis WC, editor. Methods in molecular biology. Totowa: Humana Press Inc; 1995. pp. 169–176. [DOI] [PubMed] [Google Scholar]
- McCarty K. Selective utilization of amino acids by mammalian cell cultures. Exp Cell Res. 1962;27:230–240. doi: 10.1016/0014-4827(62)90226-4. [DOI] [Google Scholar]
- Mercille S, Massie B. Induction of apoptosis in nutrient-deprived cultures of hybridoma and myeloma cells. Biotechnol Bioeng. 1994;44:1140–1154. doi: 10.1002/bit.260440916. [DOI] [PubMed] [Google Scholar]
- Miller WM, Wilke CR, Blanch HW. Transient responses of hybridoma cells to nutrient additions in continuous culture.1. Glucose pulse and step changes. Biotechnol Bioeng. 1989;33:477–486. doi: 10.1002/bit.260330413. [DOI] [PubMed] [Google Scholar]
- Morrison GR. Microchemical determination of organic nitrogen with Nessler reagent. Anal Biochem. 1971;43:527. doi: 10.1016/0003-2697(71)90283-1. [DOI] [PubMed] [Google Scholar]
- Myers RH, Montgomery DC. Response surface methodology: process and product optimization using designed experiments. New York: Wiley; 1995. [Google Scholar]
- Oh SKW, Vig P, Chua F, Teo WK, Yap MGS. Substantial overproduction of antibodies by applying osmotic-pressure and sodium-butyrate. Biotechnol Bioeng. 1993;42:601–610. doi: 10.1002/bit.260420508. [DOI] [PubMed] [Google Scholar]
- Oh SKW, Chua FKF, Choo ABH. Intracellular responses of productive hybridomas subjected to high osmotic-pressure. Biotechnol Bioeng. 1995;46:525–535. doi: 10.1002/bit.260460605. [DOI] [PubMed] [Google Scholar]
- Oyaas K, Berg TM, Bakke O, Levine DW. Hybridoma growth and antibody production under conditions of hyperosmotic stress. In: Spier RE, Griffiths JB, Stephenne J, Crooy PJ, editors. Advances in animal cell biology and technology for bioprocesses. London: Butterworth; 1989. [Google Scholar]
- Ozturk SS, Palsson BO. Effect of medium osmolarity on hybridoma growth, metabolism, and antibody-production. Biotechnol Bioeng. 1991;37:989–993. doi: 10.1002/bit.260371015. [DOI] [PubMed] [Google Scholar]
- Parampalli A, Eskridge K, Smith L, Meagher MM, Mowry MC, Subramanian A. Development of serum-free media in CHO-DG44 clls using central composite design. Cytotechnology. 2007;54:57–68. doi: 10.1007/s10616-007-9074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946;33:305–325. doi: 10.1093/biomet/33.4.305. [DOI] [Google Scholar]
- Press MF, Sauter G, Bernstein L, Villalobos IE, Mirlacher M, Zhou JY, Wardeh R, Li YT, Guzman R, Ma YL, Sullivan-Halley J, Santiago A, Park JM, Riva A, Slamon DJ. Diagnostic evaluation of HER-2 as a molecular target: an assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clin Cancer Res. 2005;11:6598–6607. doi: 10.1158/1078-0432.CCR-05-0636. [DOI] [PubMed] [Google Scholar]
- Repp R, Valerius T, Wieland G, Becker W, Steininger H, Deo Y, Helm G, Gramatzki M, Van de Winkel JG, Lang N. G-CSF-stimulated PMN in immunotherapy of breast cancer with a bispecific antibody to Fc gamma RI and to HER-2/neu (MDX-210) J Hematother. 1995;4:415–421. doi: 10.1089/scd.1.1995.4.415. [DOI] [PubMed] [Google Scholar]
- Repp R, van Ojik HH, Valerius T, Groenewegen G, Wieland G, Oetzel C, Stockmeyer B, Becker W, Eisenhut M, Steininger H, Deo YM, Blijham GH, Kalden JR, van de Winkel JGJ, Gramatzki M. Phase I clinical trial of the bispecific antibody MDX-H210 (anti-Fc gamma RI x anti-HER-2/neu) in combination with Filgrastim (G-CSF) for treatment of advanced breast cancer. Br J Cancer. 2003;89:2234–2243. doi: 10.1038/sj.bjc.6601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuveny S, Velez D, Riske F, Macmillan JD, Miller L. Production of monoclonal-antibodies in culture. Dev Biol Stand. 1985;60:185–197. [PubMed] [Google Scholar]
- Reuveny S, Velez D, Macmillan JD, Miller L. Factors affecting cell-growth and monoclonal-antibody production in stirred reactors. J Immunol Methods. 1986;86:53–59. doi: 10.1016/0022-1759(86)90264-4. [DOI] [PubMed] [Google Scholar]
- Ryu JS, Lee GM. Influence of hyperosmolar basal media on hybridoma cell growth and antibody production. Bioprocess Eng. 1997;16:305–310. doi: 10.1007/s004490050327. [DOI] [Google Scholar]
- Seewoster T, Lehmann J. Influence of targeted asparagine starvation on extra and intracellular amino acid pools of cultivated Chinese hamster ovary cells. Appl Microbiol Biotechnol. 1995;44:344–350. doi: 10.1007/BF00169927. [DOI] [PubMed] [Google Scholar]
- Selvarasu S, Kim DY, Karimi IA, Lee DY. Combined data preprocessing and multivariate statistical analysis characterizes fed-batch culture of mouse hybridoma cells for rational medium design. J Biotechnol. 2010;150:94–100. doi: 10.1016/j.jbiotec.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Shi T, Eaton AM, Ring DB. Selection of hybrid hybridomas by flow-cytometry using a new combination of fluorescent vital stains. J Immunol Methods. 1991;141:165–175. doi: 10.1016/0022-1759(91)90143-4. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, Press MF. Studies of the Her-2/Neu proto-oncogene in human-breast and ovarian-cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Waldmann TA. Immunotherapy: past, present and future. Nat Med. 2003;9:269–277. doi: 10.1038/nm0303-269. [DOI] [PubMed] [Google Scholar]
- Zhu MM, Goyal A, Rank DL, Gupta SK, Boom TV, Lee SS. Effects of elevated pCO(2) and osmolality on growth of CHO cells and production of antibody-fusion protein B1: a case study. Biotechnol Prog. 2005;21:70–77. doi: 10.1021/bp049815s. [DOI] [PubMed] [Google Scholar]