Summary: Auxin induces the expression of SAUR36 that positively regulates leaf senescence in Arabidopsis.
Abstract
Small Auxin Up RNA genes (SAURs) are early auxin-responsive genes, but whether any of them are involved in leaf senescence is not known. Auxin, on the other hand, has been shown to have a role in leaf senescence. Some of the external application experiments indicated that auxin can inhibit leaf senescence, whereas other experiments indicated that auxin can promote leaf senescence. Here, we report the identification and characterization of an Arabidopsis (Arabidopsis thaliana) leaf senescence-associated gene named SAG201, which is highly up-regulated during leaf senescence and can be induced by 1-naphthaleneacetic acid, a synthetic auxin. It encodes a functionally uncharacterized SAUR that has been annotated as SAUR36. Leaf senescence in transfer DNA insertion saur36 knockout lines was delayed as revealed by analyses of chlorophyll content, Fv/Fm ratio (a parameter for photosystem II activity), ion leakage, and the expression of leaf senescence marker genes. In contrast, transgenic Arabidopsis plants overexpressing SAUR36 (without its 3′ untranslated region [UTR]) displayed an early leaf senescence phenotype. However, plants overexpressing SAUR36 with its 3′ UTR were normal and did not exhibit the early-senescence phenotype. These data suggest that SAUR36 is a positive regulator of leaf senescence and may mediate auxin-induced leaf senescence and that the 3′ UTR containing a highly conserved downstream destabilizes the SAUR36 transcripts in young leaves.
Leaf senescence is the final phase of leaf development. Like other plant developmental processes, leaf senescence is genetically programmed; a subset of Senescence-Associated Genes (SAGs) are up-regulated at the onset of and during leaf senescence, while the vast majority of genes (including photosynthesis-related genes) are down-regulated (Gan and Amasino, 1997). Thousands of SAGs have been isolated, including genes encoding transcription factors, signal transduction components, proteinases and other catabolic enzymes, and various transporters of sugars, amino acids, and other nutrients (Gepstein et al., 2003; Guo et al., 2004; Buchanan-Wollaston et al., 2005; Guo and Gan, 2012). Some of these SAGs, such as NAC and WRKY transcription factor genes (Guo and Gan, 2006; Uauy et al., 2006; Zentgraf et al., 2010; Besseau et al., 2012; Zhang and Gan, 2012), protein kinase and phosphatase genes (Hajouj et al., 2000; Zhou et al., 2009; Xu et al., 2011; Zhang et al., 2012), lipase genes (He and Gan, 2002), and nuclease genes (Lers et al., 2006), have been shown biochemically, genetically, and/or physiologically to have a role in leaf senescence. The functions of the vast majority of SAGs (some of which encode Small Auxin Up RNAs [SAURs]) have yet to be characterized.
Leaf senescence can be regulated by various internal signals and environmental cues (Xu et al., 2011; Guo and Gan, 2012). Among the internal factors are plant hormones that promote or inhibit leaf senescence. Ethylene, abscisic acid, jasmonic acid, and salicylic acid can promote leaf senescence, and cytokinins and gibberellin inhibit senescence (Gan, 2010). However, the role of auxin in regulating leaf senescence remains elusive. In previous studies, exogenous application of auxin often delayed chlorophyll loss and protein degradation and suppressed the transcription of some SAGs in detached leaves of various plant species. Some correlative studies also showed that the total auxin levels decrease as leaf senescence progresses (Gan, 2010). These findings suggest that auxin may be a negative regulator of leaf senescence. However, the levels of free indole-3-acetic acid (IAA) in senescing leaves were shown to be higher than those in young leaves, although the levels of total IAA species decreased during Arabidopsis (Arabidopsis thaliana) leaf senescence (Quirino et al., 1999). Transcription analysis found that many genes involved in auxin biosynthesis such as Trp synthase (AtTSA1) and nitrilases (NIT1–NIT4) are up-regulated during age-dependent senescence (Quirino et al., 1999; van der Graaff et al., 2006). These results suggest that auxin may be a positive regulator of leaf senescence.
Auxin exerts its roles in cell division, elongation, and differentiation in part by altering gene expression (Kant et al., 2009; Scarpella et al., 2010). There are three major early auxin-responsive gene families, including the SAUR family (Hagen et al., 2010). SAUR genes can be readily induced by exogenous auxin (Jain et al., 2006) and have been widely used as auxin-inducible reporters. Studies have correlated some SAUR genes with auxin-mediated cell expansion based on their temporal and spatial expression and their regulation at the transcriptional, posttranscriptional, and protein levels (Esmon et al., 2006; Spartz et al., 2012). Molecular studies have also demonstrated that many SAUR genes contain a highly conserved downstream (DST) element in their 3′ untranslated region (UTR) that contributes to mRNA instability in an auxin-independent manner (Sullivan and Green, 1996; Park et al., 2012). Analysis of Arabidopsis plants expressing a SAUR15-luciferase fusion protein suggested that SAUR proteins are unstable (Zenser et al., 2003). Studies of the maize (Zea mays) ZmSAUR2 protein revealed that SAUR expression may be regulated at the protein level (Knauss et al., 2003). Recent studies showed that OsSAUR39 acts as a negative regulator of auxin synthesis and transport; overexpression of this gene in rice (Oryza sativa) confers phenotypes including reductions in lateral root development, yield, and shoot and root lengths (Kant et al., 2009). The addition of an N-terminal GFP or epitope tag dramatically increases the stability of SAUR19 to SAUR24 proteins and their ability to function as positive effectors of cell expansion through the modulation of auxin transport (Spartz et al., 2012). However, the function and underlying mechanisms of many SAUR genes remain unknown.
Here, we report our molecular genetic analyses of Arabidopsis SAUR36, which shows SAUR36 to be a positive regulator of leaf senescence, which implies that auxin, via the action of SAUR36, has a role in promoting leaf senescence.
RESULTS
The Expression of SAG201/SAUR36 Is Up-Regulated during Leaf Senescence in Arabidopsis
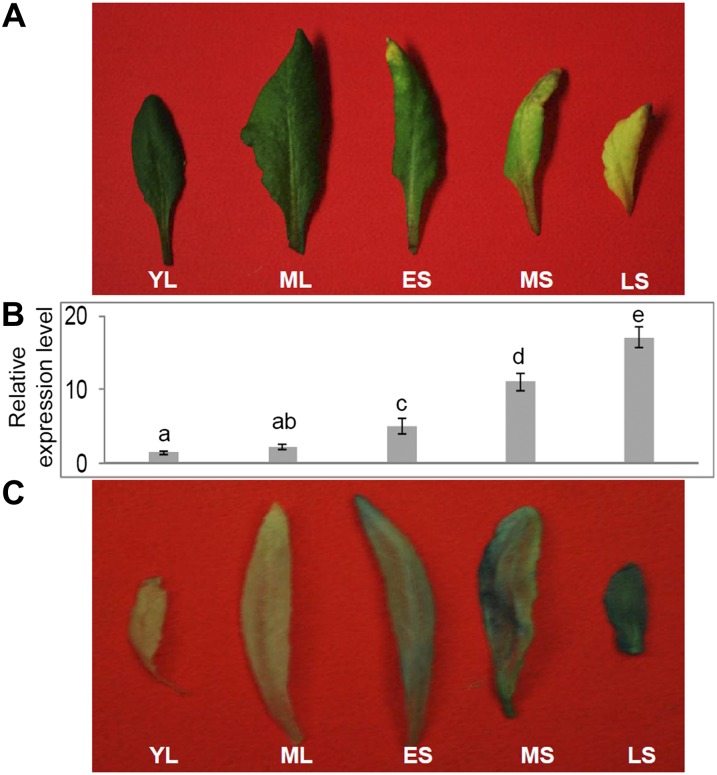
A gene, initially named SAG201, was identified from our previous transcriptomic analyses of leaf senescence in Arabidopsis (Guo et al., 2004). This gene is SAUR36. Quantitative PCR (qPCR) analyses revealed that the transcript levels of SAG201/SAUR36 were very low in expanding young leaves and fully expanded mature leaves but increased with the progression of senescence (Fig. 1, A and B).
Figure 1.
Expression of SAG201/SAUR36 during leaf senescence in Arabidopsis. A, Phenotypes of leaf senescence in wild-type plants. YL, Expanding young leaf; ML, mature leaf that is fully expanded but with no visible yellowing; ES, early senescence leaf, with about 25% leaf area yellowing; MS, middle senescence leaf, with about 50% leaf area yellowing; LS, late senescence leaf, with more than 50% leaf area yellowing. B, Relative expression of SAG201/SAUR36 during leaf senescence. The expression levels were determined using qPCR. The expression levels in young leaves were set to 1. Data represent mean values ± se of three replicates. Letters indicate significant differences by Student’s t test (P = 0.05). C, GUS staining in senescing leaves of PSAG201/SAUR36-GUS transgenic Arabidopsis plants at 40 DAE. [See online article for color version of this figure.]
We also generated transgenic Arabidopsis plants containing the promoter of SAUR36 fused with the GUS reporter gene. Histochemical staining revealed that the expression of SAUR36 was only evident in senescing leaves and not in the nonsenescent ones (Fig. 1C), which is consistent with the above qPCR data (Fig. 1B).
SAUR36 Is Induced by Exogenous Auxin
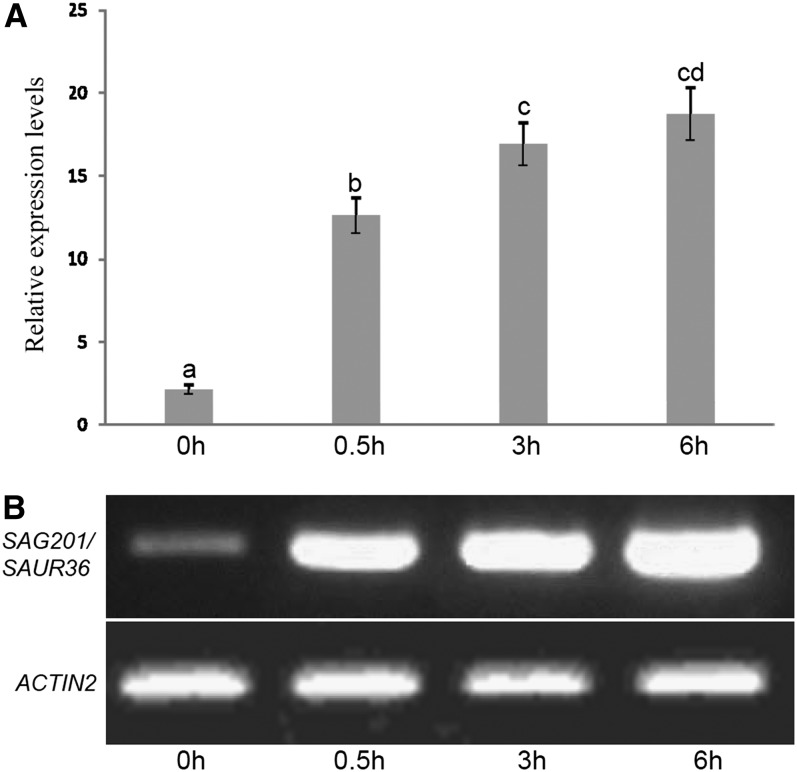
Studies have shown that some of the SAUR family genes can be rapidly induced by auxin treatment. To investigate whether SAUR36 is inducible by auxin, first we performed sequence analysis and found that there is the TGTCTC sequence in the 5′ UTR of SAUR36; the TGTCTC-containing sequence is an auxin response element (Ulmasov et al., 1995; Hagen and Guilfoyle, 2002). To confirm the inducibility of SAUR36 by auxin, 3-week-old wild-type plants were treated with α-naphthalene acetic acid (NAA; a synthetic auxin). Semiquantitative and quantitative PCR analyses revealed that the transcript levels of the gene increased readily after 30 min of NAA application and reached a high level (nearly a 20-fold increase relative to the expression at 0 h) 6 h after treatment (Fig. 2).
Figure 2.
Inducible expression of SAUR36 by NAA treatments in Arabidopsis leaves. The sixth rosette leaves of wild-type plants (30 DAE) were incubated in 3 mm MES buffer (pH 5.7) containing 20 μm NAA. The accumulation of SAG201/SAUR36 transcripts was determined using qPCR (A) and semiquantitative PCR (B). The expression levels in the 0-h leaves were set to 1. Letters indicate significant differences by Student’s t test (P = 0.05).
SAUR36 Is Knocked Out in Two T-DNA Insertion Lines
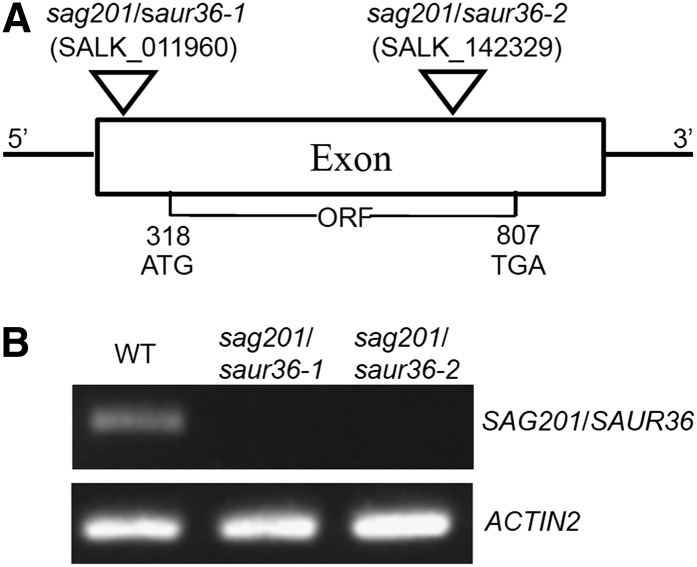
The SAUR36 gene of Arabidopsis is located on chromosome II. Like most SAUR genes in Arabidopsis (Knauss et al., 2003), the SAUR36 gene consists of only one exon (Fig. 3A) that encodes a protein with 162 amino acid residues. To investigate the biological function of the gene in leaf senescence, we obtained and identified two independent SALK transfer DNA (T-DNA) insertion lines (Columbia background) from the Arabidopsis Biological Resource Center at Ohio State University (Alonso et al., 2003). Line 1 (SALK_011960) has a T-DNA insertion in the 5′ UTR, and line 2 (SALK_118552) has a T-DNA insertion in the open reading frame (ORF; Fig. 3A). Plants homozygous for the T-DNA insertions were identified using PCR genotyping (Alonso et al., 2003). Semiquantitative PCR analyses showed that the SAUR36 transcripts in senescing leaves of two homozygous mutant lines were not detectable (Fig. 3B), suggesting that these lines are SAUR36 knockout or null mutants.
Figure 3.
Gene structure and expression of SAUR36 in two T-DNA insertion lines. A, Gene structure of SAUR36 and locations of two T-DNA insertions. B, Semiquantitative PCR analysis of SAUR36 expression in senescence leaves of the wild type (WT) and saur36-1 and saur36-2 T-DNA insertion lines.
Leaf Senescence Is Remarkably Delayed in the SAUR36 Knockout Mutant Plants
The above mutants were grown side by side in a growth chamber with corresponding wild-type plants to compare phenotypic changes in growth and development. The early stages of growth and development of both knockout lines appeared normal (Fig. 4A), but leaf senescence was significantly delayed compared with that of the wild type (Fig. 4B). The delay in leaf senescence was also obvious when the rosette leaves were excised from age-matched mutants and the wild type: the fifth leaf (numbered from the bottom; the first leaf is the oldest and the 11th leaf is the youngest) of the wild type became senescent while the third leaf of the knockout remained green (Fig. 4C).
Figure 4.
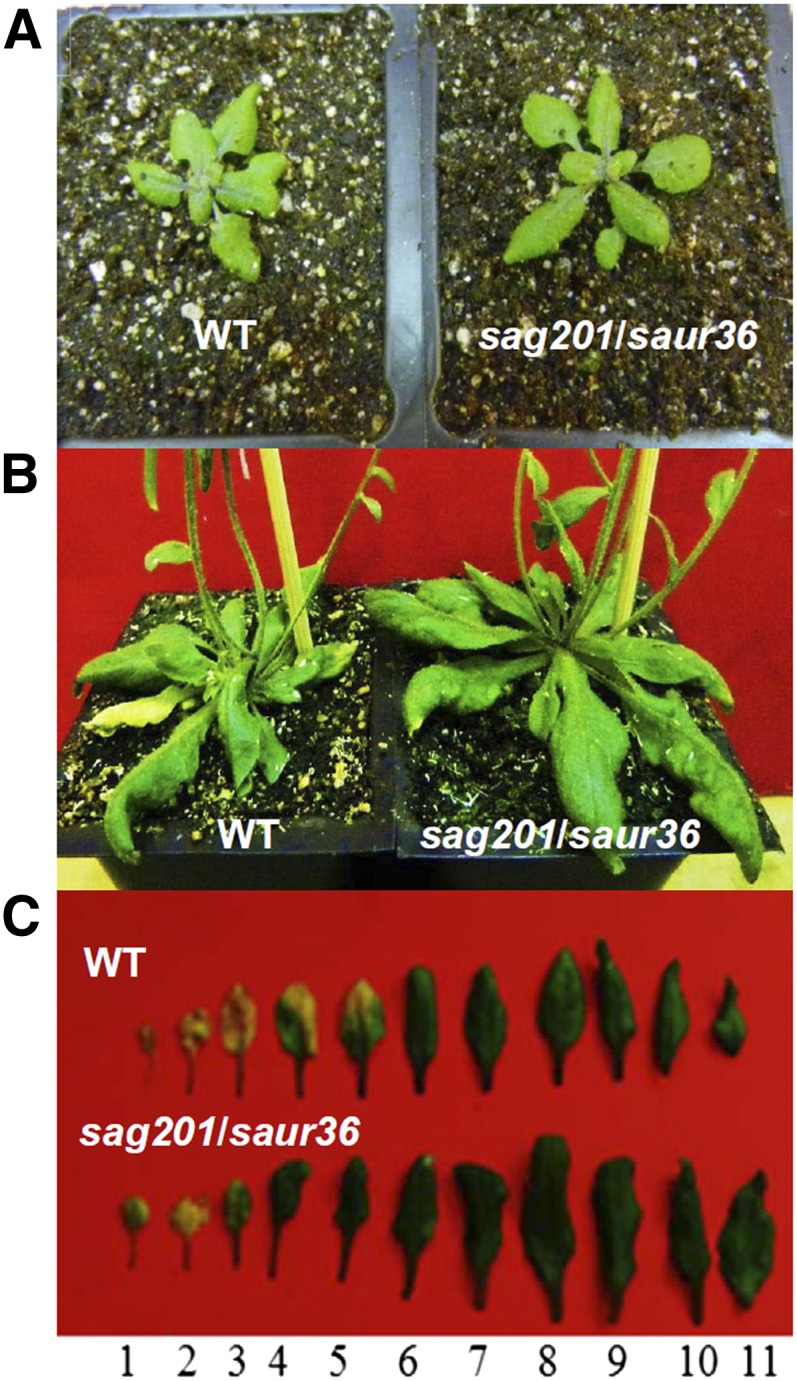
Delayed leaf senescence phenotype of the T-DNA insertion lines compared with that of the wild type. A, Phenotypes of null mutant and wild-type (WT) plants at approximately 10 DAE. Note that saur36 mutant plants are larger than the wild type. B, Phenotypes of null mutant and wild-type plants at approximately 40 DAE. Note the senescing leaves in the wild type and the larger leaves in the mutant plants. C, Phenotypes of the rosette leaves detached from age-matched plants shown in B. Leaves were numbered from bottom to top, with the first leaf being the oldest and the 11th leaf the youngest. [See online article for color version of this figure.]
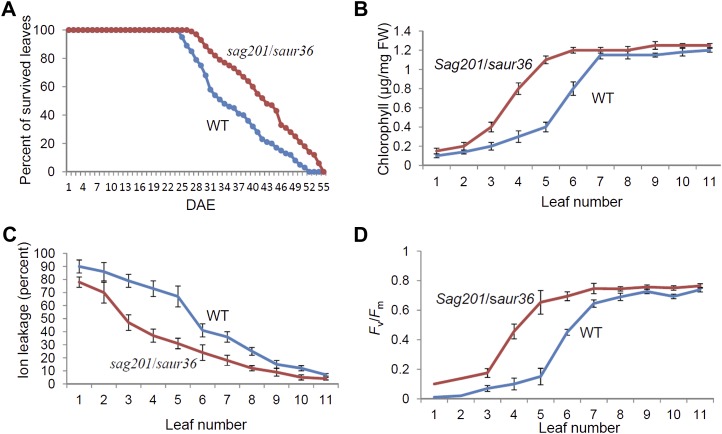
Further physiological analyses of the knockout mutant plants were performed. The leaf mortality curves (Fig. 5A) revealed that the leaves of saur36 started to senesce 3 to 4 d later than those of the wild type. Fifty percent of wild-type and null mutant leaves became senescent at 34 and 41 d after emergence (DAE), respectively, which means a 7-d delay in leaf senescence as measured by mortality analysis (Fig. 5A). The delay in leaf senescence of the null plants is also supported by changes in the total chlorophyll contents (Fig. 5B), in the ion leakage assays (Fig. 5C), and in the maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm; Fig. 5D). The Fv/Fm ratio is an indicator of the photochemical quantum efficiency of PSII and the photoreduction efficiency of the primary electron-accepting plastoquinone of PSII; a senescing leaf often loses its photosynthetic activity, as reflected by the decline in Fv/Fm ratios (He and Gan, 2002).
Figure 5.
Physiological analyses of leaves from saur36 mutant plants. A, Leaf survival curves of mutant and wild-type (WT) plants (n = 30). B to D, Chlorophyll content (B; n = 6), ion leakage (C; n = 6), and Fv/Fm ratio (D; n = 30) in individual rosette leaves of age-matched wild-type and mutant plants. Data represent mean values ± se. FW, Fresh weight. [See online article for color version of this figure.]
Leaves of saur36 Null Mutant Plants Are Much Larger
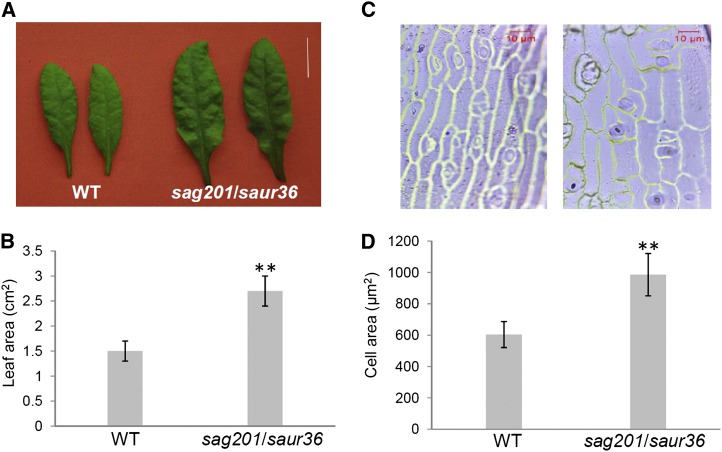
In addition to a significant delay in leaf senescence, the individual leaves of the saur36 null plants were larger compared with those of the wild type (Figs. 4 and 6A), which was confirmed by ImageJ (http://rsbweb.nih.gov/ij) leaf area measurements. The leaf area of the mutant plants was 83% greater than that of the wild type (Fig. 6A). Further microscopic analyses of the size of the epidermal cells (excluding stomatal cells) revealed that the individual cells were approximately 67% larger in the null mutant leaves than in wild-type leaves (Fig. 6B). These data suggest that the larger leaf area in the mutant plants results mainly from the larger cell size. The data also suggest that SAUR36 may inhibit cell expansion.
Figure 6.
Increased leaf and cell size of the saur36 mutants measured using ImageJ. A, Examples of fully expanded mature leaves of wild-type (WT) and mutant plants. Bar = 1 cm. B, Average areas of fully expanded mature wild-type and saur36 leaves. The sixth leaves of 35-DAE plants were used. Data represent mean values ± se (n = 6). **P ≤ 0.01 (significant difference). C, Examples of leaf epidermal cells from wild-type (left) and saur36 mutant (right) plants. D, Average cell areas of wild-type and saur36 leaves. Data represent mean values ± se (n = 30). **P ≤ 0.01 (significant difference). [See online article for color version of this figure.]
Inducible Overexpression of SAUR36 Causes Precocious Leaf Senescence
To further analyze the role of SAUR36 in leaf senescence, we performed gain-of-function analysis of SAUR36. Sequence analysis revealed that there is a DST element in the 3′ UTR of SAUR36 (Supplemental Fig. S1). Because highly conserved DST elements have been shown to be important for SAUR function (Newman et al., 1993), we generated inducible overexpression lines containing the SAUR36 ORF with [OE(+DST)] or without [OE(−DST)] its DST sequence. The glucocorticoid-inducible gene expression system (Aoyama and Chua, 1997) was used to avoid possible complications caused by constitutive expression (Guo and Gan, 2006; Zhang and Gan, 2012; Zhang et al., 2012).
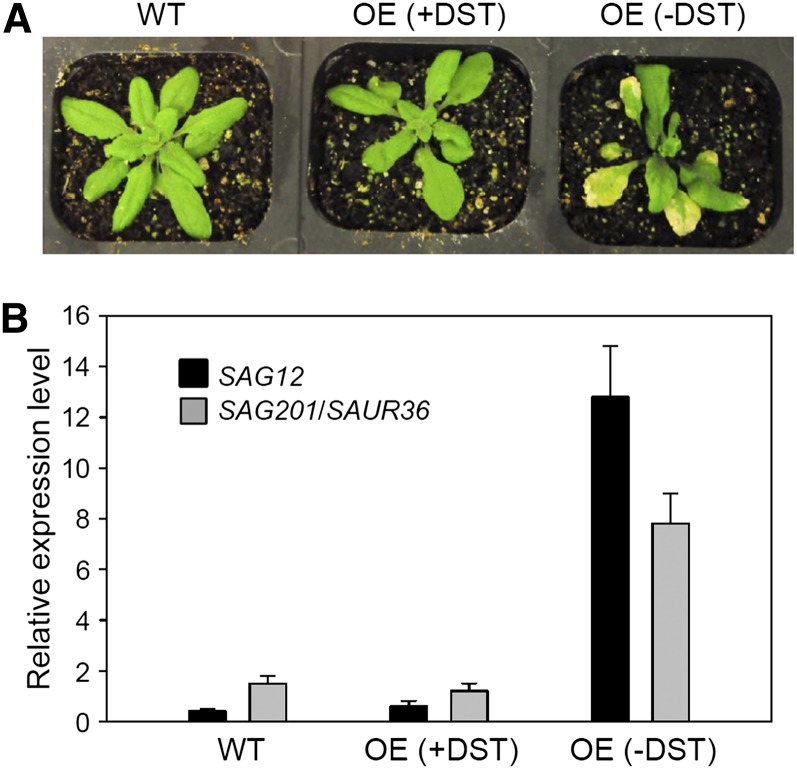
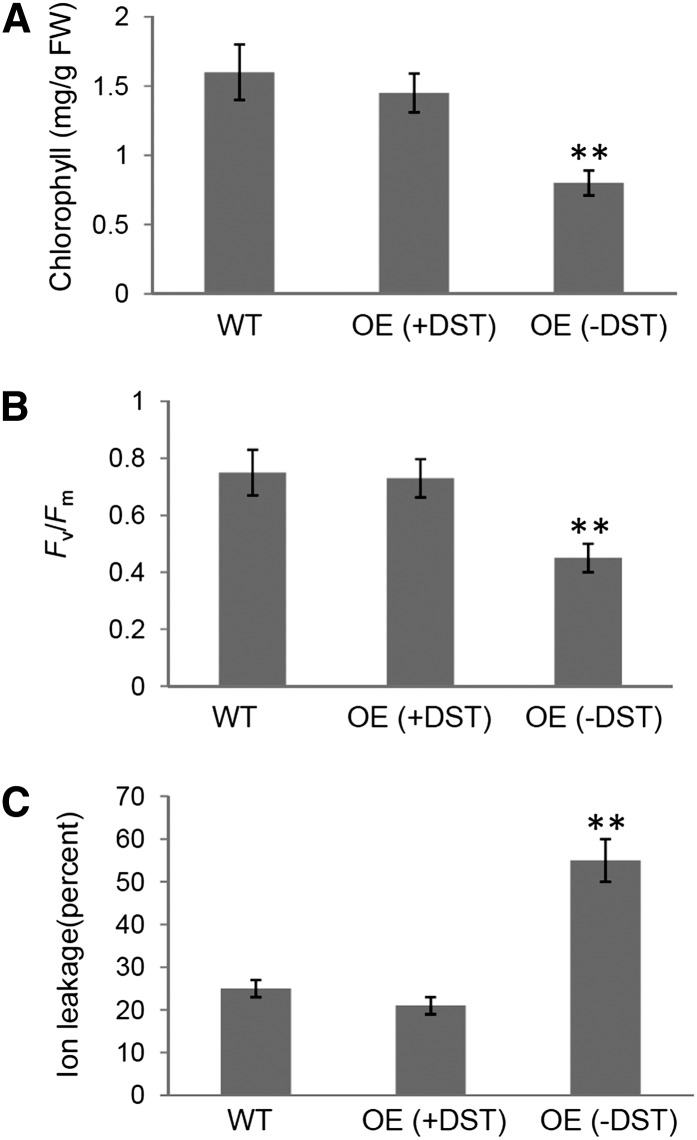
The effect of inducible overexpression of SAUR36 with or without the DST element on leaf senescence was analyzed after spraying whole plants (20 DAE) with dexamethasone (DEX; a synthetic glucocorticoid). Precocious leaf yellowing was readily observed 3 d after the DEX treatment in the OE(−DST) lines but not in the OE(+DST) plants or the wild-type plants (Fig. 7A). qPCR analysis showed that the SAUR36 transcripts accumulated to high levels in the OE(−DST) leaves but not in the OE(+DST) leaves or the wild-type plants (Fig. 7B). The DEX-treated OE(−DST) leaves displayed significantly reduced chlorophyll content, markedly declined Fv/Fm ratios, and high levels of ion leakage (Fig. 8). The senescence-specific marker gene SAG12 was also highly expressed in the OE(−DST) leaves (Fig. 7B). All these data suggest that the inducible expression of SAUR36 is sufficient to promote leaf senescence and that the DST element may cause rapid turnover of the SAUR36 transcripts.
Figure 7.
Analyses of Arabidopsis plants with induced overexpression of SAUR36 with [OE(+DST)] or without [OE(−DST)] DST. A, Phenotypes of wild-type (WT) and transgenic plants containing SAUR36 ORF with or without its DST sequence. The plants were 20 DAE, and the photograph was taken 3 d after treatment with 30 µm DEX. B, qPCR analysis of the transcript levels of SAUR36 and SAG12 in the fifth leaves of the plants shown in A. The expression level of SAUR36 in the OE(+DST) plants treated with DEX was set to 1. Data represent mean values ± se (n = 3). [See online article for color version of this figure.]
Figure 8.
Chlorophyll content (A), Fv/Fm ratio (B), and ion leakage (C) in the fifth leaves of various plants shown in Figure 7A. Data represent mean values ± se (n = 6). **P ≤ 0.01, indicating significant difference by Student’s t test. FW, Fresh weight; WT, wild type.
DISCUSSION
SUAR36 Positively Regulates Leaf Senescence in Arabidopsis
There are more than 70 SAUR family genes in the Arabidopsis genome that encode small 9- to 15-kD proteins (Hagen et al., 2010). These proteins have been shown to bind with Ca2+/calmodulin proteins in maize (Yang and Poovaiah, 2000) and Arabidopsis (Reddy et al., 2002), to regulate auxin synthesis and transport in rice (Kant et al., 2009), or to control apical hook development in Arabidopsis (Park et al., 2007), but the roles of most of the SAUR proteins in plant growth and development have yet to be investigated. In this report, we demonstrate that SAUR36 plays a role in promoting leaf senescence in Arabidopsis, because leaf senescence is substantially delayed in the saur36 null mutant plants both phenotypically and physiologically (Fig. 4), whereas the inducible expression of this gene (excluding its 3′ UTR such that the transcripts can accumulate, as seen in Fig. 7) promotes young leaves to undergo premature senescence (Figs. 7 and 8). As discussed below, it is likely that SAUR36, together with other genes, mediates auxin-promoted leaf senescence in Arabidopsis.
Auxin May Promote Leaf Senescence in Arabidopsis
Auxin has pervasive roles in regulating many aspects of plant growth and development and in response to various environmental cues (Zhao, 2008), and its involvement in senescence has long been observed (Sacher, 1973). However, how auxin affects leaf senescence is not clear. The identification and characterization of SAUR36 as a SAG reveals that SAUR36 mediates auxin-promoted leaf senescence.
There are several lines of evidence for an inhibitory role of auxin in leaf senescence. First, external application of synthetic or natural auxin can generally delay chlorophyll degradation and protein turnover in detached leaves (for review, see Gan, 2010). For example, NAA delays senescence in detached Arabidopsis leaves (Kim et al., 2011). Second, total auxin levels decrease in senescing leaves (Gan, 2010). Third, Arabidopsis plants overexpressing YUCCA6 contain high levels of auxin and display a prolonged leaf longevity phenotype in dark (Kim et al., 2011). Last, auxin treatment can down-regulate the transcript levels of SAG12 in detached senescing leaves (Noh and Amasino, 1999; Gan, 2010); SAG12 is a highly senescence-specific marker gene (Gan and Amasino, 1995). The suppression of leaf senescence by auxin appears to decline with aging (Woo et al., 2010).
However, there are also lines of evidence for a promoting role of auxin in leaf senescence. It is not unusual to observe that application of auxin promotes leaf senescence, and the endogenous auxin activity increases just prior to or at the onset of leaf senescence, as revealed in some early experiments (Atsumi and Hayashi, 1979; Gan, 2010). In fact, the levels of free IAA (the biologically active form) increase during leaf senescence in Arabidopsis and reach approximately 2-fold higher levels in leaves that are 50% yellowing compared with fully expanded green leaves, although the total auxin levels decrease (Quirino et al., 1999). Consistent with these findings, genes whose products are involved in auxin biosynthesis, such as Trp synthase (AtTSA1) and nitrilases (NIT1–NIT4), are up-regulated during leaf senescence (Quirino et al., 1999; van der Graaff et al., 2006); nitrilase converts indole-3-acetonitrile to IAA. Furthermore, several auxin response factor genes, such as ARF1, ARF2, ARF7, and ARF19, are expressed in senescing leaves in Arabidopsis. The T-DNA knockout arf2 mutant exhibits a delay in leaf senescence, and the delay in senescence becomes more striking in the arf1arf2 double mutant plants (Ellis et al., 2005). Most recently, a Senescence-Associated Receptor-Like Kinase (SARK) from soybean (Glycine max) and its ortholog in Arabidopsis have been shown to promote leaf senescence, and auxin (and ethylene) biosynthesis and signaling pathways are required for SARK to cause leaf senescence (Xu et al., 2011).
Our data reported here further support that auxin promotes leaf senescence. As discussed above, SAUR36 plays a critical role in promoting leaf senescence. Like many other SAURs that can be rapidly induced by exogenously applied auxin (Li et al., 1994; Hagen and Guilfoyle, 2002), SAUR36 is an early auxin-responsive gene that can be readily up-regulated by NAA treatment (Fig. 2). The expression of SAUR36 is up-regulated during the progression of leaf senescence (Fig. 1), which likely results from the elevated levels of the endogenous active IAA (Quirino et al., 1999). In short, auxin appears to promote leaf senescence in part by inducing the expression of SAUR36 and possibly other positive senescence-regulating genes.
How SAUR36 mediates auxin-promoted leaf senescence is not clear. The SAUR36 protein contains a nuclear localization signal, 59-RRRS-62 (Supplemental Fig. S1), predicted by the NLStradamus program (Nguyen Ba et al., 2009). A GFP fusion protein confirms that the SAUR36 protein is targeted to the nucleus (Narsai et al., 2011). Auxin can activate the 1-aminocyclopropane-1-carboxylate biosynthetic step to stimulate ethylene production, which in turn promotes senescence (Abel et al., 1995). Whether this nuclear protein functions through or is independent of ethylene has yet to be investigated.
SAUR36 May Also Be Involved in Leaf Cell Expansion
In addition to a positive role in leaf senescence, SAUR36 appears to be involved in leaf cell expansion. qPCR analysis revealed that this gene is expressed in expanding leaves at low levels prior to the up-regulation at the onset of senescence (Fig. 1B). When SAUR36 is knocked out due to T-DNA insertion, the leaves are 83% larger than wild-type leaves in terms of area (Fig. 6, A and B), which is mostly due to the formation of larger cells (Fig. 6, C and D). These data suggest that SAUR36 may inhibit cell enlargement. Early studies have also shown that some SAUR proteins play a role in auxin-modulated cell elongation (Hagen and Guilfoyle, 2002; Knauss et al., 2003; Hagen et al., 2010).
Transcriptional and Posttranscriptional Regulation of SAUR36
SAUR transcripts are unstable (McClure and Guilfoyle, 1987; Feldbrügge et al., 2002), and their high turnover rate may be due to the DST element in the 3′ UTR (Newman et al., 1993). DST is generally a 30- to 40-bp sequence consisting of three highly conserved boxes that are separated by two variable regions. The DNA sequence of the DST element and its location are highly conserved in different plant species (Newman et al., 1993). The sequences AUAGAU and GUA, which are considered invariable in DST, are present in the 3′ UTR of SAUR36 (Supplemental Fig. S1). Inducible overexpression of SAUR36 with its 3′ UTR did not cause precocious leaf senescence, because its transcripts were barely detectable (Fig. 7), presumably being rapidly degraded. However, the low levels of SAUR36 in nonsenescing leaves and the high levels in senescing leaves appeared to be regulated at the transcriptional level, because the GUS reporter directed by the SAUR36 promoter was highly detected in senescing leaves and very low in young leaves (Fig. 1C); the PSAUR36-GUS chimeric gene did not contain the DST element.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in this study. Two T-DNA insertion lines (SALK_011960 and SALK_142329, Columbia background) were obtained from the Arabidopsis Biological Resource Center (Alonso et al., 2003). As suggested by the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/tdnaprimers.2.html), a PCR-based method was used to identify homozygous T-DNA insertion mutants. The T-DNA left border primer G2325 and the gene-specific primers G3516 and G3517 for saur36-1 (SALK_011960) and G3518 and G3519 for saur36-2 (SALK_142329) were used. Plants homozygous for the T-DNA insertion were used in this study. The primers used in this research are listed in Supplemental Table S1.
Seeds were surface sterilized and then sown on one-half-strength Murashige and Skoog salts with 0.7% (w/v) phytoagar (Sigma) and appropriate antibiotics. The dishes were kept at 4°C for 2 d and then moved to a growth chamber at 22°C with 60% relative humidity under continuous light (110 μmol m−2 s−1) from a mixture of fluorescent and incandescent bulbs. The 10-d-old seedlings were transferred into Cornell mix soil (3:2:1 peat moss:vermiculite:perlite, v/v/v) and grown under the same conditions. The mutants, transgenic plants, and the wild type were grown side by side.
Plasmid Construction and Transformation of Arabidopsis
To create the PSAUR36-GUS construct, PCR products of G3556 and G3557 were cloned into pGEM-T Easy vector (Promega). The coding sequence was then released with XbaI and BamHI and subsequently cloned into pBI101 to form the GUS chimeric gene.
To make the inducible SAUR36 overexpression constructs, the SAUR36 coding regions with or without the DST element were cloned into the inducible binary vector pGL1152 (Guo and Gan, 2006).
The above binary vectors were transferred into Agrobacterium tumefaciens strain ABI. The A. tumefaciens cells containing the related binary vector constructs were used to transform Arabidopsis plants via floral dipping (Clough and Bent, 1998). Plant transformants were selected on one-half-strength Murashige and Skoog medium containing suitable antibiotics and then transferred to soil. Approximately 30 T1 transgenic lines for each transgene were selected; phenotypic analyses were performed in the T2 generation and further confirmed in the T3 generation. Homozygous plants were used in all experiments.
Chemical Induction of Gene Expression
Glucocorticoid treatments were performed as described previously (Aoyama and Chua, 1997). Twenty-day-old plants were sprayed with 30 μm DEX (synthetic glucocorticoid). The images were recorded after 3 d, and the seedlings were sampled for chlorophyll content, fluorescence, and ion leakage assays.
Histochemical GUS Staining, Chlorophyll Content, Fluorescence, and Ion Leakage Assay
Chlorophyll, fluorescence, and ion leakage analyses as well as histochemical GUS staining were performed as described previously (Zhang and Gan, 2012; Zhang et al., 2012).
Transcript Analysis
RNA extraction, complementary DNA synthesis, and qPCR analysis were carried out as described previously (He and Gan, 2002). First-strand complementary DNA was synthesized from 3 μg of total RNA (treated with RNase-free DNase; New England Biolabs) at 42°C with Promega MV-Reverse Transcriptase. For qPCR, 1 μL of each diluted sample was used as a template in a 25-μL reaction following the standard methods. All PCRs were performed on a Bio-Rad IQ-5 thermocycler with 40 cycles and an annealing temperature of 55°C. Cycle threshold values were determined by IQ-5 Bio-Rad software assuming 100% primer efficiency. Three mRNA samples from three independently harvested leaf samples were qPCR analyzed.
Auxin Treatment
The synthetic auxin NAA (Sigma) was used. The sixth leaves from approximately 30-d-old wild-type plants were incubated for various times at room temperature in 3 mm MES buffer (pH 5.7) containing 20 μm NAA.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SAG201/SAUR36 (AT2G45210), SAG12 (AT5G45890), ACTIN2 (AT3G18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide and deduced amino acid sequence of SAG201/SAUR36 and the alignment of highly conserved DST sequence.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Dr. Peter Davies (Cornell University), Dr. Yongfeng Guo (Chinese Academy of Agricultural Sciences), and William Gan (Cornell University) for critical reading of the manuscript. Dr. Xiuying Xia, Dr. Xiaohong Kou, Mr. Xingwang Liu, and other members of the Gan laboratory are thanked for useful discussion.
Glossary
- IAA
indole-3-acetic acid
- DST
downstream
- UTR
untranslated region
- qPCR
quantitative PCR
- NAA
α-naphthalene acetic acid
- ORF
open reading frame
- DAE
days after emergence
- Fv/Fm
maximum photochemical efficiency of PSII in the dark-adapted state
- DEX
dexamethasone
- T-DNA
transfer DNA
References
- Abel S, Nguyen MD, Chow W, Theologis A. (1995) ASC4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. J Biol Chem 270: 19093–19099 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Atsumi S, Hayashi T. (1979) Examination of the pronounced increase in auxin content of senescent leaves. Plant Cell Physiol 20: 861–865 [Google Scholar]
- Besseau S, Li J, Palva ET. (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63: 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574 [DOI] [PubMed] [Google Scholar]
- Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. (2006) A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA 103: 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge M, Arizti P, Sullivan ML, Zamore PD, Belasco JG, Green PJ. (2002) Comparative analysis of the plant mRNA-destabilizing element, DST, in mammalian and tobacco cells. Plant Mol Biol 49: 215–223 [DOI] [PubMed] [Google Scholar]
- Gan S-S. (2010) Hormonal control of plant senescence. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action, Ed 3 Rev. Springer Netherlands, New York, pp 597–617. [Google Scholar]
- Gan S-S, Amasino RM. (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988 [DOI] [PubMed] [Google Scholar]
- Gan S-S, Amasino RM. (1997) Making sense of senescence: molecular genetic regulation and manipulation of leaf senescence. Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp M-J, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M. (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S-S. (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27: 521–549 [Google Scholar]
- Guo Y, Gan S-S. (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S-S. (2012) Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ 35: 644–655 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ, Gray WM. (2010) Auxin signal transduction. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action, Ed 3 Rev. Springer Netherlands, New York, pp 282–307. [Google Scholar]
- Hajouj T, Michelis R, Gepstein S. (2000) Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol 124: 1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Gan S-S. (2002) A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. (2006) Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88: 360–371 [DOI] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu T, Rothstein SJ. (2009) SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol 151: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Murphy AS, Baek D, Lee S-W, Yun D-J, Bressan RA, Narasimhan ML. (2011) YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot 62: 3981–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L. (2003) The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem 278: 23936–23943 [DOI] [PubMed] [Google Scholar]
- Lers A, Sonego L, Green PJ, Burd S. (2006) Suppression of LX ribonuclease in tomato results in a delay of leaf senescence and abscission. Plant Physiol 142: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu ZB, Shi X, Hagen G, Guilfoyle TJ. (1994) An auxin-inducible element in soybean SAUR promoters. Plant Physiol 106: 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T. (1987) Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623 [DOI] [PubMed] [Google Scholar]
- Narsai R, Law SR, Carrie C, Xu L, Whelan J. (2011) In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol 157: 1342–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ. (1993) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5: 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. (2009) NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics 10: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM. (1999) Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol 41: 181–194 [DOI] [PubMed] [Google Scholar]
- Park J-E, Kim Y-S, Yoon H-K, Park C-M. (2007) Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci 172: 150–157 [Google Scholar]
- Park SH, Chung PJ, Juntawong P, Bailey-Serres J, Kim YS, Jung H, Bang SW, Kim YK, Do Choi Y, Kim JK. (2012) Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′ and 5′ untranslated regions and correlates with differential polysome association in rice. Plant Physiol 159: 1111–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RM. (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40: 267–278 [DOI] [PubMed] [Google Scholar]
- Reddy VS, Ali GS, Reddy ASN. (2002) Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J Biol Chem 277: 9840–9852 [DOI] [PubMed] [Google Scholar]
- Sacher JA. (1973) Senescence and postharvest physiology. Annu Rev Plant Physiol 24: 197–224 [Google Scholar]
- Scarpella E, Barkoulas M, Tsiantis M. (2010) Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol 2: a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan ML, Green PJ. (1996) Mutational analysis of the DST element in tobacco cells and transgenic plants: identification of residues critical for mRNA instability. RNA 2: 308–315 [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. (1995) Composite structure of auxin response elements. Plant Cell 7: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R. (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Kim J, Kim J, Lee U, Song IJ, Kim JH, Lee HY, Nam HG, Lim PO. (2010) The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot 61: 3947–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Meng T, Li P, Yu Y, Cui Y, Wang Y, Gong Q, Wang NN. (2011) A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol 157: 2131–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. (2000) Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem 275: 3137–3143 [DOI] [PubMed] [Google Scholar]
- Zenser N, Dreher KA, Edwards SR, Callis J. (2003) Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J 35: 285–294 [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Laun T, Miao Y. (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol 89: 133–137 [DOI] [PubMed] [Google Scholar]
- Zhang K, Gan S-S. (2012) An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol 158: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Xia X, Zhang Y, Gan S-S. (2012) An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J 69: 667–678 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2008) The role of local biosynthesis of auxin and cytokinin in plant development. Curr Opin Plant Biol 11: 16–22 [DOI] [PubMed] [Google Scholar]
- Zhou C, Cai Z, Guo Y, Gan S-S. (2009) An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol 150: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]