Summary: This study shows that seed-specific recombinant antibody production evokes endoplasmic reticulum stress, triggering the unfolded protein response without greatly affecting seed germination and seedling growth, however, making the PPHAS-driven expression cassette a suitable system for molecular farming.
Abstract
Among the many plant-based production systems that are being tested for molecular farming, seeds are very attractive, as they provide a stable environment in which the accumulating recombinant proteins can be stored. However, it is not known exactly how high production levels of recombinant antibodies influence the endogenous transcriptome and proteome of the developing seed. To address this question, we studied the transcriptomic status in developing Arabidopsis (Arabidopsis thaliana) seeds 13 d post anthesis of three transgenic lines, producing varying levels of recombinant VHH or single-chain Fv antibody fragments fused to the human immunoglobulin G1-derived Fc fragment under the control of the β-PHASEOLIN seed-specific promoter. Using genome-wide Tiling arrays, we demonstrated that only a small proportion of the transcriptome was significantly changed in each of the lines compared with the wild type. Strikingly, in all three lines, we found a large overlap of up-regulated genes corresponding to protein folding, glycosylation/modification, translocation, vesicle transport, and protein degradation, suggestive of a state of cellular stress called the unfolded protein response. Moreover, the gene up-regulation amplitude was similar in all three lines. We hypothesize that the production of recombinant antibodies in the endoplasmic reticulum triggers endoplasmic reticulum stress, causing a disturbance of the normal cellular homeostasis.
The use of genetically engineered crops to produce highly valuable proteins, called molecular farming, provides a promising alternative for the currently existing protein production systems that are mainly based on bacterial, yeast (Saccharomyces cerevisiae and Pichia pastoris), insect, and mammalian cells. Plants offer economical advantages for certain applications and ease in scalability and avoid most safety issues concerning viral and toxin contaminants (Fischer et al., 2004; Ma et al., 2005). Among the various plant-based production platforms, seeds are very important because they are compact organs, provide a stable environment, and have a high protein content, which is the outcome of their inherent physiological function for producing and storing proteins (Stoger et al., 2005; Lau and Sun, 2009; Boothe et al., 2010; Peters and Stoger, 2011). To evaluate the seed-specific production of recombinant proteins, the regulatory sequences of seed storage protein (SSP) genes have been used extensively. For instance, De Jaeger and coworkers (2002) demonstrated that an expression cassette using the regulatory sequences of ARCELIN5-I and β-PHASEOLIN of bean (Phaseolus vulgaris) could boost the accumulation of a single-chain Fv (scFv) up to 36.5% of the total soluble protein (TSP). Using the same cassette, a single-chain antibody fragment fused to an Fc fragment (scFv-Fc) resulted in the production of 10% bivalent functional antibodies of the TSP in Arabidopsis (Arabidopsis thaliana) seeds (Van Droogenbroeck et al., 2007). Immunolocalization studies on one of these scFv-Fc-expressing lines revealed an aberrant localization of the recombinant scFv-Fc, as well as of endoplasmic reticulum (ER) chaperone binding proteins (BiPs) and calreticulin, and of the endogenous SSP cruciferin, suggesting that the cellular homeostasis was disturbed. However, not all transformants, accumulating scFv-Fc antibodies in ER-derived vesicles, show this phenotype, indicating that also the environmental conditions of seed development and ripening affect the deposition of the recombinant protein (Van Droogenbroeck et al., 2007; Loos et al., 2011b). Similarly, interleukin-10 accumulated in compartments delimited by ribosome-associated membranes, which most likely derive from the ER (Morandini et al., 2011). Probably, the formation of these compartments does not depend on the amount of recombinant protein produced but on specific properties of the recombinant protein itself (Morandini et al., 2011). In the same study, the 65-kD isoform of Glu decarboxylase and proinsulin were, despite bearing a KDEL tag, both localized in the protein storage vacuoles, a phenomenon reported before (Petruccelli et al., 2006; Van Droogenbroeck et al., 2007; Loos et al., 2011a).
Understanding when and how SSPs are produced in developing seeds is important when using their regulatory sequences for heterologous protein production. SSPs are primarily synthesized in the late stages of seed development and accumulate in protein storage vacuoles as nutritional sources of nitrogen and sulfur, which are resorbed upon germination to give rise to the developing plant (Goldberg et al., 1994; Herman and Larkins, 1999). The synthesis of seed storage reserves in Arabidopsis occurs between 8 and 16 d post anthesis (dpa), with maximum accumulation rates between 9 and 13 dpa (Focks and Benning, 1998; Baud et al., 2002). SSP accumulation and transcription are correlated, indicative for SSP gene regulation at the transcriptional level. However, a lag phase of 1 to 2 d is observed between SSP transcription and accumulation, indicating also translational control (Li et al., 2007). The transcriptome of developing Arabidopsis seeds has been studied using EST libraries (White et al., 2000) as well as via microarray analysis (Girke et al., 2000; Ruuska et al., 2002; Spencer et al., 2007; Day et al., 2008; Le et al., 2010). Still, the coordination of gene activity during seed development is not completely understood, and the identification of, for instance, seed-specific transcription factors (TFs) will help to unravel the regulatory networks involved in the programming of seed development (Le et al., 2010). All of this is extremely valuable for the optimization of molecular farming purposes.
Generally, a higher recombinant protein production is achieved by targeting naturally secreted proteins to the secretory pathway, where posttranslational modifications, like protein folding, assembly, and glycosylation, take place, as compared with a cytoplasmic localization (Benchabane et al., 2008; Sharma and Sharma, 2009). Proteins reach their proper folding and conformation by the action of the folding-refolding machinery assisted by a set of folding chaperones, which mainly reside in the ER lumen (Ellgaard et al., 1999; Ellgaard and Helenius, 2003; van Anken and Braakman, 2005; Anelli and Sitia, 2008). When stress affects the protein-folding mechanism in the ER, or when too many secretory proteins are synthesized and transported into the ER lumen, the (re)folding system gets overloaded and the proteins cannot achieve their proper conformation anymore. In this way, unfolded or misfolded proteins accumulate, resulting in ER stress, which may compromise ER activities, such as protein synthesis and folding and cell viability, due to a decrease in essential proteins (Lu and Christopher, 2008; Moreno and Orellana, 2011).
To maintain the cellular homeostasis of the ER, signal transduction pathways are induced, which are collectively grouped into the unfolded protein response (UPR). The UPR mitigates ER stress by up-regulating the expression of genes encoding components of the protein folding-refolding machinery, such as ER-resident chaperones and folding enzymes. Also, the ER-associated degradation system can be activated (Urade, 2007, 2009; Vitale and Boston, 2008). The core chaperones activated by the UPR are the families of BiP, calnexin/calreticulin, protein disulfide isomerase (PDI), and DnaJ proteins, which all mediate multiple functions (Yoshida et al., 1998; Koizumi et al., 2001; Noh et al., 2003; Yamamoto et al., 2008). BiP is an ER-resident HSP70 family protein, which is important for overall plant viability and, among other roles, assists in the folding of glycoproteins, serves as a stress sensor, and can indirectly activate the UPR cascade (Simons et al., 1995; Hendershot et al., 1996; Fewell et al., 2001; Kimata et al., 2004; Nishikawa et al., 2005; Liebrand et al., 2012). PDI chaperones catalyze the formation and rearrangement of disulfide bonds between the correct pairs of Cys residues in nascent polypeptide chains in the ER (Gilbert, 1998; Fomenko and Gladyshev, 2003). The tandem calnexin/calreticulin consists of lectin chaperones, which are mainly involved in the ER quality control based on the recognition of certain sugar moieties in the unfolded proteins (Ellgaard and Helenius, 2001, 2003; van Anken and Braakman, 2005). Finally, the DnaJ protein Thermosensitive Male Sterile1 (TMS1) is a molecular chaperone of the HSP40 family and is suggested to work in cooperation with the HSP70 chaperones (Yamamoto et al., 2008).
Recombinant protein production is a demanding process for the host cell, as an additional foreign protein needs to be produced besides the endogenous proteome. In some cases, this burden on the cell is translated into the activation of the UPR, and this can both positively and negatively influence the recombinant protein yield. For instance, in yeast, it was found that the overexpression of heterologous 4-4-20 scFv variants activated the UPR response, which in turn led to a reduction of the scFv yield (Kauffman et al., 2002; Xu et al., 2005; Xu and Robinson, 2009). On the other hand, disrupting the endoplasmic reticulum-associated degradation (ERAD) pathway in filamentous fungi led to an enhanced heterologous protein expression by delaying protein degradation (Carvalho et al., 2011).
A lot is known about the UPR in yeast and mammals, but the plant UPR has not received much attention and its molecular components are not yet completely clear. Several ER stress-responsive genes, like BIP, PDI, and CALNEXIN, have been identified in Arabidopsis by microarray analysis of plants and plantlets in which the UPR was provoked by treating them with the chemical agents tunicamycin and dithiothreitol (DTT), which interfere with N-glycosylation or disulfide bond formation, respectively (Martínez and Chrispeels, 2003; Kamauchi et al., 2005). Although most of the induced genes were found to be orthologs of the mammalian and yeast UPR genes (Ng et al., 2000; Travers et al., 2000; Scheuner et al., 2001; Okada et al., 2002; Harding et al., 2003; Lee et al., 2003), in the plant UPR, additional genes involved in translational regulation (P58IPK) and apoptosis (BAX INHIBITOR1) were found to be up-regulated (Kamauchi et al., 2005).
Here, we analyzed the transcriptome of developing Arabidopsis seeds that accumulate fusions of an scFv antibody fragment or a VHH camel antigen-binding fragment with a human immunoglobulin Fc fragment (scFv-Fc or VHH-Fc). First, we investigated at which point during Arabidopsis seed development the antibody starts accumulating when expressed under the control of the heterologous PHASEOLIN promoter. In Arabidopsis, the 2S albumins and 12S globulins are the major SSP classes, both strictly regulated spatially and temporally (Fujiwara et al., 2002). Transcriptional and proteomic profiling of 12S SSP expression and accumulation revealed that transcription activities for all four 12S SSP genes increased from 8 to 15 dpa, followed by a very rapid increase in the 12S globulin accumulation from 12 to 23 dpa (Li et al., 2007). Then, the influence of recombinant scFv-Fc or VHH-Fc accumulation on the transcriptome was probed by performing a comparative microarray and quantitative PCR (qPCR) analysis on developing seeds. Finally, the differentially expressed genes were analyzed for their biological functions. Our results provide insight into the biological processes that are changed upon recombinant protein production in Arabidopsis seeds and suggest that seed-specific antibody production evokes ER stress triggering the UPR, already at antibody accumulation levels of 1% of total soluble seed protein.
RESULTS
Maltose-Binding Protein scFv-Fc Starts Accumulating Only after 8 dpa in Arabidopsis Seeds
Three different transgenic lines were selected to study the effect of antibody accumulation on seed homeostasis (Table I; Fig. 1). These transgenic lines were selected on the basis of two requirements. First, the selected lines accumulated similar bivalent antibody formats, and second, the amount of produced recombinant protein was different in the different seed stocks. In this way, we were able to determine whether the up- and/or down-regulated genes were specific or not for (1) one transgenic line and (2) a certain amount of produced recombinant antibody in the seeds. The lines HC26 and HC28 express a VHH-Fc antibody, which is a fusion between the antigen-binding domain of a heavy chain camel antibody (nanobody or VHH) directed against a prostate-specific antigen (VHH7) and a human IgG1-derived Fc fragment (Saerens et al., 2004; S. De Buck, J. Nolf, T. De Meyer, V. Virdi, K. De Wilde, E. Van Lerberge, B. Van Droogenbroeck, and A. Depicker, unpublished data). The transgenic line MBP39 harbored a transfer DNA (T-DNA) construct carrying the gene encoding an scFv-Fc antibody against maltose-binding protein (MBP; Van Droogenbroeck et al., 2007). In contrast to a VHH, the scFvantigen-binding element consists of two variable domains connected by a flexible linker, but similar to the VHH-Fc antibody format, the scFv-Fc contains an Fc fragment of a human IgG1 antibody (Van Droogenbroeck et al., 2009). Expression of the recombinant antibody fragments was in all three lines driven by the β-PHASEOLIN promoter (PPHAS; Hall et al., 1999). Recombinant VHH-Fc accumulated at 10% of the TSP in T3 homozygous seed stocks from line HC28 and at 5% of the TSP in T4 homozygous seed stocks from line HC26 (Table I; see “Materials and Methods”; S. De Buck, J. Nolf, T. De Meyer, V. Virdi, K. De Wilde, E. Van Lerberge, B. Van Droogenbroeck, and A. Depicker, unpublished data). T4 homozygous seed stocks from the MBP39 line accumulated the scFv-Fc at 10% of the TSP (Table I; Van Droogenbroeck et al., 2007).
Table I. Summary of the plant material used in each experiment.
See also “Materials and Methods.” References are as follows: 1, Van Droogenbroeck et al. (2007); 2, S. De Buck, J. Nolf, T. De Meyer, V. Virdi, K. De Wilde, E. Van Lerberge, B. Van Droogenbroeck, and A. Depicker, unpublished data.
Figure 1.
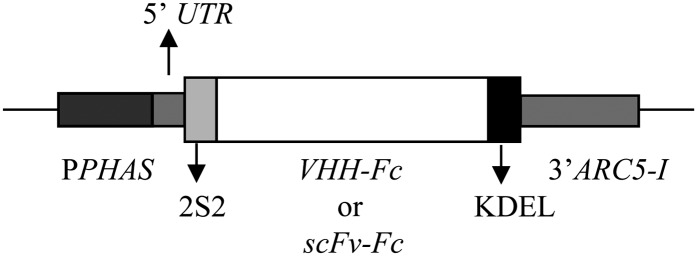
Outline of the PPHAS expression cassette for seed-specific antibody expression. PPHAS, Promoter of the β-PHASEOLIN gene from bean; 5′ UTR, 5′ untranslated region; 2S2, signal peptide of the 2S2 SSP gene from Arabidopsis; VHH-Fc, coding sequence of the VHH-Fc antibody fusion; scFv-Fc, coding sequence of the scFv-Fc antibody fusion; KDEL, ER-retention signal; 3′ARC5-I, 3′ end of the ARCELIN5-I gene. In the VHH-Fc-containing construct, the VHH-Fc coding region is flanked by attB Gateway recombination sites (De Buck et al., 2012; S. De Buck, J. Nolf, T. De Meyer, V. Virdi, K. De Wilde, E. Van Lerberge, B. Van Droogenbroeck, and A. Depicker, unpublished data). The structure is not drawn to scale.
In order to determine the ideal time point for microarray sampling, we investigated the PPHAS-driven antibody production during Arabidopsis seed development in the transgenic line MBP10 by means of an ELISA-based assay (Table I; Van Droogenbroeck et al., 2007; see “Materials and Methods”). As endogenous (storage) protein accumulation in Arabidopsis seeds starts around 7 dpa (Baud et al., 2002) and the major storage protein reserves accumulate from 8 to 16 dpa (Ruuska et al., 2002), siliques of transgenic plants expressing the anti-MBP scFv-Fc antibody were harvested at 5, 8, 11, and 14 dpa. There was no need to microdissect seeds from siliques, as it was previously shown that PPHAS is embryo specific in Arabidopsis and does not drive expression in the silique wall (Chandrasekharan et al., 2003).
At 5 dpa, no signal was detected, meaning that the anti-MBP scFv-Fc antibody accumulation was still below 0.05% of the TSP. However, a faint signal appeared at 8 dpa, corresponding to 0.05% of the TSP, and the scFv-Fc accumulation increased steadily to 1.3% of the TSP at 11 dpa and to 2.64% of the TSP at 14 dpa, demonstrating that MBP accumulates at high levels already between 11 and 14 dpa (data not shown).
Recombinant Antibody Accumulation Evokes ER Stress
Microarray Analysis Revealed the Up-Regulation of UPR Components
To evaluate possible expression changes in the transcriptome upon antibody production in developing seeds of the three antibody-expressing lines (VHH-Fc lines HC26 and HC28 and scFv-Fc line MBP29) compared with a control line, ecotype Columbia (Col-0; the wild type), plants were grown in a controlled environment and developing seeds were harvested at 13 dpa in three independent biological repeats. The extracted RNA was subjected to transcriptome analysis via hybridization to Arabidopsis Tiling 1.0R arrays (Affymetrix; www.affymetrix.com) that cover in total 29,890 genes and thus about 9,000 additional genes in comparison with the classical ATH1 arrays (Naouar et al., 2009). For details about sampling, hybridization, and data processing, see “Materials and Methods.”
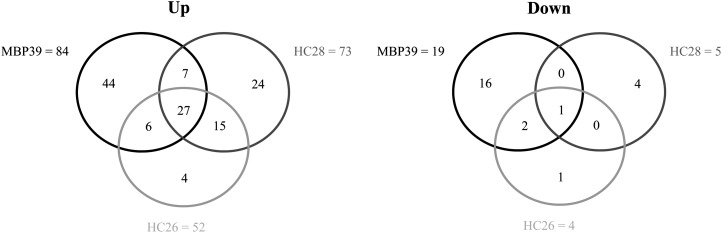
Statistical analysis of the microarray data indicated that all three antibody-accumulating lines showed a significant differential expression of a small part of the transcriptome compared with the wild type (Tables II–IV; Supplemental Tables S1–S4). Relative to the wild type, 78, 56, and 103 genes were differentially regulated in HC28, HC26, and MBP39, respectively. Most of these differentially regulated genes were up-regulated (1.8-fold cutoff): 73 for HC28, 52 for HC26, and 84 for MBP39. Strikingly, an overlap of 27 up-regulated genes was found between the three transgenic lines, as illustrated in Figure 2. In addition, 15 up-regulated genes were common to lines HC28 and HC26 (Table III; Fig. 2), seven between lines MBP39 and HC28 (Table IV; Fig. 2), and six between lines MBP39 and HC26 (Table V; Fig. 2). On the other hand, only one gene (F16N3.19), encoding a defensin-like protein, was down-regulated in all three analyzed transgenic lines (Fig. 2; Table IV). Because transcription of the SSP-encoding genes in Arabidopsis constitutes 30% of the transcriptome, the transcript levels are expected not to be under the cutoff value of 1.8; therefore, they were analyzed separately. The 2S albumins are encoded by five genes (SESA1 [At4G27140], SESA2 [At4G27150], SESA3 [At4G27160], SESA4 [At4G27170], and SESA5 [At5G54740]), and the 12S globulins are encoded by four genes (CRA1 [At5G44120], CRB [At1G03880], CRC [At4G28520], and CRA2 [located in tandem with CRA1]). In all three transgenic lines, the 2S albumins were not differentially transcribed. However, two of the 12S globulin genes (CRA1 and CRB) were reproducibly down-regulated, with a fold change varying between 0.74 and 0.82, corresponding to a 20% to 30% decrease. The transcript levels of the third and fourth globulin genes (CRC and CRA2) did not vary.
Table II. Differentially up-regulated genes in 13-dpa developing Arabidopsis seeds shared by all three lines expressing recombinant antibody fragments.
Expression of underlined genes was validated by qPCR. References are as follows: 1, Martínez and Chrispeels (2003); 2, Kamauchi et al. (2005).
| Gene |
Descriptiona |
Name |
Fold Change |
UPR Gene |
Reference |
||
|---|---|---|---|---|---|---|---|
| HC28 |
HC26 | MBP39 | |||||
| Protein folding | |||||||
| At5g28540 | Luminal binding protein1 (HSP70 family) | BIP1 | 4.76 | 4.76 | 2.6 | x | 1, 2 |
| At5g42020 | Luminal binding protein2 (HSP70 family) | BIP2 | 5.24 | 5.81 | 2.96 | x | 2 |
| At1g09080 | Luminal binding protein3 (HSP70 family) | BIP3 | 18.19 | 16.79 | 7.27 | x | 1, 2 |
| At1g77510 | PDI-like (PDIL) protein6 (TRX superfamily) | AtPDI6 | 8.91 | 9.82 | 8.56 | x | 2 |
| At2g32920 | PDIL protein9 (TRX superfamily) | AtPDI9 | 5.66 | 6.11 | 6.48 | x | 1, 2 |
| At2g47470 | PDIL protein11 (TRX superfamily) | AtPDI11 | 4 | 4.24 | 4.35 | x | 1, 2 |
| At5g61790 | Calnexin1, involved in protein folding, functions in UPR | CNX1 | 4.48 | 4.77 | 2.22 | x | 1, 2 |
| At5g07340 | Calnexin2, involved in protein folding, functions in UPR | T2I1.50 | 6.78 | 6.45 | 3.2 | x | 1, 2 |
| At3g08970 | J domain protein, localized in ER lumen (similarity to HSP40) and is induced by heat stress | TMS1 | 6.93 | 5.99 | 7.49 | ||
| Glycosylation/modification | |||||||
| At2g02810 | UDP-Gal and UDP-Glc transporter into Golgi, localized in ER, functions in UPR | UTR1 | 4.02 | 3.97 | 2.76 | x | 1, 2 |
| Translocation/protein transport | |||||||
| At1g29310 | SecY protein transport family protein | F28N24.2 | 5.71 | 8.73 | 9.01 | x | 2 |
| At5g54860 | Major facilitator superfamily protein | MBG8.12 | 3.35 | 3.78 | 3.46 | ||
| Protein degradation | |||||||
| At4g29330 | Involved in proteolysis | DER1 | 3.09 | 2.89 | 3.42 | ||
| Vesicle trafficking | |||||||
| At3g44340 | Homologous to yeast and animal Sec24 proteins | CEF | 3.23 | 3.78 | 4.78 | ||
| Stress-related protein | |||||||
| At5g47120 | BAX inhibitor1 homolog, functions as an attenuator of biotic and abiotic types of cell death | ATBI-1 | 5.29 | 4.79 | 8.24 | x | 2 |
| TF | |||||||
| At1g25310 | Maternal effect embryo arrest8, involved in embryo development ending in seed dormancy | MEE8 | 25.34 | 6.96 | 21.58 | ||
| At1g42990 | ER stress sensor | AtBZIP60 | 4.74 | 4.33 | 6.06 | x | |
| At5g64060 | NAC domain-containing protein | ANAC103 | 4.36 | 3.14 | 12.43 | ||
| Unclassified | |||||||
| At1g18830 | Transducin/WD40 repeat-like superfamily protein | F6A14.8 | 11.21 | 18.44 | 52.38 | ||
| At4g16660 | Heat shock protein70 (Hsp70) family protein | DL4355W | 7.09 | 6.31 | 4.17 | ||
| At5g23575 | Transmembrane CLPTM1 family protein | 3.88 | 4.98 | 5.1 | |||
| At5g42050 | DCD (development and cell death) domain protein | MJC20.15 | 3.2 | 3.4 | 4.93 | ||
| At1g64460 | Protein kinase superfamily protein | F1N19.4 | 2.8 | 3.21 | 3.18 | ||
| At1g64470 | Ubiquitin-like superfamily protein | F1N19.30 | 3.66 | 3.3 | 4.34 | ||
| At3g18080 | B-S glucosidase44 | BGLU44 | 3.05 | 3.18 | 3.81 | ||
| At1g78920 | Vacuolar H+-pyrophosphatase2 protein, functions as protein pump of the Golgi apparatus | AVP2 | 2.66 | 2.97 | 3.43 | ||
| At5g64510 | Located in endomembrane system, tunicamycin induced, function and biological process unknown | MUB3.3 | 7.99 | 6.8 | 5.18 | x | b |
Data relating to gene function were mainly retrieved from the TAIR database.
Table IV. Differentially up-regulated genes common to MBP39 and HC28 lines.
Reference is as follows: 1, Martínez and Chrispeels (2003).
| Gene | Descriptiona | Name | Fold Change |
UPR Gene | Reference | |
|---|---|---|---|---|---|---|
| MBP | HC28 | |||||
| Protein folding | ||||||
| At4g21180 | J domain protein, in ER membrane | ERDJ2B | 3.24 | 2.16 | x | 1 |
| Glycosylation/modification | ||||||
| At3g03640 | β-Glucosidase | BGLU25 | 15.63 | 3.52 | ||
| Protein degradation | ||||||
| At4g14800 | Proteasome complex protein | PDB2 | 2.77 | 1.82 | ||
| Vacuolar | ||||||
| At4g04910 | Protein binding | NSF | 3.8 | 1.99 | ||
| Kinase | ||||||
| At2g37050 | Leu-rich repeat protein kinase family protein | T2N18.19 | 4.32 | 2.12 | ||
| Unclassified | ||||||
| At2g31200 | Actin binding | ADF6 | 2.85 | 3 | ||
| At1g36180 | Acetyl-CoA carboxylase | ACC2 | 3.16 | 3.46 | ||
Data relating to gene function were mainly retrieved from the TAIR database.
Figure 2.
Antibody production results in a limited but shared transcriptional response in developing seeds of three antibody-producing lines. Venn diagrams group genes significantly affected by the recombinant antibody accumulation in HC26, HC28, and MBP39 transgenic lines. Both up- and down-regulated genes (1.8-fold or greater and 0.5-fold or less change, respectively) are presented.
Table III. Differentially up-regulated genes common to HC28 and HC26 lines.
References are as follows: 1, Martínez and Chrispeels (2003); 2, Kamauchi et al. (2005).
| Gene | Descriptiona | Name | Fold Change |
UPR Gene | Reference | |
|---|---|---|---|---|---|---|
| HC28 | HC26 | |||||
| Protein folding | ||||||
| At4g24190 | GRP40 ortholog, involved in protein folding | AtHsp90-7 | 7.2 | 5.71 | x | 1, 2 |
| At1g56340 | Calreticulin1 | CRT1A | 3.82 | 2.85 | x | 2 |
| At1g09210 | Calreticulin2 | CRT1B | 4.27 | 3.89 | x | 1, 2 |
| At3g54960 | PDIL protein1 (TRX superfamily) | AtPDI1 | 3.7 | 2.98 | ||
| At1g21750 | PDIL protein5 (TRX superfamily) | AtPDI5 | 3.07 | 4.2 | ||
| Glycosylation/modification | ||||||
| At1g14360 | UDP-Gal transporter, localized in ER | UTR3 | 3 | 2.88 | ||
| Translocation | ||||||
| At5g50460 | SEC61 γ-subunit | MBA10.8 | 3.39 | 3 | x | 2 |
| At4g24920 | Similar to SEC61 γ-subunit | F13M23.60 | 2.57 | 3.32 | x | 1, 2 |
| At1g52600 | Signal peptide processing | F6D8.18 | 3.07 | 2.92 | ||
| Protein degradation | ||||||
| At1g65040 | Homolog of yeast HRD1 | ATHRD1B | 3.99 | 3.05 | x | 2 |
| Unclassified | ||||||
| At2g25110 | Similar to stromal cell-derived factor2 | ATSDF2 | 3.54 | 3.33 | x | 1, 2 |
| At3g62600 | Forms a complex with SDF2 | ERDJ3B | 5.46 | 4 | ||
| Unknown | ||||||
| At3g62510 | PDI related | PDI-like1-2 | 3.22 | 5.1 | ||
| At4g34630 | T4L20.210 | 3.09 | 2.6 | |||
| At4g29520 | T16L4.30 | 5.67 | 4.61 | |||
Data relating to gene function were mainly retrieved from the TAIR database.
Table V. Differentially up-regulated genes common to MBP39 and HC26 lines.
References are as follows: 1, Martínez and Chrispeels (2003); 2, Kamauchi et al. (2005).
| Gene |
Descriptiona |
Name |
Fold Change |
UPR Gene |
Reference |
|
|---|---|---|---|---|---|---|
|
MBP |
HC26 | |||||
| Vesicle trafficking | ||||||
| At1g62020 | Coatomer α-subunit | F8K4.21 | 5.05 | 3.59 | x | 2 |
| At3g15980 | Coatomer β-subunit | MSL1.4 | 8.59 | 4.49 | ||
| At1g09180 | Member of the ARF-like GTPase family | AtSAR1 | 4.84 | 4.55 | x | 1 |
| At1g70490 | Member of the ARF-like GTPase family | ARFA1D | 11.28 | 4.78 | ||
| TF | ||||||
| At2g38470 | Member of the plant WRKY TF family, involved in abiotic stresses | AtWRKY33 | 6.1 | 2.76 | x | 1 |
| Unclassified | ||||||
| At4g14420 | DL3250C | 5.83 | 4.33 | |||
Data relating to gene function were mainly retrieved from the TAIR database.
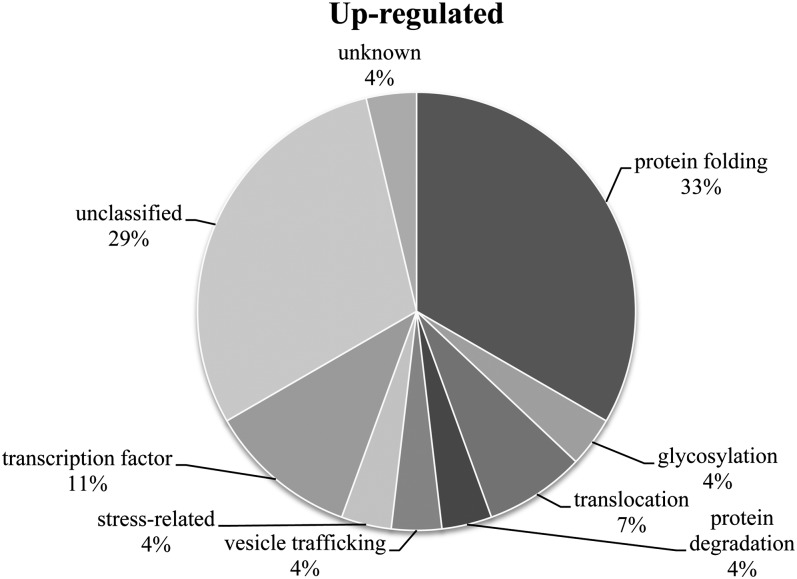
The 27 common up-regulated genes were organized into functional categories, based on their Gene Ontology (GO) annotation, collected from The Arabidopsis Information Resource (TAIR) and AmiGO databases, and analyzed for functional enrichment with BiNGO in order to gain insight in the biological process(es) they are involved in (Rhee et al., 2003; Maere et al., 2005; Carbon et al., 2009). A summary is given in Table II and is presented in Figure 3. Nine of these 27 common up-regulated genes encode protein families responsible for protein folding (BIP1, BIP2, BIP3, PDI6, PDI9, PDI11, CNX11, T2I1.50, and TMS1), and five other genes are involved in glycosylation (UTR1), protein translocation (F28N24.2 and MBG8.12), protein degradation (DER1), and vesicle trafficking (CEF; Table II). Furthermore, a stress-related protein gene and three genes encoding a TF were discovered in this set, namely ATBI-1, MEE8, AtBZIP60, and ANAC103. Finally, 10 out of the 27 listed genes could not be classified (Table II).
Figure 3.
Common up-regulated genes are mainly related to the UPR. The pie chart represents the GO annotation of the 27 common up-regulated genes upon overexpression of antibody genes in 13-dpa developing Arabidopsis seeds of the HC28, HC26, and MBP39 lines.
On all obtained data sets, we performed a BiNGO analysis, which is a tool to determine which GO categories are statistically overrepresented in a set of genes or in a biological network, based on the GO hierarchy for biological processes (Maere et al., 2005). For the differentially expressed genes that are only present in anti-MBP scFv-Fc- or VHH-Fc-expressing Arabidopsis lines, no clear overrepresentation or underrepresentation was found (Supplemental Tables S1–S4). Interestingly however, BiNGO analysis based on GO-slim categories of the 27 common up-regulated genes uncovered a molecular network implicating the UPR, protein folding, and antiapoptosis (Supplemental Fig. S1).
The Amplitude of UPR Induction Is Independent of Antibody Type and Accumulation Level
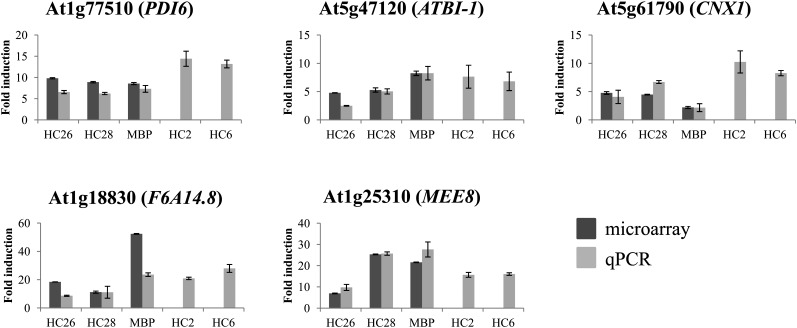
To validate the robustness of the expression data obtained by microarray analysis, we performed a qPCR analysis. For qPCR, the same RNA samples as for microarray analysis were used, and four genes were analyzed: At1g77510 (PDI6), At5g61790 (CNX1), At5g47120 (ATBI-1), and At1g18830 (F6A14.8). The qPCR data are in agreement with the microarray data (Fig. 4). Moreover, the fold induction per gene and per transgenic line is similar (Fig. 4). For instance, the expression of At1g77510 (PDI6) in the MBP39 line is 8.56- ± 0.24-fold induced as found by microarray analysis and 7.32- ± 0.79-fold induced according to the qPCR analysis, which is in the same range.
Figure 4.
The degree of gene induction is independent of the recombinant antibody accumulation level, except for MEE8. Fold induction relative to Col-0 of five differentially expressed genes is shown in five lines with different levels of recombinant antibody accumulation: 5% of the TSP in HC26, 10% in HC28 and MBP39, 1% to 2% in HC2, and 5% in HC26. The qPCR data (light gray) are plotted in comparison with the microarray data (dark gray). Two transgenic lines (HC2 and HC6) were analyzed only by qPCR. Error bars indicate sd of two or three biological repeats.
In parallel, we analyzed the expression of these four genes in developing seeds at 13 dpa from two additional lines, HC2 and HC6, which accumulate the same VHH-Fc antibody as HC26 and HC28, but at 1% to 2% and at 5% of the TSP, respectively. In these lines, the four analyzed genes were also up-regulated in the same order of magnitude as the fold inductions found for the other three transgenic lines (Fig. 4), indicating that the up-regulation of these genes upon high recombinant antibody production occurs already upon recombinant antibody accumulation levels of 1% of the total soluble seed protein (4-fold lower than the other characterized lines). Finally, the expression level of an additional gene, At1g25310 (MEE8), was also analyzed by qPCR in all five lines, because the microarray results suggested that the induction of this gene was possibly dependent on the antibody accumulation level (Table II; Fig. 4). The qPCR data are in agreement with the microarray data (Fig. 4) and show that MEE8 is induced about 2-fold higher in the lines accumulating recombinant antibody at 10% of the TSP (MBP39 and HC28) than in those at only 5% of the TSP (HC26 and HC6). Nevertheless, no difference was found in HC26 and HC6 compared with HC2, accumulating recombinant VHH-Fc antibody at 1% to 2% of the TSP.
DISCUSSION
Recombinant protein production is a demanding process for the host cell, as an additional foreign protein needs to be produced besides the endogenous proteome. Therefore, the question rises whether recombinant protein production is impinging on the cellular homeostasis in plants. Using the regulatory sequences of PHASEOLIN and ARCELIN (De Jaeger et al., 2002; Van Droogenbroeck et al., 2007), we developed an expression system with high-level antibody accumulation in Arabidopsis seed, serving as an ideal tool to evaluate how recombinant antibody accumulation influences seed homeostasis.
We performed a transcriptome analysis on 13-dpa developing seeds from several antibody-accumulating Arabidopsis lines by means of a comparative microarray-based approach. Analysis of the expression data generated by Tiling arrays revealed that, upon heterologous antibody expression, an ER stress response, called the UPR, is generated. We hypothesize that the production of recombinant antibody in the ER results in a disturbance of the normal cellular homeostasis. As protein folding is a crucial aspect in the plant’s life cycle, unfolding or misfolding of proteins can be deleterious. It is known that aberrant proteins can originate from several unfavorable conditions, such as heat or drought stress, but also as a side product of standard protein biosynthesis (Sitia and Braakman, 2003). In transformant MBP39, an aberrant localization of the SSPs was observed (Van Droogenbroeck et al., 2007). However, this aberrant localization was not always penetrant and seems dependent of the growth conditions (Loos et al., 2011b), suggesting that there is no direct link between UPR induction and aberrant seed protein localization. It is also unlikely that the format of the antibody itself (VHH-Fc or scFv-Fc) triggers the UPR response, because VHH-Fc fusions do occur in different animal species and there is a high conservation between animal and plant ER folding and assembly. However, it would be interesting to compare our results with results obtained in seed stocks accumulating full-length antibodies, because it is possible that the CH1 domain, which is not present in our antibody formats, interferes with the UPR response. Indeed, this CH1 domain controls the assembly of IgG antibodies and binds to BiP before it assembles with the light chain (Feige et al., 2009).
One comprehensive study of the Arabidopsis transcriptome during drug-induced ER stress and one study of the rice (Oryza sativa) seed transcriptome upon β-amyloid peptide accumulation have been reported (Martínez and Chrispeels, 2003; Kamauchi et al., 2005). Treatment of Arabidopsis plantlets with tunicamycin and/or DTT revealed an up-regulation of several genes, hereafter called UPR genes. Among the 27 up-regulated genes from our analyses, two (At1g25310 [MEE8] and At5g23575 [unknown]; Table II) were not present on the ATH1 chip used in the study of Martínez and Chrispeels (2003), and 12 coincided with up-regulated UPR genes identified in these studies (Table II; Martínez and Chrispeels, 2003; Kamauchi et al., 2005). The partial overlap with the data sets of Martínez and Chrispeels (2003) and Kamauchi et al. (2005) indicates that different stress inducers have somewhat different effects. This is in agreement with the observation that gene sets induced by tunicamycin and DTT are different (Martínez and Chrispeels, 2003). Nevertheless, the up-regulation of some specific UPR genes strongly suggests that recombinant protein expression in Arabidopsis seeds can also trigger an ER stress response. Similarly, upon accumulation of the β-amyloid peptides in developing rice endosperm, the expression of several BIPs, PDIs, and OsbZIP60-containing putative transmembrane domains was suggestive for an ER stress response (Oono et al., 2010).
When secretory proteins fail to (re)fold, dislocation from the ER is followed by ubiquitination and final degradation by the proteasome in a process called ERAD (Anelli and Sitia, 2008). In our data set, we found a Derlin1-like (DER1) protein. In Arabidopsis, three DER1p homologs are present, which are thought to function in the ERAD pathway, like their yeast and mammalian homologs (Kirst et al., 2005). Moreover, differential regulation of proteolysis-related genes was found in the MBP39 line and in the HC28 and HC26 lines of genes involved in ER-associated and proteasomal ubiquitin-dependent protein catabolic processes. These findings suggested that upon recombinant protein accumulation, an ERAD pathway is induced that may remove unfolded or misfolded proteins from the ER. However, it remains to be determined which proteins are misfolded upon recombinant protein production. In plants, signaling pathways from UPR to programmed cell death have been reported (Liu and Howell, 2010). It is suggested that plant apoptotic programmed cell death is initiated when the cell’s homeostasis is not restored after ER stress (Urade, 2007). ATBI-1, identified as a UPR gene (Kamauchi et al., 2005), is up-regulated in our data set, suggesting that it may function as a cross talk between apoptosis and the ER stress pathways (Sanchez et al., 2000; Lisbona et al., 2009), but its precise function is unknown.
Two translocation-related proteins, F28N24.2 and MBG8.12, are induced, and F28N2.2 is similar to the SEC61 α-subunit in yeast and mammalian cells and classified as a plant UPR protein (Kamauchi et al., 2005). The SEC61 complex is a major component of the ER translocon and functions in yeast and mammalian cells as a retrotranslocon used for the transport and subsequent disposal of misfolded ER proteins through the ERAD pathway (Pilon et al., 1997). Evidence is growing for a similar retrotranslocation system in plants, but the molecular details are not fully uncovered yet (Nakatsukasa and Brodsky, 2008). The induction of this SEC61-like protein gene suggests that the ERAD pathway in plant seeds is used to remove misfolded secretory proteins from the ER in a similar manner to that shown in yeast and mammalian cells. This might also be the case upon recombinant antibody production, which is supported by the finding that several genes involved in protein degradation were differentially regulated in our data set in all three lines (see above).
Three TF genes were up-regulated: MEE8, ATBZIP60, and ANAC103. MEE8 displays very high fold expression changes (7-fold in HC26 and more than 20-fold in the HC28 and MBP39 lines), which seems to be correlated with the recombinant antibody accumulation level, namely 5% of the TSP in HC26 and 10% of the TSP in HC28 and MBP39, this in contrast to the other 26 common up-regulated genes. Unfortunately, little information is available on the function of MEE8, but this nuclear embryonic TF seems to be involved in embryo development leading to seed dormancy (Pagnussat et al., 2005). Therefore, it is tempting to speculate that MEE8 is rather involved in a more general developmental process than specifically in the UPR. The second TF is ATBZIP60, performing a prominent role in the UPR as stress sensor. When ER stress is sensed, ATBZIP60 is proteolyzed and the soluble part is translocated to the nucleus, where it activates the promoters of the ER chaperones BiP1, BiP2, and BiP3 as well as CNX1 and CNX2 via binding to the ER stress-responsive element and to the plant unfolded protein response element, and can also activate its own transcription. Its expression was strongly up-regulated upon tunicamycin treatment of Arabidopsis seedlings (Iwata and Koizumi, 2005; Iwata et al., 2008; Lu and Christopher, 2008) and upon β-amyloid peptide accumulation in rice seeds (Oono et al., 2010). ANAC103 is the third TF that was up-regulated in our experiments and is a member of the Arabidopsis NAC family of TFs, consisting of at least 100 members (Riechmann et al., 2000; Ernst et al., 2004). NAC TFs have been implicated in a variety of plant processes, including stress responses caused by viral infection (Collinge and Boller, 2001).
Overexpression of the folding chaperone gene PDI, or co-overexpression of both BIP and PDI, resulted in a reduction of the UPR and increased the heterologous protein accumulation levels in yeast (Kauffman et al., 2002). In the same way, overexpression of the HAC1 gene, encoding a TF known as a molecular player in the UPR, increased the expression level of some heterologous proteins in Pichia pastoris, but this was dependent of the heterologous protein (Guerfal et al., 2010). Since the achieved antibody accumulation levels are already very high in our Arabidopsis lines, overexpression of one of these molecular UPR players would be unlikely to have an additional effect, but for other antibodies that do not accumulate well in Arabidopsis seeds, it could be beneficial.
Accumulation of β-amyloid peptides in rice seeds resulted in an aberrant seed phenotype caused by the decreased synthesis of SSPs at both the mRNA and protein levels (Oono et al., 2010). Here, no aberrant seed phenotype and germination capacity could be observed in the transgenic seed stocks analyzed. Interestingly, transcription levels of two of the four genes encoding the 12S globulins were decreased by 20% to 30%, most probably compensating for the 10% to 20% recombinant protein transcript levels and the induction of the UPR genes. At the same time, transcript levels of genes encoding the 2S albumin genes were not changed.
The extent of the ER stress response seemed similar for all three lines and also for the HC2 and HC6 lines, in which some selected up-regulated genes displayed comparable fold induction values. Therefore, we speculate that the threshold of antibody accumulation for the ER stress response and UPR induction is lower than 1% of TSP in seeds. Furthermore, the induced ER stress might be correlated with the type of recombinant protein being an ER-secreted glycosylated antibody. Indeed, also in maturing rice seeds, the ER stress level was primarily controlled by the properties of the expressed protein rather than the expression levels (Oono et al., 2010).
To determine whether UPR induction has a positive or negative influence on the recombinant protein yield, several experiments could be set up. For example, the assembly of antibodies in seeds is accompanied by the formation of several Cys bridges, which might be correlated with the induction of PDI-like genes. It would be interesting to investigate whether these genes would also be up-regulated upon the overexpression of proteins without disulfide bridges. Also, overexpression of the TF AtBZIP60, itself activated by IRE1, an ER-resident type I transmembrane protein (Nagashima et al., 2011), would be a valuable approach to evaluate its effect on the accumulation of recombinant proteins in seeds.
CONCLUSION
Here, we observed that upon recombinant antibody expression in developing seeds, the UPR was triggered due to a disturbance of the cellular homeostasis caused by the production of recombinant antibody in the ER. Nevertheless, the provoked ER stress did not greatly affect seed germination and seedling growth, making the PPHAS-driven expression cassette a suitable system for molecular farming in Arabidopsis seeds.
MATERIALS AND METHODS
Plant Material
Prior to the sowing for actual sampling, all Arabidopsis (Arabidopsis thaliana) seed stocks used for these experiments, including the wild type (Col-0), were sown on kanamycin-containing Murashige and Skoog medium for the transgenic lines and nonselective medium for the wild type at the same time and under the same conditions in order to exclude environmental effects possibly influencing the results. Seed stocks, harvested from plants grown simultaneously, were the starting material of the experiments. A summary of the used plant material is provided in Table I. Transformants MBP10 and MBP39 produce scFv-Fc antibodies against MBP and were earlier described by Van Droogenbroeck et al. (2007). The homozygous seed stocks of these transformants accumulate recombinant scFv-Fc at 10% of TSP. Transformants HC2, HC6, HC26, and HC28 produce VHH-Fc antibodies against the prostate-specific antigen (S. De Buck, J. Nolf, T. De Meyer, V. Virdi, K. De Wilde, E. Van Lerberge, B. Van Droogenbroeck, and A. Depicker, unpublished data). Transformants HC2 and HC26 harbor two VHH-Fc T-DNAs, and transformants HC6 and HC28 harbor four T-DNA copies (data not shown). The accumulation levels of the recombinant VHH-Fc proteins in homozygous seed stocks of these transformants are 1% to 2% (HC2), 5% (HC6 and HC26), and 10% (HC28) of TSP (S. De Buck, J. Nolf, T. De Meyer, V. Virdi, K. De Wilde, E. Van Lerberge, B. Van Droogenbroeck, and A. Depicker, unpublished data).
Activation of PPHAS via ELISA
To determine the time point at which the β-PHASEOLIN promoter is activated, seeds from both a selfed T5 homozygous anti-MBP scFv-Fc antibody (MBP10) and a wild-type seed stock were sterilized and sown on kanamycin-containing and nonselective medium, respectively. After growth for 3 weeks in the tissue culture chamber (21°C, 16-h-light/8-h-dark regime), 50 Col-0 and 50 transgenic MBP10 seedlings were transferred to soil and grown further in a growth chamber (21°C, 16-h-light/8-h-dark regime). Plants were watered with 1 L of water per tray (corresponding to about 20 mL per plant) two times per week. Once every 2 weeks, fertilization was added, supplementing 2 mL Wuxal L−1 water (corresponding to 18.75 μL per plant). From week 4 on, plants started flowering. The moment of fertilization is determined as the time point when the long stamens just exceed the height of the gynoecium and is called 0 dpa. Flowers at the 0-dpa stage were labeled with colored sewing thread. Developing siliques were harvested at 5, 8, 11, and 14 dpa and immediately frozen in liquid nitrogen. Sampling was done every 3 d until the end of the seed development. Each individual sample consisted of about 20 collected siliques. The total amount of soluble proteins extracted from complete siliques was determined by the Lowry method (Lowry et al., 1951; De Buck et al., 2012).
As determined by western blotting, the recombinant protein accumulation level in this MBP10 line was 10% of the TSP (data not shown). A standard series, ranging from 1 to 20 ng of anti-MBP scFv-Fc per well, and four dilutions (5×, 25×, 125×, and 625×) from each of the protein extracts were made. Coating was done with 1 μg of purified MBP protein per well (New England Biolabs) in 0.1 m NaHCO3 in a volume of 100 μL overnight at 4°C. The plate was washed with 1× phosphate-buffered saline + 0.1% Tween 20 and incubated for 1 h with blocking buffer (1× phosphate-buffered saline + 2% skim milk). The samples, blank, and standards were incubated for 2 h at room temperature. After washing, the anti-MBP scFv-Fc was detected with anti-human IgG (Fc specific)-alkaline phosphatase antibody produced in goat (1:3,000; Sigma). Alkaline phosphatase activity was detected by addition of the substrate p-nitrophenyl phosphate disodium salt hexahydrate (5 mg; Sigma), dissolved in alkaline phosphatase buffer (48.5 mL diethanolamine per 500 mL, 1 mm MgCl2·6H2O, pH 9.8), and the absorbance was measured at 405-nm wavelength after 30 min of incubation using the VERSAmax Microplate reader. Background correction was done by subtracting the value for the blank from the optical density value of each sample.
Sampling for Microarray Experiments
From every seed stock, more than 100 seeds were sown on kanamycin-selective Murashige and Skoog medium, except for Col-0, which was sown on nonselective medium. Seedlings were transferred to soil after 3 weeks and grown in a 16-h-day/8-h-night regime at 21°C. Plants were watered with 1 L of water per aratray (±50 plants per tray, corresponding to 20 mL of water per plant) two times per week. Once every 2 weeks, fertilization was added (2 mL Wuxal L−1 water, corresponding to 18.75 μL per plant). As soon as flowering started, from week 4 onward, flowers were labeled at the stage of anthesis (0 dpa) with colored yarn thread. Sampling was done in triplicate (biological repeats), with additional samples preserved as backups. Each biological repeat was collected with about 3 d in between, but always at the age of 13 dpa. About 30 siliques were harvested per biological repeat, and the developing seeds were dissected from the siliques, kept on dry ice or liquid nitrogen, and stored at −70°C.
RNA Extraction from Developing Seeds
The procedure was followed as described by Ruuska and Ohlrogge (2001). The final RNA pellets were dissolved in 30 μL of RNase-free water, and RNA was stored at −70°C.
Microarray Analysis and Data Processing
The Arabidopsis Tiling 1.0R chip description file used here was created by Naouar et al. (2009) and consists of 29,890 genes.
Total RNA quantity was assessed with the Nanodrop ND-1000 UV-VIS spectrophotometer (www.nanodrop.com), and RNA quality was determined with the Agilent 2100 BioAnalyzer (www.agilent.com). Reverse transcription, RNA labeling (7 μg of total RNA), hybridization to GeneChip Arabidopsis Tiling 1.0R, and scanning were carried out according to the manufacturer’s instructions (Affymetrix; www.affymetrix.com) at the VIB Microarray Facility (www.microarrays.be).
The raw probe-level data (cell intensity files) were preprocessed using the Robust Multichip Average statistical algorithm, translating them in expression data (Irizarry et al., 2003). The required tools are implemented in R/Bioconductor (www.r-project.org; www.bioconductor.org) using the affy package (Gautier et al., 2004). The Robust Multichip Average preprocessing comprised three steps: (1) background correction, eliminating a noise component; (2) quantile normalization, normalizing every slide to obtain the same cumulative frequency distribution; and (3) summarization, using the median polish algorithm (Irizarry et al., 2003). Of the obtained expression values, P values were calculated and converted into false-discovery rates on the basis of the Bayes t statistic for contrasts (Smyth, 2004, 2005), which is implemented in the Bioconductor package LIMMA (Smyth, 2005). Genes with values of P < 0.001 and q < 0.05 were retained for further analysis. The resulting data set was log transformed. Array element annotations were compiled by TAIR (www.Arabidopsis.org; Rhee et al., 2003).
qPCR
To determine the mRNA levels of some differentially expressed genes according to the microarray results, qPCR analyses were performed on RNA extracted from 13-dpa developing seeds of MBP39, HC28, HC26, HC2, and HC6 transgenic lines and wild-type Col-0. Primers for five different genes were designed using Beacon Designer 7.0 (Premier Biosoft International; Table VI) and diluted to a concentration of 500 ng μL−1. As an internal standard, EF1A was used (Baud et al., 2007).
Table VI. Primers used for qPCR amplification.
| Gene | Accession No. | Direction | Primer Sequence (5′–3′) |
|---|---|---|---|
| PDI6 | At1g77510 | Forward | TGGACGCAACCGCAAATG |
| Reverse | TGGCTTCTTCTCACTGTTCTTC | ||
| MEE8 | At1g25310 | Forward | TGAAGAAGAAACGGCGGGAAC |
| Reverse | ACAGTCTCCAGTCGTCACATATTC | ||
| CNX1 | At5g61790 | Forward | GTGGTTGTATTCTTCTCGCTCTTC |
| Reverse | CTTCCTTCTTCTCCGCCTCATC | ||
| ATBI-1 | At5g47120 | Forward | TTCTCAGCAGCAGCAATGTTAG |
| Reverse | TGTGTCCACCACCATGTATCC | ||
| F6A14.8 | At1g18830 | Forward | GCTCCACCACCAACTGTTCAG |
| Reverse | ATCTTCTGCCTCACGCTTCTTG | ||
| EF1A | At5g60390 | Forward | ATTGCCACACCTCTCACATTGC |
| Reverse | CCATACCAGCGTCACCATTCTTC |
First-strand complementary DNA was generated from 1 μg of total RNA using the iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer’s instructions. Reactions were RNase treated, diluted 20 times, and stored at −20°C until further use. Sample loading on 384-well plates was done using the Janus robotic platform (JANUS Mini Format; Perkin-Elmer). The analysis was performed with an iCycler (Bio-Rad) using SYBR Green (Eurogentec) in a 5-μL volume according to the manufacturer’s recommendations. PCR was performed in triplicate technical repeats and with cycling conditions of 10 min of preincubation at 95°C followed by 45 cycles at 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s. The LightCycler480 software (release 1.5.0; Roche) was used to calculate melting temperature and cycle threshold values. Relative gene expression was analyzed by means of the comparative threshold cycle method, also known as the 2−ΔΔCt method (Livak and Schmittgen, 2001; Pfaffl, 2001; Schmittgen and Livak, 2008).
Analysis of Germination and Plant Development upon High-Level Antibody Accumulation
For each line, at least 64 seeds were sown on selective medium and the wild type on medium without any antibiotic selection to determine germination percentage. The transgenic lines were also sown at a later time point on nonselective medium to determine the speed of germination and to observe seedling growth.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Antibody accumulation affects protein folding and secretion and triggers an antiapoptosis and UPR.
Supplemental Table S1. Differentially expressed genes only present in the MBP39 line.
Supplemental Table S2. Differentially expressed genes only present in the HC28 line.
Supplemental Table S3. Differentially expressed genes only present in the HC26 line.
Supplemental Table S4. Down-regulated genes present in the HC28 and/or HC26 and/or MBP39 lines.
Acknowledgments
We thank the VIB Microarray Facility for performing the Tiling 1.0R arrays; Marnik Vuylsteke, Stephanie De Bodt, Laurens Pauwels, and Naira Naouar for help in setup and analysis of the microarrays; Stephen Depuydt, Laurens Pauwels, Alain Goossens, Geert De Jaeger, Geert Angenon, and Lieve Gheysen for critical reading of the manuscript; Jonah Nolf, Els Van Lerberge, and Hannes Peeters for practical assistance; and Annick Bleys for help in preparing the manuscript. We also thank the colleagues involved in the European Cooperation in Science and Technology action FA0804 for helpful discussions.
Glossary
- dpa
d post anthesis
- SSP
seed storage protein
- scFv
single-chain Fv
- TSP
total soluble protein
- ER
endoplasmic reticulum
- BiP
binding protein
- UPR
unfolded protein response
- PDI
protein disulfide isomerase
- ERAD
endoplasmic reticulum-associated degradation
- DTT
dithiothreitol
- qPCR
quantitative PCR
- MBP
maltose-binding protein
- Col-0
ecotype Columbia
- GO
Gene Ontology
- TAIR
The Arabidopsis Information Resource
- TF
transcription factor
- T-DNA
transfer DNA
References
- Anelli T, Sitia R. (2008) Protein quality control in the early secretory pathway. EMBO J 27: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin J-P, Miquel M, Lepiniec L, Rochat C. (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Wuillème S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C. (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52: 405–419 [DOI] [PubMed] [Google Scholar]
- Benchabane M, Goulet C, Rivard D, Faye L, Gomord V, Michaud D. (2008) Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol J 6: 633–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothe J, Nykiforuk C, Shen Y, Zaplachinski S, Szarka S, Kuhlman P, Murray E, Morck D, Moloney MM. (2010) Seed-based expression systems for plant molecular farming. Plant Biotechnol J 8: 588–606 [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho NDSP, Arentshorst M, Kooistra R, Stam H, Sagt CM, van den Hondel CAMJJ, Ram AFJ. (2011) Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus niger. Appl Microbiol Biotechnol 89: 357–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Bishop KJ, Hall TC. (2003) Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J 33: 853–866 [DOI] [PubMed] [Google Scholar]
- Collinge M, Boller T. (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46: 521–529 [DOI] [PubMed] [Google Scholar]
- Day RC, Herridge RP, Ambrose BA, Macknight RC. (2008) Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol 148: 1964–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck S, Virdi V, De Meyer T, De Wilde K, Piron R, Nolf J, Van Lerberge E, De Paepe A, Depicker A. (2012) Production of camel like antibodies in plants. In S Muyldermans, D Saerens, eds, Single Domain Antibodies: Methods and Protocols. Methods in Molecular Biology,Vol 911. Springer, New York, pp 305–324 [DOI] [PubMed] [Google Scholar]
- De Jaeger G, Scheffer S, Jacobs A, Zambre M, Zobell O, Goossens A, Depicker A, Angenon G. (2002) Boosting heterologous protein production in transgenic dicotyledonous seeds using Phaseolus vulgaris regulatory sequences. Nat Biotechnol 20: 1265–1268 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. (2001) ER quality control: towards an understanding at the molecular level. Curr Opin Cell Biol 13: 431–437 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. (2003) Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. (1999) Setting the standards: quality control in the secretory pathway. Science 286: 1882–1888 [DOI] [PubMed] [Google Scholar]
- Ernst HA, Olsen AN, Larsen S, Lo Leggio L. (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. (2009) An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell 34: 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell SW, Travers KJ, Weissman JS, Brodsky JL. (2001) The action of molecular chaperones in the early secretory pathway. Annu Rev Genet 35: 149–191 [DOI] [PubMed] [Google Scholar]
- Fischer R, Stoger E, Schillberg S, Christou P, Twyman RM. (2004) Plant-based production of biopharmaceuticals. Curr Opin Plant Biol 7: 152–158 [DOI] [PubMed] [Google Scholar]
- Focks N, Benning C. (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenko DE, Gladyshev VN. (2003) Genomics perspective on disulfide bond formation. Antioxid Redox Signal 5: 397–402 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Nambara E, Yamagishi K, Goto DB, Naito S. (2002) Storage proteins. The Arabidopsis Book 1: e0020, doi/ 10.1199/tab.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gilbert HF. (1998) Protein disulfide isomerase. Methods Enzymol 290: 26–50 [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. (1994) Plant embryogenesis: zygote to seed. Science 266: 605–614 [DOI] [PubMed] [Google Scholar]
- Guerfal M, Ryckaert S, Jacobs PP, Ameloot P, Van Craenenbroeck K, Derycke R, Callewaert N. (2010) The HAC1 gene from Pichia pastoris: characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb Cell Fact 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TC, Chandrasekharan MB, Li G. (1999) Phaseolin: its past, properties, regulation and future. In PR Shewry, R Casey, eds, Seed Proteins. Kluwer, Dordrecht, The Netherlands, pp 209–240 [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633 [DOI] [PubMed] [Google Scholar]
- Hendershot L, Wei J, Gaut J, Melnick J, Aviel S, Argon Y. (1996) Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc Natl Acad Sci USA 93: 5269–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EM, Larkins BA. (1999) Protein storage bodies and vacuoles. Plant Cell 11: 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamauchi S, Nakatani H, Nakano C, Urade R. (2005) Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J 272: 3461–3476 [DOI] [PubMed] [Google Scholar]
- Kauffman KJ, Pridgen EM, Doyle FJ, III, Dhurjati PS, Robinson AS. (2002) Decreased protein expression and intermittent recoveries in BiP levels result from cellular stress during heterologous protein expression in Saccharomyces cerevisiae. Biotechnol Prog 18: 942–950 [DOI] [PubMed] [Google Scholar]
- Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. (2004) A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol 167: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst ME, Meyer DJ, Gibbon BC, Jung R, Boston RS. (2005) Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol 138: 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ. (2001) Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol 127: 949–962 [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Sun SSM. (2009) Plant seeds as bioreactors for recombinant protein production. Biotechnol Adv 27: 1015–1022 [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A-H, Iwakoshi NN, Glimcher LH. (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang B-C, Xu Y, Zhu Y-X. (2007) Systematic studies of 12S seed storage protein accumulation and degradation patterns during Arabidopsis seed maturation and early seedling germination stages. J Biochem Mol Biol 40: 373–381 [DOI] [PubMed] [Google Scholar]
- Liebrand TWH, Smit P, Abd-El-Haliem A, de Jonge R, Cordewener JHG, America AHP, Sklenar J, Jones AME, Robatzek S, Thomma BPHJ, et al. (2012) Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of Cf receptor-like proteins involved in pathogen resistance of tomato. Plant Physiol 159: 1819–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, et al. (2009) BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1α. Mol Cell 33: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-X, Howell SH. (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loos A, Van Droogenbroeck B, Hillmer S, Grass J, Kunert R, Cao J, Robinson DG, Depicker A, Steinkellner H. (2011a) Production of monoclonal antibodies with a controlled N-glycosylation pattern in seeds of Arabidopsis thaliana. Plant Biotechnol J 9: 179–192 [DOI] [PubMed] [Google Scholar]
- Loos A, Van Droogenbroeck B, Hillmer S, Grass J, Pabst M, Castilho A, Kunert R, Liang M, Arcalis E, Robinson DG, et al. (2011b) Expression of antibody fragments with a controlled N-glycosylation pattern and induction of endoplasmic reticulum-derived vesicles in seeds of Arabidopsis. Plant Physiol 155: 2036–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Lu D-P, Christopher DA. (2008) Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol Genet Genomics 280: 199–210 [DOI] [PubMed] [Google Scholar]
- Ma JK-C, Barros E, Bock R, Christou P, Dale PJ, Dix PJ, Fischer R, Irwin J, Mahoney R, Pezzotti M, et al. (2005) Molecular farming for new drugs and vaccines: current perspectives on the production of pharmaceuticals in transgenic plants. EMBO Rep 6: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ. (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandini F, Avesani L, Bortesi L, Van Droogenbroeck B, De Wilde K, Arcalis E, Bazzoni F, Santi L, Brozzetti A, Falorni A, et al. (2011) Non-food/feed seeds as biofactories for the high-yield production of recombinant pharmaceuticals. Plant Biotechnol J 9: 911–921 [DOI] [PubMed] [Google Scholar]
- Moreno AA, Orellana A. (2011) The physiological role of the unfolded protein response in plants. Biol Res 44: 75–80 [DOI] [PubMed] [Google Scholar]
- Nagashima Y, Mishiba K-i, Suzuki E, Shimada Y, Iwata Y, Koizumi N. (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL. (2008) The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Rätsch G, Inzé D, Kuiper M, et al. (2009) Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Ng DTW, Spear ED, Walter P. (2000) The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol 150: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S-i, Brodsky JL, Nakatsukasa K. (2005) Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J Biochem 137: 551–555 [DOI] [PubMed] [Google Scholar]
- Noh S-J, Kwon CS, Oh D-H, Moon JS, Chung W-I. (2003) Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311: 81–91 [DOI] [PubMed] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. (2002) Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J 366: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Wakasa Y, Hirose S, Yang L, Sakuta C, Takaiwa F. (2010) Analysis of ER stress in developing rice endosperm accumulating β-amyloid peptide. Plant Biotechnol J 8: 691–718 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H-J, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie L-F, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Peters J, Stoger E. (2011) Transgenic crops for the production of recombinant vaccines and anti-microbial antibodies. Hum Vaccin 7: 367–374 [DOI] [PubMed] [Google Scholar]
- Petruccelli S, Otegui MS, Lareu F, Tran Dinh O, Fitchette A-C, Circosta A, Rumbo M, Bardor M, Carcamo R, Gomord V, et al. (2006) A KDEL-tagged monoclonal antibody is efficiently retained in the endoplasmic reticulum in leaves, but is both partially secreted and sorted to protein storage vacuoles in seeds. Plant Biotechnol J 4: 511–527 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Römisch K. (1997) Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J 16: 4540–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB. (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Ohlrogge JB. (2001) Protocol for small-scale RNA isolation and transcriptional profiling of developing Arabidopsis seeds. Biotechniques 31: 752–758 [DOI] [PubMed] [Google Scholar]
- Saerens D, Kinne J, Bosmans E, Wernery U, Muyldermans S, Conrath K. (2004) Single domain antibodies derived from dromedary lymph node and peripheral blood lymphocytes sensing conformational variants of prostate-specific antigen. J Biol Chem 279: 51965–51972 [DOI] [PubMed] [Google Scholar]
- Sanchez P, de Torres Zabala M, Grant M. (2000) AtBI-1, a plant homologue of Bax inhibitor-1, suppresses Bax-induced cell death in yeast and is rapidly upregulated during wounding and pathogen challenge. Plant J 21: 393–399 [DOI] [PubMed] [Google Scholar]
- Scheuner D, Song BB, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. (2001) Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Sharma AK, Sharma MK. (2009) Plants as bioreactors: recent developments and emerging opportunities. Biotechnol Adv 27: 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JF, Ferro-Novick S, Rose MD, Helenius A. (1995) BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol 130: 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R, Braakman I. (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426: 891–894 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2005) Limma: linear models for microarray data. In RC Gentleman, VJ Carey, S Dudoit, R Irizarry, W Huber, eds, Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Spencer MWB, Casson SA, Lindsey K. (2007) Transcriptional profiling of the Arabidopsis embryo. Plant Physiol 143: 924–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger E, Ma JK-C, Fischer R, Christou P. (2005) Sowing the seeds of success: pharmaceutical proteins from plants. Curr Opin Biotechnol 16: 167–173 [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- Urade R. (2007) Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J 274: 1152–1171 [DOI] [PubMed] [Google Scholar]
- Urade R. (2009) The endoplasmic reticulum stress signaling pathways in plants. Biofactors 35: 326–331 [DOI] [PubMed] [Google Scholar]
- van Anken E, Braakman I. (2005) Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol 40: 191–228 [DOI] [PubMed] [Google Scholar]
- Van Droogenbroeck B, Cao JY, Stadlmann J, Altmann F, Colanesi S, Hillmer S, Robinson DG, Van Lerberge E, Terryn N, Van Montagu M, et al. (2007) Aberrant localization and underglycosylation of highly accumulating single-chain Fv-Fc antibodies in transgenic Arabidopsis seeds. Proc Natl Acad Sci USA 104: 1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Droogenbroeck B, De Wilde K, Depicker A. (2009) Production of antibody fragments in Arabidopsis seeds. In L Faye, V Gomord, eds, Recombinant Proteins from Plants. Humana Press, New York, pp 89–101 [DOI] [PubMed] [Google Scholar]
- Vitale A, Boston RS. (2008) Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, de Ilárduya OM, Jaworski JG, Ohlrogge JB, Benning C. (2000) A new set of Arabidopsis expressed sequence tags from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol 124: 1582–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Raden D, Doyle FJ, III, Robinson AS. (2005) Analysis of unfolded protein response during single-chain antibody expression in Saccharomyces cerevisiae reveals different roles for BiP and PDI in folding. Metab Eng 7: 269–279 [DOI] [PubMed] [Google Scholar]
- Xu P, Robinson AS. (2009) Decreased secretion and unfolded protein response up-regulation are correlated with intracellular retention for single-chain antibody variants produced in yeast. Biotechnol Bioeng 104: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Maruyama D, Endo T, Nishikawa S-i. (2008) Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol 49: 1547–1562 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. (1998) Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins: involvement of basic leucine zipper transcription factors. J Biol Chem 273: 33741–33749 [DOI] [PubMed] [Google Scholar]