Summary: Analyses of gene regulation and cell chemistry in acclimated fed batch cultures of Phaeodactylum tricornutum identify genes and metabolic pathways of carbon metabolism, and the subcellular localization and timing of genes and processes during light/dark cycles.
Abstract
The regulation of carbon metabolism in the diatom Phaeodactylum tricornutum at the cell, metabolite, and gene expression levels in exponential fed-batch cultures is reported. Transcriptional profiles and cell chemistry sampled simultaneously at all time points provide a comprehensive data set on carbon incorporation, fate, and regulation. An increase in Nile Red fluorescence (a proxy for cellular neutral lipids) was observed throughout the light period, and water-soluble glucans increased rapidly in the light period. A near-linear decline in both glucans and lipids was observed during the dark period, and transcription profile data indicated that this decline was associated with the onset of mitosis. More than 4,500 transcripts that were differentially regulated during the light/dark cycle are identified, many of which were associated with carbohydrate and lipid metabolism. Genes not previously described in algae and their regulation in response to light were integrated in this analysis together with proposed roles in metabolic processes. Some very fast light-responding genes in, for example, fatty acid biosynthesis were identified and allocated to biosynthetic processes. Transcripts and cell chemistry data reflect the link between light energy availability and light energy-consuming metabolic processes. Our data confirm the spatial localization of processes in carbon metabolism to either plastids or mitochondria or to glycolysis/gluconeogenesis, which are localized to the cytosol, chloroplast, and mitochondria. Localization and diel expression pattern may be of help to determine the roles of different isoenzymes and the mining of genes involved in light responses and circadian rhythms.
The ecological success of diatoms has drawn attention from phycologists for a long time, and their usefulness in many areas (e.g. as feed organisms in aquaculture or raw material producers in bioenergy) has made this group of microalgae known to a broader audience. The recent completion of the genome sequence of Phaeodactylum tricornutum (Bowler et al., 2008) has provided a useful model organism for this group, and knowledge of its physiology can now be extended to the genetic and metabolic levels, resulting in a deeper understanding of some of the factors that regulate major cell processes in diatoms (Parker et al., 2008; Fernie et al., 2012). The transcriptome of P. tricornutum has been studied in a few contexts, such as silicon metabolism (Sapriel et al., 2009) and light acclimation (Nymark et al., 2009).
Carbon metabolism in autotrophic algae starts with photosynthesis and carbon acquisition, followed by incorporation of carbon into different organic metabolites. In diatoms, chrysolaminaran (1,3-β-d-glucan) is one of the main sinks for carbon fixed during light periods, and it is also incorporated into glucans through gluconeogenesis (Myklestad and Granum, 2009). In darkness, glucans provide energy for nutrient assimilation and carbon skeletons for the synthesis of other biomolecules through glycolysis and the tricarboxylic acid (TCA) cycle. Triosephosphate exported from the chloroplast might serve as a precursor for hexose sugars (Glc and Fru) or might be transformed into pyruvate and enter the TCA cycle in the mitochondrion. The metabolism of storage carbohydrates is a dynamic process, and water-soluble glucans are located in vacuoles (Granum and Myklestad, 2002; Kroth et al., 2008). Cells use monomeric Glc in various metabolic pathways, and therefore, polymeric 1,3-β-d-glucans have to be broken down into monomers. The synthesis of polymers (that must be reduced to monomers again) is considered a way to reduce the large oscillations in cellular Glc levels that would otherwise occur over the light/dark cycles (Vårum et al., 1986).
Another fraction of the fixed carbon is incorporated via acetyl-CoA into fatty acids. Short fatty acids with few double bonds are combined with glycerol on the endoplasmic reticulum (ER) and stored as neutral triacylglycerols in lipid droplets in the cytosol or in chloroplasts (Fan et al., 2011). Other fatty acids are elongated and desaturated and combined with phosphorus or carbohydrates (glycans) into more complex phospholipids or glycolipids to provide polar membrane material. Fatty acids store more energy in smaller molecules, but the processes of lipid synthesis and catabolism are slower compared with carbohydrate metabolism (Lancelot and Mathot, 1985). It has been shown that other factors such as nutrient depletion (Reitan et al., 1994; Breteler et al., 2005) or excess light energy (Norici et al., 2011) may increase cellular lipid content in diatoms. Along with the mapping and identification of protein coding regions of microalgae genomes, genetic engineering is a rapidly developing field that may provide a tool to enhance lipid synthesis and storage in algae for use in biofuel production (Radakovits et al., 2010).
Cell metabolism occurs in cyclic patterns, and some cycles are cued by external geophysical zeitgebers such as light/dark periods (e.g. photosynthetic carbon reduction) while other cycles (e.g. the TCA cycle) are cued by internal biological clocks. The internal molecular clocks are related to gene transcription and translation regulated in feedback loops (Tessmar-Raible et al., 2011), and the circadian control of many cell processes has been studied in the green alga Chlamydomonas reinhardtii (Schulze et al., 2010). Control points can be at the translational level (Mittag, 2003) or the posttranslational level (Zhang et al., 2011) and sometimes are regulated as a feedback signal system. As an example, oscillations of sugar content contributed to sugar-responsive transcripts in plant cells (Haydon et al., 2011). In general, major cellular processes in microalgae, such as photosynthesis, nutrient uptake and incorporation, and cell growth and division, show diel regulation that follow the light/dark periods (Sanchez et al., 2009; Radakovits et al., 2010). However, this general pattern may be altered in species with high growth rates, where some cells undergo more than one cell division per day (Granum et al., 2009). Several studies have investigated diel regulation of the transcriptome in cyanobacteria, and 25% to 90% of the expressed genes were found to exhibit oscillating expression profiles (Stöckel et al., 2008; Zinser et al., 2009; Shi et al., 2010; Straub et al., 2011). Biochemical pathways involved in carbon metabolism appeared to be tightly regulated, with processes such as photosynthesis and carbon assimilation dominating during the day and catabolic utilization of energy reserves occurring mainly during the night (Dron et al., 2012). Similar analyses of eukaryotic plankton are rare, but about 94% of the expressed genes in the prasinophyte Ostreococcus tauri were differentially expressed during a light/dark cycle (Monnier et al., 2010).
In this work, we describe the transcriptional regulation of glucan and lipid metabolism in P. tricornutum acclimated to light/dark cycles and surplus nutrient supply, based on a time series of cell chemistry measurements and simultaneous data from microarray analysis. The fate of the incorporated carbon was followed through gluconeogenesis or glycolysis and from acetyl-CoA into the TCA cycle or to fatty acid synthesis and elongation. Analysis of data from the transcriptional level was integrated with cell chemistry measurements, focusing on the uptake and metabolism of major carbon pools in the cells. Our results show that genes encoding enzymes involved in processes such as the TCA cycle and fatty acid biosynthesis are highly coordinated over the light/dark cycle, and the transcriptional data indicate activity that may ultimately result in the synthesis of carbon skeletons in the chloroplast during daytime, and the breakdown of carbon-rich compounds in the cytosol and mitochondria and cell division during the night.
RESULTS AND DISCUSSION
Two samples of 50 mL were removed from each culture at selected time points over a course of 26.5 h; one sample was used for cell chemistry measurements, and the other sample was subjected to RNA extraction. The sampling points were (hours after light on): T1 = 0.5, T2 = 6, T3 = 10.5, T4 = 15.5, T5 = 16.5 (darkness), T6 = 20 (darkness), T7 = 23.5 (darkness), and (the next day) T8 = 27. The chronologically last sample (T8) corresponds to a time point between T1 and T2 when considering the light/dark cycle (discussed below). In the analyses of gene expression, the latest point in darkness (T7) was used as a reference.
Cell Chemistry
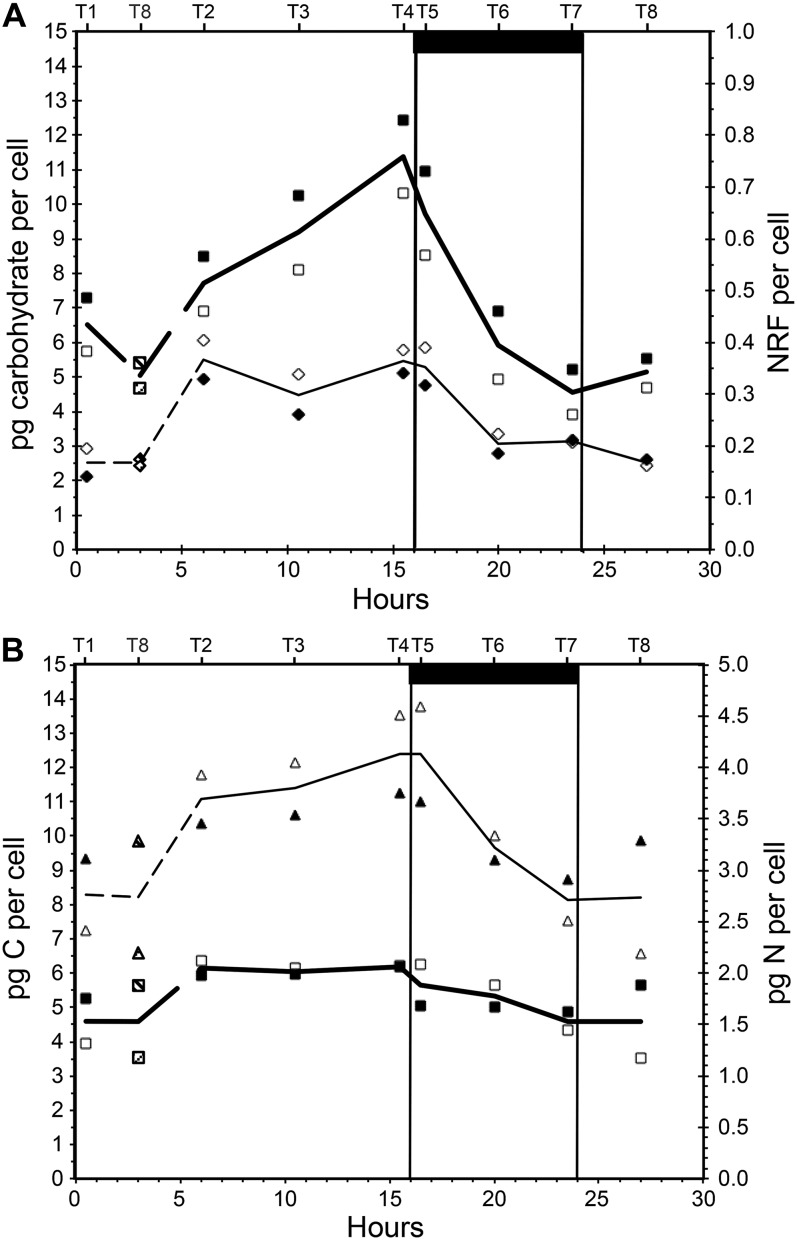
Four cell-specific chemical variables are show in Figure 1. Our predictions were that when cultures are in steady state, sampling is independent of chronology and only dependent on the point of time with respect to the light/dark cycle. As seen from our data, the T8 measurements fit well between T1 and T2 and thus support steady state. Steady state was also confirmed by stable biomass during a period of at least 5 d before sampling, and this phenomenon is well known (Droop, 1974). As a consequence, T8 is plotted in accordance with its position in the light/dark cycle, and not chronology, in the remaining figures.
Figure 1.
Variation in cell chemistry over a 16/8-h light/dark cycle in P. tricornutum. A, Water-soluble carbohydrates (pg cell−1; squares) and cell specific NRF (diamonds, right axis). B, Cellular content of carbon (pg cell−1; triangles) and nitrogen (pg cell−1; squares, right axis). White and black symbols represent replicate cultures, and lines show averages of the two replicate data series. The bottom x axis shows hours from light on in the morning, and the top x axis shows sampling times labeled T1 to T8. The last sampling point in the series (T8, sampled at 27 h after start) corresponds to a point between T1 and T2 (0.5 and 6 h after start) and has been indicated in the graphs (dashed lines and shaded symbols). The dark bar at top and vertical lines indicate the period when the light was off.
Total water-soluble carbohydrates in the cells increased from 5 to 11.5 pg cell−1 (average of two cultures) during the light period, and the rate of increase was very high toward the end of the light period (Fig. 1A). The same pattern has been observed in other diatoms (Vårum et al., 1986; Granum et al., 2009). A few hours into the light period, cellular carbon increased rapidly from 8 to 12.4 pg cell−1 and thus increased approximately 50% during the light period (Fig. 1B). Cellular nitrogen increased from 1.5 to 2 pg cell−1 during the first hours of the light period and was stable until it decreased during the dark period (Fig. 1B). The observed increase in both cellular carbon and nitrogen (per cell) during the light was followed by a decrease in the dark, and there was no accumulation of nitrogen or carbon in the cells over a light/dark cycle. Hockin and coworkers (2012) studied the proteome and transcriptome of Thalassiosira pseudonana and found that the response of carbon metabolism to nitrogen starvation in this diatom is more similar to cyanobacteria than other photosynthetic eukaryotes. Cell numbers were stable during the light period, on average 7.41 × 105 mL−1 (±2.22%; average of four measurements), and increased steadily during the dark period. On the last sampling (T8, 27 h after the start), the cell numbers were 1.02 × 106, which is an increase of 38%. These numbers indicate that cell division took place during the dark and a few hours into the next light period, and biodilution may partially explain the observed decrease in nitrogen per cell in the dark. Optical density, on the other hand, increased 35% during the light period and decreased rapidly in the dark (data not shown). The increase in optical density during the day may reflect a cell volume increment rather than an increase in cell numbers.
Nile Red fluorescence (NRF) was used as a proxy for neutral lipids (Greenspan and Fowler, 1985; Chen et al., 2009). The fluorescence was stable and low from the middle of the dark period to the first 3 h into the light period. Over the next hours, the signal intensity nearly doubled and remained so until the beginning of the dark period, when it decreased to a level close to the level at the beginning of the light period (Fig. 1A). This observation is in accordance with previous studies in the centric diatom Cyclotella meneghiniana (Sicko-Goad et al., 1988) and the eustigmatophyte Nannochloropsis sp. (Sukenik and Carmeli, 1990). Nile Red is also used to stain proteins (Demeule et al., 2007), and although excitation/emission wavelengths differ from those for lipid analyses, it may be a source of interference with the lipid measurements. Larsen et al. (2011) quantified lipids and proteins in a biofilm using Nile Red and other fluorochromes and found that oleic acid fluoresced almost twice as much as palmitic and stearic acids when the lipids were precipitated in water. P. tricornutum is rich in palmitic and palmitoleic acids (Domergue et al., 2003), and Nile Red as a proxy for neutral lipids may be biased due to the specific fatty acid composition. Gravimetric measurements of total neutral lipids (Bligh and Dyer, 1959) sampled 27 h after the start of the experimental period showed that P. tricornutum contained 10.1% ± 3.5% (dry weight) neutral lipids.
Gene Expression
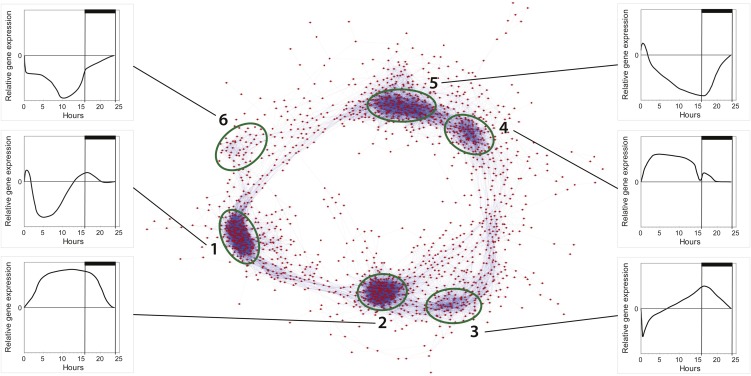
Global transcriptome analyses using whole-genome oligonucleotide microarrays for P. tricornutum showed that 4,567 genes were significantly regulated (P > 0.05) during at least one of the eight time points in the study, equivalent to about 44% of the whole transcriptome. We generated a coexpression network based on Pearson correlation values between the gene pairs of the 1,936 most responsive genes in the experiment (Fig. 2). Six clusters of coregulated genes were identified; together, the clusters formed a circle similar to the coexpression network observed in a diurnal experiment performed on the cyanobacterium Cyanothece sp. (Stöckel et al., 2008).
Figure 2.
Coexpression network of diel cycling genes in P. tricornutum. A network representation of the 1,936 most responsive genes in the experiment was made using Cytoscape and Pearson correlation values (r > 0.95) for each gene pair. In total, 44,003 gene pairs are included. Six clusters of tightly coregulated genes are distinguished, and graphs showing the general expression pattern of a given cluster (normalized to time point T7, 23.5 h) are presented for each cluster (labeled 1–6). [See online article for color version of this figure.]
Cluster 1 mainly contains genes involved in ribosome biogenesis and the processing of pre-ribosomal RNAs. A hallmark of this cluster is that the genes are down-regulated around midday. Cluster 2 contains genes with highest expression late in the day and during the night and includes genes associated with many cellular processes, among them cell division and the TCA cycle. Cluster 3 is composed mainly of genes that are down-regulated after the transition from dark to light and includes genes involved in nitrogen metabolism (urease accessory protein, urease and cyanate hydratase) as well as genes central to the glyoxylate cycle (malate synthase and isocitrate lyase). Cluster 3 also includes genes coupled to mitochondrial β-oxidation such as β-ketoacyl-CoA thiolase (KCT3) and multifunctional fatty acid oxidation complex subunit α (Phatr2_35240). Cluster 4 includes genes coupled to various processes with maximum transcriptional activity during midday. Notably, many of them encode heat shock proteins, chaperones, and proteins involved in protein modification/degradation. Cluster 5 includes genes that are down-regulated during nighttime and encode proteins involved in many cellular processes such as carotenoid, chlorophyll, and fatty acid biosynthesis. Cluster 6 is mainly composed of genes transcribed from the chloroplast as well as nucleus-encoded ribosomal proteins.
Carbon Fixation
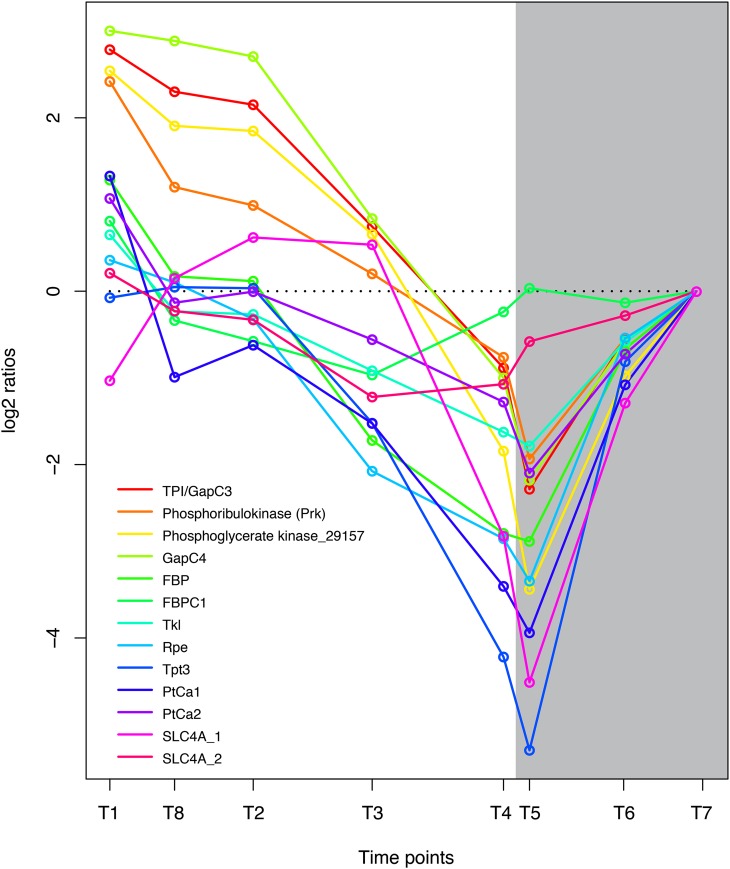
Glycerate 3-phosphate is one of the primary products of CO2 assimilation and is phosphorylated to 1,3-bisphosphoglycerate by phosphoglycerate kinase. Four central enzymes in carbon fixation were highly coregulated and showed highest expression early in the light period and reduced activity in the dark period: phosphoribulokinase (Prk), phosphoglycerate kinase (Phatr2_29157), glyceraldehyde-3-phosphate dehydrogenase (GapC4), and the recently described Stramenopile protein triosephosphate isomerase/glyceraldehyde-3-phosphate dehydrogenase (TPI/GapC3; Fig. 3). Prk converts d-ribulose 5-phosphate to d-ribulose 1,5-bisphosphate, which acts as the acceptor for CO2 during the production of glycerate 3-phosphate by Rubisco. 3-Phospho-d-glycerate is in turn phosphorylated to 1,3-diphosphoglycerate by phosphoglycerate kinase (Phatr2_29157). The Stramenopile-specific fusion protein TPI/GapC3 might be one of the proteins responsible for the reduction of 1,3-diphosphoglycerate to glyceraldehyde 3-phosphate (catalyzed by the glyceraldehyde phosphate dehydrogenase [GAPDH] domain) as well as the isomerization of glyceraldehyde 3-phosphate to dihydroxyacetone phosphate (catalyzed by the N-terminal triosephosphate isomerase domain; Liaud et al., 2000). TPI/GapC3 is predicted to encode a chloroplast-localized protein by WoLF PSORT(Horton et al., 2007); however, previous work by Liaud et al. (2000) suggested that this protein was localized to the mitochondria. Triosephosphate is transported out of the chloroplast by triosephosphate transporters. There are three triosephosphate transporters in P. tricornutum; of these, the plastidic Tpt3, which is related to Arabidopsis (Arabidopsis thaliana) TPT/APE2, is most light responsive. The triosephosphate exported from the chloroplast may serve as a precursor for hexose sugars (Glc and Fru) or may be transformed into pyruvate and enter the TCA cycle in the mitochondrion (glycolysis). d-Ribulose 5-phosphate is regenerated through the sequential action of several enzymes of the Calvin cycle, including fructose 1,6-bisphosphatase (FBP, FBPC1), transketolase (Tkl), and the plastidic ribulose phosphate 3-epimerase (Rpe). The expression pattern of these genes was closely related to the other carbon fixation enzymes, although it was not induced to the same degree by light (Fig. 3). The sedoheptulose bisphosphatase was not significantly regulated at any of the time points.
Figure 3.
Coordinated regulation of genes coupled to carbon fixation in P. tricornutum. Expression levels at sampling points (hours after light on) T1 = 0.5, T8 = 27 (day 2), T2 = 6, T3 = 10.5, T4 = 15.5, T5 = 16.5 (darkness), T6 = 20 (darkness), and T7 = 23.5 (darkness) are normalized to time point T7. GenBank accession numbers are as follows: TPI/GapC3, EEC50801.1; Prk, ACI65926.1; Phatr2_29157, EEC46310; GapC4, EEC50484.1; FBP, EEC44418.1; FBPC1, EEC51753.1; Tkl, EEC50321.1; Rpe, EEC51500.1; Tpt3, EEC51811.1; PtCa1, EEC51057.1; PtCa2, EEC48791.1; SLC4A_1, EEC48911.1; SLC4A_2, EEC51950.1. [See online article for color version of this figure.]
Diatoms possess organic carbon-concentrating mechanisms that provide a higher CO2 concentration around Rubisco than that in the surrounding medium (Roberts et al., 2007); however, the details of this process are still unknown. Genes that may be coupled to carbon-concentrating mechanisms in diatoms, such as the plastidic HCO3− transporters SLC4A_1 and SLC4A_2 and the carbonic anhydrases PtCa1 and PtCa2, were strongly down-regulated at dark (Fig. 3). Interestingly, four genes previously linked to a proposed C4 pathway in diatoms (Reinfelder et al., 2000) appear to have higher transcriptional activity late in the day and during the night. Phosphoenolpyruvate carboxylases PEPCase_1 and PEPCase_2, which convert phosphoenolpyruvate and HCO3− to oxaloacetate, show a similar expression pattern (Supplemental Table S1). The same is also observed for phosphoenolpyruvate carboxykinase (PEPCK1), which decarboxylates oxaloacetate, and the NADP-dependent malic enzyme (Phatr2_51970), which decarboxylates malate. The mitochondrial pyruvate carboxylase (PYC1), which carboxylates pyruvate to oxaloacetate, is closely coregulated with PEPCK1, while the plastid-localized PYC2 shows lowest transcriptional activity late at night. The transcriptional activity of pyruvate-phosphate dikinase is also highest late at night. However, regulation of the alleged C4 enzymes may be an indirect effect of aerating the culture with 1% to 2% CO2 (v/v) and could be connected to the regulation of pH homeostasis at least during the night (Haimovich-Dayan et al., 2013).
Glucan Biosynthesis and Glycolysis
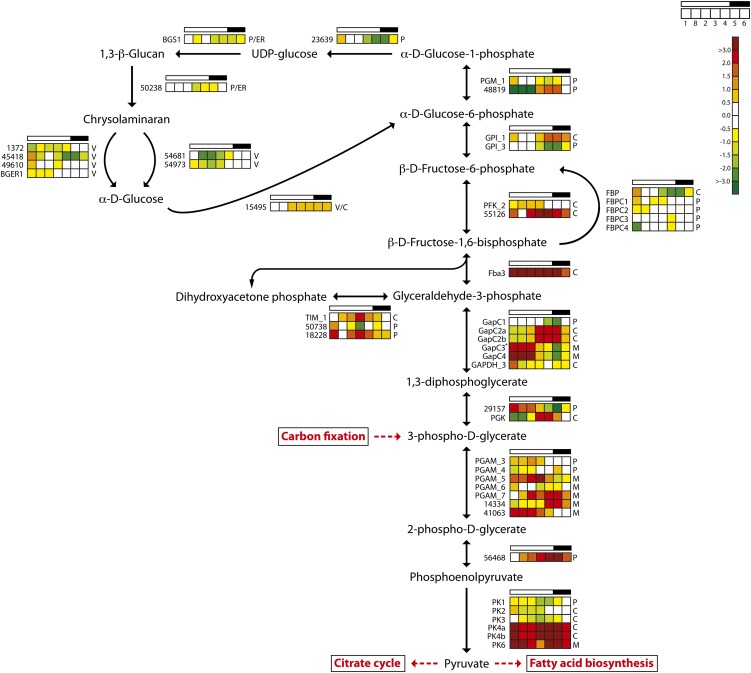
One of the key enzymes in gluconeogenesis is fructose-1,6-bisphosphate aldolase (Fba), which reversibly converts d-glyceraldehyde 3-phosphate into d-Fru-1,6-bisP. The evolutionary origin and functional diversification of the various Fba found in diatoms are described by Allen et al. (2012). Fba3 is a class II gene encoding a cytosolic enzyme; a minimum expression level was observed late at night, and it showed a strong light dependency, with about 16 times higher expression during the light period (Fig. 4). This suggests that Fba3 has an important role in the biosynthesis of hexose sugars and storage polysaccharides. Blue light induction of Fba3 has been reported previously (Tachibana et al., 2011). The other class II gene (FbaC2, with a putative plastid localization) showed less diel regulation.
Figure 4.
Light/dark cycle regulation of genes encoding glycolytic and glucan biosynthesis enzymes in P. tricornutum. Colored squares indicate expression levels at time points (hours after light on) T1 = 0.5, T8 = 27 (day 2), T2 = 6, T3 = 10.5, T4 = 15.5, T5 = 16.5 (darkness), and T6 = 20 (darkness), and the data are normalized to time point T7 = 23.5 (darkness). Light and dark periods are indicated in the bar above the squares. The color scale indicates log2-transformed gene expression ratios. +GAPC3 is a protein fusion with TPI. BGER, β-Glucan elicitor receptor; BGS, 1,3-β-glucan synthase; C, cytosolic localization; M, mitochondrial localization; P, plastid localization; PFK, phosphofructokinase; PGAM, phosphoglycerate mutase; PGK, phosphoglycerate kinase; PK, pyruvate kinase; TIM, triosephosphate isomerase; V, vacuolar localization. [See online article for color version of this figure.]
The first irreversible step of hexose sugar synthesis is the production of Fru-6-P catalyzed by the cytosolic FBP. There are five FBP enzymes in P. tricornutum, but only two of the enzymes, FBP and FBPC1, are light induced and show clear diel expression patterns (Fig. 3). FBPC1 most likely encodes a plastidic form, while FBP is a cytosolic enzyme and the best candidate gene responsible for sugar biosynthesis (Kroth et al., 2008). Fru-6-P is converted to Glc-6-P by glucose-6-phosphate isomerase (GPI), and the two GPIs observed here (GPI_1 and GPI_3) showed opposite regulation (Fig. 4). GPI_3, which carries a chloroplast targeting signal, is closely coregulated with enzymes of the Calvin cycle and is most likely coupled to gluconeogenesis. GPI_1 may be a key enzyme of glycolysis; it has a predicted cytosolic localization and showed highest transcriptional activity at night. The conversion of Glc-6-P to Glc-1-P is catalyzed by phosphoglucomutase (PGM; Fig. 4). There are five PGM enzymes in P. tricornutum, but only two of these genes, PGM_1 (eukaryotic type) and Phatr2_48819 (cyanobacterial type), show clear diel expression patterns. Both proteins have chloroplast-targeting signals. PGM_1 is related to Arabidopsis PGM, which is essential for starch biosynthesis, and displays a gene expression pattern related to carbon fixation/Calvin cycle genes, suggesting that it has functions connected to gluconeogenesis. In contrast, the cyanobacteria-type PGM is strongly down-regulated by light and has low expression during the first 6 h of the day, with maximum transcriptional activity in the evening and during the night (Fig. 4).
Recently the presence of a functional Entner-Doudoroff pathway (EDP), which converts Glc-6-P to pyruvate and glyceraldehyde-3-phosphate, was reported in P. tricornutum (Fabris et al., 2012). The putative key enzyme of this pathway, 2-keto-3-deoxyphosphogluconate aldolase (EDA; Phatr2_34120), peaked at the beginning of the light period and reached the expression minimum at the beginning of the dark period (Supplemental Table S1). Both enzymes of the P. tricornutum EDP have a putative mitochondrial localization; however, the EDA expression pattern is opposite to most of the genes encoding mitochondria- and cytosol-localized glycolytic enzymes. The role of the EDP may be to feed pyruvate to the TCA cycle early in the light period, when pyruvate production through the “classic” glycolytic pathway is low.
Several of the genes encoding putatively plastid- or ER-localized enzymes in the gluconeogenesis/1,3-β-glucan pathway, such as GPI_3, PGM1, Phatr2_23639, BGS1, and a putative 1,6-β-branching enzyme, Phatr2_50238 (Fabris et al., 2012), showed a coordinated diel expression pattern, with a peak at the beginning and a minimum at the end of the light period (Fig. 4). In contrast, both endo-1,3-β-glucosidases that were differentially expressed in the experiment (Phatr2_54681 and Phatr2_54973) reached maximum (yet low) expression during the dark period. Both the endo-1,3-β-glucanases and the exo-1,3-β-glucosidases identified on the P. tricornutum genome have a putative localization to the ER/Golgi or vacuoles (Kroth et al., 2008). Based on the spatial and temporal separation of the different components, a possible pattern of glucan biosynthesis and metabolism emerges. During the day, carbon fixed through photosynthesis is shuttled through the gluconeogenesis pathway in chloroplasts to produce the nucleotide sugar UDP-Glc. UDP-Glc is made available to BGS1 through ER connected to the plastid, leading to the synthesis of chrysolaminaran that may be transported through the ER/Golgi to storage vacuoles. At night, the expression of biosynthetic enzymes decreases and the expression of endo-1,3-β-glucanases and exo-1,3-β-glucosidases increases, shifting the equilibrium toward the degradation of chrysolaminaran to Glc. P. tricornutum encodes one glucokinase (Phatr2_15495) that may be cytosolic or localized to the vacuole (Fig. 4). A Glc transporter (Phatr2_12520) with a predicted vacuolar localization (WoLF PSORT) showed highest expression during the dark period (Supplemental Table S1) and may act to shuttle Glc to the cytosol. A cytosolic glucokinase could convert Glc to Glc-6-P, which may enter the glycolytic pathway via the cytosolic GPI_1 that was, in fact, up-regulated during the dark period.
TCA Cycle
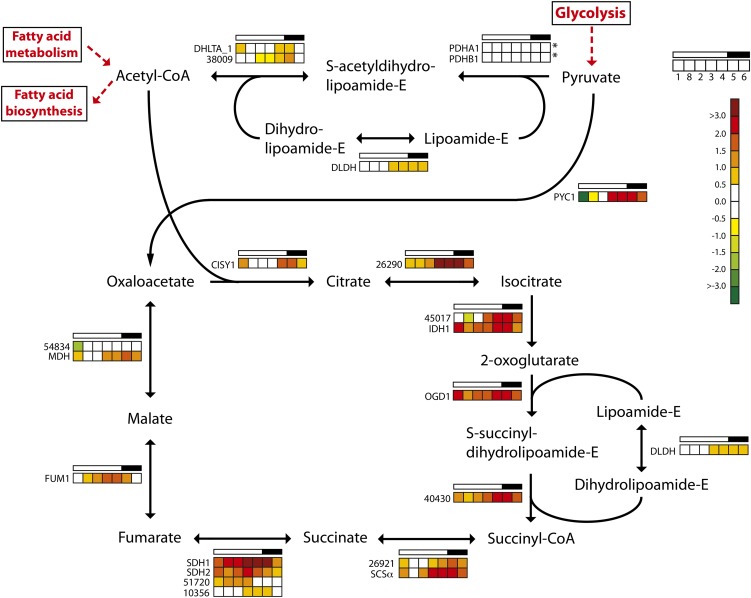
The TCA cycle is located to the mitochondria, although some of the involved isoenzymes have a putative cytosolic localization (Kroth et al., 2008; Millar et al., 2011). Pyruvate produced through glycolysis enters the TCA cycle in two ways: either through conversion to acetyl-CoA by the mitochondrial pyruvate dehydrogenase complex or through conversion to oxaloacetate by pyruvate carboxylase (Fig. 5). The genes PDHA1, PDHB1, DHLTA_1, Phatr2_38009, and DLDH encode components of the mitochondrial pyruvate dehydrogenase complex and show low and relatively stable expression throughout the diel cycle. In contrast, the majority of the genes encoding TCA cycle enzymes displayed a coordinated, cyclic expression pattern. The expression levels of these genes increased throughout the light period, reaching maximum levels around the light/dark transition, and decreased in the dark period. Thus, the TCA cycle is up-regulated at the onset of night in order to provide the cells with ATP, carbon skeletons, and other metabolites. A major sink for the carbon skeletons is in nitrogen assimilation, which occurs mainly in the light. Similar regulation of TCA pathway genes has been observed in cyanobacteria (Stöckel et al., 2008; Shi et al., 2010; Straub et al., 2011).
Figure 5.
Light/dark cycle regulation of genes encoding TCA cycle enzymes in P. tricornutum. Colored squares indicate expression levels at time points (hours after light on) T1 = 0.5, T8 = 27 (day 2), T2 = 6, T3 = 10.5, T4 = 15.5, T5 = 16.5 (darkness), and T6 = 20 (darkness), and the data are normalized to time point T7 = 23.5 (darkness). Light and dark periods are indicated in the bar above the squares. The color scale indicates log2-transformed gene expression ratios. Asterisks indicate that the gene did not show significantly differential expression (P < 0.05) at any time point. CISY, Citrate synthase; DHLTA, dihydrolipoamide acetyltransferase; DLDH, dihydrolipoyl dehydrogenase; FUM, fumarase; IDH, isocitrate dehydrogenase; MDH, malate dehydrogenase; OGD, 2-oxoglutarate dehydrogenase E1 component; PDH, pyruvate dehydrogenase; PYC, pyruvate carboxylase; SCSα, succinyl-CoA ligase α-subunit; SDH, succinate dehydrogenase flavoprotein. [See online article for color version of this figure.]
Fatty Acid Biosynthesis
De novo biosynthesis of fatty acids occurs in the plastids of all plants and algae. Plastidial acetyl-CoA acts as a precursor for fatty acid biosynthesis and is provided from different sources: (1) from the conversion of pyruvate by the pyruvate dehydrogenase complex, which is present both in mitochondria and chloroplasts; and (2) from the conversion of acetate by acetyl-CoA synthetase. Nannochloropsis sp. incorporated acetate into lipids during the light period; in the dark, acetate was incorporated into nonlipid components at a lower rate (Sukenik and Carmeli, 1990).
Recently, a plastidial sodium-dependent pyruvate transporter was reported in plants (Furumoto et al., 2011). Both P. tricornutum (Phatr2_34802) and T. pseudonana (Thaps3_711) encode an ortholog with high similarity to the plant pyruvate transporters from Flaveria trinervia and Arabidopsis. Phatr2_34802 contains 35% identical and 48% conserved residues compared with the Arabidopsis pyruvate transporter BASS2 (At2g26900). A phylogenetic analysis based on a peptide alignment of the plant pyruvate transporters and related transporters showed that Phatr2_34802 and Thaps3_711 cluster together with Arabidopsis and F. trinervia BASS2 transporters with high significance (Supplemental Fig. S1A). Phatr2_34802 contains a putative bipartite N-terminal presequence indicative of a plastid localization. P. tricornutum also encodes an acetyl-CoA transporter (Phatr2_54049) with unclear localization; however, the TargetP localization server (Emanuelsson et al., 2007) predicts a chloroplast transit peptide. All three genes encoding the plastidic pyruvate dehydrogenase complex in P. tricornutum showed similar expression profiles throughout the diel cycle (Supplemental Fig. S1B). The genes were unregulated or moderately up-regulated at the beginning of the light period, but expression levels decreased throughout the day and reached a minimum around the light/dark transition.
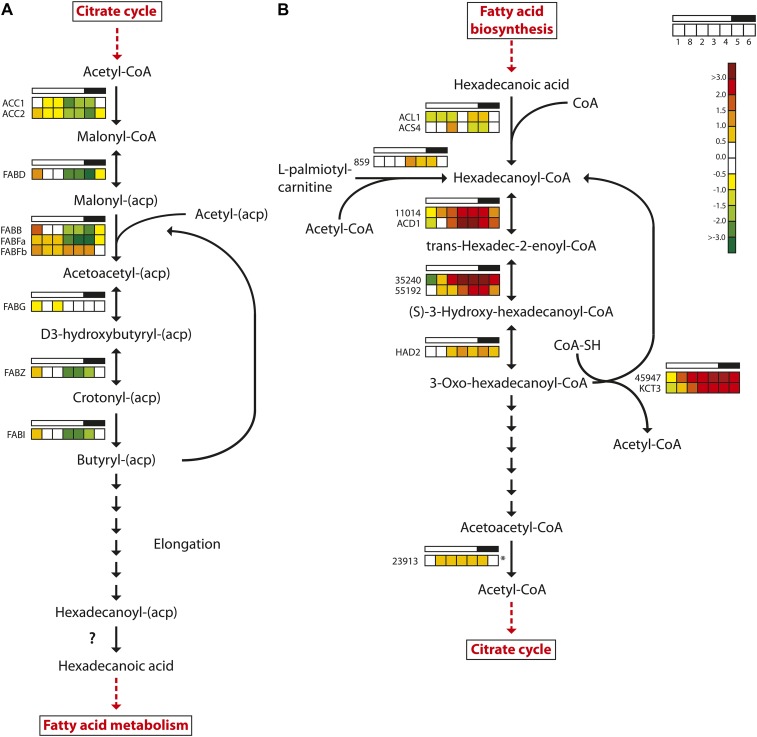
A similar expression profile was observed for the majority of the genes encoding enzymes involved in fatty acid biosynthesis (Fig. 6). Of the nine fatty acid biosynthesis genes that were regulated in the experiment, eight reached maximum expression levels after the onset of light and minimum expression levels toward the end of the light period. FABFb, encoding one of three 3-oxoacyl-[acyl carrier protein] synthases catalyzing the condensation reaction in fatty acid synthesis in P. tricornutum, showed a divergent expression pattern. It peaked twice during the diel cycle: after the onset and at the end of the light period. FABFb expression reached the lowest levels during the dark period. The two other 3-oxoacyl-[acyl carrier protein] synthase genes (FABB and FABFa) showed the “consensus” expression pattern of fatty acid biosynthesis genes; furthermore, their maximum expression levels were 10 to 20 times higher than that of FABFb (Supplemental Table S1). Several genes encoding desaturases involved in the biosynthesis of unsaturated fatty acids (PtFAD2, PtFAD6, PTD5b, and PTD6) had similar expression patterns, with maximum expression levels after the onset of light (Supplemental Table S1). These genes were among the most responsive to the dark/light transition. We were not able to identify any clear candidate gene product responsible for the final hydrolysis generating the fatty acid. Daylight appears to be anticipated, as expression increased throughout the dark period. The spike in expression at the beginning of the light period was still pronounced for most of the fatty acid biosynthesis genes. It could act as a response to a sudden flux/increase in carbon fixed by photosynthesis as the light is turned on. However, no increase in neutral lipid levels was observed until 3 h after the onset of light. One explanation for this apparent lag could be that increased lipid production during this period is channeled into the biosynthesis of polar lipids or free fatty acids, which are not detected by the Nile Red assay applied here.
Figure 6.
Light/dark cycle regulation of genes encoding fatty acid metabolism enzymes in P. tricornutum. A, Fatty acid biosynthesis. B, Fatty acid β-oxidation. Colored squares indicate expression levels at time points (hours after light on) T1 = 0.5, T8 = 27 (day 2), T2 = 6, T3 = 10.5, T4 = 15.5, T5 = 16.5 (darkness), and T6 = 20 (darkness), and the data are normalized to time point T7 = 23.5 (darkness). Light and dark periods are indicated in the bar above the squares. The color scale indicates log2-transformed gene expression ratios. The question mark indicates that no candidate gene for the enzyme activity has been identified. The asterisk indicates that the gene did not show significantly differential expression (P < 0.05) at any time point. ACC, Acetyl-CoA carboxylase; FABB/FABF, 3-oxoacyl-[acyl carrier protein] synthase; FABD, malonyl-CoA transacylase; FABG, 3-oxoacyl-(acyl carrier protein) reductase; FABI, enoyl-ACP reductase; FABZ, 3R-hydroxyacyl-[acyl carrier protein] dehydrase; HAD, 3-hydroxyacyl-CoA dehydrogenase; KCT, β-ketoacyl-CoA thiolase. [See online article for color version of this figure.]
Fatty Acid β-Oxidation
Fatty acid degradation in plants takes place in the peroxisomes via the β-oxidation cycle. In diatoms, however, there appear to be two fatty acid degradation pathways, similar to mammalian cells, where one pathway is located in the peroxisomes and the other is located in the mitochondria. Two key enzymes involved in mitochondrial β-oxidation, the multifunctional fatty acid oxidation complex subunit α (Phatr2_35240) and β-ketoacyl-CoA thiolase (KCT3; also known as the mitochondrial trifunctional protein β-subunit), were both down-regulated during the late dark period/beginning of the light period and showed highest expression late in the light period and in the dark period (Fig. 6). Five other genes predicted to encode enzymes involved in mitochondrial β-oxidation show a similar diel regulation pattern: acyl-CoA dehydrogenase (Phatr2_11014), catalyzing the first step in β-oxidation; enyol-CoA hydratase small chain (Phatr2_55192), catalyzing the second step in mitochondrial fatty acid β-oxidation; 3-ketoacyl-CoA thiolase (Phatr2_45947); the short-chain acyl-CoA dehydrogenase; and 3-hydroxyacyl-CoA dehydrogenase. The observed expression pattern suggests that the mitochondrial β-oxidation pathway provides acetyl-CoA that can be used in the TCA cycle during the night. Metabolic energy and carbon can be stored as fat that is later used for the synthesis of Glc and other carbohydrates via the glyoxylate cycle. Malate synthase and isocitrate lyase (Phatr2_14401) are two central enzymes in the glyoxylate cycle, and they enable the synthesis of hexoses using acetyl-CoA as an intermediate. In addition to its role as an intermediate in gluconeogenesis, acetyl-CoA is also a substrate in the glyoxylate cycle. Both genes were strongly down-regulated when the light was turned on (Supplemental Table S1). PEPCK1, which catalyzes the final step in the production of phosphoenolpyruvate used for gluconeogenesis, showed a similar expression pattern.
The 3,5-Δ-2,4-dienoyl-CoA isomerase (DEI1) is predicted to encode a peroxisome-localized enzyme involved in fatty acid β-oxidation. DEI1 showed increased transcriptional activity during the daytime and decreased activity late in the evening and during the night. None of the other enzymes predicted to be connected to the peroxisomal β-oxidation pathway, such as acyl-CoA oxidase, acyl-CoA dehydrogenase (Phatr2_42907), and peroxisomal 2,4-dienoyl-CoA reductase, showed increased transcriptional activity late in the evening or at night.
Cell Division
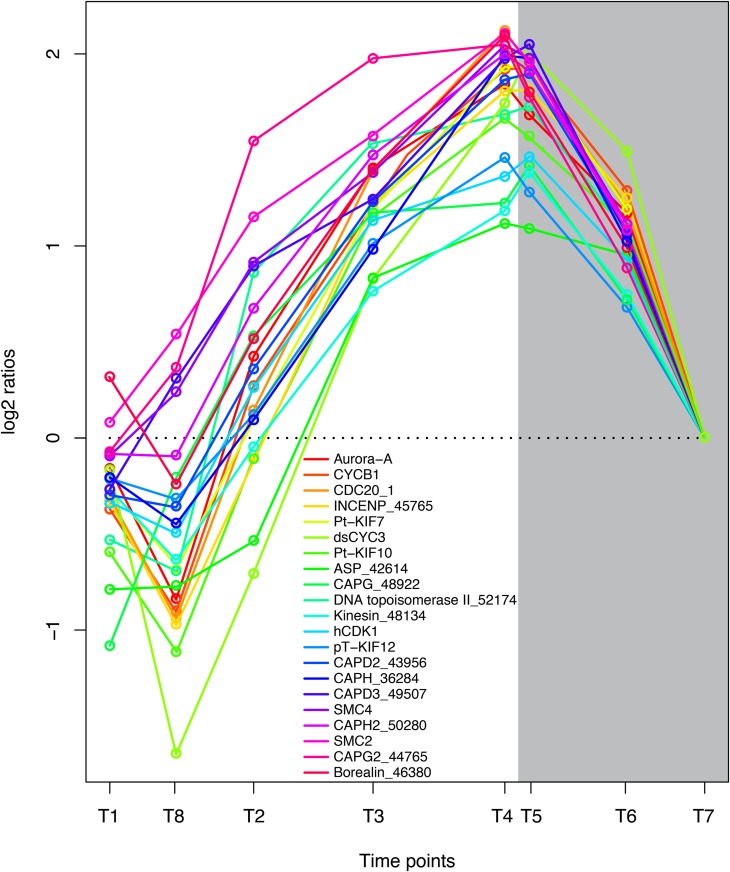
The goal of the previously described metabolic processes is to enable cell division, and more than 100 transcripts encoding proteins that are involved in DNA replication, cell cycle regulation, and cell division showed a high degree of coregulation during the time course of the experiment. Genes encoding proteins with key functions in mitosis had the lowest expression early in the day and increased to a maximum late in the evening and early night (Fig. 7), showing that cell division is more frequent late in the evening and during the night. These observations are in correspondence with previous results (Nelson and Brand, 1979; Vaulot et al., 1986; Bowler et al., 2010). Chromosomes undergo a major structural reorganization during mitosis, and the compaction of interphase chromatin into mitotic chromosomes is regulated by a multisubunit ATPase complex, the condensin complex. The condensation of chromosomes is a continuous process that is initiated in early prophase and is required for accurate chromosome segregation during anaphase (Swedlow and Hirano, 2003; Moser and Swedlow, 2011). Another protein with important functions during chromosome condensation is topoisomerase II (Uemura et al., 1987). The P. tricornutum ortholog of topoisomerase II (Phatr2_ 52174) and all components of the condensin complex I/II showed highly coregulated gene expression, with highest expression levels observed late in the evening and during early night. In addition, we observed that genes encoding proteins coupled to the spindle checkpoint are tightly coregulated with the condensin complex genes. Aurora kinases are reported to phosphorylate condensin subunits during mitosis (Nakazawa et al., 2011). P. tricornutum Aurora-A, the INCENP ortholog (Phatr_45765), Borealin/CDCA8-related protein (Phatr2_46380), and CDC20 ortholog (Phatr2_12783) showed the same diel pattern of regulation as the condensin complex genes (Fig. 7). These genes might serve as useful markers for cells undergoing mitosis.
Figure 7.
Coordinated regulation of genes involved in cell division and mitosis. Expression levels at sampling points (hours after light on) T1 = 0.5, T8 = 27 (day 2), T2 = 6, T3 = 10.5, T4 = 15.5, T5 = 16.5 (darkness), T6 = 20 (darkness), and T7 = 23.5 (darkness) are normalized to time point T7. Protein names and GenBank accession numbers are as follows: Aurora-A, Aurora kinase A (EEC48936.1); CYCB1, cyclin B1 (EEC47805.1); CDC20_1, cell division cycle20 (EEC47990.1); Phatr2_45765, INCENP protein (EEC48380.1); Pt-KIF7, kinesin family-like protein (EEC45857.1); dsCYC3, cyclin protein (EEC51859.1); Pt-KIF10, kinesin family-like protein (EEC45010.1); Phatr2_42614, abnormal spindle-like protein (EEC51580.1); CAPG_48922, non-SMC condensin I complex subunit G (EEC44841.1); Phatr2_52174, DNA topoisomerase II (EEC47151.1); Phatr2_48134, kinesin motor domain protein (EEC45795.1); hCDK1, mitogen-activated protein kinase (EEC47173.1); pT-KIF12, kinesin family-like protein (EEC51889.1); CAPD2_43956, non-SMC condensin I complex subunit D2 (ACI65548.1); CAPH_36284, non-SMC condensin I complex subunit H (EEC47398.1); CAPD3_49507, condensin II complex subunit D3 (EEC44317.1); SMC4, condensin complex subunit SMC4 (ACI65666.1); CAPH2_50280, condensin II complex subunit H2 (EEC43358.1); SMC2, condensin complex subunit SMC2 (EEC44274.1); CAPG2_44765, condensin II complex subunit G2 (EEC49642.1); Borealin_46380, borealin-related protein (EEC47609.1). Genes that previously have not been assigned gene symbols or gene functions are marked with an underscore and corresponding Phatr2 identifier (Phatr2; version 2.0 of the P. tricornutum genome). [See online article for color version of this figure.]
CONCLUSION
This time-series study of carbon metabolism, storage, and regulation in the diatom P. tricornutum acclimated to a 16/8-h light/dark period highlights more than 4,500 transcripts that were differentially regulated, many of which are associated with carbohydrate and lipid metabolism and are responsive to light. Orthologous genes not previously described in algae, such as a pyruvate transporter, have been identified. Examples of concerted transcriptional activities regulating steps of the same process/metabolic pathway are suggested (e.g. fast-responding genes in fatty acid biosynthesis). More than 100 transcripts related to cell cycle regulation, DNA replication, and cell division show a high degree of coregulation.
Our data confirm the spatial regulation of different pathways in carbon metabolism, with some processes taking place in either plastids or mitochondria. Enzymes involved in glycolysis/gluconeogenesis, however, are localized to the cytosol, chloroplast, and mitochondria. Our data also suggest a clear temporal separation of the processes that capture carbon (light period) and mitosis (dark period) in P. tricornutum acclimated to a regular light/dark cycle of 16/8 h. In general, pathways localized to chloroplasts, such as carbon fixation, glucan biosynthesis, and lipid biosynthesis, are up-regulated at the beginning of the light period. Pathways localized to the mitochondria, such as the TCA cycle and lipid β-oxidation, peak toward the end of the light period. The localization combined with the diel expression pattern identified in this study may be of help in dissecting the roles of different isoenzymes. Furthermore, this data set provides a valuable source to aid the work of mining genes involved in light responses and circadian rhythms.
MATERIALS AND METHODS
Cultures and Growth Conditions
Phaeodactylum tricornutum (originating from the CCMP 632 strain) was grown in axenic cultures at 20°C and illuminated with 150 μmol photons m−2 s−1 provided by fluorescent tubes (Philips Master TL-D 36W/840) in cycles of 16/8-h photoperiods. Growth medium was made from natural seawater filtered through 0.2-μm Tuffryn membrane capsules (Pall Gelman) before nutrient amendment and autoclaving. Nutrient amendment was according to the f/2 recipe of Guillard (1975), but phosphorus was increased to 1.1 of the f/2 recipe to prevent phosphorus limitation throughout the experimental period.
Three cultures of each treatment were grown in 2,000-mL Nalgene optically clear flasks and aerated with a mixture of air and 1% to 2% CO2 (v/v) to prevent settling of cells and carbon limitation. Without the addition of CO2, its concentration and thus also pH could vary dependent on the presence/absence of light, with biased light/dark responses as a consequence. Cultivation was performed in exponential fed-batch modus: fresh medium was added to the cultures every 1 min by micropumps (Bio-Chem Valve), and medium doses increased exponentially based on information on actual culture volume and the desired dilution rate. The maximum specific growth rate was determined from measurements of optical density at 750 nm (OD750) and cell numbers in batch cultures prior to the fed-batch study, and an average maximum specific growth rate of 1.4 d−1 was observed in different treatments (unlimited/nitrogen or phosphorus limitation). Both experimental cultures were filled and harvested several times until a stable optical density (i.e. biomass) for more than 5 consecutive days confirmed steady-state growth. Steady state was used as a criterion for when sampling was initiated. An increase in cell numbers of 30% was observed during the dark phase of the experimental period, indicating that the effective growth rate was slightly higher in the experimental fed-batch cultures, 1.55 ± 0.12 d−1, based on a regression of cell numbers versus time in the two replicate cultures. Both cultures were checked to confirm axenity by flow cytometric analyses of samples stained with SYBR Green I during the cultivation period.
Sampling and Analyses
At each sampling point during a 26.5-h period, two samples of 50 mL were removed from each culture. The sampling started 0.5 h after the light was turned on in the morning on the first day and was finalized after 27 h. The sampling points were as follows (given as hours after light on): T1 = 0.5, T2 = 6, T3 = 10.5, T4 = 15.5, T5 = 16.5 (darkness), T6 = 20 (darkness), T7 = 23.5 (darkness), and T8 = 27 (next day). T8 represents the chronological midpoint between T1 and T2 in the figures to emphasize the cyclicity of cell processes in acclimatized fed-batch cultures. In the analyses of gene expression, sample T7 was used as a reference. One 50-mL sample was used for measurements of OD750 and NRF and analyses of particulate carbon and nitrogen as well as water-soluble carbohydrates (data analyzed at all time points). Flow cytometric analyses of cell numbers were performed on samples from time points 1, 2, 4, 6, and 8 h. The other 50-mL sample was harvested by centrifugation for RNA extraction.
Biomass and Cell Chemistry
OD750 was measured in replicate aliquots of 2.5 mL from each culture in a Merck Spectroquant Pharo 100 spectrophotometer. The same samples were transferred to a Turner Designs Aquafluor spectrofluorometer for measurement of autofluorescence before the samples were stained with Nile Red (0.25 mg mL−1 in acetone) and incubated for at least 15 min in darkness at room temperature prior to measurements of Nile Red fluorescence (Chen et al., 2009). Excitation/emission wavelengths are centered around 525/575 nm on this instrument, and the measured fluorescence is considered as representative of neutral lipids. The measured NRF was corrected for the autofluorescence from the cells and made biomass specific by normalization to cell counts.
Cells were counted in a Becton Dickinson FACScan flow cytometer equipped with an argon-ion laser that provides excitation light of 488 nm. Subsamples of cells fixed with glutaraldehyde were analyzed by collecting signals of side scatter and autofluorescence (detector FL3 with a 650-nm/LP filter), and Fluoresbrite Carboxylate YG 1-μm Microspheres (Polysciences) were used as an internal reference.
Samples for analysis of particulate carbon and nitrogen were collected on precombusted Whatman GF/F filters and stored at −23°C until analysis. Duplicate samples and blank filter pieces were bored out and treated with fuming HCl (37%) before they were packed into tin capsules and dried at 60°C. The samples were analyzed on an ECS 4010 Costech Instruments element analyzer.
Total water-soluble carbohydrates were analyzed spectrophotometrically using a modified phenol-sulfuric method where absorbance readings at 485 nm are converted to carbohydrate content using Glc as a standard (Granum and Myklestad, 2002).
Samples for gravimetric determination of total neutral (chloroform-extractable) lipids were collected by centrifugation of 1.25 L of culture from each treatment when the experimental sampling was finished and the cultures were refilled. The samples were lyophilized before they were blown with nitrogen and stored at −80°C until analysis. Lipid extraction was based on the method of Bligh and Dyer (1959).
Sample Preparation and RNA Isolation
From each time point, a sample of 50-mL culture volume was collected from each culture flask. The samples were centrifuged at 4,000g for 10 min at 15°C and resuspended in 1 mL of f/2 medium. The resuspended cells were transferred to 2-mL tubes and centrifuged at 18,000g for 1 min, the supernatant was removed, and the remaining cell pellet was flash frozen in liquid nitrogen and stored at 80°C. Samples harvested during the dark period were protected from stray light by covering tubes with aluminum foil, and as little light as possible was allowed in the laboratory during sampling and centrifugation. Total RNA was isolated from the samples using the Spectrum Plant Total RNA kit (Sigma-Aldrich) as described by Nymark et al. (2009).
DNA Microarray Experiments
The RNA from each sample (83–200 ng) was reverse transcribed, amplified, and labeled using the Low Input Quick Amp Labeling Kit, One-Color (Agilent; 5190-2305). A total of 1,650 ng of copy RNA from each sample was fragmented and hybridized on 4x44K P. tricornutum whole-genome 60-mer oligonucleotide microarrays (Agilent Technologies) in an Agilent G2545A Hybridization Rotary Oven (10 rpm, 65°C, 17.5 h). Hybridization was performed with the Gene Expression Hybridization Kit (Agilent; 5188-5242). The slides were washed with buffers 1 2 from the Gene Expression Wash Buffer Kit (Agilent; 5188-5327) and scanned twice at 5-μm resolution on a laser scanner (G2505 B; Agilent Technologies) using the “dynamic range expander” option in the scanner software. The resulting images were processed using Agilent Feature Extraction software version 9.5. Microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE42514.
Statistical Analysis
The single-color scan data (feature extraction files) were analyzed, and genes with statistically significant differential expression were identified using the Limma package (version 3.2.3; Smyth et al., 2005) and R (version 2.10.1). Spots identified as feature outliers were excluded from analysis, and weak or not detected spots were given reduced weight (0.5). The data were normalized using the quantile method, and no background subtraction was performed. A design matrix was created, and contrasts between time point T7 (23.5 h; the last dark time point) against each of the time points (T1 = 0.5 h, T2 = 6 h, T3 = 10.5 h, T4 = 15.5 h, T5 = 16.5 h, T6 = 20 h, T7 = 23.5 h, and T8 = 27 h) were computed. The method of Benjamini and Hochberg (1995) was used to estimate the false discovery rate. Genes with adjusted P < 0.05 were considered to be statistically significantly differentially expressed. Genes selected for further analysis must have average adjusted P < 0.05 in at least one time point. It should be noted that each gene could be represented with one to five oligonucleotide probes.
Annotation of Genes and Pathways
The genes were annotated based on data from the P. tricornutum genome database at the Department of Energy Joint Genome Institute (Bowler et al., 2008; http://genome.jgi-psf.org/Phatr2/Phatr2.home.html). Where no name was given, we used the protein identifier. The biochemical pathways were drawn based on data from the Kyoto Encyclopedia of Genes and Genomes (Kanehisa et al., 2012) and DiatomCyc (Fabris et al., 2012) databases using Adobe Illustrator.
Network Analysis
A table with log2 ratios for all genes with positive detection in at least one of the time points (10,365 in total) was used to calculate Pearson correlation values for each gene pair. To reduce the number of genes included in the analysis, a filtered list of genes where only the most responsive genes was represented was made using the following filtering criteria: the gene must have an absolute log2 > 1.5 and adjusted P < 0.05 for at least one of the time points. The resulting list of 1,991 genes was used to extract gene pairs with Pearson correlation values above 0.95, in total 44,024 gene pairs. The gene pair list was imported into Cytoscape (Cline et al., 2007), and an unweighted network was made using the force-directed algorithm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Light/dark cycle regulation of genes encoding a pyruvate transporter and the plastidic pyruvate dehydrogenase complex in P. tricornutum.
Supplemental Table S1. Diel regulation of genes in P. tricornutum. List of 4,567 genes at sampling times T1 = 07:30 am, T2 = 1 pm, T3 = 5:30 pm, T4 = 10:30 pm, T5 = 11:30 pm (darkness), T6 = 3 am (darkness), and T7 = 6:30 am.
Acknowledgments
We thank Leila Alipanah for assisting with the harvesting of cultures and Torfinn Sparstad for help with the microarray hybridization.
Glossary
- TCA
tricarboxylic acid
- ER
endoplasmic reticulum
- NRF
Nile red fluorescence
- PGM
phosphoglucomutase
- GPI
glucose-6-phosphate isomerase
- EDP
Entner-Doudoroff pathway
- OD750
optical density at 750 nm
References
- Allen AE, Moustafa A, Montsant A, Eckert A, Kroth PG, Bowler C. (2012) Evolution and functional diversification of fructose bisphosphate aldolase genes in photosynthetic marine diatoms. Mol Biol Evol 29: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bligh EG, Dyer WJ. (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244 [DOI] [PubMed] [Google Scholar]
- Bowler C, De Martino A, Falciatore A. (2010) Diatom cell division in an environmental context. Curr Opin Plant Biol 13: 623–630 [DOI] [PubMed] [Google Scholar]
- Breteler WCMK, Schogt N, Rampen S. (2005) Effect of diatom nutrient limitation on copepod development: role of essential lipids. Mar Ecol Prog Ser 291: 125–133 [Google Scholar]
- Chen W, Zhang CW, Song LR, Sommerfeld M, Hu Q. (2009) A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J Microbiol Methods 77: 41–47 [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeule B, Gurny R, Arvinte T. (2007) Detection and characterization of protein aggregates by fluorescence microscopy. Int J Pharm 329: 37–45 [DOI] [PubMed] [Google Scholar]
- Domergue F, Spiekermann P, Lerchl J, Beckmann C, Kilian O, Kroth PG, Boland W, Zähringer U, Heinz E. (2003) New insight into Phaeodactylum tricornutum fatty acid metabolism: cloning and functional characterization of plastidial and microsomal Δ12-fatty acid desaturases. Plant Physiol 131: 1648–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron A, Rabouille S, Claquin P, Le Roy B, Talec A, Sciandra A. (2012) Light-dark (12:12) cycle of carbon and nitrogen metabolism in Crocosphaera watsonii WH8501: relation to the cell cycle. Environ Microbiol 14: 967–981 [DOI] [PubMed] [Google Scholar]
- Droop MR. (1974) Nutrient status of algal cells in continuous culture. J Mar Biol Assoc U K 54: 825–855 [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fabris M, Matthijs M, Rombauts S, Vyverman W, Goossens A, Baart GJ. (2012) The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J 70: 1004–1014 10.1111/j.1365-1313X.2012.04941.x [DOI] [PubMed] [Google Scholar]
- Fan JL, Andre C, Xu CC. (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett 585: 1985–1991 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Obata T, Allen AE, Araújo WL, Bowler C. (2012) Leveraging metabolomics for functional investigations in sequenced marine diatoms. Trends Plant Sci 17: 395–403 10.1016/j.tplants.2012.1002.1005 [DOI] [PubMed] [Google Scholar]
- Furumoto T, Yamaguchi T, Ohshima-Ichie Y, Nakamura M, Tsuchida-Iwata Y, Shimamura M, Ohnishi J, Hata S, Gowik U, Westhoff P, et al. (2011) A plastidial sodium-dependent pyruvate transporter. Nature 476: 472–475 [DOI] [PubMed] [Google Scholar]
- Granum E, Myklestad SM. (2002) A simple combined method for determination of beta-1,3-glucan and cell wall polysaccharides in diatoms. Hydrobiologia 477: 155–161 [Google Scholar]
- Granum E, Roberts K, Raven JA, Leegood RC. (2009) Primary carbon and nitrogen metabolic gene expression in the diatom Thalassiosira pseudonana (Bacillariophyceae): diel periodicity and effects of inorganic carbon and nitrogen. J Phycol 45: 1083–1092 [DOI] [PubMed] [Google Scholar]
- Greenspan P, Fowler SD. (1985) Spectrofluorometric studies of the lipid probe, Nile red. J Lipid Res 26: 781–789 [PubMed] [Google Scholar]
- Guillard RRL. (1975) Culture of phytoplankton for feeding marine invertebrates. In WL Smith, MH Chanley, eds, Culture of Marine Invertebrate Animals. Plenum Press, New York, pp 29–60
- Haimovich-Dayan M, Garfinkel N, Ewe D, Marcus Y, Gruber A, Wagner H, Kroth PG, Kaplan A. (2013) The role of C(4) metabolism in the marine diatom Phaeodactylum tricornutum. New Phytol 197: 177–185 10.1111/j.1469-8137.2012.04375.x [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Bell LJ, Webb AAR. (2011) Interactions between plant circadian clocks and solute transport. J Exp Bot 62: 2333–2348 [DOI] [PubMed] [Google Scholar]
- Hockin NL, Mock T, Mulholland F, Kopriva S, Malin G. (2012) The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol 158: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroth PG, Chiovitti A, Gruber A, Martin-Jezequel V, Mock T, Parker MS, Stanley MS, Kaplan A, Caron L, Weber T, et al. (2008) A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 3: e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancelot C, Mathot S. (1985) Biochemical fractionation of primary production by phytoplankton in Belgian coastal waters during short-term and long-term incubations with C-14 bicarbonate. 1. Mixed diatom population. Mar Biol 86: 219–226 [Google Scholar]
- Larsen J, Hatzakis NS, Stamou D. (2011) Observation of inhomogeneity in the lipid composition of individual nanoscale liposomes. J Am Chem Soc 133: 10685–10687 [DOI] [PubMed] [Google Scholar]
- Liaud MF, Lichtlé C, Apt K, Martin W, Cerff R. (2000) Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol Biol Evol 17: 213–223 [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Soole KL, Day DA. (2011) Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol 62: 79–104 [DOI] [PubMed] [Google Scholar]
- Mittag M. (2003) The function of circadian RNA-binding proteins and their cis-acting elements in microalgae. Chronobiol Int 20: 529–541 [DOI] [PubMed] [Google Scholar]
- Monnier A, Liverani S, Bouvet R, Jesson B, Smith JQ, Mosser J, Corellou F, Bouget FY. (2010) Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genomics 11: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser SC, Swedlow JR. (2011) How to be a mitotic chromosome. Chromosome Res 19: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myklestad S, Granum E. (2009) Biology of (1,3)-β-glucans and related glucans in protozoans and chromistans. In A Bacic, GB Fincher, BA Stone, eds, Chemistry, Biochemistry, and Biology of (1-3)-β-Glucans and Related Polysaccharides. Academic Press, Boston, pp 353–385
- Nakazawa N, Mehrotra R, Ebe M, Yanagida M. (2011) Condensin phosphorylated by the Aurora-B-like kinase Ark1 is continuously required until telophase in a mode distinct from Top2. J Cell Sci 124: 1795–1807 [DOI] [PubMed] [Google Scholar]
- Nelson DM, Brand LE. (1979) Cell-division periodicity in 13 species of marine phytoplankton on a light-dark cycle. J Phycol 15: 67–75 [Google Scholar]
- Norici A, Bazzoni AM, Pugnetti A, Raven JA, Giordano M. (2011) Impact of irradiance on the C allocation in the coastal marine diatom Skeletonema marinoi Sarno and Zingone. Plant Cell Environ 34: 1666–1677 [DOI] [PubMed] [Google Scholar]
- Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM. (2009) An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 4: e7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MS, Mock T, Armbrust EV. (2008) Genomic insights into marine microalgae. Annu Rev Genet 42: 619–645 [DOI] [PubMed] [Google Scholar]
- Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. (2010) Genetic engineering of algae for enhanced biofuel production. Eukaryot Cell 9: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinfelder JR, Kraepiel AM, Morel FM. (2000) Unicellular C4 photosynthesis in a marine diatom. Nature 407: 996–999 [DOI] [PubMed] [Google Scholar]
- Reitan KI, Rainuzzo JR, Olsen Y. (1994) Effect of nutrient limitation on fatty acid and lipid content of marine microalgae. J Phycol 30: 972–979 [Google Scholar]
- Roberts K, Granum E, Leegood RC, Raven JA. (2007) Carbon acquisition by diatoms. Photosynth Res 93: 79–88 [DOI] [PubMed] [Google Scholar]
- Sanchez S, Bouget FY, Sanchez F, Garnier L. (2009) Functional genomics approaches to study the involvement of transcription factors in the microalgae Ostreococcus tauri circadian clock. Comp Biochem Physiol A Mol Integr Physiol 153: S207 [Google Scholar]
- Sapriel G, Quinet M, Heijde M, Jourdren L, Tanty V, Luo G, Le Crom S, Lopez PJ. (2009) Genome-wide transcriptome analyses of silicon metabolism in Phaeodactylum tricornutum reveal the multilevel regulation of silicic acid transporters. PLoS ONE 4: e7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze T, Prager K, Dathe H, Kelm J, Kiessling P, Mittag M. (2010) How the green alga Chlamydomonas reinhardtii keeps time. Protoplasma 244: 3–14 [DOI] [PubMed] [Google Scholar]
- Shi T, Ilikchyan I, Rabouille S, Zehr JP. (2010) Genome-wide analysis of diel gene expression in the unicellular N(2)-fixing cyanobacterium Crocosphaera watsonii WH 8501. ISME J 4: 621–632 [DOI] [PubMed] [Google Scholar]
- Sicko-Goad L, Simmons MS, Lazinsky D, Hall J. (1988) Effect of light cycle on diatom fatty acid composition and quantitative morphology. J Phycol 24: 1–7 [Google Scholar]
- Smyth GK, Michaud J, Scott HS. (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Stöckel J, Welsh EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB. (2008) Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc Natl Acad Sci USA 105: 6156–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Quillardet P, Vergalli J, de Marsac NT, Humbert JF. (2011) A day in the life of Microcystis aeruginosa strain PCC 7806 as revealed by a transcriptomic analysis. PLoS ONE 6: e16208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukenik A, Carmeli Y. (1990) Lipid synthesis and fatty acid composition in Nannochloropsis sp (Eustigmatophyceae) grown in a light-dark cycle. J Phycol 26: 463–469 [Google Scholar]
- Swedlow JR, Hirano T. (2003) The making of the mitotic chromosome: modern insights into classical questions. Mol Cell 11: 557–569 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Allen AE, Kikutani S, Endo Y, Bowler C, Matsuda Y. (2011) Localization of putative carbonic anhydrases in two marine diatoms, Phaeodactylum tricornutum and Thalassiosira pseudonana. Photosynth Res 109: 205–221 [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K, Raible F, Arboleda E. (2011) Another place, another timer: marine species and the rhythms of life. Bioessays 33: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. (1987) DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50: 917–925 [DOI] [PubMed] [Google Scholar]
- Vårum KM, Østgaard K, Grimsrud K. (1986) Diurnal rhythms in carbohydrate metabolism of the marine diatom Skeletonema costatum (Grev) Cleve. J Exp Mar Biol Ecol 102: 249–256 [Google Scholar]
- Vaulot D, Olson RJ, Chisholm SW. (1986) Light and dark control of the cell cycle in two marine phytoplankton species. Exp Cell Res 167: 38–52 [DOI] [PubMed] [Google Scholar]
- Zhang L, Weng WY, Guo JH. (2011) Posttranscriptional mechanisms in controlling eukaryotic circadian rhythms. FEBS Lett 585: 1400–1405 [DOI] [PubMed] [Google Scholar]
- Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML, Wright MA, Rector T, Steen R, McNulty N, et al. (2009) Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE 4: e5135. [DOI] [PMC free article] [PubMed] [Google Scholar]