Summary: The primary metabolic pathway gene NtMTHFR negatively regulates the secondary metabolism pathway nicotine demethylation gene to potentially recycle methyl groups from alkaloids.
Abstract
Methylenetetrahydrofolate reductase (MTHFR) is a key enzyme of the tetrahydrofolate (THF)-mediated one-carbon (C1) metabolic network. This enzyme catalyzes the reduction of 5,10-methylene-THF to 5-methyl-THF. The latter donates its methyl group to homocysteine, forming methionine, which is then used for the synthesis of S-adenosyl-methionine, a universal methyl donor for numerous methylation reactions, to produce primary and secondary metabolites. Here, we demonstrate that manipulating tobacco (Nicotiana tabacum) MTHFR gene (NtMTHFR1) expression dramatically alters the alkaloid profile in transgenic tobacco plants by negatively regulating the expression of a secondary metabolic pathway nicotine N-demethylase gene, CYP82E4. Quantitative real-time polymerase chain reaction and alkaloid analyses revealed that reducing NtMTHFR expression by RNA interference dramatically induced CYP82E4 expression, resulting in higher nicotine-to-nornicotine conversion rates. Conversely, overexpressing NtMTHFR1 suppressed CYP82E4 expression, leading to lower nicotine-to-nornicotine conversion rates. However, the reduced expression of NtMTHFR did not affect the methionine and S-adenosyl-methionine levels in the knockdown lines. Our finding reveals a new regulatory role of NtMTHFR1 in nicotine N-demethylation and suggests that the negative regulation of CYP82E4 expression may serve to recruit methyl groups from nicotine into the C1 pool under C1-deficient conditions.
Tetrahydrofolate (THF)-mediated one-carbon (C1) metabolism generates and provides C1 units in different oxidation states for various anabolic pathways, including alkaloid biosynthesis in plants (Hanson et al., 2000). Methylenetetrahydrofolate reductase (MTHFR) catalyzes the reduction of 5,10-methylene-THF into the most reduced C1 derivative, 5-methyl-THF (Guenther et al., 1999; Roje et al., 1999; Gelling et al., 2004). The latter then serves as the methyl group donor for Met synthesis from homocysteine (Guenther et al., 1999; Hanson et al., 2000). More than 80% of synthesized Met is further converted to S-adenosyl-methionine (AdoMet), which is the universal methyl group donor in reactions leading to modifications of various primary and secondary metabolites (Giovanelli et al., 1985; Hanson et al., 2000; Roje, 2006). In plants, MTHFRs have been cloned and characterized from Arabidopsis (Arabidopsis thaliana) and maize (Zea mays; Roje et al., 1999). In contrast to the mammalian enzymes, which are NADPH dependent and inhibited by AdoMet, plant MTHFRs are NADH dependent and AdoMet insensitive. In addition, the reaction that they catalyze is likely reversible in the cytosol (Roje et al., 1999). To date, MTHFRs from alkaloid-producing plants have not been cloned and studied in relation to their role in alkaloid metabolism, despite the importance of methyl group availability in the biosynthesis of multiple alkaloids.
In nature, about 20% of plant species produce alkaloids, which are indispensable compounds for plant defense (Baldwin et al., 2001) as well as for human health (Verpoorte and Memelink, 2002; Cragg and Newman, 2005; Ziegler and Facchini, 2008). Biosynthesis of numerous alkaloids, such as nicotine, ricinine, and morphine, in these plants requires methyl groups that are drawn from the THF-mediated C1 metabolism via AdoMet (Hanson et al., 2000; Ziegler and Facchini, 2008). Since MTHFR is a key enzyme of the THF-mediated C1 metabolic pathway, controlling the flux of C1 units toward methyl group biogenesis, we hypothesized that perturbing MTHFR expression would have an effect on alkaloid biosynthesis as a consequence of altered methyl group availability. On the other hand, it has been reported that the above three alkaloids can undergo enzymatic demethylation (Dawson, 1945; Skurský et al., 1969; Miller et al., 1973). Isotope-labeling studies in Nicotiana plumbaginifolia suspension cells have shown that methyl groups released from nicotine by N-demethylation find their way into many primary metabolites, such as Met and Ser, suggesting that they reenter the C1-folate pool (Mesnard et al., 2002; Bartholomeusz et al., 2005). Although it is well documented that C1 units from the THF-mediated pathway are required for alkaloid biosynthesis in plants (Hanson et al., 2000), it is unknown whether any molecular mechanisms are involved in regulating alkaloid N-demethylation in response to the C1 demand.
Tobacco (Nicotiana tabacum) plants are an ideal model system to study the role of MTHFR in regulating alkaloid metabolism, which is well studied in this model plant species (Bush et al., 1999). It has been well established that the methyl group for nicotine biosynthesis is derived from the THF-mediated C1 metabolic pathway via AdoMet (Mizusaki et al., 1972; Hibi et al., 1994). In commercial tobacco plants, nicotine accounts for 90% of the total alkaloid content, which is about 2% to 5% of dry leaf weight (Saitoh et al., 1985). Nicotine is synthesized in the roots (Dawson, 1941) by the condensation of two precursors, nicotinic acid and N-methylpyrrolinium cation, which are produced independently by the pyridine-nucleotide cycle and the methylpyrroline pathway, respectively (Wagner et al., 1986; Bush et al., 1999; Kajikawa et al., 2011). In the methylpyrroline pathway, the biosynthesis of N-methylputrescine from putrescine requires AdoMet as a cosubstrate (Mizusaki et al., 1972; Hibi et al., 1994). The synthesized nicotine is translocated through the xylem and transported to the leaves (Morita et al., 2009; Shoji et al., 2009; Hildreth et al., 2011), where it accumulates. In the leaf, nicotine can undergo N-demethylation to produce nornicotine (Griffith et al., 1955; Chakrabarti et al., 2008). Nornicotine is the second most abundant alkaloid in most tobacco cultivars (Saitoh et al., 1985), and its conversion is mainly controlled by the nicotine N-demethylase gene CYP82E4 (Siminszky et al., 2005; Xu et al., 2007). It is believed that CYP82E4-mediated oxidative N-demethylation of nicotine produces an undefined unstable intermediate, which then breaks down spontaneously to nornicotine and formaldehyde (Mesnard et al., 2002; Bartholomeusz et al., 2005; Siminszky et al., 2005; Molinié et al., 2007). Mesnard et al. (2002) suggested that the formaldehyde released from nicotine is recycled back into the THF-mediated C1 pool.
Based on the available information concerning nicotine biosynthesis and N-demethylation in tobacco, one would expect that impaired MTHFR function in tobacco plants would reduce the production of Met and AdoMet by reducing 5-methyl-THF availability and that reduced AdoMet availability would limit nicotine biosynthesis in roots. In addition, if the methyl groups released from nicotine are recycled back to the THF-mediated C1 metabolic pathway in a regulated manner in response to a perceived C1 deficiency, then reduced MTHFR expression in tobacco plants may stimulate nicotine N-demethylation as a mechanism to replenish the THF-mediated C1 pool in leaves.
To monitor nicotine N-demethylation, the nicotine-to-nornicotine conversion rate (NCR) has been widely used as an index in tobacco breeding and physiological studies (Jack et al., 2007). NCR is calculated as a percentage of nornicotine content versus the sum of nicotine and nornicotine contents. Many studies have shown that NCRs are relatively constant at around or below 3.0% in most commercial tobacco cultivars, but absolute nicotine and nornicotine contents are far more variable in individual plants (Bush, 1981; Jack et al., 2007). Any tobacco lines with NCRs greater than 3.0% are defined as “converters” and often have high nicotine N-demethylase activity (Jack et al., 2007). Therefore, NCR is a better parameter than nicotine or nornicotine content for measuring nicotine N-demethylation.
To investigate the role of MTHFR in alkaloid metabolism, we cloned NtMTHFR1 and NtMTHFR2 from tobacco, altered NtMTHFR1 expression, and studied the effects of its altered expression on nicotine biosynthesis and N-demethylation by analyzing NCRs and by monitoring the expression of the major nicotine N-demethylase gene CYP82E4. Contents of folates, amino acids, and AdoMet were quantified to find out if the biosynthesis of these metabolites had been affected in the transgenic plants. We discovered that altered NtMTHFR1 expression reshapes the alkaloid profile in the transgenic tobacco plants by negatively regulating CYP82E4 expression. Our finding provides the first molecular evidence that NtMTHFR1 negatively regulates nicotine N-demethylation. Based on the stable levels of folate, Met, and AdoMet in the transgenic plants, the possible regulatory mechanism that acts to recycle the C1 units from a nonessential metabolite (nicotine) into the core THF-mediated C1 pathway in response to a perceived deficiency in the methyl group supply is discussed.
RESULTS
Cloning of Full-Length Nicotiana spp. MTHFR cDNAs and Functional Assay for NtMTHFR1
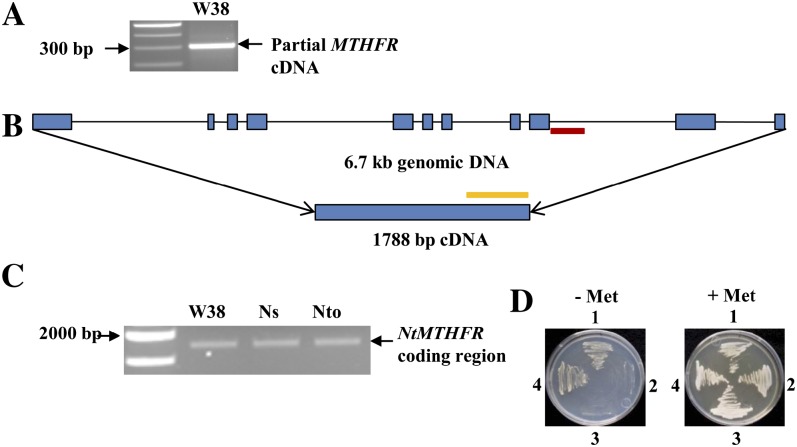
Since no complementary DNA (cDNA) sequences were available for tobacco MTHFR, we first cloned partial MTHFR cDNAs by PCR using degenerate primers based on conserved regions of Arabidopsis, maize, and rice (Oryza sativa) MTHFRs. A PCR product of approximately 300 bp was amplified from tobacco cv Wisconsin 38 (W38; Fig. 1A) and then subcloned. Of the six clones sequenced, four were identical and were designated as NtMTHFR1. The other two clones shared 89% and 98% nucleotide identity with NtMTHFR1. Based on the partial sequence of NtMTHFR1, we carried out two rounds of inverse PCR and obtained a 7,156-bp genomic sequence encompassing the whole NtMTHFR1 gene. After aligning its genomic sequence with the Arabidopsis MTHFR2, we found that this gene consists of 11 exons and 10 introns with a coding region of 1,788 bp (Fig. 1B).
Figure 1.
PCR amplification of the partial and full-length coding regions of NtMTHFRs and functional complementation of a yeast mthfr mutant by NtMTHFR1. A, A 300-bp fragment was amplified from tobacco cDNAs using degenerate primers targeting MTHFR. B, The NtMTHFR1 genomic DNA contains 11 exons (blue rectangles) and 10 introns (black solid lines). Regions used for the RNAi sense and antisense arms, as well as the spacer, are shown in orange and red, respectively. C, Full-length coding regions of MTHFRs amplified from cDNAs from W38, N. sylvestris (Ns), and N. tomentosiformis (Nto) using a pair of primers targeting the MTHFR. D, Functional complementation of a yeast mthfr mutant by NtMTHFR1. Yeast strains DAY4 (wild type; 1), RRY3 (null mutant met12met13; 2), RRY3 with the pVT103-U vector alone (3), and RRY3 with pVT103-U expressing NtMTHFR1 (4) were grown in minimal medium in the presence (+) or absence (−) of Met. [See online article for color version of this figure.]
Modern tobacco is an amphidiploid, containing both ancestral Nicotiana tomentosiformis and Nicotiana sylvestris genomes (Burk, 1973). To clone the MTHFR full-length cDNAs of W38 as well as of the two progenitor species, reverse transcription PCR with a pair of primers (NtMF/NtMR) targeting specifically the 1,788-bp coding region (Fig. 1B) was used. Only one PCR band was observed for each sample (Fig. 1C). After subcloning, eight colonies for each sample were sequenced to test if the PCR band contains more than one type of sequence. We identified only one unique MTHFR sequence in N. sylvestris (NsMTHFR1), two in W38 (NtMTHFR1 and NtMTHFR2), and three in N. tomentosiformis (NtoMTHFR1, NtoMTHFR2, and NtoMTHFR3). Six out of eight NtMTHFR sequences from W38 belong to NtMTHFR1, which suggests that the expression levels of NtMTHFR1 are higher than NtMTHFR2. All six unique Nicotiana spp. MTHFR sequences were deposited in GenBank with accession numbers HQ113127 through HQ113132. Alignment results showed that all Nicotiana spp. MTHFR coding regions have 1,788 bp, while those of Arabidopsis and rice have 1,785 bp, and that of maize has 1,782 bp (Supplemental Fig. S1). NtMTHFR1 and NtMTHFR2 from W38 were found to differ by 4% at the nucleotide level. NtMTHFR1 is highly homologous to NsMTHFR1, with only two base-pair differences, whereas NtMTHFR2 and NtoMTHFR1 are identical, suggesting that NtMTHFR1 was inherited from N. sylvestris whereas NtMTHFR2 was inherited from N. tomentosiformis. Alignment of the deduced amino acid sequences of NtMTHFR1 and NtMTHFR2 with other plant MTHFRs (Supplemental Fig. S2) showed that they are 80% and 81% identical to Arabidopsis AtMTHFR2, 83% identical to rice OsMTHFR, and 83% and 82% identical to maize ZmMTHFR. The unique ATP synthase motif PA/SVNG/AER/KSDS (Supplemental Fig. S2) present in other plant MTHFRs (Kasap et al., 2007) is also conserved in those from the three Nicotiana spp.
Arabidopsis and maize MTHFRs have been shown to encode functional MTHFRs by a yeast complementation test (Roje et al., 1999). To confirm whether NtMTHFR1 encodes a functional MTHFR, its coding region was cloned into a yeast expression vector, pVT103-U, and expressed in the yeast mutant RRY3 (met12met13), a Met auxotroph strain lacking MTHFR activity (Raymond et al., 1999). RRY3 carrying NtMTHFR1 grew well in the medium lacking Met and had similar growth to that of the wild-type strain DAY4 (Fig. 1D). Neither RRY3 nor RRY3 carrying the pVT103-U vector alone could grow in the medium lacking Met. These results show that NtMTHFR1 can complement the loss of MTHFR activity in mutant RRY3, which provides evidence that NtMTHFR1 indeed encodes a functional MTHFR.
NCRs Are Altered in NtMTHFR1 Overexpression and RNA Interference Knockdown Lines
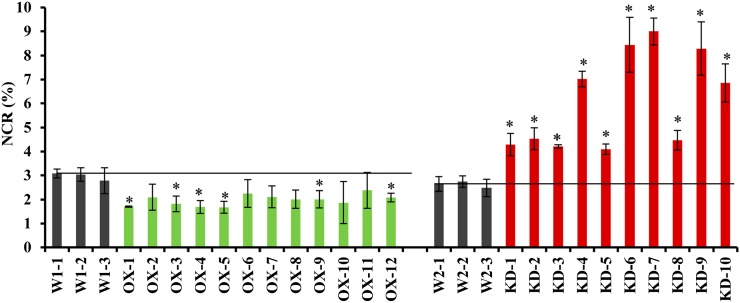
We created NtMTHFR1 overexpression (OX) and knockdown (KD) tobacco lines to study the effect of its altered expression on nicotine metabolism. The construction of NtMTHFR1 overexpression and RNA interference (RNAi) cassettes is detailed in Supplemental Figure S3. The NtMTHFR1 RNAi construct was designed such that it knocked down both NtMTHFR1 and NtMTHFR2, since they share 96% sequence identify in the region used for constructing the RNAi cassette. Both OX and KD lines, each with its own set of three vector-alone transgenic control lines, were generated by Agrobacterium tumefaciens-mediated transformation of W38 leaf explants. After confirming the presence of nptII by PCR, segregation of the transgene was determined by screening T1 seedlings for kanamycin resistance. Transgenic lines with a single transgene locus (segregation ratio of 3:1) were used in subsequent analyses to minimize gene dosage effects (Supplemental Table S1). Previously harvested T0 leaves of 12 OX lines (labeled as OX-1 to OX-12) and 10 KD lines (labeled as KD-1 to KD-10) were selected and subjected to nicotine and nornicotine analyses as described previously (Shi et al., 2003).
We observed 3- to 4-fold higher total alkaloid contents in KD and their vector control lines compared with OX and their vector control lines (Supplemental Table S2). This is because the plants were grown at different times of the year: the KD plants were grown from April to July, while the OX plants were grown from November to February. Long days are known to favor alkaloid production (Tso et al., 1970). However, the NCRs in vector control lines used in both OX and KD experiments were similar (2.96% and 2.62%) regardless of the growing season, which is in agreement with the NCRs of less than 3.0% that were reported for commercial tobacco lines (Jack et al., 2007). As in traditional tobacco breeding programs, NCRs were used in this study to indicate the alteration of the alkaloid profile in transgenic tobacco lines.
All KD lines showed significantly higher NCRs (P < 0.05) compared with the vector control lines (Fig. 2). Among them, five lines (KD-4, KD-6, KD-7, KD-9, and KD-10) exhibited about 3-fold higher NCRs compared with the control lines (Fig. 2). Higher NCRs in these lines resulted from a 30% to 50% reduction in nicotine accumulation (KD-1, KD-2, and KD-3) but also, surprisingly, from a 40% to 400% increase in nornicotine accumulation (KD-5, KD-8, KD-9, and KD-10) or from the combination of the two (KD-4, KD-6, and KD-7) compared with the control lines (Supplemental Table S2). These results suggest that the suppression of NtMTHFR favors nicotine N-demethylation in leaves instead of the expected lower nicotine production because of the limited supply of methyl groups from the C1 metabolic pool. KD lines, despite these changes in alkaloid contents, showed no significant phenotype change in terms of plant height, leaf number, and days to flowering compared with the vector control lines when grown under greenhouse conditions (Supplemental Table S1).
Figure 2.
NCRs in OX and KD lines. NCR (%) was calculated as nornicotine/(nicotine + nornicotine) × 100%. Control lines (shown in black) had average NCRs of 2.96% (OX experiment) and 2.62% (KD experiment), which were then used to draw baselines across the figure for comparison with OX (shown in green) and KD (shown in red) lines. Data represent averages of two independent measurements ± sd. Each independent measurement consisted of three replicates. Asterisks indicate significant differences (P < 0.05) of NCRs between an individual transgenic line and the average of three control lines. [See online article for color version of this figure.]
All OX lines showed NCRs in the range of 1.7% to 2.4%, corresponding to a 20% to 40% reduction compared with the control lines (Fig. 2). Six (OX-1, OX-3, OX-4, OX-5, OX-9, and OX-12) out of 12 lines had significantly reduced NCRs (P < 0.05). Lower NCRs in OX plants were mainly due to the 25% to 50% reduction in nornicotine accumulation (Supplemental Table S2). Like KD lines, OX lines also showed no obvious differences in phenotype when compared with the control plants (Supplemental Table S1). The alteration of NCRs in KD and OX lines indicates that perturbation of the primary pathway gene NtMTHFR1 indeed affects nicotine metabolism.
NtMTHFR1 Negatively Affects CYP82E4 Expression
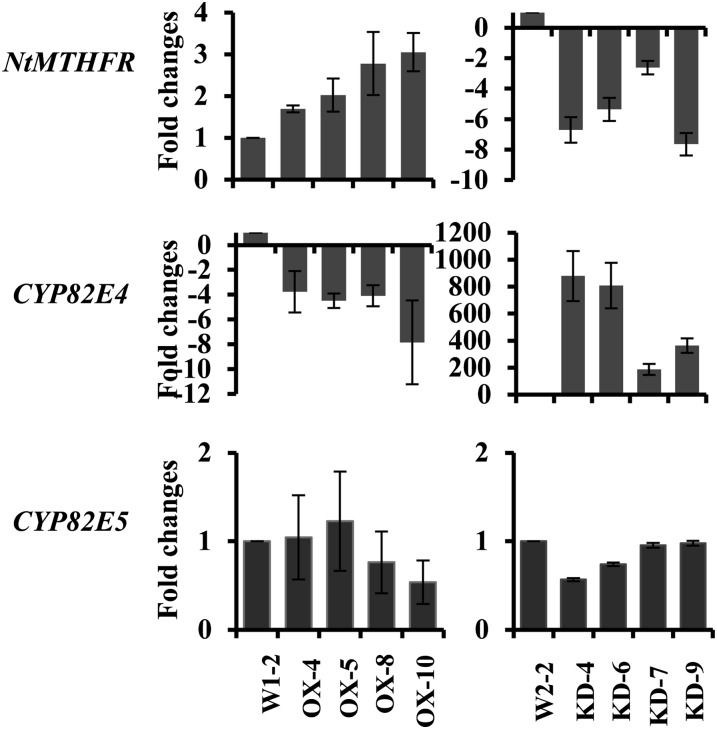
Five cytochrome P450 genes, CYP82E2, CYP82E3, CYP82E4, CYP82E5, and CYP82E10, have been characterized in tobacco (Lewis et al., 2010). CYP82E4 is the main enzyme responsible for the nicotine N-demethylation in leaves (Siminszky et al., 2005; Xu et al., 2007). CYP82E5 and CYP82E10 have minor N-demethylase activity in green leaves and roots, respectively (Gavilano and Siminszky, 2007; Lewis et al., 2010), whereas CYP82E2 and CYP82E3 lack the N-demethylase activity (Siminszky et al., 2005). Therefore, we selected CYP82E4 and CYP82E5 to study whether the transcript levels of NtMTHFR1 correlate with their expressions in leaves. We used quantitative real-time (qRT)-PCR with gene-specific primers to quantify the expression levels of NtMTHFR1, CYP82E4, and CYP82E5 in four OX lines (OX-4, OX-5, OX-8, and OX-10) with low NCRs (around 1.7%–2%) and four KD lines (KD-4, KD-6, KD-7, and KD-9) with high NCRs (around 7%–9%) compared with the vector-alone control lines (W1-2 for the OX lines and W2-2 for the KD lines). To determine the levels of NtMTHFR in these lines, we also carried out immunoblotting using an antibody raised against human MTHFR.
The results showed that all four KD lines had a 3- to 8-fold reduction in NtMTHFR1 expression (Fig. 3). An immunoblot of crude protein extracts of the transgenic lines showed an immunoreactive band of 64 kD, close to the size of 65.5 kD predicted from the amino acid sequence (Supplemental Fig. S4). The intensity of this band was reduced in all four KD lines compared with the vector control line W2-2 (Supplemental Fig. S4). We found a concurrent several-hundred-fold increase in the expression level of CYP82E4 in these lines compared with the control line W2-2 (Fig. 3), suggesting that the reduced NtMTHFR1 expression strongly stimulates the expression of CYP82E4. The qRT-PCR analysis of the four OX lines showed a roughly 2- to 3-fold increase in NtMTHFR1 transcript abundance compared with the control line W1-2 (Fig. 3), but this increase in transcripts did not produce a corresponding increase in the intensity of the protein band on the immunoblot compared with the control line W1-2 (Supplemental Fig. S4). One explanation for this may be that the antibody we used cross reacts with both NtMTHFR1 and NtMTHFR2 and that NtMTHFR2 expression was reduced in the OX lines in response to the overexpression of NtMTHFR1. Nevertheless, CYP82E4 expression was reduced by 4- to 8-fold in these OX lines (Fig. 3). The expression of CYP82E5 was not affected in either OX or KD lines (Fig. 3). These results provide strong evidence that a negative correlation exists between the expression of NtMTHFR1 and CYP82E4 and also suggest that high NCRs in KD lines and low NCRs in OX lines are due to the up- and down-regulation of CYP82E4 expression, respectively.
Figure 3.
qRT-PCR analysis of changes in transcript levels of NtMTHFR1, CYP82E4, and CYP82E5 in OX and KD lines. Data shown are fold changes calculated as transcript levels in OX and KD lines compared with control lines (W1-2 for OX lines and W2-2 for KD lines). The control line value is defined as 1. Negative values indicate reduced expression. Data represent averages of three independent assays ± sd. Each assay was done in triplicate.
Elevated CYP82E4 Transcript Levels in KD Lines Were Likely Not Due to Leaf Senescence
Leaf senescence and curing are the two major triggers of nicotine N-demethylation (Wersman and Matzinger, 1968). It has been reported that CYP82E4 is expressed at very low levels in green healthy leaves and is strongly induced during senescence (Siminszky et al., 2005; Gavilano et al., 2006; Chakrabarti et al., 2008). Therefore, we investigated whether the increased CYP82E4 expression could have been triggered by leaf senescence.
Leaf yellowing, which is a visible indicator of leaf senescence (Lim et al., 2007), was not observed in any of the leaves of the analyzed 16-leaf-stage transgenic plants or the wild-type control. All the plants looked green and healthy and appeared to follow the same developmental pattern. The third leaf from the top, which was used for the CYP82E4 analysis, was just fully expanded in all the plants analyzed.
To further investigate whether the changes in CYP82E4 transcripts in the KD lines could have been due to early stages of leaf senescence, in which the senescing symptoms are not yet visible to the human eye, we also quantified ethylene production, which is induced at the initiation of leaf senescence (Trobacher, 2009). Ethylene production in the third leaves from the top (just fully expanded) of 16-leaf-stage plants was quantified. The analyzed leaves were enclosed in bags either still attached to or excised from the plant. None of the 12 attached leaves from four OX, KD, and wild-type lines produced detectable levels of ethylene after being sealed for 24 or 48 h. Since none of the attached leaves produced detectable ethylene, the experiment was only performed with one plant from each line. For excised leaves, ethylene production was measured in three individual plants from each line representing three biological replicates. The ethylene concentrations after 24 h were 0.069 to 0.088, 0.059 to 0.093, and 0.073 to 0.097 μg g−1 fresh leaf for the wild-type, OX, and KD lines, respectively (Supplemental Fig. S5); the differences were not significant.
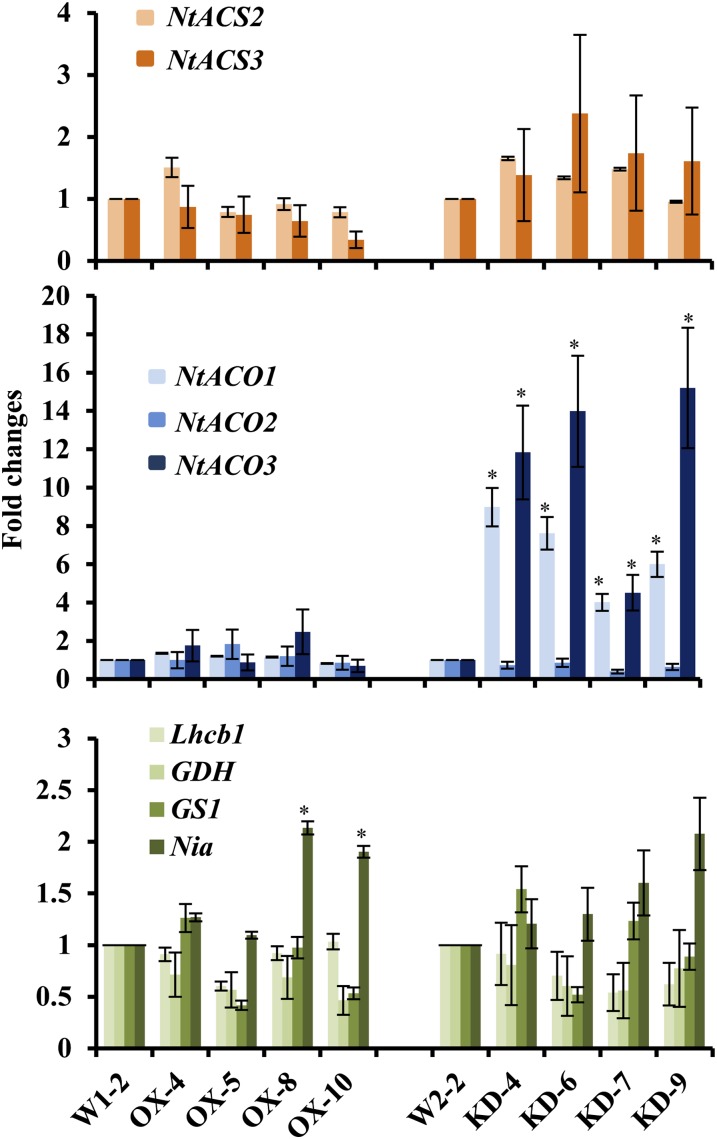
Furthermore, we examined the expression levels of 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS) and ACC oxidase (ACO), the hallmark genes for leaf senescence on the ethylene biosynthesis pathway. ACS is involved in the rate-limiting step of ethylene biosynthesis from AdoMet to ACC, while ACO is involved in the oxidation of ACC to ethylene (Lin et al., 2009). In Nicotiana glutinosa, the ACO1 and ACO3 transcripts accumulate during senescence and are dramatically induced by ethylene treatment (Kim et al., 1998). The expression of ACS genes is regulated developmentally and environmentally (Kende, 1993). Expression of the three ACS genes (NtACS1, NtACS2, and NtACS3; Zhang et al., 2009) and three ACO genes (NtACO1, NtACO2, and NtACO3; Zhang et al., 2009) was analyzed by qRT-PCR with gene-specific primers. Since NtACS1 was not detected in any of the samples, it is not listed in Figure 4. Both NtACO1 and NtACO3 had 3- to 10-fold higher expression levels, while NtACO2, NtACS2, and NtACS3 did not change significantly in KD lines compared with the vector control W2-2 (Fig. 4). No transcripts of these five genes were changed in any of the OX lines (Fig. 4). Since ethylene production was not increased in any of the KD plants analyzed, it is unlikely that senescence is responsible for the induction of the NtACO1 and NtACO3 genes. Finding the mechanism behind this induction is beyond the scope of this paper.
Figure 4.
Expression levels of ethylene biosynthesis genes ACS and ACO and five senescence-related genes, CP1, GS1, GDH, Lhcb1, and Nia, in selected OX and KD lines. The expression level of each gene was quantified by SYBR Green-based qRT-PCR. Data shown are fold changes calculated as transcript levels in OX and KD lines compared with control lines (W1-2 for OX lines and W2-2 for KD lines). The control line value is defined as 1. Data represent averages of three independent assays ± sd. Each assay was done in triplicate. *P < 0.05. [See online article for color version of this figure.]
In parallel, we examined the expression levels of three senescence-inducible genes encoding Cys Protease1 (CP1; Beyene et al., 2006; Niewiadomska et al., 2009), cytosolic Gln Synthetase1 (GS1; Pageau et al., 2006), and Glu Dehydrogenase (GDH; Pageau et al., 2006), and the two senescence-suppressible genes encoding chlorophyll a/b-binding protein (Lhcb1; Niewiadomska et al., 2009) and nitrate reductase (Nia; Pageau et al., 2006). Expression of CP1, a senescence-induced marker gene orthologous to the gene encoding SAG12 protease of Arabidopsis (Gan and Amasino, 1995), was not detected in any of the OX, KD, and vector control lines. The expression level of GS1 and GDH in OX and KD lines was similar to that in control lines (Fig. 4). Meanwhile, the two senescence-suppressible genes Lhcb1 and Nia were not suppressed in either OX or KD lines. To the contrary, the expression level of Nia in two OX lines and one KD line was slightly higher than that in the vector control.
In conclusion, the KD plants analyzed did not show visible signs of senescence or increased ethylene production. Of the nine genes known to change expression during senescence, only two (NtACO1 and NtACO3) were induced as anticipated for the senescing plants; these genes are also known to be induced by other signals, such as wounding and viral infection (Kim et al., 1998). Therefore, the presented results taken together support the hypothesis that the highly induced expression of CYP82E4 in young green leaves of the KD lines is likely not triggered by senescing signals.
Folate Contents in OX and KD Lines
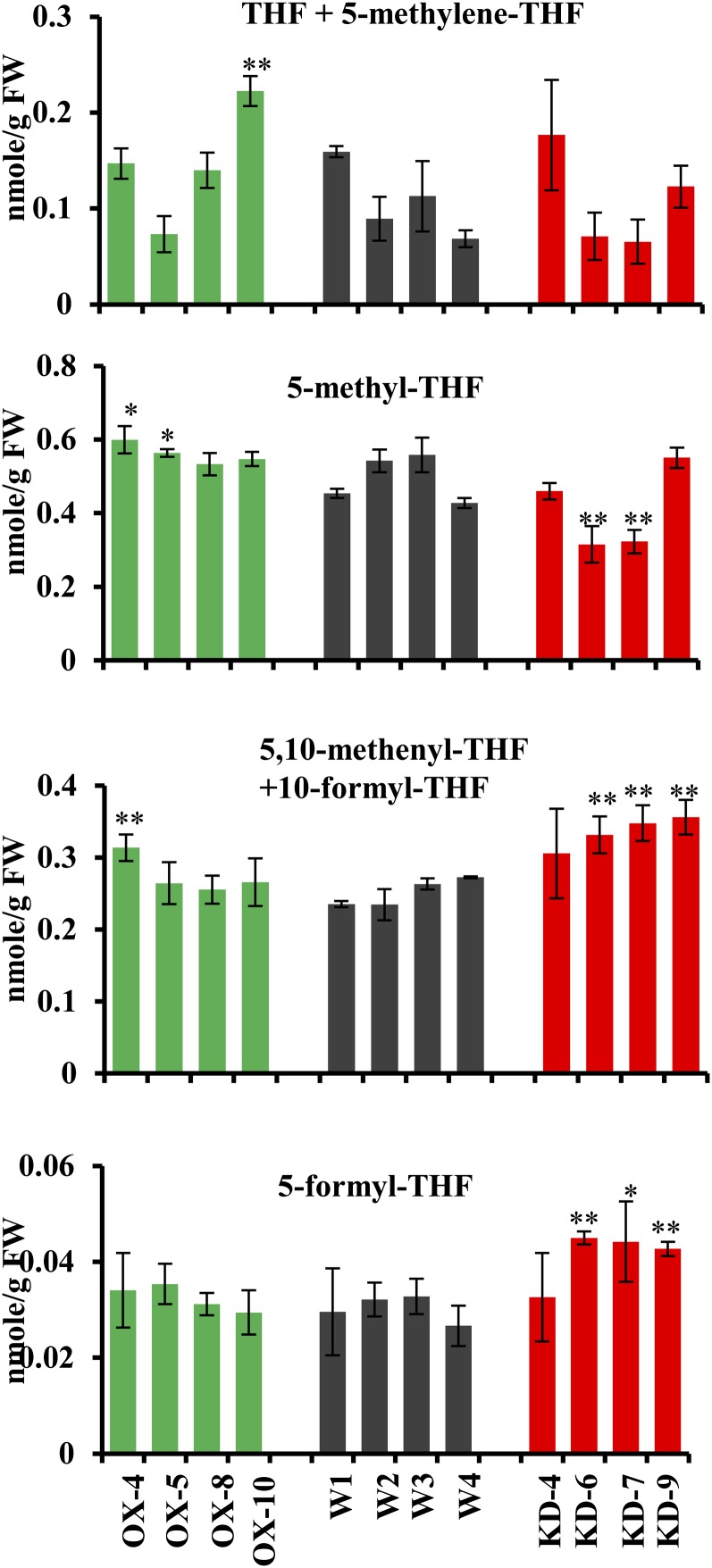
Interconvertible C1-substituted folates, varying in oxidation state from the most oxidized 10-formyl-THF to the most reduced 5-methyl-THF, are required for many anabolic reactions in plants and other organisms (Hanson et al., 2000). Despite the importance of the individual folate species in multiple essential cellular processes, little is known about the molecular mechanisms regulating folate biosynthesis and the balance of the individual folate species in plants and other organisms in response to metabolic demands. Because MTHFR catalyzes the reduction of 5,10-methylene-THF to 5-methyl-THF, we hypothesized that the cellular contents of 5-methyl-THF would decrease and those of the more oxidized folate species would either remain constant or increase, as a result of the reduced expression of NtMTHFR in KD lines and vice versa in OX lines. Therefore, folates were measured in the OX and KD lines as described earlier with five heterozygous T1 transgenic plants from each line using a published procedure (Quinlivan et al., 2006). Of the six folate derivatives on the THF-mediated C1 pathway, 5-methyl-THF and 5-formyl-THF could be separated and quantified individually. During the extraction, 5,10-methylene-THF decomposes to THF and formaldehyde, while 10-formyl-THF is spontaneously reduced to 5,10-methenyl-THF; therefore, these folates were quantified in pairs.
A significant decrease in 5-methyl-THF was observed in two KD lines compared with the wild-type control (Fig. 5). A significant increase (P < 0.01) in 5,10-methenyl-THF + 10-formyl-THF and 5-formyl-THF was observed in three out of four KD lines, while the ratio of 5-methyl-THF to 5,10-methenyl-THF + 10-formyl-THF was reduced in all four KD lines compared with the wild-type control (Fig. 5). These results are consistent with our hypothesis that the suppression of NtMTHFR can decrease cellular contents of 5-methyl-THF and increase those of the oxidized folate species. In the case of OX lines, the contents of all folate species remained constant (Fig. 5). These data suggest that, in the OX plants, a 2- to 3-fold increase in NtMTHFR1 transcripts does not affect the accumulation of 5-methyl-THF. This is consistent with the immunoblotting results showing no increase in the MTHFR band intensity in OX lines despite the increase in NtMTHFR1 transcripts compared with the wild type (Fig. 3; Supplemental Fig. S4). Overall, the total folate contents did not differ significantly between OX, KD, and wild-type lines, suggesting that altered MTHFR expression did not affect de novo folate biosynthesis.
Figure 5.
Folate levels in leaves of OX, KD, and wild-type lines. Folates were extracted from the third leaf collected from 16-leaf-stage T1 and wild-type plants. Four OX lines, four KD lines, and four wild-type plants were analyzed. Equal amounts of leaf tissues from five T1 heterozygous plants of each line were pooled for folate analysis. The bars represent folate levels as averages ± sd of three biological replicates. FW, Fresh weight. *P < 0.05, **P < 0.01. [See online article for color version of this figure.]
Contents of Amino Acids and AdoMet in OX and KD Lines
The THF-mediated C1 pathway is required for the biosynthesis of the amino acids Met, Gly, and Ser and also of the universal methyl group donor AdoMet in plants (Cossins and Chen, 1997). 5,10-Methylene-THF and THF are required for the interconversion of Gly and Ser. 5-Methyl-THF is the methyl donor in the biosynthesis of Met and its adenylated derivative AdoMet (Hanson et al., 2000; Hanson and Roje, 2001). Met biosynthesis is regulated at multiple levels within the metabolic network for the biosynthesis of Asp family amino acids, which include Asp, Lys, Met, Thr, and Ile, so altered Met levels have a potential to affect fluxes throughout the whole network (Galili, 2011). Since suppression of NtMTHFR affected the contents of individual folate species (Fig. 5) and may have affected the flux of C1 units into the downstream products as discussed earlier, we hypothesized that the levels of AdoMet and Asp family amino acids may be affected in the transgenic plants.
Therefore, the aforementioned leaf tissues used for folate analyses were analyzed for amino acid and AdoMet contents. The results show that, while there is some variation in amino acid and AdoMet levels from line to line in both transgenic and wild-type lines, the overall amino acid and AdoMet profiles of both OX and KD transgenic plants and the wild-type control are similar (Supplemental Table S3; Supplemental Fig. S6). Thus, these data indicate that perturbing NtMTHFR expression in tobacco does not affect the pool sizes of amino acids or AdoMet in leaves.
DISCUSSION
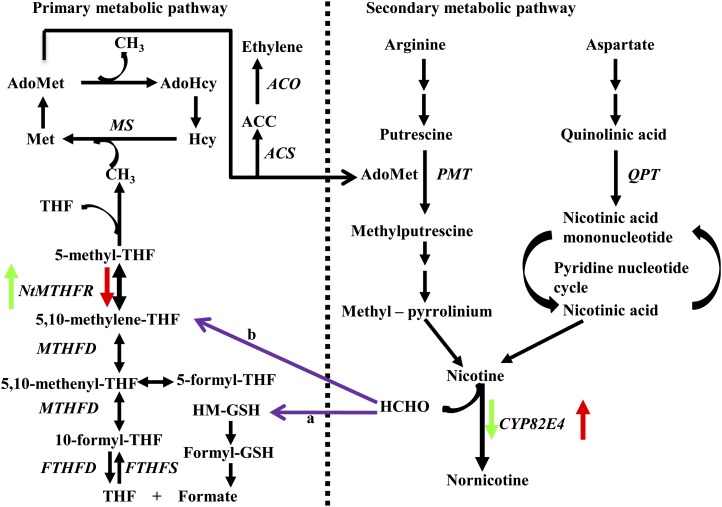
MTHFR-driven biosynthesis of 5-methyl-THF directs C1 units from the central THF-mediated C1 metabolism toward the biosynthesis of Met and AdoMet. AdoMet is required in the first committed step of nicotine biosynthesis for the methylation of putrescine to N-methylputrescine. Therefore, suppression of NtMTHFR1 was expected to restrict the C1 flux into the nicotine biosynthesis pathway, thereby decreasing the accumulation of the nicotine precursor N-methylputrescine as well as that of nicotine and its demethylated product nornicotine. Surprisingly, we observed that knocking down NtMTHFR1 induced the expression of CYP82E4, resulting in increased conversion of nicotine into nornicotine, whereas overexpressing NtMTHFR1 reduced the expression of CYP82E4, leading to reduced conversion of nicotine to nornicotine (Fig. 2; Supplemental Table S2). When the altered alkaloid profiles were analyzed in detail together with the CYP82E4 transcripts in OX and KD lines, we found that the nornicotine content in these lines was not determined by the content of its precursor nicotine but by the level of CYP82E4. This observation is consistent with previous reports that nicotine N-demethylation is mainly controlled by CYP82E4 (Siminszky et al., 2005; Xu et al., 2007). The results of gene expression studies together with the altered alkaloid profiles in both OX and KD lines suggest that NtMTHFR1 negatively regulates the expression of the nicotine metabolic pathway gene CYP82E4, affecting nicotine metabolism in tobacco (Fig. 6).
Figure 6.
THF-mediated C1 metabolism, the biosynthesis of ethylene and nicotine, and the predicted relationship between NtMTHFR and CYP82E4. The arrows indicate the direction of metabolite change under the condition when the NtMTHFR1 was overexpressed (green) or suppressed (red). Purple arrows indicate two possible routes (a and b) for the entry of formaldehyde released from nicotine into the THF-mediated C1 pool. Genes involved in the pathways and not abbreviated in the text are as follows: FTHFD, 10-formyltetrahydrofolate deformylase; FTHFS, 10-formyltetrahydrofolate synthetase; MS, Met synthase; PMT, putrescine methyltransferase. Hcy, Homo-Cys; HM-GSH, S-hydroxymethylglutathione; HCHO, formaldehyde. [See online article for color version of this figure.]
It is known that CYP82E4 is strongly induced during senescence but that its expression level is very low in young and healthy green leaves (Siminszky et al., 2005; Gavilano et al., 2006; Chakrabarti et al., 2008). Our data show no evidence of early senescence as a trigger for the strong induction of CYP82E4 in young leaves of the MTHFR KD lines. This conclusion is supported by analyses of ethylene release and of expression levels of ethylene biosynthesis and senescence-related genes in KD and wild-type lines. First, there was no significant difference in ethylene levels and the contents of Met and AdoMet between KD lines and wild-type plants (Supplemental Table S3; Supplemental Figs. S5 and S6). Second, of the nine senescence-related genes analyzed, only NtACO1 and NtACO3 were induced in the KD lines (Fig. 4). Leaf senescence is known to induce both ACS and ACO enzymes sharply, but the formation of ACC is often considered to be the rate-limiting step on the pathway (Kim et al., 1998; Wang et al., 2002; Lin et al., 2009). The lack of ACS induction, therefore, is consistent with the lack of increase in ethylene production in the analyzed transgenic plants. These results, together with the lack of any visible senescing symptoms in the analyzed plants, support the hypothesis that the just-expanded third leaves used for studying CYP82E4 and MTHFR expression were not undergoing senescence and that the induced expression of CYP82E4 in the KD lines was unrelated to early senescence.
Previous isotope-labeling experiments have shown that the methyl group derived from nicotine is recycled back into the folate pool (Mesnard et al., 2002; Bartholomeusz et al., 2005). These studies, together with the altered alkaloid profiles in the transgenic lines described here, lead us to speculate that C1 units may mediate the regulation of NtMTHFR1 on CYP82E4 (Fig. 6). Since maintaining the C1 pool at an appropriate level is critical for many physiological functions (Cossins and Chen, 1997; Hanson et al., 2000) and thus essential to an organism’s survival and development, it is possible that nicotine demethylation can be induced to serve as a physiological source of C1 units in times of restricted supply from other sources. In tobacco, nicotine accounts for 2% to 5% of dry leaves (Saitoh et al., 1985). Its N-demethylation could potentially produce a large amount of C1 units, which could tremendously affect C1 fluxes and, hence, plant metabolism.
The full chain of events that lead to an altered alkaloid profile in tobacco plants remains to be determined. We speculate that the following sequence of events could be responsible for the results observed. Suppression of NtMTHFR led to a reduction in the level of 5-methyl-THF (Fig. 5). In response, conversion of nicotine to nornicotine was induced in the KD plants in an attempt to compensate for the perceived deficiency in methyl group supply. This is consistent with the higher NCRs observed in the KD lines (Fig. 2). So far, the molecular mechanisms of how plant cells via the folate status regulate CYP82E4 expression and of how they adjust the flux of C1 units through the folate pathway in response to C1 demand are unclear. The facts that KD lines had unchanged pools of Met and AdoMet (Supplemental Table S3; Supplemental Fig. S6) and higher pools of oxidized folate species (Fig. 5) suggest that the influx of C1 units into the C1 metabolism is increased in these lines compared with the wild-type control. Increased expression of CYP82E4 (Fig. 3) supports the formaldehyde released from nicotine (Mesnard et al., 2002) as a possible source of the C1 units. It is plausible that the observed increase in expression of CYP82E4 in KD lines is part of the cellular response attempting to correct the perceived imbalance in the ratio of 5-methyl-THF to 5,10-methenyl-THF + 10-formyl-THF. It is likely that MTHFR is operating below its Km value for 5,10-methylene-THF in plants. The Km value for the monoglutamylated 5,10-methylene-THF is 287 µm for Arabidopsis MTHFR (Roje et al., 2002), which is well above the 5,10-methylene-THF concentration (in the nanomolar range) in the cytosol (Goyer et al., 2005). The Km values for the physiologically more relevant polyglutamylated substrates have not been determined yet for the plant MTHFRs. Those for the MTHFR purified from pig liver were 33 µm for the monoglutamylated substrate and 1.9 µm for the hexaglutamylated substrate (Matthews et al., 1987). A similar drop in Km values for polyglutamylated substrates in plant MTHFRs would still render these values in the micromolar range, while the substrate concentration is in the nanomolar range (Goyer et al., 2005). Thus, these enzymes are expected to operate at folate substrate concentrations well below the corresponding Km values. Therefore, increased influx of C1 units into the core C1 metabolism could increase the flux of C1 units through the folate pathway, including the MTHFR-mediated reaction, by increasing the substrate concentration and thus the turnover rate. The lack of effect of increased NtMTHFR1 expression on the levels of 5-methyl-THF, AdoMet, and amino acids in OX lines may be due to MTHFR operating very close to the reaction equilibrium as part of a mechanism to regulate the conversion of 5,10-methylene-THF to 5-methyl-THF by the downstream demand for methyl groups.
The route(s) by which C1 units released by nicotine N-demethylation reenter the C1 metabolic pool is unclear at present. Two possible routes (Fig. 6) have been proposed (Hanson et al., 2000; Mesnard et al., 2002; Robins et al., 2007), the first one via formate (route a) and the second one via the direct reaction of formaldehyde with THF to form 5,10-methylene-THF (route b). The increase in 5,10-methenyl-THF and 10-formyl-THF in the KD lines suggests that the C1 units released from nicotine are likely entering the core C1 pool at the level of formate via the glutathione-mediated route a. The entry via route b is also possible, because formaldehyde can spontaneously react with THF to form 5,10-methylene-THF, which can then be enzymatically converted to 5,10-methenyl-THF and 10-formyl-THF (Hanson and Roje, 2001). Determining the route responsible for recycling of the C1 units released by nicotine N-demethylation into the core C1-mediated THF metabolism will require further study. Nevertheless, the data presented here suggest that nicotine N-demethylation in tobacco plants responds to the folate status of the cell.
Regardless of the molecular mechanism responsible for the induction of CYP82E4 and the entry route of formaldehyde into the THF-mediated C1 pathway, our study provides evidence in support of the regulatory role of MTHFR in nicotine N-demethylation and implies a possible cross talk between the THF-mediated C1 pathway and the secondary metabolism. It is known that many plants contain high levels of alkaloids and that many alkaloids, such as opiates and tropanes, contain an N-methyl group (Miller et al., 1973). If these methyl-rich alkaloids act as sources of C1 units for anabolic reactions in plants, we need to reevaluate their roles in plant growth. Moreover, considering that many plants are rich in methylated compounds such as lignin, alkaloids, and betaines (Hanson et al., 2000), our findings also provide valuable information for future metabolic engineering of secondary metabolism and open up possibilities for new strategies for metabolic engineering of alkaloids and other secondary metabolites in medicinal plants. At present, metabolic engineering of alkaloids focuses primarily on manipulating key genes involved in secondary metabolic pathways (Wu and Chappell, 2008) and the associated transcription factors (Grotewold, 2008). To reduce the content of certain alkaloids by interrupting the rate-limiting gene is relatively easy (Xie et al., 2004; Gavilano et al., 2006), but increasing the level of certain alkaloids by overexpressing one or several key genes remains a real challenge because of the emergence of new rate-limiting steps (Verpoorte and Memelink, 2002). For future metabolic engineering studies, genes such as NtMTHFR1, whose major functions are in the primary metabolism but that are involved in the regulation of secondary metabolism, could be good targets.
MATERIALS AND METHODS
Plant Growth
Tobacco (Nicotiana tabacum ‘Wisconsin 38’; NCR of 2.83%) and its progenitor species Nicotiana sylvestris and Nicotiana tomentosiformis were grown in a greenhouse under natural light and controlled temperature (25°C). T0 and T1 transgenic plants were also grown in a greenhouse under the same conditions. Transgenic seeds were germinated on Murashige and Skoog medium plates with 150 mg L−1 kanamycin to examine the segregation ratio.
Isolation of RNA and Genomic DNA, and Preparation of cDNA
Genomic DNA and total RNA were prepared using Qiagen DNeasy and RNeasy Plant Mini kits, respectively. All RNA samples were treated with DNase I (Qiagen). They were then quantified spectrophotometrically by NanoDrop (NanoDrop Technologies) and visualized using agarose gel electrophoresis. The first-strand cDNAs were synthesized using MultiScribe Reverse Transcriptase combined with a random primer (ABI).
MTHFR Cloning
All the procedures for cloning MTHFRs from W38, N. sylvestris, and N. tomentosiformis are provided in Supplemental Information S1.
NtMTHFR1 Functional Complementation in Yeast Mutant RRY3
The coding region of NtMTHFR1 was isolated from the PCR clone by digestion with SmaI and SacI and was cloned into the BamHI-blunted and SacI sites in the pVT103-U vector containing an ADH1 promoter and a URA3 selection marker (Vernet et al., 1987). Wild-type yeast strain DAY4 and the double mutant strain RRY3 (met12met13; Raymond et al., 1999) were kindly provided by Dr. Dean R. Appling. Recombinant plasmid and the pVT103-U vector alone were transformed into RRY3. Plasmid preparation, yeast transformation, and growth test on Met-free medium were carried out as described by Raymond et al. (1999). Wild-type strain DAY4 was used as a control.
NtMTHFR1 OX and KD Lines
All the procedures for the construction of the NtMTHFR1 OX lines and RNAi cassettes (Supplemental Fig. S3) are provided in Supplemental Information S1. Each construct was transformed into Agrobacterium tumefaciens strain LBA4404 by the heat-shock method. Both OX and KD lines with a single transgenic locus were generated, each with its own set of three transgenic control plants carrying pBI121 vector alone, by A. tumefaciens-mediated transformation as described previously (Musa et al., 2009). T0 transgenic plants of OX lines were grown from November to February, while those of KD lines were grown from April to July, in Durham, North Carolina. When plants reached the 16-leaf stage, the fifth and sixth leaves from the top were collected and lyophilized for determining nicotine and nornicotine contents, and the third leaf was harvested for isolating DNA, RNA, and proteins. The presence of nptII was validated by PCR as described previously (Darlington et al., 2009). For determining the single-locus lines, T1 seedlings were germinated on Murashige and Skoog medium containing 150 mg L−1 kanamycin.
To generate plant materials for metabolite analysis, kanamycin-resistant T1 transgenic plants of the four OX lines and the four KD lines that were used for examining gene expression were grown together along with four wild-type W38 lines. Ten plants from each line were planted. The third expanded leaf from the top of each plant was harvested when the plant was at the 16-leaf stage. Harvested leaf was used to isolate DNA for TaqMan probe-based quantitative PCR (qPCR) assay (see below) to distinguish heterozygous and homozygous plants. After confirming their zygosity, five heterozygous plants of each line were pooled in equal weight for further metabolite analysis.
qRT-PCR and qPCR
Expression levels of NtMTHFR1, CYP82E4, CYP82E5, NtACS1, NtACS2, NtACS3, NtACO1, NtACO2, NtACO3, CP1, GS1, GDH, Lhcb1, and Nia in selected T0 transgenic plants were measured by SYBR Green-based qRT-PCR as described previously (Hung et al., 2010). Four OX lines (OX-4, OX-5, OX-8, and OX-10) and four KD lines (KD-4, KD-6, KD-7, and KD-9) were analyzed. Vector-only transgenic line W1-2 was used as a control for the OX lines, while line W2-2 was used as a control for the KD lines. Primers used for CYP82E4 (DQ131886) or CYP82E5 (EU182719) expression measurement were specifically designed to exclude other members of the nicotine N-demethylase family. Published sequences of the three reported NtACSs and NtACOs (Zhang et al., 2009), three senescence-inducible genes, CP1 (Y881011; Beyene et al., 2006; Niewiadomska et al., 2009), GS1 (X95933; Pageau et al., 2006), and GDH (AY366369; Pageau et al., 2006), and two senescence-suppressible genes, Lhcb1 (AY219853; Niewiadomska et al., 2009) and Nia (X14058; Pageau et al., 2006), were used to design gene-specific primers. For the T1 zygosity test, the TaqMan probe-based qPCR assay described by Yuan et al. (2007) was adapted to target nptII with some modifications. The assay was done in duplex with the target gene and one copy control. Intron 7 of the tobacco quinolinate phosphoribosyltransferase gene (QPT1; AJ748262) was used to design a specific set of primers and probe for the QPT1 intron 7 only as one copy control. Probes targeting nptII and QPT1 intron 7 were labeled with fluorescein amidite and tetrachloro fluorescein (Biosearch Technologies), respectively. Sequences of primers and probes used for expression analyses are listed in Supplemental Table S4.
Protein Extraction, SDS-PAGE, and Immunoblot Analysis
Total leaf protein isolation and immunoblot analysis were carried out as described by Hung et al. (2010). About 100 mg of leaf tissues was used for protein extraction. About 12.5 µg of protein for each sample was separated on a 12.5% SDS-PAGE gel and electrotransferred onto a polyvinylidene difluoride membrane. Human anti-MTHFR (1:1,000; Novus Biologicals) was used as a primary antibody to detect tobacco MTHFR. Secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories; 1:15,000) was used. After capturing signal, the blot was stained with Amido Black to confirm sample loading.
Measurement of Nicotine and Nornicotine
Nicotine and nornicotine contents were measured twice in triplicate assays following the method described by Shi et al. (2003) using a Perkin-Elmer Autosystem XL Gas Chromatograph. NCR was calculated as: nornicotine/(nicotine + nornicotine) × 100%.
Ethylene Measurement
Ethylene production was measured using a gas chromatograph (GC-8A; Shimadzu). All the procedures for sampling and the ethylene measurement are detailed in Supplemental Information S1.
Folate Analysis
Tissues from the third leaf were pulverized in liquid nitrogen and stored in −80°C until analysis. Folates were extracted using a published procedure (Díaz de la Garza et al., 2004) with some modifications. Three biological samples of each line were used as three replicates. All the procedures for the folate analysis are detailed in Supplemental Information S1.
Analysis of Amino Acids and AdoMet
Approximately 100 mg of pulverized leaf tissue was used to measure amino acids and AdoMet. Three biological samples of each line were used as three replicates. All the procedures for the extraction, derivatization, and detection of amino acids and AdoMet are provided in Supplemental Information S1.
Statistical Analysis
For selecting single-locus insertion lines, a χ2 test was used. For the comparison of NCRs between transgenic and control lines, the mean values of three vector-only control lines and each individual transgenic line were subjected to one-way ANOVA using R language (http://www.r-project.org). For the comparison of ethylene, folates, AdoMet, amino acids, and genes between the wild type and the OX or KD lines, Student’s t test was used.
Sequence data from this article for six Nicotiana spp. MTHFR genes can be found in GenBank under accession numbers HQ113127 through HQ113132.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple DNA sequence alignment of six Nicotiana spp. MTHFR sequences.
Supplemental Figure S2. Multiple amino acid sequence alignment of six putative Nicotiana spp. MTHFRs.
Supplemental Figure S3. Diagram of two cassettes used for overexpression and knockdown of NtMTHFR1.
Supplemental Figure S4. Immunoblotting of MTHFR in selected OX and KD lines.
Supplemental Figure S5. Ethylene concentrations produced from excised leaves of OX, KD, and wild-type lines.
Supplemental Figure S6. AdoMet levels in leaves of OX, KD, and wild-type lines.
Supplemental Table S1. Genetic analysis and growth comparison of OX and KD transgenic lines.
Supplemental Table S2. Nicotine and nornicotine contents in OX and KD transgenic lines.
Supplemental Table S3. Amino acid contents in OX, KD, and wild-type lines.
Supplemental Table S4. Primer and probe sequence information for qRT-PCR and qPCR.
Supplemental Table S5. Gradient elution system used for analysis of THF, 5-methyl-THF, and 5,10-methenyl-THF.
Supplemental Table S6. Gradient elution system used for analysis of AccQ-amino acids (Waters).
Supplemental Table S7. Gradient elution system used for analysis of AdoMet.
Supplemental Information S1. MTHFR cloning, construction of NtMTHFR1 OX lines and RNAi cassettes, and analysis of ethylene, folates, amino acids, and AdoMet.
Acknowledgments
We thank Dr. Zhenming Pei (Duke University), Dr. Susan L. Peacock (North Carolina Central University), and Dr. Weihua Fan (BASF Plant Science) for critical reading of the manuscript. We thank Drs. John M. Dole and Weiwen Gou for ethylene measurement and Mr. Jeffory K. Barton for taking care of plants (all North Carolina State University). We are grateful to Dr. Dean R. Appling (University of Texas at Austin) for yeast strains and Dr. Reed B. Wickner (National Institute of Diabetes and Digestive and Kidney Diseases) for the yeast vector.
Glossary
- THF
tetrahydrofolate
- C1
one-carbon
- AdoMet
S-adenosyl-methionine
- NCR
nicotine-to-nornicotine conversion rate
- W38
Wisconsin 38
- OX
overexpression
- KD
knockdown
- RNAi
RNA interference
- qRT
quantitative real-time
- ACC
1-aminocyclopropane-1-carboxylate
- qPCR
quantitative PCR
- cDNA
complementary DNA
- RT
reverse transcription
- RT
real-time
References
- Baldwin IT, Halitschke R, Kessler A, Schittko U. (2001) Merging molecular and ecological approaches in plant-insect interactions. Curr Opin Plant Biol 4: 351–358 [DOI] [PubMed] [Google Scholar]
- Bartholomeusz TA, Bhogal RK, Molinié R, Felpin FX, Mathé-Allainmat M, Meier AC, Dräger B, Lebreton J, Roscher A, Robins RJ, et al. (2005) Nicotine demethylation in Nicotiana cell suspension cultures: N′-formylnornicotine is not involved Phytochemistry 66: 2432–2440 [DOI] [PubMed] [Google Scholar]
- Beyene G, Foyer CH, Kunert KJ. (2006) Two new cysteine proteinases with specific expression patterns in mature and senescent tobacco (Nicotiana tabacum L.) leaves. J Exp Bot 57: 1431–1443 [DOI] [PubMed] [Google Scholar]
- Burk LG. (1973) Partial self-fertility in a theoretical amphiploid progenitor of N. tabacum. J Hered 64: 348–350 [Google Scholar]
- Bush LP. (1981) Physiology and biochemistry of tobacco alkaloids. Recent Adv Tob Sci 7: 75–106 [Google Scholar]
- Bush LP, Hempfling WP, Burton H. (1999) Biosynthesis of nicotine and related alkaloids. In JW Gorrod, P Jacob, eds, Analytical Determination of Nicotine and Related Compounds and Their Metabolites. Elsevier Science, New York, pp 13–44 [Google Scholar]
- Chakrabarti M, Bowen SW, Coleman NP, Meekins KM, Dewey RE, Siminszky B. (2008) CYP82E4-mediated nicotine to nornicotine conversion in tobacco is regulated by a senescence-specific signaling pathway. Plant Mol Biol 66: 415–427 [DOI] [PubMed] [Google Scholar]
- Cossins EA, Chen L. (1997) Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45: 437–452 [DOI] [PubMed] [Google Scholar]
- Cragg GM, Newman DJ. (2005) Plants as a source of anti-cancer agents. J Ethnopharmacol 100: 72–79 [DOI] [PubMed] [Google Scholar]
- Darlington DE, Hung CY, Xie JH. (2009) Developing an Agrobacterium tumefaciens-mediated genetic transformation for a selenium-hyperaccumulator Astragalus racemosus. Plant Cell Tissue Organ Cult 99: 157–165 [Google Scholar]
- Dawson RF. (1941) The localization of the nicotine synthetic mechanism in tobacco plant. Science 94: 396–397 [DOI] [PubMed] [Google Scholar]
- Dawson RF. (1945) On the biosynthesis of nornicotine and anabasine. J Am Chem Soc 67: 503–504 [Google Scholar]
- Díaz de la Garza R, Quinlivan EP, Klaus SM, Basset GJ, Gregory JF, III, Hanson AD. (2004) Folate biofortification in tomatoes by engineering the pteridine branch of folate synthesis. Proc Natl Acad Sci USA 101: 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G. (2011) The aspartate-family pathway of plants: linking production of essential amino acids with energy and stress regulation. Plant Signal Behav 6: 192–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988 [DOI] [PubMed] [Google Scholar]
- Gavilano LB, Coleman NP, Burnley LE, Bowman ML, Kalengamaliro NE, Hayes A, Bush LP, Siminszky B. (2006) Genetic engineering of Nicotiana tabacum for reduced nornicotine content. J Agric Food Chem 54: 9071–9078 [DOI] [PubMed] [Google Scholar]
- Gavilano LB, Siminszky B. (2007) Isolation and characterization of the cytochrome P450 gene CYP82E5v2 that mediates nicotine to nornicotine conversion in the green leaves of tobacco. Plant Cell Physiol 48: 1567–1574 [DOI] [PubMed] [Google Scholar]
- Gelling CL, Piper MDW, Hong SP, Kornfeld GD, Dawes IW. (2004) Identification of a novel one-carbon metabolism regulon in Saccharomyces cerevisiae. J Biol Chem 279: 7072–7081 [DOI] [PubMed] [Google Scholar]
- Giovanelli J, Mudd SH, Datko AH. (1985) Quantitative analysis of pathways of methionine metabolism and their regulation in Lemna. Plant Physiol 78: 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer A, Collakova E, Díaz de la Garza R, Quinlivan EP, Williamson J, Gregory JF, III, Shachar-Hill Y, Hanson AD. (2005) 5-Formyltetrahydrofolate is an inhibitory but well tolerated metabolite in Arabidopsis leaves. J Biol Chem 280: 26137–26142 [DOI] [PubMed] [Google Scholar]
- Griffith RB, Valleau WD, Stokes GW. (1955) Determination and inheritance of nicotine to nornicotine conversion in tobacco. Science 121: 343–344 [DOI] [PubMed] [Google Scholar]
- Grotewold E. (2008) Transcription factors for predictive plant metabolic engineering: are we there yet? Curr Opin Biotechnol 19: 138–144 [DOI] [PubMed] [Google Scholar]
- Guenther BD, Sheppard CA, Tran P, Rozen R, Matthews RG, Ludwig ML. (1999) The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia. Nat Struct Biol 6: 359–365 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Gage DA, Shachar-Hill Y. (2000) Plant one-carbon metabolism and its engineering. Trends Plant Sci 5: 206–213 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Roje S. (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52: 119–137 [DOI] [PubMed] [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y. (1994) Gene expression in tobacco low-nicotine mutants. Plant Cell 6: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildreth SB, Gehman EA, Yang H, Lu RH, Ritesh KC, Harich KC, Yu S, Lin J, Sandoe JL, Okumoto S, et al. (2011) Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proc Natl Acad Sci USA 108: 18179–18184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Sun YH, Chen JJ, Darlington DE, Williams AL, Burkey KO, Xie JH. (2010) Identification of a Mg-protoporphyrin IX monomethyl ester cyclase homologue, EaZIP, differentially expressed in variegated of Epipremnum aureum ‘Golden Pothos’ is achieved through a unique method of comparative study using tissue regenerated plants. J Exp Bot 61: 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A, Fannin N, Bush LP. (2007) Implications of reducing nornicotine accumulation in burley tobacco. Appendix A: the LC protocol. Rec Adv Tob Sci 33: 58–79 [Google Scholar]
- Kajikawa M, Shoji T, Kato A, Hashimoto T. (2011) Vacuole-localized berberine bridge enzyme-like proteins are required for a late step of nicotine biosynthesis in tobacco. Plant Physiol 155: 2010–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasap M, Sazci A, Ergul E, Akpinar G. (2007) Molecular phylogenetic analysis of methylenetetrahydrofolate reductase family of proteins. Mol Phylogenet Evol 42: 838–846 [DOI] [PubMed] [Google Scholar]
- Kende H. (1993) Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 44: 283–307 [Google Scholar]
- Kim YS, Choi D, Lee MM, Lee SH, Kim WT. (1998) Biotic and abiotic stress-related expression of 1-aminocyclopropane-1-carboxylate oxidase gene family in Nicotiana glutinosa L. Plant Cell Physiol 39: 565–573 [DOI] [PubMed] [Google Scholar]
- Lewis RS, Bowen SW, Keogh MR, Dewey RE. (2010) Three nicotine demethylase genes mediate nornicotine biosynthesis in Nicotiana tabacum L.: functional characterization of the CYP82E10 gene. Phytochemistry 71: 1988–1998 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lin ZF, Zhong SL, Grierson D. (2009) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Matthews RG, Ghose C, Green JM, Matthews KD, Dunlap RB. (1987) Folylpolyglutamates as substrates and inhibitors of folate-dependent enzymes. Adv Enzyme Regul 26: 157–171 [DOI] [PubMed] [Google Scholar]
- Mesnard F, Roscher A, Garlick AP, Girard S, Baguet E, Arroo RR, Lebreton J, Robins RJ, Ratcliffe G. (2002) Evidence for the involvement of tetrahydrofolate in the demethylation of nicotine by Nicotiana plumbaginifolia cell-suspension cultures. Planta 214: 911–919 [DOI] [PubMed] [Google Scholar]
- Miller RJ, Jolles C, Rapoport H. (1973) Morphine metabolism and normorphine in Papaver somniferum. Phytochemistry 12: 597–603 [Google Scholar]
- Mizusaki S, Tanabe Y, Noguchi M, Tamaki E. (1972) N-Methylputrescine oxidase from tobacco roots. Phytochemistry 11: 2757–2762 [Google Scholar]
- Molinié R, Kwiecień RA, Paneth P, Hatton W, Lebreton J, Robins RJ. (2007) Investigation of the mechanism of nicotine demethylation in Nicotiana through 2H and 15N heavy isotope effects: implication of cytochrome P450 oxidase and hydroxyl ion transfer. Arch Biochem Biophys 458: 175–183 [DOI] [PubMed] [Google Scholar]
- Morita M, Shitan N, Sawada K, Van Montagu MC, Inzé D, Rischer H, Goossens A, Oksman-Caldentey KM, Moriyama Y, Yazaki K. (2009) Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc Natl Acad Sci USA 106: 2447–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa TA, Hung CY, Darlington DE, Sane DC, Xie JH. (2009) Overexpression of human erythropoietin in tobacco does not affect plant fertility or morphology. Plant Biotechnol Rep 3: 157–165 [Google Scholar]
- Niewiadomska E, Polzien L, Desel C, Rozpadek P, Miszalski Z, Krupinska K. (2009) Spatial patterns of senescence and development-dependent distribution of reactive oxygen species in tobacco (Nicotiana tabacum) leaves. J Plant Physiol 166: 1057–1068 [DOI] [PubMed] [Google Scholar]
- Pageau K, Reisdorf-Cren M, Morot-Gaudry JF, Masclaux-Daubresse C. (2006) The two senescence-related markers, GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilization, are differentially regulated during pathogen attack and by stress hormones and reactive oxygen species in Nicotiana tabacum L. leaves. J Exp Bot 57: 547–557 [DOI] [PubMed] [Google Scholar]
- Quinlivan EP, Hanson AD, Gregory JF. (2006) The analysis of folate and its metabolic precursors in biological samples. Anal Biochem 348: 163–184 [DOI] [PubMed] [Google Scholar]
- Raymond RK, Kastanos EK, Appling DR. (1999) Saccharomyces cerevisiae expresses two genes encoding isozymes of methylenetetrahydrofolate reductase. Arch Biochem Biophys 372: 300–308 [DOI] [PubMed] [Google Scholar]
- Robins RJ, Molinie R, Kwiecien RA, Paneth P, Lebreton J, Bartholomeusz TA, Roscher A, Drager B, Mesnard F. (2007) Progress in understanding the N-demethylation of alkaloids by exploiting isotopic techniques. Phytochem Rev 6: 51–63 [Google Scholar]
- Roje S. (2006) S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67: 1686–1698 [DOI] [PubMed] [Google Scholar]
- Roje S, Chan SY, Kaplan F, Raymond RK, Horne DW, Appling DR, Hanson AD. (2002) Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the methylenetetrahydrofolate reductase reaction in vivo. J Biol Chem 277: 4056–4061 [DOI] [PubMed] [Google Scholar]
- Roje S, Wang H, McNeil SD, Raymond RK, Appling DR, Shachar-Hill Y, Bohnert HJ, Hanson AD. (1999) Isolation, characterization, and functional expression of cDNAs encoding NADH-dependent methylenetetrahydrofolate reductase from higher plants. J Biol Chem 274: 36089–36096 [DOI] [PubMed] [Google Scholar]
- Saitoh F, Nona M, Kawashima N. (1985) The alkaloid contents of sixty Nicotiana species. Phytochemistry 24: 477–480 [Google Scholar]
- Shi H, Kalengamaliro NE, Krauss MR, Hempfling WP, Gadani F. (2003) Stimulation of nicotine demethylation by NaHCO(3) treatment using greenhouse-grown burley tobacco. J Agric Food Chem 51: 7679–7683 [DOI] [PubMed] [Google Scholar]
- Shoji T, Inai K, Yazaki Y, Sato Y, Takase H, Shitan N, Yazaki K, Goto Y, Toyooka K, Matsuoka K, et al. (2009) Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol 149: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminszky B, Gavilano L, Bowen SW, Dewey RE. (2005) Conversion of nicotine to nornicotine in Nicotiana tabacum is mediated by CYP82E4, a cytochrome P450 monooxygenase. Proc Natl Acad Sci USA 102: 14919–14924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurský L, Burleson D, Waller GR. (1969) Interconversion of ricinine and N-demethylricinine in senescent and green castor plant leaves. J Biol Chem 244: 3238–3242 [PubMed] [Google Scholar]
- Trobacher CP. (2009) Ethylene and programmed cell death in plants. Botany 87: 757–769 [Google Scholar]
- Tso TC, Kasperbauer MJ, Sorokin TP. (1970) Effect of photoperiod and end-of-day light quality on alkaloids and phenolic compounds of tobacco. Plant Physiol 45: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet T, Dignard D, Thomas DY. (1987) A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52: 225–233 [DOI] [PubMed] [Google Scholar]
- Verpoorte R, Memelink J. (2002) Engineering secondary metabolite production in plants. Curr Opin Biotechnol 13: 181–187 [DOI] [PubMed] [Google Scholar]
- Wagner R, Feth F, Wagner KG. (1986) The regulation of enzyme activities of the nicotine pathway in tobacco. Physiol Plant 68: 667–672 [DOI] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR. (2002) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl) 14: S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wersman EA, Matzinger DF. (1968) Time and site of nicotine conversion in tobacco. Tob Sci 12: 226–228 [Google Scholar]
- Wu SQ, Chappell J. (2008) Metabolic engineering of natural products in plants: tools of the trade and challenges for the future. Curr Opin Biotechnol 19: 145–152 [DOI] [PubMed] [Google Scholar]
- Xie JH, Song W, Maksymowicz W, Jin W, Cheah K, Chen WX, Carnes C, Ke J, Conkling MA. (2004) Biotechnology: a tool for reduced risk tobacco products. The nicotine experience from test tube to cigarette pack. Recent Adv Tob Sci 30: 17–37 [Google Scholar]
- Xu DM, Shen YX, Chappell J, Cui MW, Nielsen M. (2007) Biochemical and molecular characterizations of nicotine demethylase in tobacco. Physiol Plant 129: 307–319 [Google Scholar]
- Yuan JS, Burris J, Stewart NR, Mentewab A, Stewart CN., Jr (2007) Statistical tools for transgene copy number estimation based on real-time PCR. BMC Bioinformatics (Suppl 7) 8: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZJ, Zhang HW, Quan RD, Wang XC, Huang RF. (2009) Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol 150: 365–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Facchini PJ. (2008) Alkaloid biosynthesis: metabolism and trafficking. Annu Rev Plant Biol 59: 735–769 [DOI] [PubMed] [Google Scholar]