Abstract
Summary: By aligning QTLs from many populations and several species, this study increases knowledge of the genetic control of agriculturally and developmentally important traits in the Saccharinae clade of grasses, expedites translation of knowledge from better-studied to less-studied species, and facilitates investigation of fundamental questions across the genomes and paleoduplicated “subgenomes” of divergent taxa.
CSGRqtl (http://helos.pgml.uga.edu/qtl/) is a comparative genomic database that facilitates the cross utilization of information among members of the Saccharinae clade of grasses, and between Saccharinae and other taxa. CSGRqtl is developed as a specific data mining resource for Saccharinae crops, weeds, and models, complementing and supplementing another database, Gramene, which includes a variety of mapping data for a broad spectrum of grass taxa. To facilitate data comparisons, a plant trait ontology defined by Gramene is applied to categorize Saccharinae quantitative trait loci (QTLs). Using the sorghum (Sorghum bicolor) genome sequence as a central reference, CSGRqtl provides approximate physical positions for QTL likelihood peaks. In order to facilitate QTL mapping and further study of the functions and evolution of candidate genes that may underlie QTLs, CSGRqtl integrates gene annotations, genetic markers, and paleoduplicated regions, and provides a series of query functionalities to navigate different data components on the basis of QTL alignments. The goals of CSGRqtl are to provide both for practical needs of crop improvement by serving as a toolbox for QTL visualization and manipulation, and to facilitate investigation of fundamental questions about similarities and differences in the genetic control of traits across paleoduplicated “subgenomes” and across the genomes of divergent taxa.
The Saccharinae clade of grasses has a rich history of contributions to humanity, with the promise of still greater contributions as a result of recent invigorated interest and research activity in several members of this clade. Sorghum ranks fifth in importance among the world’s grain crops, is a versatile source of food, fodder, and fuel, and is the most drought-tolerant of the world’s top five cereal crops, a trait essential in the U.S. southern plains and the arid countries of sub-Saharan and northeastern Africa, where it is used heavily. A close relative, Sorghum halepense (2n = 40), is of greatest importance as one of the world’s most noxious weeds, having spread from its west Asian center of diversity across much of Asia, Africa, Europe, North and South America, and Australia. Including the world’s leading sugar crop and arguably also the leading bioethanol crop, the Saccharum (sugarcane) genus includes a complex polyploid series with cultivated forms being interspecific hybrid aneuploids. Among the highest yielding of biomass crops with 2 to 3 times the yield of other leading candidates in the U.S. Midwest (Heaton et al., 2008), the genus Miscanthus is an attractive candidate for producing cellulosic biomass in temperate latitudes (Lewandowski et al., 2000, 2003).
Knowledge of the various Saccharinae taxa varies widely, from a rich history of genetic, quantitative trait locus (QTL) and physical data aligned with a high-quality reference genome for sorghum (Paterson et al., 2009), to scattered EST and genomic survey sequence data for Saccharum spp. and Miscanthus spp., to nothing at all for many others. We hypothesize that the rich body of existing information about the locations of agriculturally important genes/QTLs in well-studied Saccharinae and other grasses will have useful predictive value in accelerating the identification of diagnostic DNA markers for traits important to the “domestication” (early improvement) of grasses such as Miscanthus spp. that are of relatively recent interest. We have shown that genes/QTLs for domestication traits often correspond across divergent grasses (Lin et al., 1995; Paterson et al., 1995a, 1995b; Ming et al., 2002; Hu et al., 2003) and that meta-QTL data from diverse populations shed valuable light on the genetic control of traits (Feltus et al., 2006; Rong et al., 2007).
Here, we describe a new element of an ongoing effort to provide online resources to facilitate the study and improvement of the important Saccharinae clade. The primary goal of this new resource is the anchoring of published QTLs for this clade to the sorghum genome. Genetic map alignments translate the wealth of genomic information from sorghum to Saccharum spp., Miscanthus spp., and other taxa. In addition, genome alignments facilitate comparison of the Saccharinae QTL sets to those of other taxa that enjoy comparable resources, exemplified herein by rice (Oryza sativa; Gramene).
QTL ALIGNMENT
The availability of sequence-tagged markers such as RFLP probes and simple sequence repeat (SSR) primers in the National Center for Biotechnology Information probe database (http://www.ncbi.nlm.nih.gov/probe) provides alignable information to convert genetic positions (in centimorgan) of markers to physical positions (bp). Subsequently, QTLs are anchored to the sorghum genome by identifying two flanking markers (Fig. 1). Maps based largely on Random Amplification of Polymorphic DNA, Amplified Fragment Length Polymorphism, and Diversity Arrays Technology markers do not provide alignable information and were not included.
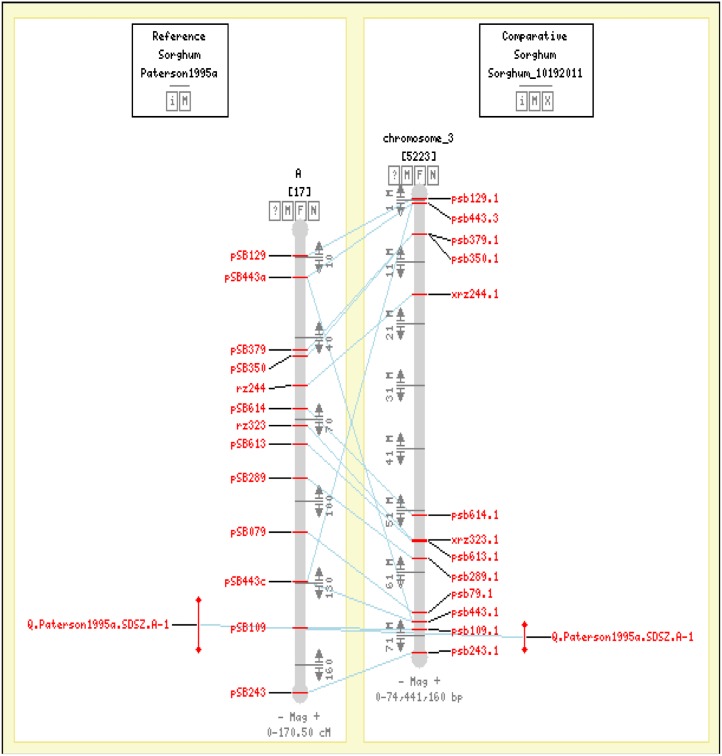
Figure 1.
CMap viewer displaying a QTL affecting seed size in linkage group A (Paterson et al., 1995a) aligned to sorghum chromosome 3. Two flanking markers, pSB443 and pSB243, are identified to anchor the QTL. [See online article for color version of this figure.]
After marker sequences are prepared, BLASTN (Altschul et al., 1990) is applied to anchor markers to the sorghum genome. Hits with E ≤ 1e-10 and E ≤ 50 for RFLP sequences and SSR primers, respectively, are postprocessed to assemble into loci. All hits with distances of 5,000 bp or less are assembled into one RFLP locus. For SSRs, one forward primer hit is combined with one reverse primer hit if the distance between the two hits is 1,000 bp or less.
Striking discrepant loci are removed, based on the order of markers in the original source. Colinearity between genetic and physical positions is determined by ColinearScan 1.0.1 (Wang et al., 2006).
A QTL region is delineated by two flanking markers nearest to the likelihood peak that have alignment information.
QTL CORRESPONDENCE
Numerous studies have shown nonrandom correspondences of QTL locations across taxa (Paterson et al., 1995a), and such comparisons may quickly advance knowledge in less well-studied species such as Miscanthus spp. by taking advantage of better studied species such as sorghum. As an example, the wealth of rice QTL information in Gramene (Ware et al., 2002; http://www.gramene.org/qtl/) and genomic synteny data in the Plant Genome Duplication Database (Tang et al., 2008; http://chibba.agtec.uga.edu/duplication/) allow us to convert genetic marker correspondence to genomic region correspondence and, in turn, to investigate alignments among large populations of QTLs. A total of 8,686 rice QTLs affecting 238 traits are anchored to the rice genome (Michigan State University release 7). Among trait ontology accessions assigned to QTLs in sorghum and rice, 24 are in common and include some widely studied traits such as seed size, plant height, and flowering time. Colinearity provides a bridge to globally investigate overlaps among QTLs in the sorghum and rice genomes for specific traits (Fig. 2). Identification of QTL correspondence sheds light on the evolution of the underlying phenotype and also aids in the development of practical tools such as DNA markers that may be diagnostic across populations and taxa.
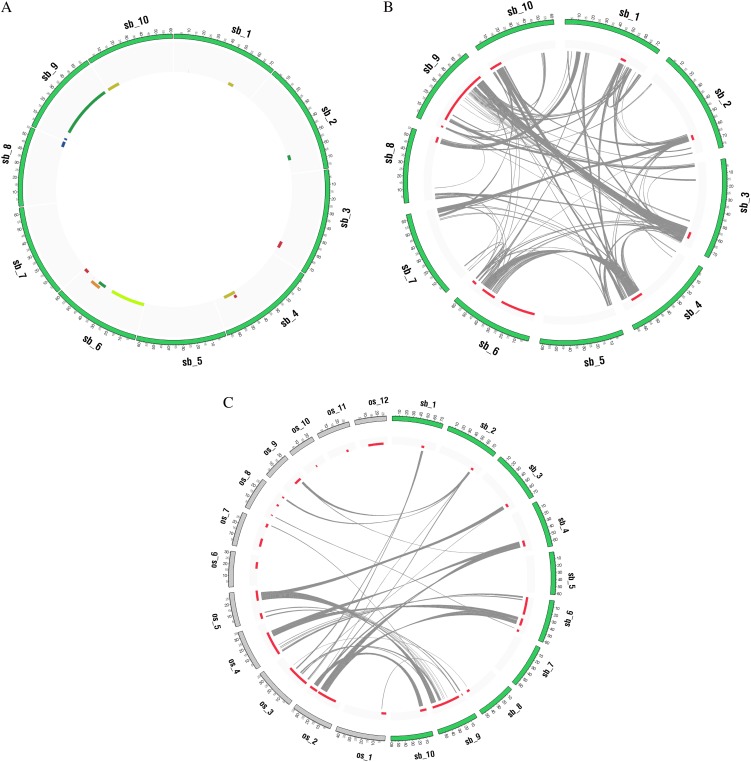
Figure 2.
A, The distribution of QTLs underlying days to flower in the sorghum (green)/rice (gray) genomes and QTL correspondence by intergenomic/intragenomic synteny. B, The distribution of QTLs in the sorghum genome. QTLs from different studies are indicated by colors. C, The paralogs for genes bounded by nonoverlapping QTLs in the sorghum genome. The QTL correspondence between sorghum and rice is established by intergenomic synteny.
TOOLS
CSGRqtl contains a number of analysis tools to allow a user to query and visualize the background database.
(1) Text-based search. Searching for a trait returns a set of QTLs underlying this trait. A circular plot created by Circos (Krzywinski et al., 2009) gives a genome-wide overview of QTL distribution (Fig. 2A). The stacking regions yield potential QTL hotspots in the genome. Inputting a sorghum gene identifier or annotation results in a list of QTLs containing genes that match queries. A plot is created to depict the approximate positions of genes and QTLs in the genome.
(2) Trait ontology browser. Each QTL is allocated a proper trait accession, based on the Gramene Plant Trait Ontology. By a trait ontology browser, the hierarchy of trait ontology is displayed, and QTLs belonging to each trait accession are listed.
(3) QTL correspondence. In order to describe connections between paleoduplicated regions and QTLs in rice and sorghum, CSGRqtl provides circular plots to show nonoverlapping QTLs divided/narrowed by intergenomic/intragenomic synteny (Fig. 2, B and C) and allows users to download orthologs/paralogs for genes bounded by nonoverlapping QTLs for each trait.
(4) CMap database. CMap version 0.16 (Youens-Clark et al., 2009) is downloaded from the Generic Components for Model Organism Database project (http://www.gmod.org). Using CMap, a user can view alignments between genetic maps and the sorghum genome sequence (Fig. 1). The database also provides alignable information such as RFLP probe and SSR primer sequences for each marker anchored. The CMap resource will greatly expedite the process of marker screening for QTL mapping.
(5) Genome browser for gene annotations in QTL regions. To associate sorghum QTL data with gene annotations, a sorghum genome browser is implemented using Generic Genome Browser version 2.39 (Stein et al., 2002). Gene models are from standard sorghum genome annotation version 1.4. A total of 209,828 sorghum ESTs from the National Center for Biotechnology Information are also anchored on the genome. G/C content, six-frame translation, and restriction sites are also available for each genomic region. For a QTL region, a user can get information about all annotated genes in that region (Fig. 3). All QTLs can be easily accessed given any genomic region.
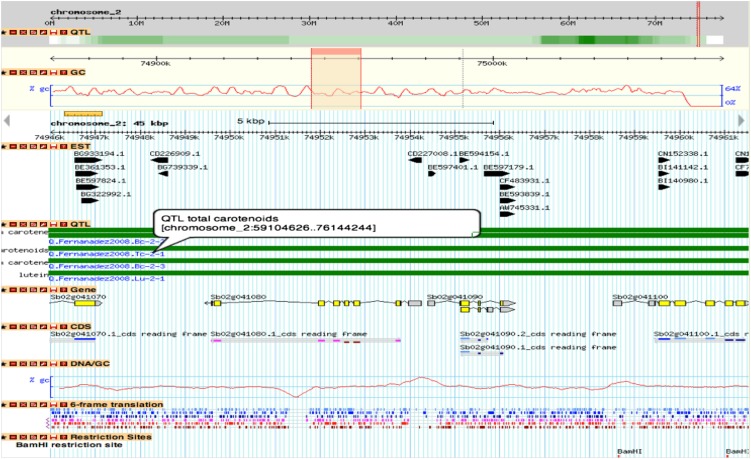
Figure 3.
Overview of a sorghum genomic region on chromosome 2. The first drill down shows a heat map of QTL density across all of sorghum chromosome 2. The second drill down displays GC content of a 200-kb region indicated by a narrow red window in the first drill down. The third and following drilldowns list various annotation information in a 15-kb region indicated by another red window within the 200-kb region. For any particular region, all QTLs are shown as green bars in the QTL track. The name and genomic position of each QTL is indicated on the left side of the track or by mouse hovering. The literature source from which the QTL is derived is shown at left bottom of each QTL bar. More gene annotations will be available by mouse hovering on each gene track.
APPLICATION EXAMPLES
Analysis of Genetic Complexity of Traits
Most Saccharinae traits are complex or quantitative, the result of collective actions of multiple genes, so the determination of “saturation” in terms of QTL discovery is a major challenge. CSGRqtl provides the user with a repository compiling QTL mapping results from different parental combinations and in different environments that yields a more complete picture of genetic complexity of a trait than any one study alone. For example, flowering time in sorghum is commonly thought to be controlled by six genes, Maturity1 (Ma1) to Ma6 (Quinby, 1974; Rooney and Aydin, 1999). A total of 14 flowering QTL confidence intervals published in six studies fall into more than 11 nonoverlapping regions in the sorghum genome, strongly suggesting that genetic control of sorghum flowering involves more than six genes. Similarly, 51 plant height QTLs published in seven studies fall into 13 blocks, implicating far more than the classically suggested four genes, dwarf1 (dw1) to dw4, in genetic control of sorghum height (Quinby, 1974).
The lengths of QTL likelihood intervals for a particular trait depend on the quality of genetic mapping and the richness of marker sequence information. For flowering, QTL genomic regions are relatively small on chromosomes 1, 2, 3, 4, 6, 8, and 10, compared with a larger region on chromosome 9. Concentrations of QTL data in a specific genomic region provide a good a priori hypothesis for the user to combine further evidence, such as results from genome-wide association studies, to identify specific candidate genes with reduced risk of false-positive signals.
CSGRqtl also offers multiple means for users to mine prospective causal genes. As an example, some users may have a priori information that a specific gene family might be essential, such as auxins for plant height development. CSGRqtl will assist the user in queries such as identifying any plant height QTL intervals bounding genes with the auxin annotation.
QTL Distributions in Relation to Patterns of Whole-Genome Duplication
Combination of paleoduplications and QTL intervals can be broadly applied in evolutionary questions, such as the exploration of whether paralogous/orthologous genes resulting from genome duplication/speciation events continue to function in related ways or now function differently. In the current database, we made use of QTL data from rice to do such comparisons. The divergence of the ancestors of rice and sorghum occurred shortly after a whole-genome duplication (rho) event occurred approximately 70 million years ago (Paterson et al., 2003, 2004). For flowering time, by aligning 11 QTL intervals in sorghum to 17 QTL intervals in rice, we were able to identify 2,556 genes present in the corresponding regions of both species, which are candidates that may underlie the flowering trait of the common ancestor of rice and sorghum. In addition to intergenomic comparison, CSGRqtl indicates that there are 3,505 sorghum paralogous gene pairs in which 466 have both copies encompassed by flowering QTLs. For plant height QTLs, 8,619 genes are present in both rice and sorghum in the regions of overlap between 13 QTL intervals. Among 3,505 paralogous gene pairs in sorghum, 757 have both copies located in the sorghum QTL regions. While these numbers remain large, they provide a subset of positional/evolutionary candidates that might be further narrowed by a range of other approaches such as association genetics or expression profiling.
DATA AVAILABILITY
All of the data in CSGRqtl are freely available.
FUTURE ENHANCEMENTS
CSGRqtl is a Saccharinae QTL repository and a data-mining tool for QTL gene identification to which we intend to add data as they become available. In addition to sorghum and Saccharum spp., we plan to compile QTLs for closely related species such as Miscanthus spp. that are the subject of much research. While the sorghum genome is a valuable reference, as additional Saccharinae genomes are sequenced we may introduce additional reference genomes and alignments among these genomes. In much the same manner as illustrated for rice, orthologous connections between sorghum and other cereal species such as maize (Zea mays) offer the opportunity to further explore comparative genomics information toward QTL identification and other goals.
Glossary
- QTL
quantitative trait locus
- SSR
simple sequence repeat
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Feltus FA, Hart GE, Schertz KF, Casa AM, Kresovich S, Abraham S, Klein PE, Brown PJ, Paterson AH. (2006) Alignment of genetic maps and QTLs between inter- and intra-specific sorghum populations. Theor Appl Genet 112: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Heaton EA, Dohleman FG, Long SP. (2008) Meeting US biofuel goals with less land: the potential of Miscanthus. Glob Change Biol 14: 2000–2014 [Google Scholar]
- Hu FY, Tao DY, Sacks E, Fu BY, Xu P, Li J, Yang Y, McNally K, Khush GS, Paterson AH, et al. (2003) Convergent evolution of perenniality in rice and sorghum. Proc Natl Acad Sci USA 100: 4050–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski I, Clifton-Brown JC, Scurlock JMO, Huisman W. (2000) Miscanthus: European experience with a novel energy crop. Biomass Bioenergy 19: 209–227 [Google Scholar]
- Lewandowski I, Scurlock JMO, Lindvall E, Christou M. (2003) The development and current status of perennial rhizomatous grasses as energy crops in the US and Europe. Biomass Bioenergy 25: 335–361 [Google Scholar]
- Lin YR, Schertz KF, Paterson AH. (1995) Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 141: 391–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, DelMonte TA, Hernandez E, Moore PH, Irvine JE, Paterson AH. (2002) Comparative analysis of QTLs affecting plant height and flowering among closely-related diploid and polyploid genomes. Genome 45: 794–803 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Peterson DG, Estill JC, Chapman BA. (2003) Structure and evolution of cereal genomes. Curr Opin Genet Dev 13: 644–650 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lin YR, Li Z, Schertz KF, Doebley JF, Pinson SR, Liu SC, Stansel JW, Irvine JE. (1995a) Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269: 1714–1718 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Schertz KF, Lin YR, Liu SC, Chang YL. (1995b) The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) Pers. Proc Natl Acad Sci USA 92: 6127–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinby JR. (1974) Sorghum Improvement and the Genetics of Growth. Texas A&M University Press, College Station
- Rong J, Feltus FA, Waghmare VN, Pierce GJ, Chee PW, Draye X, Saranga Y, Wright RJ, Wilkins TA, May OL, et al. (2007) Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics 176: 2577–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney WL, Aydin S. (1999) Genetic control of a photoperiod-sensitive response in Sorghum bicolor (L.) Moench. Crop Sci 2: 397–400 [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. (2002) The generic genome browser: a building block for a model organism system database. Genome Res 12: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. (2008) Synteny and collinearity in plant genomes. Science 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Wang X, Shi X, Li Z, Zhu Q, Kong L, Tang W, Ge S, Luo J. (2006) Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinformatics 7: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware D, Jaiswal P, Ni J, Pan X, Chang K, Clark K, Teytelman L, Schmidt S, Zhao W, Cartinhour S, et al. (2002) Gramene: a resource for comparative grass genomics. Nucleic Acids Res 30: 103–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youens-Clark K, Faga B, Yap IV, Stein L, Ware D. (2009) CMap 1.01: a comparative mapping application for the Internet. Bioinformatics 25: 3040–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data in CSGRqtl are freely available.