Summary: The dual-targeting ability of proteins arose early in evolution, and although it is conserved in many cases, acquisition and loss still occur.
Abstract
The dual-targeting ability of a variety of proteins from Physcomitrella patens, rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana) was tested to determine when dual targeting arose and to what extent it was conserved in land plants. Overall, the targeting ability of over 80 different proteins from rice and P. patens, representing 42 dual-targeted proteins in Arabidopsis, was tested. We found that dual targeting arose early in land plant evolution, as it was evident in many cases with P. patens proteins that were conserved in rice and Arabidopsis. Furthermore, we found that the acquisition of dual-targeting ability is still occurring, evident in P. patens as well as rice and Arabidopsis. The loss of dual-targeting ability appears to be rare, but does occur. Ascorbate peroxidase represents such an example. After gene duplication in rice, individual genes encode proteins that are targeted to a single organelle. Although we found that dual targeting was generally conserved, the ability to detect dual-targeted proteins differed depending on the cell types used. Furthermore, it appears that small changes in the targeting signal can result in a loss (or gain) of dual-targeting ability. Overall, examination of the targeting signals within this study did not reveal any clear patterns that would predict dual-targeting ability. The acquisition of dual-targeting ability also appears to be coordinated between proteins. Mitochondrial intermembrane space import and assembly protein40, a protein involved in oxidative folding in mitochondria and peroxisomes, provides an example where acquisition of dual targeting is accompanied by the dual targeting of substrate proteins.
Gene transfer to the host nucleus followed the endosymbiotic events that led to the formation of mitochondria and plastids in plant cells (Adams et al., 2000; Dyall et al., 2004; Kleine et al., 2009; Keeling, 2010). This resulted in a reduction of the coding capacity of these organelles to approximately 5% of the original endosymbiont’s genome (Pfannschmidt, 2010). Therefore, the majority of organellar proteins are encoded in the nucleus, translated in the cytosol, and imported into their respective organelles. This process of protein targeting required that new machinery, not present in the original endosymbionts, be acquired to specifically recognize and translocate thousands of proteins across their respective organelle membranes (Dolezal et al., 2006). Studies into mitochondrial and plastid protein import revealed that targeting and import is specific for each organelle (Rudhe et al., 2002; Glaser and Whelan, 2007). This specificity is believed to be due to a number of factors: the nature of the targeting signals, the presence of cytosolic “targeting” factors, and the presence of protein receptors on the organelle surface, all of which contribute to maintain the specificity of protein import (Chew and Whelan, 2004). The molecular mechanisms of how these features maintain specificity is largely unknown and further complicated with the growing identification of proteins that can be targeted to multiple organelles.
The initial report that glutathione reductase (GR) from Pisum sativum was targeted to both mitochondria and plastids revealed that targeting could occur to two organelles and that protein targeting was not location specific (Creissen et al., 1995). Since this initial report, 107 proteins have now been reported to be dual targeted to mitochondria and plastids in a variety of plants (Carrie et al., 2009a; Carrie and Small, 2013). The dual targeting of proteins can occur by a variety of mechanisms (Peeters and Small, 2001; Yogev and Pines, 2011), such as ambiguous targeting signals, where a single targeting signal has the ability to target a protein to two distinct locations, or alternative transcription/translation, where altered targeting signals are produced for each respective organelle (Peeters and Small, 2001; Yogev and Pines, 2011). Dual targeting by ambiguous signals is of particular interest, as it questions how two distinct organelle import machineries can recognize these ambiguous signals and yet distinguish between organelle-specific signals. In addition to dual targeting to mitochondria and plastids, dual targeting of proteins to mitochondria and peroxisomes has also been reported (Carrie et al., 2008, 2009b). The mechanism of targeting differs in that proteins dual targeted to mitochondria and peroxisomes contain two signals, which results in the same protein being imported into two distinct locations.
To date, 72 proteins in Arabidopsis have been shown to be dual targeted (Carrie et al., 2009a; Carrie and Small, 2013); however, as many as 500 are predicted to be dual targeted because they contain ambiguous signals (Mitschke et al., 2009). The necessity for dual targeting remains largely unknown, though it may be related to gene copy number and restriction of genome size (Morgante et al., 2009), or be required for the coordination of organelle function (Chew et al., 2003; Carrie et al., 2009a). There is only a limited amount of information available regarding the extent of dual targeting of orthologous proteins between species. Dual targeting of four proteins (Met aminopeptidase, monodehydroascorbate reductase [MDHAR], glutamyl-transfer RNA synthetase, and tyrosyl-transfer RNA synthetase) has been shown to be conserved in rice and Arabidopsis (Morgante et al., 2009). Other studies have found that organellar seryl-transfer RNA is dual targeted in Arabidopsis and maize (Zea mays; Rokov-Plavec et al., 2008). In addition, the MutS HOMOLOG1 was found to be dual targeted in a number of dicot plants (Xu et al., 2011).
This study was undertaken to determine if dual-targeted proteins in Arabidopsis were also dual targeted in rice and P. patens. In addition, if differences were observed in the dual-targeting ability of proteins from within these three species, we investigated the targeting ability of Picea glauca orthologs and therefore the dual-targeting ability of proteins spanning 500 million years of land plant evolution. Overall, we posed the following questions to gain a better understanding of the dual targeting of proteins to mitochondria and plastids or mitochondria and peroxisomes. When did dual targeting arise in plant evolution? Is dual targeting of proteins conserved? Is acquisition or loss of protein dual targeting still occurring? Is dual targeting of proteins associated with a gain of function in organelles?
RESULTS
In land plants, 107 proteins have been experimentally reported to be dual targeted to mitochondria and plastids, or mitochondria and peroxisomes (Supplemental Tables S1 and S2; Carrie et al., 2009a; Carrie and Small, 2013). To determine when dual targeting arose in land plant evolution and if dual-targeting ability is conserved, we investigated whether the orthologs of some dual-targeted proteins in Arabidopsis were also dual targeted in P. patens and rice. As P. patens diverged from Arabidopsis approximately 450 million years ago (Fig. 1A), it represents the earliest land plant with a complete genome sequence, allowing identification of all orthologs within gene families of Arabidopsis dual-targeted proteins. It should be noted that for many dual-targeted proteins, location-specific orthologs also exist. Therefore, identification of all gene family members is required to ensure that all orthologous proteins are being identified when testing for targeting ability. We also identified the orthologs from Chlamydomonas reinhardtii (Merchant et al., 2007) and Chlorella variabilis (Blanc et al., 2010) and included them in the phylogenetic analysis to obtain a more robust characterization of orthologs. The predicted protein that showed the highest level of sequence identity to the Arabidopsis dual-targeted protein was defined as the closest ortholog, and its targeting ability was tested (Supplemental Data Set S1). In addition, we tested the targeting of P. patens proteins in Arabidopsis cell suspensions, onion (Allium cepa) epidermal cells, and P. patens protonemal tissues, as there have been no previous reports of targeting P. patens proteins in nonhomologous plant tissues. We also tested the mitochondrial targeting signals from alternative oxidase (AOX) and the α-subunit of the mitochondrial processing peptidase; the plastid targeting signals of the small subunit of Rubisco and PSI subunit 2; and the peroxisomal targeting signals from thiolase and malate synthase in Arabidopsis cell suspensions, onion epidermal cells, and P. patens tissue to define the fluorescence characteristics of these organelles in the various tested tissues (Fig. 1B). This demonstrated that the mitochondrial, plastid, and peroxisomal red fluorescent protein (RFP) markers previously used in Arabidopsis to define these organelles can also be used with P. patens (Carrie et al., 2009b).
Figure 1.
Experimental design to investigate dual targeting of proteins in land plants. A, Tree diagram showing the approximate time (in millions of years ago) that the major group of plants diverged. The four species of land plants used in this study, Arabidopsis, rice, P. glauca, and P. patens, are shown. Additionally, C. reinhardtii and C. variabilis were included in the phylogenetic analysis. B, Verifying the P. patens-derived organelle-specific markers for mitochondria, plastids (rbcS [for small subunit of Rubisco]), and peroxisomes (thiolase and malate synthase) fused to GFP. Accession numbers are indicated. The biolistic transformation of each P. patens-derived organelle marker was carried out with previously published organelle marker sets, with the mitochondrial, plastid, and peroxisomal (SRL [for Cucurbita spp. malate synthase]) targeting signal fused to RFP (Carrie et al., 2009b). In addition, cytochrome oxidase IV-mCherry (Nelson et al., 2007) and P. patens AOX-mCherry were used as mitochondrial markers in onion epidermal cells and P. patens protonemal tissues, respectively. Targeting was tested in Arabidopsis cell suspensions, onion epidermal cells, and P. patens protonemal tissues. Scale bar indicates 20 µm. Mitochondria (M), plastids (Pl), and peroxisomes (Px) are indicated, respectively.
Dual Targeting Arose Early and Is Conserved during Land Plant Evolution
A number of orthologs to dual-targeted proteins in Arabidopsis were also found to be dual targeted from rice and P. patens. DNA topoisomerase represents an example of a protein that is dual targeted to mitochondria and plastids in Arabidopsis (Carrie et al., 2009b) and was similarly found to be dual targeted to mitochondria and plastids from rice and P. patens (Fig. 2). Analysis of topoisomerase genes revealed that there are three in Arabidopsis, four in rice, and five in P. patens (Fig. 2A). While the targeting predictions of the proteins differed using various prediction programs (Fig. 2B), it was apparent that topoisomerases were dual targeted in all three species when tested by biolistic transformation (Fig. 2C). While dual targeting of the rice topoisomerase was readily apparent in Arabidopsis cell suspensions and onion epidermal cells, dual targeting of the P. patens topoisomerase was only apparent in onion epidermal cells and in P. patens protonemal tissues, where the mitochondria were elongated and cylinder like in shape (Fig. 2C). This elongated shape was similar to that observed with the control mitochondrial markers (Fig. 1B). Thus, while dual targeting of topoisomerases was conserved, the dual targeting of the P. patens topoisomerase was not observed in all tested cell types.
Figure 2.
Dual targeting of DNA topoisomerase to mitochondria and plastids. A, Phylogenetic analysis of genes encoding DNA topoisomerases (Top) from Arabidopsis (At), rice (Os), P. patens (Pp), P. glauca (Pg), C. reinhardtii (Cr), and C. variabilis (Cv) using MEGA 5 (see “Materials and Methods”). B, Table of Top proteins from Arabidopsis, rice, P. patens, P. glauca, C. reinhardtii, and C. variabilis with genomic loci numbers, targeting predictions (see “Materials and Methods”), and experimental localization based on GFP targeting. C, GFP images of the targeting ability of tested Top proteins. Dual targeting of AtTopIA1 and OsTopI was evident in Arabidopsis cell suspensions and onion epidermal cells. In contrast, dual targeting of PpTop was only evident in onion epidermal cells and P. patens protonemal tissues. Mitochondria (M) and plastids (Pl) are indicated, respectively. Scale bar indicates 20 µm. Cyto, Cytosol; N, nuclear; S, secretory pathway.
Analysis of a large number of other proteins revealed that while many were observed to be dual targeted, the ability for dual targeting differed between cell types. In the case of DNA helicase (Hel; Fig. 3), previously shown to be dual targeted in Arabidopsis (Carrie et al., 2009b), the tested P. patens ortholog (PpHel1) and the only rice ortholog (OsHel) were also determined to be dual targeted in all tested tissues (Fig. 3). Thus, it differed from the P. patens topoisomerase that was not observed to be dual targeted in Arabidopsis cell suspensions. However, in the case of DNA polymerase (Pol; Fig. 4), while the two Arabidopsis orthologs AtPolγ1 (AtPol1) and AtPol2 and the rice ortholog OsPol1 were observed to be dual targeted to mitochondria and plastids (Carrie et al., 2009b), the P. patens orthologs showed a different pattern (Fig. 4). Of the four P. patens DNA polymerase orthologs, three (PpPol2, PpPol3, and PpPol1) branched with the Arabidopsis proteins. However, PpPol1 was only targeted to plastids in all tested tissues (Fig. 4C). In contrast, while PpPol2 was predominantly targeted to mitochondria in Arabidopsis cell suspensions, plastid and mitochondrial targeting was evident in onion epidermal cells (Fig. 4C). As in Arabidopsis, mitochondrial targeting was dominant in P. patens protonemal tissues, with plastid targeting only weakly observed (Fig. 4C).
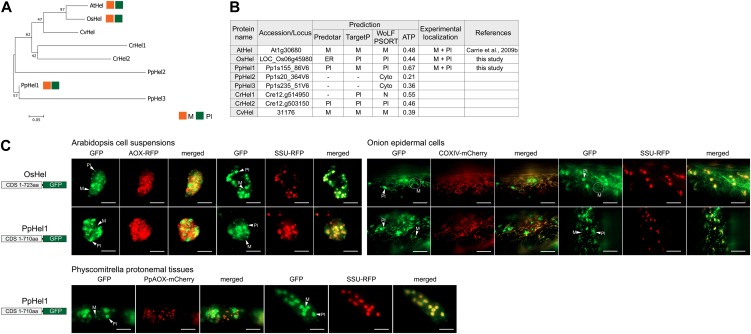
Figure 3.
Dual targeting of DNA helicase to mitochondria and plastids. A, Phylogenetic analysis of genes encoding DNA helicase (Hel) from Arabidopsis (At), rice (Os), P. patens (Pp), C. reinhardtii (Cr), and C. variabilis (Cv) using MEGA 5 (see “Materials and Methods”). There was no ortholog found in P. glauca. B, Table of Hel proteins from Arabidopsis, rice, P. patens, C. reinhardtii, and C. variabilis with genomic loci numbers, targeting predictions, and experimental localization based on GFP tagging. C, GFP images of the targeting ability of Hel proteins tested. Dual targeting of OsHel and PpHel1 was evident in Arabidopsis cell suspensions, onion epidermal cells, and P. patens protonemal tissues. Mitochondria (M) and plastids (Pl) are indicated, respectively. Scale bar indicates 20 µm. Cyto, Cytosol; ER, endoplasmic reticulum; N, nuclear.
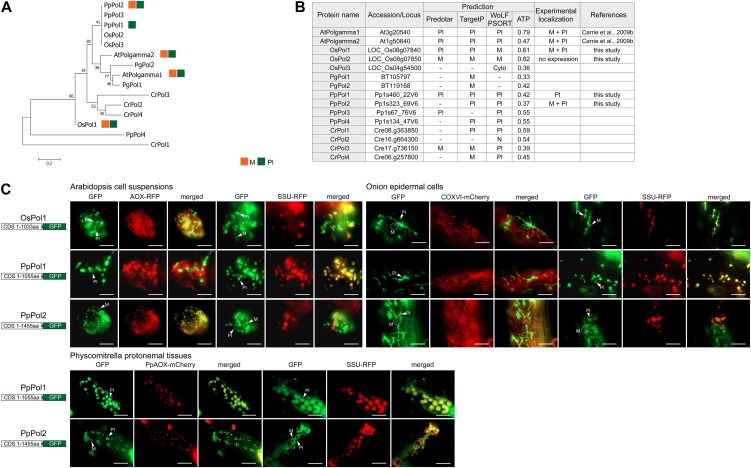
Figure 4.
Dual targeting of DNA polymerase to mitochondria and plastids. A, Phylogenetic analysis of genes encoding DNA polymerases (Pol) from Arabidopsis (At), rice (Os), P. patens (Pp), P. glauca (Pg), and C. reinhardtii (Cr) using MEGA 5 (see “Materials and Methods”). There was no ortholog found in C. variabilis. B, Table of the Pol proteins from Arabidopsis, rice, P. patens, P. glauca, and C. reinhardtii with genomic loci numbers, predictions using a variety of prediction programs, and experimental localization based on GFP tagging. C, GFP images of targeting ability of tested Pol proteins. Dual targeting of OsPol1 was evident in all three tested cells types. Only plastid targeting could be detected for PpPol1 in all three tissues, while plastid and mitochondrial targeting was detected for PpPol2 in onion epidermal cells and P. patens protonemal tissues. Mitochondria (M) and plastids (Pl) are indicated, respectively. Scale bar indicates 20 µm. Cyto, Cytosol; N, nuclear.
In an analysis of 38 Arabidopsis proteins that were previously reported to be dual targeted to mitochondria and plastids, rice contained an ortholog that was dual targeted in 28 out of 38 cases [Supplemental Fig. S1; Supplemental Table S1, shaded in green and yellow; the following proteins were not dual targeted to mitochondria and plastids in rice compared with Arabidopsis: ascorbate peroxidase (APX), glutamine synthetase, cochaperone GrpE protein, methylenetetrahydrofolate reductase, organellar single-stranded DNA binding protein, rhodanase-like protein, 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase, sulfiredoxin, thiazole biosynthetic enzyme, and AAA-type ATPase family protein], while nine out of 13 cases of rice and P. patens were dual targeted in the tested tissues [Supplemental Fig. S1; Supplemental Table S1, shaded in green; the following proteins were not dual targeted to mitochondria and plastids in rice and P. patens compared with Arabidopsis: APX, cochaperone GrpE protein, DNA ligase, and Arabidopsis 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase].
The dual targeting of proteins to mitochondria and peroxisomes, or plastids and peroxisomes has also been reported, with 11 cases identified to date (Supplemental Table S2). The dual targeting of orthologs from rice and P. patens of 6-phosphogluconolactonase (dual targeted to plastids and peroxisomes; Reumann et al., 2007) and Ala:glyoxylate aminotransferase (dual targeted to mitochondria and peroxisomes; Carrie et al., 2009b) was tested to determine if dual targeting was also conserved when it involved another organelle, the peroxisomes. The tested orthologs were found to be targeted to both plastids and peroxisomes, or mitochondria and peroxisomes (Supplemental Fig. S2) in all tissues.
Overall, in the analysis of four Arabidopsis proteins that were dual targeted to mitochondria and peroxisomes, or plastids and peroxisomes, three out of four (Supplemental Table S2; the following protein was not dual targeted to mitochondria and peroxisomes in rice compared with Arabidopsis: malonyl-CoA decarboxylase) orthologs from rice tested in Arabidopsis suspension and onion were dual targeted, and two out of three proteins from rice and P. patens were dual targeted to mitochondria and peroxisomes or plastids and peroxisomes (Supplemental Table S2).
Dual Targeting Is Gained and Lost during Land Plant Evolution
While a variety of proteins were observed to be dual targeted, P. patens DNA polymerase hinted that targeting to one organelle may be stronger or more efficient than targeting to another (Fig. 4) and that this may be associated with gene duplication. Therefore, a detailed study was carried out to investigate the dual-targeting ability of a number of proteins that are known to be encoded by small gene families. Enzymes of the ascorbate-GR cycle were chosen, as GR, APX, and MDHAR contain orthologs that are dual targeted in Arabidopsis and are part of multigene families (Fig. 5A; Chew et al., 2003). GR orthologs from rice (OsGR1) and P. patens (PpGR1) were similarly observed to be dual targeted in Arabidopsis, onion, and P. patens tissue (Fig. 5D). In the case of APX, four genes encode APX in P. patens, five in P. glauca, seven in Arabidopsis, and eight in rice (Fig. 6A). The Arabidopsis APX, designated as a plastid stromal isoform (AtSAPX), has been previously reported to be dual targeted (Chew et al., 2003), and this was confirmed in the current study (Fig. 6). However, examination of P. patens APX1 revealed that it was only targeted to plastids in all three tested tissue types (Fig. 6B). Examination of the rice APX proteins OsAPX5, OsAPX6, OsAPX7, and OsAPX8, which grouped with and displayed the highest sequence identity to AtSAPX (Fig. 6, A and C), revealed that OsAPX5 and OsAPX6 were targeted to mitochondria, while OsAPX7 and OsAPX8 were targeted to plastids (Fig. 6B). To gain insight into when dual targeting of APX was gained (or lost), the targeting of the most orthologous APX from P. glauca, PgAPX1, was examined and found to be dual targeted (Fig. 6). Thus, it is proposed that the dual-targeting ability of APX arose following the split between P. patens and P. glauca, and was subsequently lost in rice following monocot divergence. In rice, gene duplication resulted in two genes, with each encoding organelle-specific proteins (Fig. 6A). The gene family of MDHAR has multiple members identified with four genes in P. patens, three in P. glauca, five in Arabidopsis, and six in rice (Fig. 7A). AtMDHAR6, previously shown to be dual targeted (Chew et al., 2003), has two orthologs in P. patens, one of which showed targeting to both mitochondria and plastids. The detection of MDHAR dual-targeted orthologs in all four species, including P. glauca (Fig. 7), suggests that as with GR, dual-targeting ability arose early in land plant evolution and has been conserved. However, PpMDHAR1 and OsMDHAR1.2 are not dual targeted. This suggests that dual targeting of these isoforms may have been lost after gene duplication in these organisms, so that only a single dual-targeted isoform is conserved in each organism.
Figure 5.
Dual targeting of the enzymes of the ascorbate-glutathione cycle. A, Overview of the ascorbate-glutathione cycle. B, Phylogenetic analysis of the genes encoding GR from Arabidopsis (At), rice (Os), P. glauca (Pg), P. patens (Pp), C. reinhardtii (Cr), and C. variabilis (Cv) using MEGA 5. C, Table of GR proteins from Arabidopsis, rice, P. patens, P. glauca, C. reinhardtii, and C. variabilis with genomic loci numbers, targeting predictions, and experimental localization based on GFP tagging. D, GFP images of targeting ability of tested GR proteins. Dual targeting of GR from Arabidopsis, rice, and P. patens was detected in all tested tissues. Mitochondria (M) and plastids (Pl) are indicated, respectively. Scale bar indicates 20 µm. DHAR, Dehydroascorbate reductase; ASC, ascorbate; DHA, dehydroascorbate; GSH, glutathione; GSSG, glutathione disulfide; M, mitochondria; Pl, plastids; Cyto, cytosol.
Figure 6.
Dual targeting of APX to mitochondria and plastids. A, Phylogenetic analysis of genes encoding APX from Arabidopsis (At), rice (Os), P. glauca (Pg), P. patens (Pp), and C. reinhardtii (Cr) using MEGA 5 (see methods). There was no ortholog found in C. variabilis. B, GFP images of targeting ability of tested APX proteins. Dual targeting of AtSAPX was evident in Arabidopsis cell suspensions and onion epidermal cells. Four rice APX proteins were tested. OsAPX5 and OsAPX6 showed targeting to mitochondria, while OsAPX7 and OsAPX8 showed targeting to plastids. PgAPX1 showed targeting to both mitochondria and plastids in onion epidermal cells, while PpAPX1 showed targeting to plastids only in all tested tissues. C, Table of the APX proteins from Arabidopsis, rice, and P. patens with genomic loci numbers, targeting predictions, and experimental localization based on GFP targeting. Mitochondria (M) and plastids (Pl) are indicated, respectively. Scale bar indicates 20 µm. Cyto, Cytosol; ER, endoplasmic reticulum.
Figure 7.
Dual targeting of MDHAR to mitochondria and plastids. A, Phylogenetic analysis of genes encoding MDHAR from Arabidopsis (At), rice (Os), P. glauca (Pg), P. patens (Pp), and C. reinhardtii (Cr) using MEGA 5 (see “Materials and Methods”). There is no ortholog found in C. variabilis. B, GFP images of targeting ability of the tested MDHAR proteins. Dual targeting of AtMDHAR6 was evident in Arabidopsis cell suspensions and onion epidermal cells. While OsMDHAR1.1 displayed dual targeting to mitochondria and plastids, OsMDAHR1.2 only showed mitochondrial targeting ability. Likewise for both P. patens and P. glauca, of the two proteins tested for each species, only one (PgMDHAR2 and PpMDHAR2 for P. glauca and P. patens, respectively) displayed dual-targeting ability. C, Table of the MDHAR proteins from Arabidopsis, rice, P. glauca, P. patens, and C. reinhardtii with genomic loci numbers, targeting predictions, and experimental localization based on GFP tagging. Mitochondria (M) and plastids (Pl) are indicated respectively. Scale bar indicates 20 µm. Cyto, Cytosol; ER, endoplasmic reticulum; EX, extracellular; S, secretory pathway.
Hexokinase (HXK) proteins represent another example of the acquisition of dual-targeting ability throughout land plant evolution. While the targeting of Arabidopsis orthologs has not previously been studied, a recent study (Nilsson et al., 2011) in P. patens identified isoforms that were dual targeted to mitochondria and plastids. Of the 11 genes encoding HXK (PpHXK1–PpHXK11) in this study (Fig. 8A), at least six isoforms are dual targeted (Fig. 9). Compared with the previous study, some differences have been observed (Nilsson et al., 2011), in that PpHXK5 and PpHXK9 were found to be dual targeted while PpHXK8 was not (Fig. 8B). However, none of the four HXK orthologs from Arabidopsis (AtHXK1–AtHXK4), nor one HXK-like (AtHKL1), displayed dual-targeting ability, with four orthologs targeted to mitochondria and one targeted to plastids (Fig. 8C). Phylogenetic analysis of the dual-targeted HXK isoforms from P. patens reveals that five out of the six isoforms branch together (Fig. 8A). This suggests that a single gene in P. patens encoding a dual-targeted HXK underwent gene duplication, as all proteins in this group display dual-targeting ability (Fig. 8D). On the other hand, PpHKK5 also displays dual-targeting ability (Fig. 8D), most likely acquired throughout evolution, as its closest orthologs (PpHXK1 and PpHXK6; Fig. 8A) are not dual targeted (Fig. 8D). Because P. patens HXKs are more similar to each other than to HXKs in other land plants and because they have “undergone concerted evolution” (Nilsson et al., 2011), this finding is consistent with the proposal that dual targeting arose after P. patens diverged from the lineage that gave rise to other land plants.
Figure 8.
Dual targeting of HXK to mitochondria and plastids. A, Phylogenetic analysis of genes encoding HXK from Arabidopsis (At), Rice (Os), P. glauca (Pg), P. patens (Pp), C. reinhardtii (Cr), and C. variabilis (Cv) using MEGA 5 (see “Materials and Methods”). B, Table of the HXK proteins from Arabidopsis, rice, P. glauca, P. patens, C. reinhardtii, and C. variabilis with genomic loci numbers, targeting predictions, and experimental localization based on GFP tagging. C, GFP tagging of several HXK proteins from Arabidopsis showed that no dual targeting was evident for any HXK protein in Arabidopsis. D and E, GFP tagging of several HXK proteins from P. patens revealed that PpHXK2, PpHXK3, PpHXK5, PpHXK7, PpHXK9, and PpHXK11 displayed dual-targeting ability. Mitochondria (M) and plastids (Pl) are indicated respectively. Scale bar indicates 20 µm. Cyto, Cytosol; ER, endoplasmic reticulum; EX, extracellular; S, secretory pathway.
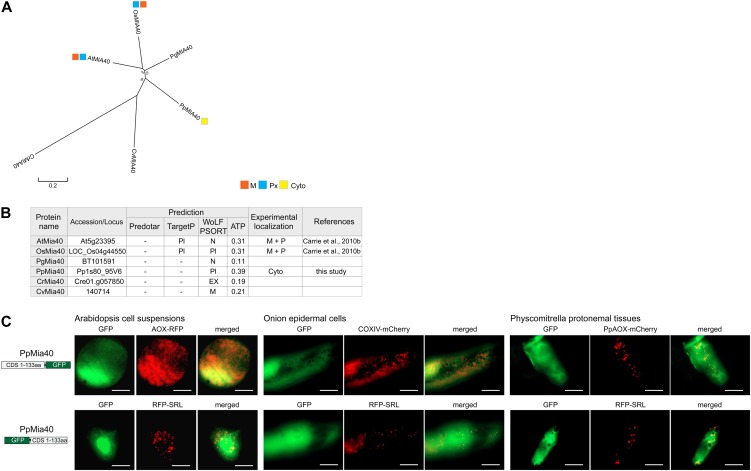
Figure 9.
Targeting ability of Mia40 in P. patens. A, Phylogenetic analysis of genes encoding Mia40 in Arabidopsis (At), rice (Os), P. glauca (Pg), P. patens (Pp), C. reinhardtii (Cr), and C. variabilis (Cv) using MEGA 5 (see “Materials and Methods”). B, Table of the Mia40 proteins from Arabidopsis, rice, P. patens, P. glauca, C. reinhardtii, and C. variabilis with genomic loci numbers, targeting prediction, and experimental localization based on GFP tagging. C, GFP tagging of P. patens Mia40, displaying cytosolic targeting in all tested tissues. Scale bar indicates 20 µm. M, Mitochondria; Px, peroxisomes; Pl, plastids; Cyto, cytosol; EX, extracellular; N, nuclear; Cyto, cytosol.
Acquisition of Dual-Targeting Ability May Allow Organelles to Gain Additional Functions
In the case of peroxisomes, Mitochondrial intermembrane space import and assembly protein40 (Mia40) represents an example of a protein that has acquired dual-targeting ability during land plant evolution. Mia40 was first identified in Arabidopsis as an ortholog to the essential yeast (Saccharomyces cerevisiae) protein Mia40 (Carrie et al., 2010b). However, subsequent work on the Arabidopsis Mia40 showed that it differed from the yeast protein. Firstly, the Arabidopsis Mia40 was shown to be a nonessential protein and was dual targeted to both the mitochondrial intermembrane space and the peroxisomal matrix (Carrie et al., 2010b). It was also demonstrated that the rice Mia40 was similarly dual targeted to mitochondria and peroxisomes (Carrie et al., 2010b). Further analyses of the protein sequences from other plant Mia40 proteins showed that in higher plants, Mia40 contains a known peroxisomal targeting sequence (peroxisomal targeting signal type1 [PTS1]), while Mia40 from lower plant species does not contain this PTS1 sequence (Supplemental Fig. S3). Thus, the P. patens Mia40 (PpMia40) was selected for subcellular localization assay. GFP analysis of PpMia40 revealed that it did not target to either mitochondria or peroxisomes and instead appeared to reside in the cytosol, as evidenced by a diffuse GFP signal in all tested tissue types (Fig. 9C). While the lack of peroxisomal targeting is expected due to the lack of a PTS1 sequence at the C terminus, the lack of mitochondrial targeting is surprising considering that all other known Mia40 proteins have been shown to be located within mitochondria. In addition, the PpMia40 sequence was observed to contain the conserved Cys residues and is highly similar to the Arabidopsis Mia40 in the enzymatically active domain. PpMia40 is missing 28 amino acids from the N-terminal end when compared with Arabidopsis Mia40 (Supplemental Fig. S3). In an attempt to deduce the targeting ability of Mia40 over a wider range of plants, Mia40 sequences from 11 plant species, ranging from C. reinhardtii to Arabidopsis, were examined for predicted targeting ability (Supplemental Fig. S3; Fig. 10), and also the proposed substrates for Mia40, copper chaperone for superoxide dismutase1 (Ccs1), and copper zinc superoxide dismutase1 (CSD1), CSD2, and CSD3 (Fig. 10). It was observed that mitochondrial targeting of Mia40, as well as dual targeting of Mia40, arose relatively late in plant evolution (Fig. 10). It was notable that P. patens, Volvox carteri, C. variabilis, and, to a lesser extent, C. reinhardtii lacked the first 30 to 40 amino acids compared with the predicted mitochondrial targeted Mia40 from other plants (Supplemental Fig. S3). However, the peroxisomal acquisition of Mia40 is also accompanied by the peroxisomal targeting ability of Ccs1 and CSD3. Note that as previously demonstrated, Essential for respiration and vegetative growth1 can carry out the oxidative folding of proteins in mitochondria alone (Carrie et al., 2010b). It is therefore likely that Ccs1- and CSD-like proteins can be assembled in mitochondria without a mitochondrial Mia40 in P. patens and P. glauca (Fig. 10).
Figure 10.
The evolution of targeting of Mia40 and putative substrate proteins. In plants such as Arabidopsis, G. max, and P. trichocarpa, Mia40 is dual targeted to mitochondria and peroxisomes. This dual targeting is also accompanied by the targeting of Mia40 substrates to the same organelles. However, in plants such as C. reinhardtii, V. carteri, C. variabillis, P. patens, and P. glauca, Mia40 is not predicted to be targeted to any organelle and was confirmed for P. patens in the current study (Fig. 9). This finding is accompanied by either the absence of substrate proteins or the absence of targeting of substrate proteins to peroxisomes. Monocot species such as rice, Brachypodium distachyon, and maize appear to be an intermediate, as they contain a dual-targeted Mia40 but lack substrate proteins in the peroxisome. This suggests how dual targeting of one protein may facilitate the acquisition of whole metabolic pathways between different organelles. See Supplemental Figure S3 for sequence alignment of Mia40 from various plants and the predicted targeting signals highlighted.
DISCUSSION
This study used GFP tagging of proteins from several land plants to determine when dual targeting of proteins arose in land plant evolution and if it was conserved. While it is desirable to use a variety of approaches to determine the location of a protein (Millar et al., 2009), for the analysis of 96 proteins from Arabidopsis, rice, and P. patens, and three from P. glauca, GFP tagging is the only realistic approach to determine targeting ability. The use of various other approaches to determine the presence of a protein, either by immunodetection or mass spectrometry, was not feasible due to large gene families, thus the requirement of isoform-specific antibodies. Mass spectrometric approaches are only feasible for highly purified organelles and may not identify proteins that are relatively low in abundance. Even in Arabidopsis, the most intensively studied plant in subcellular proteomics, only 13 of the 72 dual-targeted proteins that have been determined by GFP tagging were also detected in two organelles via proteomic studies (Heazlewood et al., 2007). The in vivo targeting assay using GFP tagging offers a realistic approach to assess targeting because, if observed, targeting to mitochondria, plastids, peroxisomes, or a single organelle indicates that the protein is in an import-competent state.
It was observed that in many cases, dual targeting was conserved. If dual targeting ability arose early in evolution, it remained conserved from P. patens to Arabidopsis and rice, as 11 out of 16 tested proteins were confirmed to be dual targeted in all three species (Supplemental Tables S1 and S2, shaded in green). Similarly, if dual targeting arose later in plant evolution, it remained conserved, with two out of five tested proteins confirmed to be dual targeted from rice and Arabidopsis. Loss of dual targeting could be concluded with confidence, as was observed with APX. Loss of dual targeting in rice was accompanied by gene duplication followed by neofunctionalization, meaning that the duplicated genes encoded proteins that were targeted to single locations. A similar scenario also appears to have occurred with MDHAR isoforms in rice and P. patens. It was also observed that dual-targeting ability was acquired, as seven proteins were dual targeted from Arabidopsis alone. The dual-targeting ability of HXKs from P. patens is likely to be a derived feature rather than it being lost from Arabidopsis.
While dual targeting of proteins was conserved in many cases, we observed differences in the targeting ability of proteins to mitochondria and plastids with the different tissues/cells used (e.g. P. patens DNA polymerase2). This does not appear to be coordinated with differences in the mitochondrial protein import apparatus of P. patens, rice, and Arabidopsis. The outer mitochondrial membrane protein import receptor64, which is derived from an ancestral gene encoding a plastid outer envelope protein, is only present in rice and Arabidopsis (Chew et al., 2004; Carrie et al., 2010a), yet dual targeting occurs in P. patens. The observed differences may be due to a variety of reasons: first, there are isoforms of the protein import components present in plastids and mitochondria (Soll and Schleiff, 2004; Lister et al., 2007; Jarvis, 2008), and for plastids it has been proposed that these different isoforms may import different sets of proteins (Jarvis, 2008). Thus, the difference in import between systems may reflect the different abundance of various isoforms in various cells and/or the fact that there is coevolution or specialization of precursor proteins to bind to specific isoforms of protein import components. An analysis of the Translocase of the outer membrane20 import family of proteins in Arabidopsis suggested that different Translocase of the outer membrane20 isoforms may display some preference or differences for binding different precursor proteins (Lister et al., 2007; Duncan et al., 2013). Furthermore, although targeting signals are generally considered to be well conserved across wide phylogenetic gaps, an analysis of mitochondrial targeting signals from rice and Arabidopsis revealed differences in length and amino acid composition. This finding suggests that subtle differences between species may affect the efficiency of targeting in difference cell types (Huang et al., 2009). Another reason for the differences between various tested cells is that even within a single species the extent of dual targeting varies in cells from different tissues (Carrie et al., 2009b). Finally, it has previously been reported that while some proteins are dual targeted, targeting to a single organelle is only observed in a single cell (Beardslee et al., 2002). The reasons for this, however, are unknown. Therefore, the observed variation in dual targeting between systems with some proteins may be due to a range of factors.
Analysis of the targeting signals of dual-targeted proteins does not reveal any specific motifs or residues that are associated with dual targeting. In this study, protein isoforms that display very high levels of sequence similarity differed in dual-targeting ability. In the case of rice MDHAR, one splicing isoform, OsMDAHR1.1, was dual targeted (Fig. 7), yet another isoform with only four different amino acids, OsMDHAR1.2, was not dual targeted (Fig. 7; Supplemental Fig. S4). With PpMDHAR1 or PpMDHAR2, four amino acid differences in the predicted targeting region result in PpMDAHR1 not being targeted to mitochondria or plastids, and result in PpMDAHR2 being dual targeted (Fig. 7; Supplemental Fig. S5). Overall, it appears that small changes in protein sequences result in large differences in targeting; it is likely not an all-or-nothing situation. The threshold of GFP detection may result in a more dramatic difference that occurs in vivo, as observed with DNA helicases and DNA polymerase where the ability to detect dual targeting differed between cell types (Figs. 3 and 4).
One of the incentives for this study was to gain a better understanding of the purpose of dual targeting. As outlined above, dual targeting appears to be conserved once it arises, and is therefore maintained under positive selection. However, the functional characterization of several dual-targeted proteins has demonstrated that the effects of the inactivation of genes encoding dual-targeted proteins are only observed in a single organelle. For example, inactivation of a dual-targeted RNA polymerase targeted to mitochondria and plastids only resulted in changes to mitochondrial transcript abundance (Kühn et al., 2009). For MutS HOMOLOG1, a dual-targeted protein that maintains genome stability in plastids and mitochondria (Xu et al., 2011), suppression of its protein abundance resulted in responses that were plastid in origin (Xu et al., 2012). In the case of the dual-targeted mitochondrial carrier protein Arabidopsis Brittle1 (AtBT1), which has been shown to be additionally targeted to plastids (Bahaji et al., 2011b), complementation of a transfer DNA mutant using a mitochondrial specific form of AtBT1 resulted in the restoration of a normal growth phenotype (Bahaji et al., 2011a). These studies indicate that dual-targeted proteins have a predominantly central role in a single organelle; however, the current study indicates that dual targeting is conserved, which suggests that dual-targeted proteins may function in both organelles and be under a certain form of positive selection. These apparently conflicting conclusions may be reconciled by the fact that while other location-specific isoforms can restore the normal phenotype under a given set of conditions, this does not indicate that the underlying complex molecular interactions (transcriptome, proteome, metabolome) are restored to normal. Consequently, these hemicomplemented plants provide a valuable resource to explore more cellular or condition-specific functions of dual-targeted proteins, which drive the conservation of dual targeting.
The evolutionary history of the targeting of Mia40 makes for an interesting case. Mia40 is believed to be found in most eukaryotic species but is best studied in yeast model systems (Chacinska et al., 2004). In yeast, Mia40 is located within the mitochondrial intermembrane space, where it plays an essential role in the oxidation and folding of intermembrane space proteins (Chacinska et al., 2004). It was originally believed that this was the role of Mia40 in all eukaryotic species. However, in Arabidopsis it was demonstrated that a mia40 knockout is not lethal and that it was also targeted to peroxisomes (Carrie et al., 2010b). The role for Arabidopsis Mia40 is believed to involve the oxidation and folding of both the mitochondrial and peroxisomal-located Ccs1, CSD1, and CSD3 (Carrie et al., 2010b). It is interesting to note that not all plant Mia40 proteins are targeted to peroxisomes. It appears that the dual targeting of Mia40 has arisen later in plant evolution. The interesting point is that a dual-targeted Mia40 is accompanied by Ccs1 and CSD proteins with a clear PTS1 sequence in Arabidopsis (Fig. 10; Supplemental Fig. S3). Therefore, in lower plant species such as P. patens and P. glauca, Mia40 is not dual targeted, and their peroxisomes are not predicted to contain Ccs1 or CSD proteins (Supplemental Fig. S3; Fig. 10). In higher plants such as Arabidopsis and Populus trichocarpa, not only does Mia40 contain a PTS1 sequence, Ccs1 and CSD proteins do as well. Rice and other monocots appear to be an intermediate of the above case, as rice has a dual-targeted Mia40 but no predicted peroxisomal-targeted Ccs1 or CSD proteins (Fig. 10). This suggests that, over time, biochemical pathways involving several proteins may become dual targeted.
It appears that as observed in Arabidopsis, dual targeting of proteins is widespread in various land plant lineages. Overall, it appears that dual targeting is also well conserved; however, with gene duplication and amplification, dual targeting may be lost with the resulting individual genes encoding proteins that are location specific. Because dual targeting of proteins appears well conserved and because acquisition of dual targeting may result in additional biochemical pathways within organelles, as observed with Mia40, at least one role for dual targeting is the development of metabolic complexity and functional diversification. In other words, the same protein targeted to two locations can result in two different functions without the introduction of any change in the genome per se. Once this occurs and these pathways are established, gene duplication followed by neofunctionalization may result in proteins that are targeted to only one organelle. Phylogenetic analysis of the plastid and mitochondrial proteome suggests that they are composed of proteins that arose from a variety of evolutionary sources (Gray et al., 2001; Martin, 2010; Suzuki and Miyagishima, 2010; Szklarczyk and Huynen, 2010). In fact, proteins originating from the endosymbionts that gave rise to mitochondria and plastids do not represent the majority of proteins in these organelles. Thus, dual targeting may represent an ancient means to develop metabolic/proteomic complexity in plant cells, a process that is still ongoing.
MATERIALS AND METHODS
Bioinformatic Analyses
The tree showing the evolutionary relationship between the six plant species used in this study was determined according to their time of divergence (in millions of years) as reported by previous studies (Bowman et al., 2007; Rensing et al., 2008; Carrie et al., 2010b).
Protein sequences of all published Arabidopsis (Arabidopsis thaliana) dual-targeted proteins (Supplemental Tables S1 and S2) were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org) and orthologs identified using BLASTp (Altschul et al., 1990) against rice (Oryza sativa), Physcomitrella patens, and Chlamydomonas reinhardtii protein databases using Phytozome (http://www.phytozome.net). Orthologs from Picea glauca were identified using tBLASTn (Altschul et al., 1990) against EST sequences from the National Center for Biotechnology Information database (http://blast.icbi.nlm.nih.gov/blast.cgi). BLASTp searches for the Chlorella variabilis NC64 genome (Blanc et al., 2010) were done at http://genome.jgi-psf.org/ChlNC64A_1/ChlNC64A_1.home.html. Candidate proteins with a similarity percentage above 50% and containing the same functional domains were included in this study. The ortholog with the highest percentage similarity and identity to the known Arabidopsis dual-targeted protein was selected for GFP targeting (Supplemental Data Set S1, indicated in red). When the ortholog did not exhibit dual-targeting ability, the ortholog demonstrating the next-highest percentage similarity and identity was chosen. Subcellular localizations were predicted using Predotar (Small et al., 2004), TargetP (Emanuelsson et al., 2007), and WoLF PSORT (Horton et al., 2007). The Ambiguous Targeting Predictor was also used to predict the probability of a protein being dual targeted (Mitschke et al., 2009). Multiple sequence alignments of protein family members were conducted using MAFFT (Katoh et al., 2005). Multiple Align Show (http://www.bioinformatics.org/sms/multi_align.html) was used to visualize multiple sequence alignment. MEGA 5 (Tamura et al., 2011) was used to construct phylogenetic trees using the statistical method of neighboring-joining with 1,000 bootstrap replications. MatGAT 2 (http://bitincka.com/ledion/matgat) was used to determine the percentage identity and similarity matrices.
Construction of GFP Fusion Vectors
RNA extraction from Arabidopsis, rice, and P. patens was carried out using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. P. glauca RNA was obtained from Dr. Olivier Keech (Umeå Plant Science Center). Reverse transcription was carried out using the SuperScript III First-Strand Synthesis System (Invitrogen). The translational start sites for all P. glauca genes were confirmed by 5′ RACE using CapFishing full-length complementary DNA (cDNA) premix kit (Seegene). Full-length cDNA was amplified using gene-specific primers flanked by Gateway recombination cassettes (see Supplemental Table S3) according to the manufacturer’s instructions. A number of genes (see Supplemental Table S3) were cloned directly from cDNA clones obtained from the Knowledge-based Oryza Molecular biological Encyclopedia (Kikuchi et al., 2003) or the RIKEN BioResource Center (Nishiyama et al., 2003). The targeting signals of Glycine max AOX, Pisum sativum small subunit of Rubisco, and Cucurbita spp. malate synthase were fused to RFP and used as mitochondrial, plastid, and peroxisomal markers, respectively (Carrie et al., 2007, 2009b). Cytochrome oxidase IV-mCherry (Nelson et al., 2007) was used as the mitochondrial marker in onion (Allium cepa) epidermal cells. P. patens AOX-mCherry was generated by replacing cytochrome oxidase IV targeting signal (1–29 amino acids) with full-length P. patens AOX (1–365 amino acids) in front of mCherry fluorescence protein, and used as the mitochondrial marker in P. patens protonemal tissues.
Biolistic Transformation and Microscopy
Biolistic cotransformation of the GFP and RFP fusion vectors was performed on Arabidopsis cell suspensions and onion epidermal cells, as previously described (Carrie et al., 2009b). Briefly, 5 µg of GFP and RFP/mCherry plasmids were coprecipitated onto gold particles and bombarded onto 4-d-old Arabidopsis cell suspensions and freshly peeled onion epidermal cells, using the PDS-1000/He biolistic transformation system (Bio-Rad). For putative P. patens proteins, transformation was also performed on 7-d-old protonemal tissues. Following incubation for 12 to 24 h at 22°C (25°C for P. patens) in the dark, GFP and RFP/mCherry expression was visualized at 100× magnification using a BX61 Olympus microscope (Olympus) with excitation wavelengths of 460/480 nm (GFP) and 535/555 nm (RFP/mCherry), and emission wavelengths of 495 to 540 nm (GFP) and 570 to 625 nm (RFP/mCherry). Images were captured using Cell imaging software (Olympus), as previously described (Carrie et al., 2009b).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtTopIA1 (At4g31210), AtGR2 (At3g54660), AtSAPX (At4g08390), AtMDHAR6 (At1g63940), AtHXK1 (At4g29130), AtHXK2 (At2g19860), AtHXK3 (At1g47840), AtHXK4 (At3g20040), and AtHKL1 (At1g50460). Accession numbers for rice and P. patens can be found in Supplemental Tables S1 and S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Targeting ability of orthologs of known Arabidopsis dual-targeted proteins to mitochondria and plastids.
Supplemental Figure S2. Targeting ability of orthologs of known Arabidopsis dual targeted proteins to mitochondria and peroxisomes or plastids and peroxisomes.
Supplemental Figure S3. Sequence alignment of orthologs of Mia40 proteins from different plant species.
Supplemental Figure S4. Sequence alignment of the two isoforms of rice MDHAR1 (LOC_Os08g05570.1 and LOC_Os08g05570.2).
Supplemental Figure S5. Sequence alignment of PpMDHAR1 and PpMDHAR2.
Supplemental Table S1. List of all known dual-targeted proteins to mitochondria and plastids from Arabidopsis and their orthologs in other plants.
Supplemental Table S2. List of all known dual-targeted proteins to mitochondria and peroxisomes, or plastids and peroxisomes, from Arabidopsis and their orthologs in other plants.
Supplemental Table S3. List of primers and cDNA clones used in this study.
Supplemental Data Set S1. Percentage of similarity and identity scores generated by MatGAT for protein sequences in gene families.
Acknowledgments
We would like to thank Dr. Olivier Keech for the kind donation of P. glauca RNA. J.W. and C.C. designed the experiments; L.X., S.R.L., and C.C. performed the experimental procedures; L.X., C.C., and M.W.M. carried out the data analysis; and all authors contributed to the writing of the manuscript.
Glossary
- AOX
alternative oxidase
- RFP
red fluorescent protein
- GR
glutathione reductase
- APX
ascorbate peroxidase
- MDHAR
monodehydroascorbate reductase
- cDNA
complementary DNA
References
- Adams KL, Daley DO, Qiu YL, Whelan J, Palmer JD. (2000) Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408: 354–357 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bahaji A, Muñoz FJ, Ovecka M, Baroja-Fernández E, Montero M, Li J, Hidalgo M, Almagro G, Sesma MT, Ezquer I, et al. (2011a) Specific delivery of AtBT1 to mitochondria complements the aberrant growth and sterility phenotype of homozygous Atbt1 Arabidopsis mutants. Plant J 68: 1115–1121 [DOI] [PubMed] [Google Scholar]
- Bahaji A, Ovecka M, Bárány I, Risueño MC, Muñoz FJ, Baroja-Fernández E, Montero M, Li J, Hidalgo M, Sesma MT, et al. (2011b) Dual targeting to mitochondria and plastids of AtBT1 and ZmBT1, two members of the mitochondrial carrier family. Plant Cell Physiol 52: 597–609 [DOI] [PubMed] [Google Scholar]
- Beardslee TA, Roy-Chowdhury S, Jaiswal P, Buhot L, Lerbs-Mache S, Stern DB, Allison LA. (2002) A nucleus-encoded maize protein with sigma factor activity accumulates in mitochondria and chloroplasts. Plant J 31: 199–209 [DOI] [PubMed] [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et al. (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22: 2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K. (2007) Green genes-comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Carrie C, Giraud E, Duncan O, Xu L, Wang Y, Huang S, Clifton R, Murcha M, Filipovska A, Rackham O, et al. (2010b) Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J Biol Chem 285: 36138–36148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Giraud E, Whelan J. (2009a) Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J 276: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J. (2009b) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, Eubel H, Meyer E, Day DA, Millar AH, Whelan J. (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 582: 3073–3079 [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Millar AH, Smith SM, Whelan J. (2007) Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol Biol 63: 97–108 [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Whelan J. (2010a) An in silico analysis of the mitochondrial protein import apparatus of plants. BMC Plant Biol 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Small I. (2013) A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim Biophys Acta 1833: 253–259 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuán Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N. (2004) Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J 23: 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew O, Lister R, Qbadou S, Heazlewood JL, Soll J, Schleiff E, Millar AH, Whelan J. (2004) A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett 557: 109–114 [DOI] [PubMed] [Google Scholar]
- Chew O, Whelan J. (2004) Just read the message: a model for sorting of proteins between mitochondria and chloroplasts. Trends Plant Sci 9: 318–319 [DOI] [PubMed] [Google Scholar]
- Chew O, Whelan J, Millar AH. (2003) Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278: 46869–46877 [DOI] [PubMed] [Google Scholar]
- Creissen G, Reynolds H, Xue Y, Mullineaux P. (1995) Simultaneous targeting of pea glutathione reductase and of a bacterial fusion protein to chloroplasts and mitochondria in transgenic tobacco. Plant J 8: 167–175 [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313: 314–318 [DOI] [PubMed] [Google Scholar]
- Duncan O, Murcha MW, Whelan J. (2013) Unique components of the plant mitochondrial protein import apparatus. Biochim Biophys Acta 1833: 304–313 [DOI] [PubMed] [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ. (2004) Ancient invasions: from endosymbionts to organelles. Science 304: 253–257 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Glaser E, Whelan J. (2007) Import of nuclear-encoded mitochondrial proteins. In Logan DC, ed, Plant Mitochondria. Blackwell, Oxford, pp 97–128 [Google Scholar]
- Gray MW, Burger G, Lang BF. (2001) The origin and early evolution of mitochondria. Genome Bio 2: reviews1018.1–reviews1018.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Whelan J, Millar AH. (2009) Refining the definition of plant mitochondrial presequences through analysis of sorting signals, N-terminal modifications, and cleavage motifs. Plant Physiol 150: 1272–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. (2010) The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci 365: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al. (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. (2009) DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol 60: 115–138 [DOI] [PubMed] [Google Scholar]
- Kühn K, Richter U, Meyer EH, Delannoy E, de Longevialle AF, O’Toole N, Börner T, Millar AH, Small ID, Whelan J. (2009) Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21: 2762–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Carrie C, Duncan O, Ho LH, Howell KA, Murcha MW, Whelan J. (2007) Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19: 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. (2010) Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans R Soc Lond B Biol Sci 365: 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Carrie C, Pogson B, Whelan J. (2009) Exploring the function-location nexus: using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke J, Fuss J, Blum T, Höglund A, Reski R, Kohlbacher O, Rensing SA. (2009) Prediction of dual protein targeting to plant organelles. New Phytol 183: 224–235 [DOI] [PubMed] [Google Scholar]
- Morgante CV, Rodrigues RA, Marbach PA, Borgonovi CM, Moura DS, Silva-Filho MC. (2009) Conservation of dual-targeted proteins in Arabidopsis and rice points to a similar pattern of gene-family evolution. Mol Genet Genomics 281: 525–538 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nilsson A, Olsson T, Ulfstedt M, Thelander M, Ronne H. (2011) Two novel types of hexokinases in the moss Physcomitrella patens. BMC Plant Biol 11: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Fujita T, Shin IT, Seki M, Nishide H, Uchiyama I, Kamiya A, Carninci P, Hayashizaki Y, Shinozaki K. (2003) Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proc Natl Acad Sci USA 100: 8007–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N, Small I. (2001) Dual targeting to mitochondria and chloroplasts. Biochim Biophys Acta 1541: 54–63 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. (2010) Plastidial retrograde signalling—a true “plastid factor” or just metabolite signatures? Trends Plant Sci 15: 427–435 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O. (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokov-Plavec J, Dulic M, Duchêne AM, Weygand-Durasevic I. (2008) Dual targeting of organellar seryl-tRNA synthetase to maize mitochondria and chloroplasts. Plant Cell Rep 27: 1157–1168 [DOI] [PubMed] [Google Scholar]
- Rudhe C, Chew O, Whelan J, Glaser E. (2002) A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J 30: 213–220 [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. (2004) Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Soll J, Schleiff E. (2004) Protein import into chloroplasts. Nat Rev Mol Cell Biol 5: 198–208 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Miyagishima SY. (2010) Eukaryotic and eubacterial contributions to the establishment of plastid proteome estimated by large-scale phylogenetic analyses. Mol Biol Evol 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Szklarczyk R, Huynen MA. (2010) Mosaic origin of the mitochondrial proteome. Proteomics 10: 4012–4024 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Arrieta-Montiel MP, Virdi KS, de Paula WB, Widhalm JR, Basset GJ, Davila JI, Elthon TE, Elowsky CG, Sato SJ, et al. (2011) MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 23: 3428–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Santamaria RdlR, Virdi KS, Arrieta-Montiel MP, Razvi F, Li S, Ren G, Yu B, Alexander D, Guo L, et al. (2012) The chloroplast triggers developmental reprogramming when MUTS HOMOLOG1 is suppressed in plants. Plant Physiol 159: 710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O, Pines O. (2011) Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim Biophys Acta 1808: 1012–1020 [DOI] [PubMed] [Google Scholar]