Summary: Analysis of all the expressed genes in white lupin roots and leaves shows that acclimation to phosphorous deficiency involves changes in root development and modifications in metabolism.
Abstract
Phosphorus, in its orthophosphate form (Pi), is one of the most limiting macronutrients in soils for plant growth and development. However, the whole-genome molecular mechanisms contributing to plant acclimation to Pi deficiency remain largely unknown. White lupin (Lupinus albus) has evolved unique adaptations for growth in Pi-deficient soils, including the development of cluster roots to increase root surface area. In this study, we utilized RNA-Seq technology to assess global gene expression in white lupin cluster roots, normal roots, and leaves in response to Pi supply. We de novo assembled 277,224,180 Illumina reads from 12 complementary DNA libraries to build what is to our knowledge the first white lupin gene index (LAGI 1.0). This index contains 125,821 unique sequences with an average length of 1,155 bp. Of these sequences, 50,734 were transcriptionally active (reads per kilobase per million reads ≥ 3), representing approximately 7.8% of the white lupin genome, using the predicted genome size of Lupinus angustifolius as a reference. We identified a total of 2,128 sequences differentially expressed in response to Pi deficiency with a 2-fold or greater change and P ≤ 0.05. Twelve sequences were consistently differentially expressed due to Pi deficiency stress in three species, Arabidopsis (Arabidopsis thaliana), potato (Solanum tuberosum), and white lupin, making them ideal candidates to monitor the Pi status of plants. Additionally, classic physiological experiments were coupled with RNA-Seq data to examine the role of cytokinin and gibberellic acid in Pi deficiency-induced cluster root development. This global gene expression analysis provides new insights into the biochemical and molecular mechanisms involved in the acclimation to Pi deficiency.
Phosphorus is an essential macronutrient for all living organisms, including plants. Plants take up phosphorus in orthophosphate (Pi) forms: H2HPO4− and HPO42−. Although the Pi content of soil is generally high, Pi availability is often limited due to slow diffusion and high fixation in soils (Shen et al., 2011), making Pi availability one of the most limiting factors for plant growth and productivity worldwide (Marschner, 1995; Vance et al., 2003; Shen et al., 2011). Mined rock phosphate, a nonrenewable resource, is the primary source of Pi fertilizer. It is predicted that easily accessed global Pi reserves may be depleted in 50 years (Cordell et al., 2009). In addition, the application of Pi fertilizer causes severe environmental problems, such as eutrophication of water systems (Cordell et al., 2009). Thus, a fuller understanding of the strategies used by plants to acquire and efficiently utilize Pi is important for the breeding or engineering of crop plants with greater capacity to acquire, store, and recycle soil Pi (Vance et al., 2003; Cordell et al., 2009; Yang and Finnegan, 2010; Chiou and Lin, 2011; Gaxiola et al., 2011; Lambers et al., 2011).
A variety of adaptive strategies to cope with Pi deficiency have evolved in plants. These include morphological, physiological, biochemical, and molecular responses, such as changes in root and shoot development (Hermans et al., 2006; Lynch, 2011), the optimization of internal Pi use (Plaxton and Tran, 2011), and the expression of secreted acid phosphatases, organic anions, and high-affinity phosphate transporters (TPs; Raghothama, 1999; Vance et al., 2003; Misson et al., 2005; Plaxton and Tran, 2011). Hundreds of plant genes are differentially regulated in response to Pi deficiency, as demonstrated by microarray and EST analyses (Hammond et al., 2003; Hernández et al., 2007; Nilsson et al., 2010). The complex network of regulatory genes necessary to sense and respond to Pi deficiency has only recently been addressed. Regulatory components identified so far include transcription factors (TFs), SPX (for SYG1, Pho81, XPR1) subfamily proteins, plant hormones, noncoding RNAs, and protein modifiers, including proteins involved in SUMOylation, phosphorylation, dephosphorylation, protein translocation, and epigenetic modifications (Nilsson et al., 2010; Smith et al., 2010; Yang and Finnegan, 2010; Cheng et al., 2011a, 2011b; Chiou and Lin, 2011; Hammond and White, 2011; Kuo and Chiou, 2011).
While 80% of plants are capable of establishing symbiotic associations with mycorrhizal fungi that aid Pi acquisition (Burleigh et al., 2002), other effective adaptations to low Pi can be found. One of the most studied is the formation of cluster roots, which are closely spaced tertiary lateral rootlets that resemble bottle brushes (Neumann and Martinoia, 2002; Vance et al., 2003; Shane and Lambers, 2005; Neumann, 2010; Lambers et al., 2011). White lupin (Lupinus albus), which forms cluster roots, has become a model plant for the study of Pi acquisition due to its exceptional ability to acquire nutrients unavailable to most other plants (Neumann and Martinoia, 2002; Vance et al., 2003; Tian et al., 2009; Zhu et al., 2009; Cheng et al., 2011a, 2011b). Cluster roots increase the root surface area for enhanced Pi absorption and exude organic anions (Johnson et al., 1996; Massonneau et al., 2001; Sas et al., 2001; Wang et al., 2007) and acid phosphatases (Gilbert et al., 1999; Miller et al., 2001) that release Pi from sparingly soluble inorganic and organic compounds. In addition, high-affinity phosphate TPs (Liu et al., 2001) help with the acquisition of solubilized Pi. Although other plants form cluster roots, mainly from the family Proteaceae (Lambers et al., 2011), white lupin’s response to Pi deficiency is currently the most thoroughly evaluated (Neumann and Martinoia, 2002; Vance et al., 2003; Zhou et al., 2008; Neumann, 2010; Cheng et al., 2011a, 2011b; Rodriguez-Medina et al., 2011).
Lupin (Lupinus spp.) has proven to be more than a model for Pi deficiency; in Australia, the seeds are used in animal feed, and their high-protein/low-oil content make them ideal for human consumption (Gao et al., 2011). To facilitate crop improvement, genetic maps have been released for both white lupin (Phan et al., 2007) and a narrow-leafed lupin, Lupinus angustifolius (Nelson et al., 2006). Recently, a 12× bacterial artificial chromosome (BAC) library for L. angustifolius was developed, and 13,985 BAC end sequences (BES; representing 1% of the lupin genome) were deposited to the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov; Gao et al., 2011). Gene-coding regions were identified on 2,700 BES (Gao et al., 2011), representing approximately 0.5 Mb of sequence. The BAC library and BES will be useful in assembling the L. angustifolius genome sequence, currently under way.
Traditionally, genomic studies have focused on model systems such as Arabidopsis (Arabidopsis thaliana), Medicago truncatula, and maize (Zea mays). International consortiums and vast federal funding were required to develop genomic tools for these species, which in turn can be used as platforms to study closely related organisms. The emergence of next-generation sequencing has facilitated the identification of transcribed genes in a number of nonmodel species. For many species, next-generation sequencing technology has allowed the development of a transcriptome sequence prior to the sequencing of the genome, as in alfalfa (Medicago sativa; Yang et al., 2011), lentil (Lens culinaris; Kaur et al., 2011), pea (Pisum sativum; Franssen et al., 2011), pigeon pea (Cajanus cajan; Dubey et al., 2011), sweet potato (Ipomoea batatas; Wang et al., 2010b), eucalyptus (Eucalyptus spp.; Mizrachi et al., 2010), carrot (Daucus carota; Iorizzo et al., 2011), watermelon (Citrullis vulgaris; Guo et al., 2011), rubber tree (Hevea brasiliensis; Xia et al., 2011), pitcher plant (Sarracenia spp.; Srivastava et al., 2011), and cucurbits (Cucurbitaceae spp.; Blanca et al., 2011). Beyond identifying and sequencing the transcriptome, the Illumina RNA-Seq platform allows researchers to examine the expression pattern of transcripts in specific tissues of interest. Although often used in species with a sequenced genome and high-quality gene predictions, the development of tools such as ABySS (Birol et al., 2009), SOAPdeNovo (Li et al., 2009), Trinity (Grabherr et al., 2011), and Velvet/Oases (Zerbino and Birney, 2008) allows researchers to assemble de novo transcriptomes from Illumina sequences and study gene expression patterns. This approach has proven successful in a variety of plant species, including chickpea (Cicer arietinum; Garg et al., 2011), sweet potato (Wang et al., 2010b), eucalyptus (Mizrachi et al., 2010), alfalfa (Yang et al., 2011), and now lupin. De novo RNA-Seq experiments are particularly useful in species with complex genomes (such as tetraploids) and outcrossing species where genomic sequencing is considered cost prohibitive (such as alfalfa).
In this study, we present, to our knowledge, the first white lupin gene index (LAGI 1.0) and whole-genome expression profiles of transcripts in plants grown under Pi-sufficient and Pi-deficient conditions. While the predominant focus of our report is on Pi deficiency-induced cluster roots, we also present data on leaves. Utilizing our newly developed gene index and expression patterns, we identified 2,128 genes whose expression is modified due to the Pi status of the plant. We present evidence for novel Pi deficiency-induced metabolism and hormone signaling.
RESULTS
RNA-Seq Using the Illumina GA-IIx Platform and the White Lupin Gene Index 1.0
We developed complementary DNA (cDNA) libraries derived from roots and leaves collected from white lupin plants grown under Pi-sufficient or Pi-deficient conditions: Pi-sufficient roots (PsR), Pi-deficient cluster roots (PdCR), Pi-sufficient leaves (PsL), and Pi-deficient leaves (PdL) collected 16 d after emergence (DAE). Analyses of internal Pi concentrations confirmed that the cotyledon provides ample Pi throughout early stages of plant development and there is no difference in the Pi content of Pi-sufficient- and Pi-deficient-grown plants until at least 10 DAE (Supplemental Fig. S1). A detailed time-course experiment may yield valuable insight to Pi deficiency acclimation; however, that is beyond the scope of this project. At 16 DAE, Pi-deficient plants have been Pi deficient for approximately 6 d and have developed functional cluster roots with all zones (Neumann et al., 2000) present. No visible anthocyanin accumulated in PdL, nor did we observe any other differences in the growth of Pi-sufficient and Pi-deficient aerial tissues. These results are consistent with previous studies, which showed that Pi-sufficient and Pi-deficient plants at 25 DAE exhibit differences in Pi concentrations but show no change in overall growth responses (Schulze et al., 2006). Our analyses comparing Pi-sufficient and Pi-deficient transcript expression patterns at 16 DAE provide insight into gene expression profiles as plants experience Pi deficiency. The experiment was replicated three times (three biological replicates), resulting in a total of 12 cDNA libraries for RNA-Seq analysis. A total of 367,973,656 Illumina EST reads were generated from 12 cDNA libraries. After filtering homopolymers and short reads (less than 76 bp), a total of 277,224,180 reads (76 bp in size) were selected for further analysis. The Illumina reads generated in this study are available at the NCBI Sequence Read Archive browser (http://ncbi.nlm.nih.gov/sra, accession no. GSE31132).
The de novo assembled transcripts were combined with 8,441 publicly available Sanger ESTs to build the first white lupin gene index (LAGI 1.0), which is publicly available on the white lupin species resource page at http://lupal.comparative-legumes.org/#tabs-3. LAGI 1.0 contains a total of 125,821 unique sequences with an average length of 1,155 bp and a GC content of 39.6%. Unique sequence lengths ranged from 100 to 15,514 bp (Supplemental Fig. S2, sequence distribution). The total base count of the sequences in LAGI 1.0 is 145,286,614 bp. The newly built white lupin gene index increases the number of publicly available white lupin sequences by 14-fold. Putative functions for LAGI 1.0 sequences were assigned using BLASTX searches against the Arabidopsis and soybean (Glycine max) proteomes (for details, see “Materials and Methods”; annotation file available at http://lupal.comparative-legumes.org/#tabs-3). Putative functions were assigned for 63% of the sequences. We also assigned Gene Ontology functional classes and MapMan functional classifications (Thimm et al., 2004; annotation file available at http://lupal.comparative-legumes.org/#tabs-3). Annotations for genes and pathways analyzed in greater detail throughout this paper were manually curated to ensure accurate analysis and interpretation.
Transcript profiles of lupin roots and leaves under Pi-sufficient or Pi-deficient conditions were measured. We analyzed the reads per kilobase per million reads (RPKM)-normalized gene expression counts for each sequence in LAGI 1.0 from 11 samples {(Pi sufficient and Pi deficient) × (roots and leaves) × (three biological replicates [except PdL; two biological replicates])}. Previous studies have suggested that expression of transcripts with low RPKM scores may be statistical artifacts (Zhao et al., 2010; Burke and Strand, 2012). Based on the length of the sequenced reads, an RPKM of 3 reflects 2-fold coverage of the sequence, assuming equal distribution of reads across the sequence. Sequences with RPKM < 3 exhibit low transcript abundance under the conditions tested and may not provide reliable expression data. With this in mind, we consider transcripts with RPKM < 3 as silent. Of the 125,821 sequences assembled, 50,734 had RPKM ≥ 3 in at least one library (transcript expression file available at http://lupal.comparative-legumes.org/#tabs-3) and are considered transcriptionally active. A total of 2,128 transcripts are differentially expressed (2-fold change, P ≤ 0.05, RPKM ≥ 3) in response to Pi deficiency (Fig. 1; Supplemental Tables S1 and S2). Additionally, we identified 8,371 transcripts exhibiting tissue-specific expression patterns (RPKM ≥ 3 in a single tissue, RPKM < 3 in all other tissues; Supplemental Table S3). We found 3,042 transcripts uniquely expressed in PdL and 3,086 transcripts uniquely expressed in PdCR.
Figure 1.
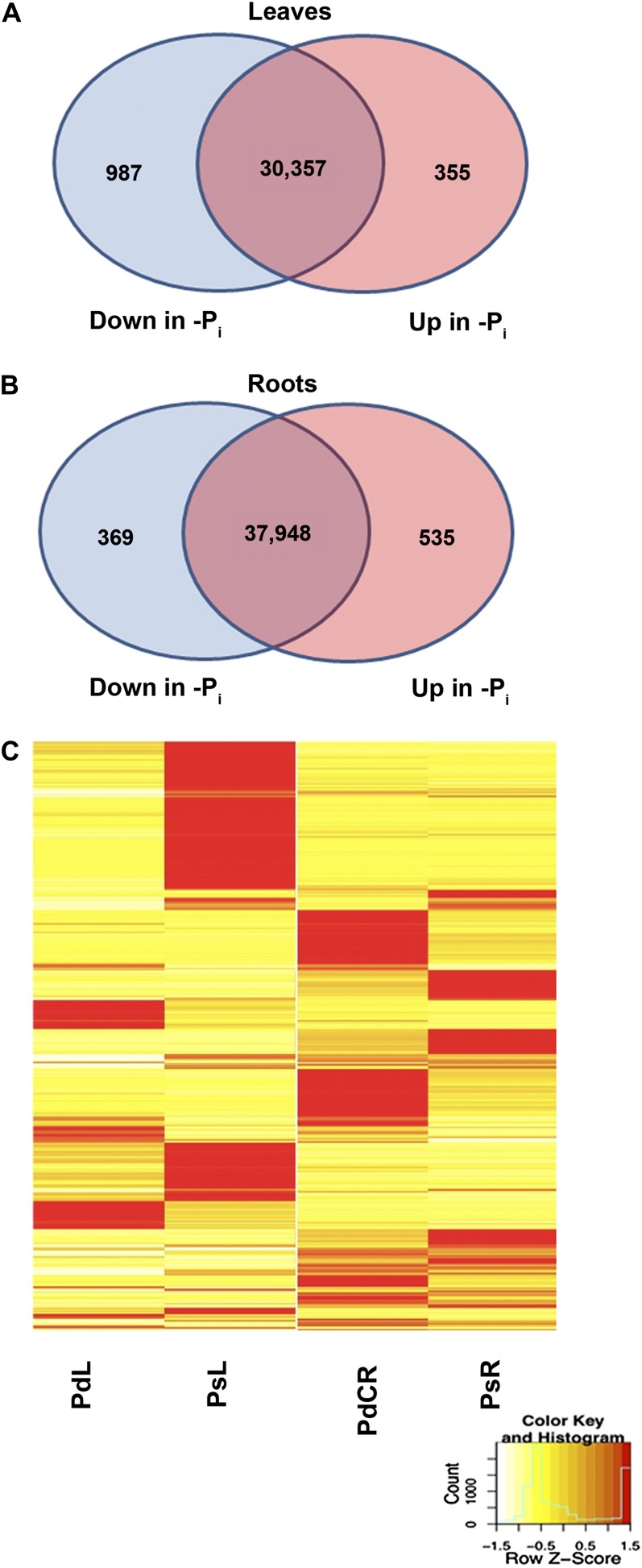
Transcripts differentially expressed due to Pi. A total of 2,128 transcripts were identified as differentially expressed between Pi-sufficient and Pi-deficient tissues. To be considered differentially expressed, the transcript must have RPKM ≥ 3 in at least one tissue, 2-fold or greater change between tissues, and P ≤ 0.05. A, A total of 1,342 transcripts differentially expressed due to Pi in leaves. B, A total of 904 transcripts differentially expressed due to Pi in roots. C, Heat map of Z scores (number of sd) illustrating expression profiles of 2,128 transcripts differentially expressed due to Pi. Red indicates high expression, yellow indicates intermediate expression, and white indicates low expression. See also Supplemental Tables S1 and S2.
The 1,000 most highly expressed transcripts in leaves and roots (regardless of Pi status) were also identified (Supplemental Table S4). In leaves, the 1,000 most highly expressed transcripts are involved in processes including photosynthesis, tetrapyrrole synthesis, glycolysis, amino acid metabolism, protein, and chloroplast synthesis. Other transcripts highly abundant in leaves are involved in lipid metabolism, carbohydrate catabolism, flavonoid biosynthesis, and protein transport. In roots, the 1,000 most highly expressed transcripts are involved in carbohydrate metabolism, cell wall modification, lignin biosynthesis, sugar/nutrient signaling, and transport. Other highly expressed transcripts in roots are also involved in the cytosolic branch of glycolysis, the tricarboxylic acid cycle, amino acid synthesis, and a modified glyoxylate-formate pathway (Fig. 2). Lastly, transcripts involved in hormone homeostasis are highly expressed in both root and leaf tissues.
Figure 2.
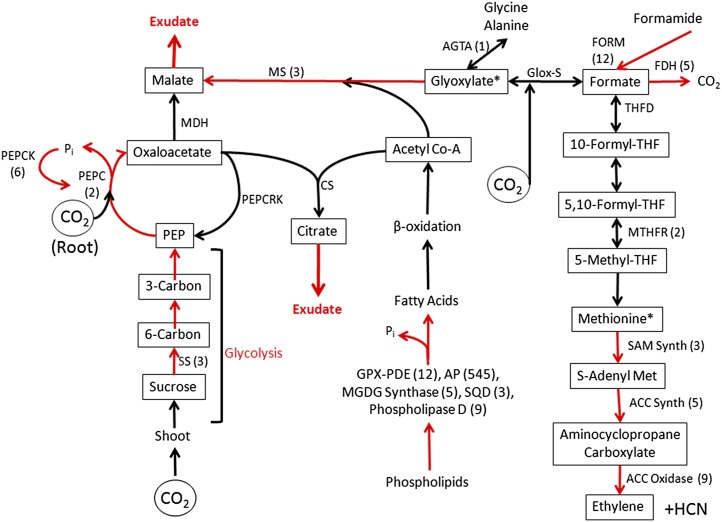
Adjustments in root metabolism promote acclimation to Pi deficiency. Shown are modifications in white lupin cluster root metabolism that facilitate acclimation to Pi deficiency as evidenced by transcript expression. Increased expression in PdCR was confirmed by qPCR as indicated by fold change in parentheses. Red arrows represent genes known to be up-regulated due to Pi deficiency. Increased Suc metabolism via glycolysis and organic acid production provide carbon for malate and citrate exudation into the rhizosphere. Organic acids lost through exudation are replenished through anaplerotic pathways involving phosphoenolpyruvate carboxylase (PEPC) and a glyoxylate-like cycle malate synthase (MS). One-carbon metabolism is enhanced through a formate and THF pathway. The THF pathway contributes to Met and ethylene production. Increased expression of transcripts involved in phospholipid degradation releases Pi for recycling and carbon for acetyl-CoA synthesis. Acetyl-CoA and glyoxylate provide carbon for malate synthesis through malate synthase. Formate may be carboxylated to glyoxylate by a putative glyoxylate synthase (Glox-S). Additional abbreviations are as follows: Suc synthase (SS); phosphoenolpyruvate carboxylase kinase (PEPCK); phosphoenolpyruvate carboxylase carboxykinase (PEPCRK); malate dehydrogenase (MDH); citrate synthase (CS); formamidase (FORM); formate dehydrogenase (FDH); Ala glyoxylate transaminase (AGTA); THF deformylase (THFD); methylene-THF reductase (MTHFR); S-adenosyl-Met synthase (SAM Synth); aminocyclopropane synthase (ACC Synth); aminocyclopropane oxidase (ACC Oxidase); phospholipase A (PLA1); glycerophosphodiester-phosphodiesterase (GPX-PDE); acid phosphatase (AP); monogalactosyldiacylglycerol synthase (MGDG synthase); sulfoquinovosyltransferase (SQD).
We identified putative housekeeping genes that showed little variation in expression but were expressed at relatively high levels. To identify housekeeping genes, we first selected genes with an average RPKM-normalized transcript count greater than 10. Next, we selected the top 10% of genes (1,250) with the lowest coefficient of variation (sd/mean; Supplemental Table S5; Severin et al., 2010). These housekeeping genes may be useful as reference genes in quantitative PCR (qPCR) or other experiments to normalize gene expression levels across different conditions (Czechowski et al., 2005).
Differential Expression of Transcripts Due to Pi
A total of 2,128 transcripts are differentially expressed in response to Pi deficiency: 1,342 and 904 genes in PdL versus PsL and PdCR versus PsR comparisons, respectively (Fig. 1; Supplemental Tables S1 and S2). In the 1,342 differentially expressed transcripts identified in leaves, 987 transcripts were up-regulated in PsL while 355 were up-regulated in PdL. Of the 904 transcripts differentially expressed between PdCR and PsR, 396 were up-regulated in PsR and 535 were up-regulated in PdCR.
The uptake of Pi from soil and the formation of cluster roots in response to Pi deficiency results in the altered expression of transcripts encoding TPs and TFs. Of the 2,128 transcripts differentially expressed in response to Pi deficiency, 110 were identified as TPs and 155 as TFs (Tables I and II; Supplemental Tables S6 and S7). Both TFs and TPs differentially expressed due to Pi deficiency exhibit distinct expression patterns between leaves and roots, with no overlapping differential expression. We identified nine Pi TPs differentially expressed in response to Pi deficiency (Table I; Supplemental Table S6); eight of them are differentially expressed in roots, while one is differentially expressed in leaves. In addition, the majority of potassium TPs differentially expressed in response to Pi deficiency are expressed in roots. Our RNA-Seq data revealed eight multidrug and toxin efflux (MATE) transcripts differentially expressed due to Pi deficiency: four in leaves and four in roots (Table I; Supplemental Table S6). The four MATE transcripts differentially expressed in leaves exhibited reduced expression under Pi deficiency stress, while expression of the MATE transcripts in roots was induced in PdCR, as seen in previous experiments. Root TPs were highly responsive to the Pi status of the plant.
Table I. TPs differentially expressed in roots and leaves due to Pi deficiency.
TPs with altered expression patterns between Pi-sufficient and Pi-deficient roots and leaves (PdCR versus PsR and PdL versus PsL) were used. The numbers of individual transcripts annotated as a member of each TP family exhibiting increased or decreased expression (2-fold or greater change) in Pi-deficient tissues compared with Pi-sufficient tissues are shown. ABC, ATP-binding cassette; PIP, plasma membrane intrinsic protein; TIP, tonoplast intrinsic protein.
|
PdCR |
PdL |
|||
|---|---|---|---|---|
| TP Family | Increase | Decrease | Increase | Decrease |
| ABC | 4 | 2 | ||
| Amino acid | 4 | 1 | ||
| ATP/ADP antiporter | 1 | 1 | ||
| Calcium | 3 | 4 | ||
| Divalent cations | 5 | 4 | ||
| Lipid transfer | 1 | 5 | 1 | |
| MATE | 4 | 4 | ||
| Miscellaneous | 1 | |||
| Nucleotide | 3 | |||
| Membrane H+-ATPases | 8 | |||
| Phosphate | 8 | 1 | ||
| PIP | 2 | 7 | ||
| Porins | 3 | |||
| Potassium | 5 | 1 | ||
| Sugar | 8 | 3 | ||
| Sulfate | 6 | |||
| TIP | 1 | |||
| Unspecified | 5 | 6 | ||
Table II. TFs differentially expressed in roots and leaves due to Pi deficiency.
TFs with altered expression patterns between Pi-sufficient and Pi-deficient roots and leaves (PdCR versus PsR and PdL versus PsL) were used. The numbers of individual transcripts within a TF family exhibiting increased or decreased expression (2-fold or greater change) in Pi-deficient tissues compared with Pi-sufficient tissues are shown.
Additionally, some 23 TF families show differential expression due to Pi deficiency stress (Table II; Supplemental Table S7). The bHLH and AP2_EREB TF families are the two largest families responding to Pi deficiency stress identified in the root tissues. We identified 14 members of the bHLH TF family exhibiting increased expression in PdCR. The 17 transcripts in the AP2_EREB TF family exhibit decreased expression in PdCR compared with PsR (Table II; Supplemental Table S7). In the leaves, the MYB TF family, with 33 transcripts showing increased expression in PdL, is the largest TF family represented. Members of these TF families have been identified in the Pi deficiency response of Arabidopsis and rice (Rubio et al., 2001; Yi et al., 2005; Chen et al., 2007; Devaiah et al., 2007, 2009; Hernández et al., 2007).
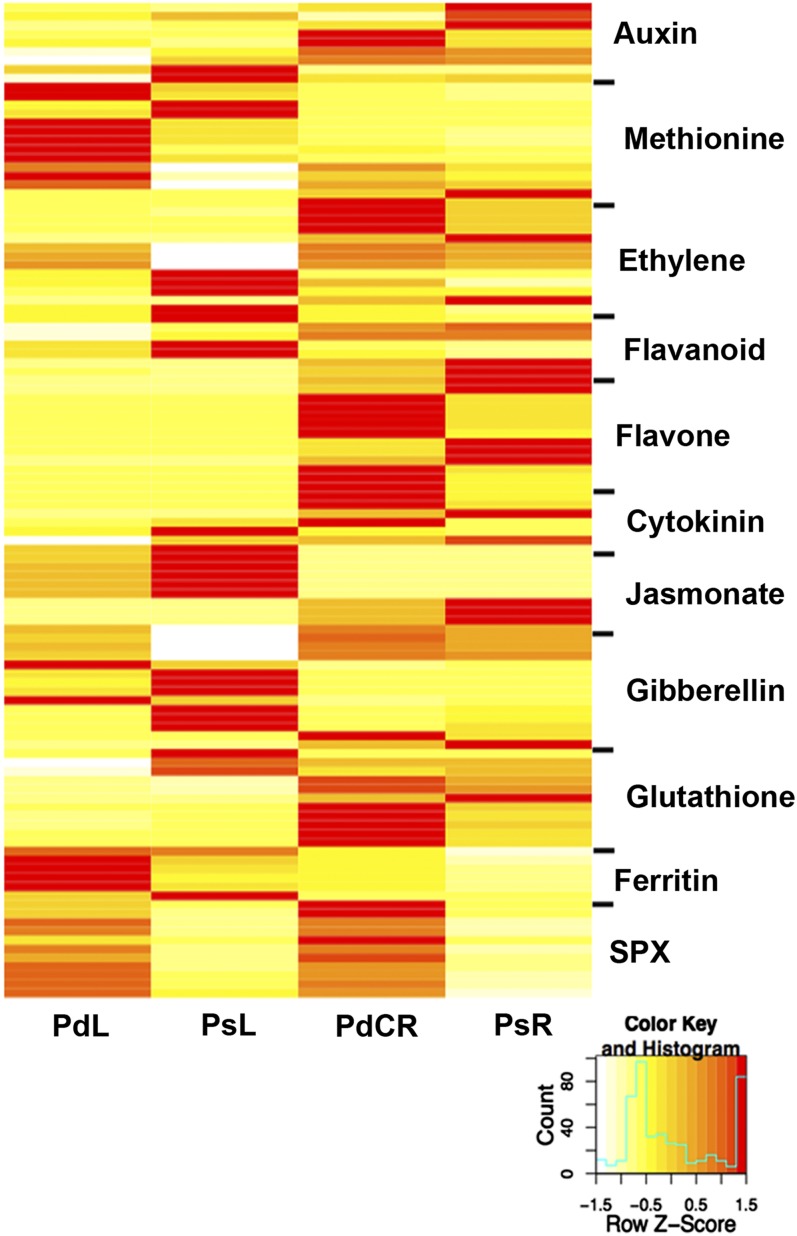
As noted in previous studies, abiotic stress, including Pi deficiency, frequently results in the generation of harmful reactive oxygen species (ROS; Misson et al., 2005). To further understand how Pi-deficient white lupin plants acclimate to Pi deficiency, we evaluated our transcriptome data for differential expression of glutathione S-transferase (GST), glutathione peroxidase, lactoyl-glutathione lyase (glyoxylase), ferritins, and NADPH-oxidase transcripts (Fig. 3; Supplemental Table S8). As compared with PsR and PsL, PdCR showed increased abundance of transcripts encoding GST, glutathione peroxidase, and glyoxylase. Interestingly, PdL had extremely high expression of ferritin and ascorbate peroxidase transcripts compared with other treatments and tissues. We did not detect any NADPH oxidases that showed increased transcript abundance based upon Pi status.
Figure 3.
Heat map of expression profiles of transcripts involved in acclimation to Pi deficiency. Expression, represented by Z scores, is shown for selected transcripts differentially expressed due to Pi deficiency. To be considered differentially expressed, the transcript must have RPKM ≥ 3 in at least one tissue, 2-fold or greater change between tissues, and P ≤ 0.05. Red indicates high expression, yellow indicates intermediate expression, and white indicates low expression. Transcripts have been grouped by function to clarify the role of each group in the Pi deficiency response. For more details, see Supplemental Table S8.
Anthocyanin formation frequently accompanies and is a phenotypic marker for Pi deficiency (Sánchez-Calderón et al., 2006). The lack of visually detectable anthocyanin accumulation in the leaves of our Pi-deficient plants likely reflects the early (6 d of Pi-deficient growth conditions) Pi deficiency status of our plants. With this in mind, we queried the database for differentially expressed transcripts related to flavonoids and anthocyanins, including isoflavonoid mevalonyl transferase, anthocyanin transferase, chalcone reductase, flavonoid monooxygenase, and isoflavone synthesis (Fig. 3; Supplemental Table S8). Unexpectedly, transcripts for the flavonoid genes show enhanced expression in PsR and PsL rather than in Pi-deficient plants. Flavone-related transcripts exhibiting differential expression were found only in roots: three transcripts up-regulated in PdCR and three transcripts up-regulated in PsR.
We identified 82 SPX domain-containing transcripts in the LAGI 1.0 assembly (Supplemental Table S9). Of these, 11 show greater than 2-fold increased expression under Pi-deficient growth conditions (all 11 were induced in PdCR, and three of the 11 transcripts were also induced in PdL; Fig. 3; Supplemental Table S8). Interestingly, SPX domain-containing proteins differentially expressed due to Pi status in lupin all belong to the same family, SPX; no additional domains are present in these proteins.
Under Pi-deficient conditions, more carbon is partitioned to roots relative to shoots (Cakmak et al., 1994; Jiang et al., 2007; Morcuende et al., 2007). We as well as others have shown that carbon metabolism is altered in Pi-deficient plants (Johnson et al., 1994; Hernández et al., 2007; Müller et al., 2007). In particular, carbon metabolism in roots is directed toward organic acid synthesis accompanied by the exudation of malate and citrate into the rhizosphere (Johnson et al., 1994; Neumann et al., 2000; Massonneau et al., 2001). Our RNA-Seq data are consistent with previous reports showing transcripts involved in phospholipid degradation increase, which would contribute to an increased acetyl-CoA pool for malate and citrate synthase (Fig. 2). Moreover, upon further analysis, we noted that PdCR had increased abundance of transcripts related to formate, folate, glyoxylate, Met, and ethylene (Fig. 2). Reconstruction of metabolic pathways based upon expression profiles of transcripts expressed in formate, glyoxylate, and Met biosynthesis provides evidence for previously unrecognized Pi stress-induced pathways and reinforces the interpretation that carbon metabolism is modified in Pi-deficient roots and is driven toward malate and citrate. In addition, carbon for organic acids is replenished in part via a one-carbon pathway. Many transcripts in formate and tetrahydrofolate (THF) metabolism (Fig. 2) were more abundant in PdCR- than in either PdL- or Pi-sufficient tissues. Additionally, three primary transcripts within the Yang ethylene cycle are up-regulated in PdCR. The increased expression of transcripts involved in glycolysis, organic acid formation, and ethylene production in PdCR was confirmed with qPCR (Fig. 2).
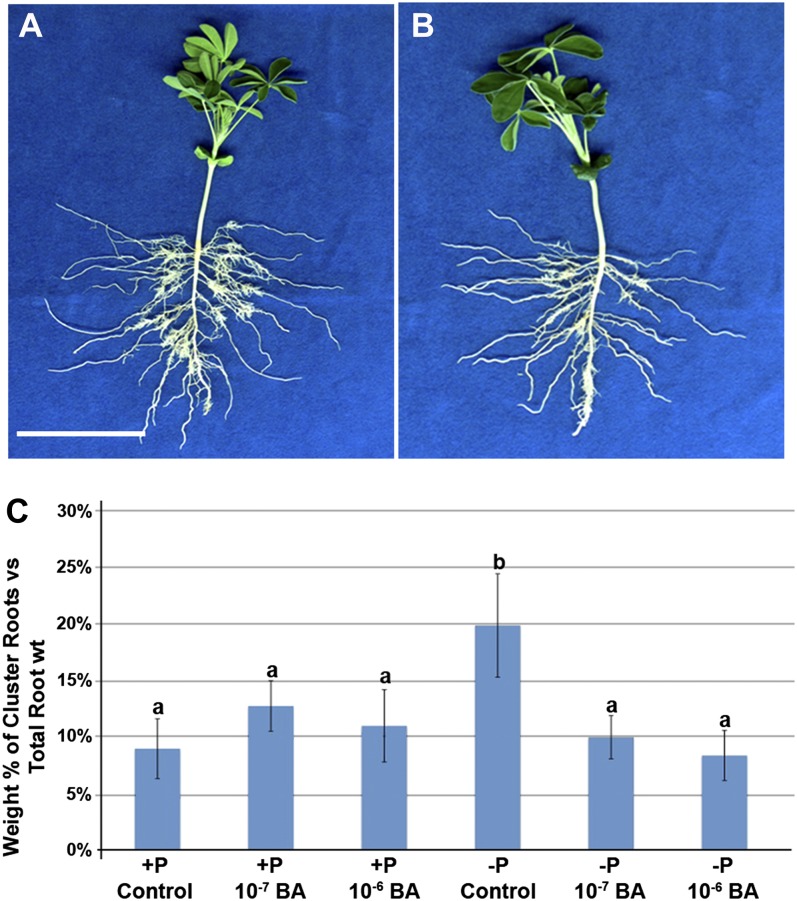
To better understand the role of cytokinin (CK) and GA in cluster root development, we manipulated the availability of these hormones and measured the responses of genes in known pathways using qPCR. In our RNA-Seq data, we identified two transcripts encoding cytokinin oxidase (CKX) up-regulated 3-fold in PdCR (Fig. 3; Supplemental Table S8). CKX is the primary enzyme in CK degradation. Preliminary studies silencing the expression of CKX by RNA interference (RNAi) resulted in an interruption of the cluster rootlet developmental pattern along the primary root (Supplemental Fig. S3). The first cluster rootlet to emerge within the interrupted zone was elongated relative to the other rootlets. The addition of synthetic CK, benzyladenine (BA), inhibits Pi deficiency-induced cluster root formation (Fig. 4), as described previously by Neumann et al. (2000). Moreover, added CK increased the expression of cytokinin receptors (CRE) and CKX. The expression patterns for CRE and CKX were confirmed with qPCR. Additionally, lupin roots transformed with the GUS gene driven by the CRE and CKX promoters confirmed high GUS expression in young and emerging rootlets of Pi-deficient plants (Supplemental Figs. S4 and S5). The expression of both CRE and CKX GUS proteins decreased in mature rootlets. In Pi-deficient conditions, CKX-driven GUS expression is localized to the meristem and elongation zone of emerging rootlets, while in Pi-sufficient conditions, GUS expression appears more diffuse. Under Pi-sufficient conditions, CRE-driven GUS expression is almost completely lacking, illustrating the importance of CK in Pi deficiency acclimation. The expression patterns of additional transcripts in the CK activation and degradation pathway were measured using qPCR in control and CK (BA)-treated plants in both Pi-sufficient and Pi-deficient normal and cluster roots (Supplemental Fig. S6). The homolog of Lonely Guy (LAGI01_56645) mirrors the CKX expression pattern, with expression significantly higher in PdCR in both normal and BA-treated plants.
Figure 4.
CK application impairs Pi stress-induced cluster root formation. All root samples were collected at 14 DAE of shoots from quartz/sand growth medium. CK-treated plants were given 10−6 or 10−7 m BA at 3, 6, and 8 DAE. A, Pi-deficient control plant. Bar = 10 cm. B, Pi-deficient plant treated with 10−6 m BA. Note the reduction in cluster roots on the BA-treated plant. C, BA application reduces the percentage of root weight of cluster roots from 20% in Pi-deficient control plants (b) to approximately 10% (±se), comparable to those of Pi-sufficient plants (a). Columns with the same lowercase letter are not significantly different (P = 0.05).
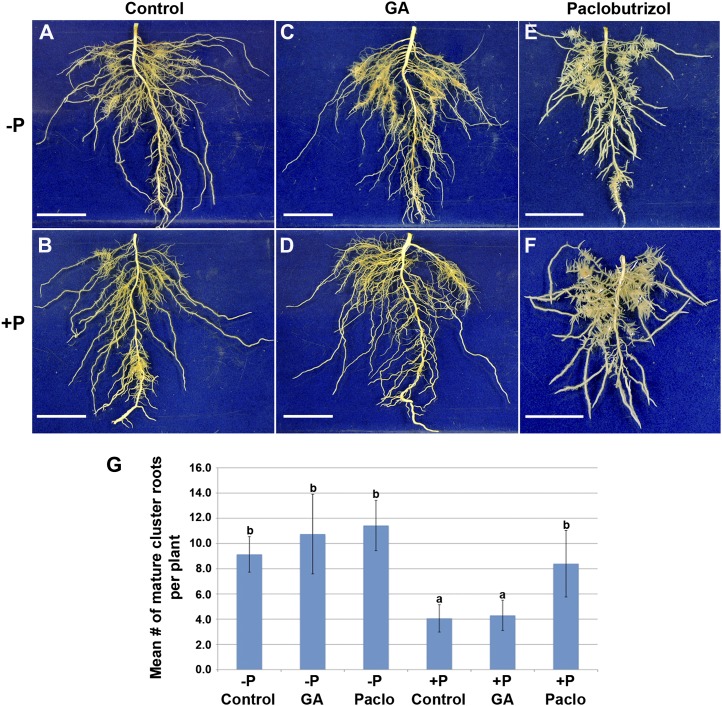
To alter the availability of GA, we treated plants with either GA3 or paclobutrazol (Paclo), an inhibitor of GA biosynthesis (Fig. 5). The application of Paclo mimicked Pi-deficient stress, causing a dramatic induction of cluster roots and increased rootlet density in Pi-sufficient plants (Fig. 5). By comparison, addition of GA had no effect on cluster root numbers of Pi-deficient plants, but the density of rootlets within a mature zone was reduced (Fig. 5; Supplemental Fig. S7). Neither GA3 nor Paclo appeared to affect root hair development (data not shown). To further investigate the role of GA on cluster root development and the Pi deficiency response, we used qPCR to measure the expression levels of genes known to be up-regulated under Pi-deficient growth conditions (Table III). The expression levels of acid phosphatase, MATE, and GA receptor increased in both Pi-sufficient and Pi-deficient growth conditions with the application of exogenous GA. Not surprisingly, exogenous GA application appears to inhibit the expression of ent-kaurenoic acid oxidase (KAO), a gene important in GA biosynthesis. The expression of KAO increases in both Pi-sufficient and Pi-deficient conditions when the GA biosynthesis pathway is blocked by the application of Paclo. Lastly, there appears to be no correlation between CK degradation and GA availability. Overall, these qPCR results illustrate the importance of GA homeostasis in the lupin Pi deficiency response.
Figure 5.
Effects of GA3 and Paclo on white lupin cluster root development. All root samples were collected at 14 DAE of shoots from quartz/sand growth medium. GA-treated plants were treated with 10−6 m GA at 3, 6, 9, and 12 DAE. Paclo-treated plants were given 1 mg of Paclo (per each 6-L pot) at 5 DAE. A and B, Roots of Pi-sufficient and Pi-deficient control plants. C and D, Roots of Pi-sufficient and Pi-deficient plants treated with 10−6 m GA. E and F, Roots of Pi-sufficient and Pi-deficient plants treated with Paclo. Bars = 5 cm. G, Mean number of cluster roots per plant (±se). Note that Paclo-treated Pi-sufficient plants have increased cluster root formation mimicking that of Pi-deficient plants. GA had no effect on mean cluster root number but does appear to decrease rootlet density (Supplemental Fig. S7). Columns with the same lowercase letter are not significantly different (P = 0.05).
Table III. qPCR expression profiles of transcripts involved in root GA3 signaling.
Transcripts corresponding to genes known to be involved in GA signaling and affected by GA were identified from the LAGI 1.0 assembly. Expression levels of these transcripts were measured using qPCR in normal and cluster (CR) roots of plants grown under Pi-sufficient and Pi-deficient conditions, control plants, plants provided with exogenous GA (+GA), and plants treated with Paclo to block GA biosynthesis. Expression ratios were calculated using the method described by Schmittgen and Livak (2008). Expression values are presented compared with Pi-sufficient normal root expression levels. Transcripts with statistically significant changes in expression compared with Pi-sufficient normal roots are denoted with asterisks. GPX-PDE, Glycerophosphodiester phosphodiesterase.
| LAGI 1.0 Identifier | Transcript Name | Pi-DeficientCR | Pi-Deficient + GA CR | Pi-Deficient + PacloCR | Pi-Sufficient CR | Pi-Sufficient + GA CR | Pi-Sufficient + Paclo CR |
|---|---|---|---|---|---|---|---|
| LAGI01_35427 | Acid phosphatase | 32* | 85* | 28* | 3* | 6* | 3* |
| LAGI01_48446 | GPX-PDE | 5* | 10* | 5* | 2* | 2* | 2* |
| LAGI01_46294 | Low-phosphate root1 | 26* | 18* | 18* | 10* | 10* | 6* |
| LAGI01_21605 | MATE | 40* | 46* | 4* | 16* | 35* | −2.5* |
| LAGI01_36029 | GA3 receptor | 2* | 4* | 1 | 3* | 6 | 2* |
| LAGI01_20605 | Phosphate transporter1 | 3* | 6* | 2* | 2* | 2* | 1 |
| LAGI01_30270 | CKX | 2* | 2* | 2* | 2* | 3* | 3* |
| LAGI01_21906 | KAO | 3* | 3* | 5* | 4* | 3* | 6* |
| LAGI01_45033 | Fe(II) oxoglutarate | 3* | 7* | 2* | 2* | 2* | 3* |
We have previously shown that auxin is involved in cluster root development (Gilbert et al., 2000; Cheng et al., 2011b). The RNA-Seq data further reflect auxin homeostasis involvement in cluster root formation. We found transcripts related to auxin metabolism and sensing to be highly expressed in PdCR (Fig. 3; Supplemental Table S8). Similarly, sequences involved in ethylene biosynthesis were up-regulated in PdCR. However, in leaves, Pi deficiency results in the down-regulation of transcripts involved in both ethylene and jasmonic acid signaling (Fig. 3; Supplemental Table S8).
RNA-Seq Expression Validation by qPCR
Transcripts known to be involved in the phosphate starvation response (such as purple acid phosphatase and low phosphate root1) were confirmed by qPCR to have similar expression patterns to those measured by RNA-Seq (Table IV; Supplemental Table S10). Additionally the expression profiles of transcripts involved in root carbon metabolism were also measured by qPCR. Finally, transcripts involved in hormone signaling pathways were identified in the LAGI 1.0 assembly and used to design primers for qPCR analysis to measure expression profiles in plants treated with GA3, Paclo, or CK (Table III; Supplemental Fig. S6). In total, 65 transcripts were evaluated by qPCR, and 96% were consistent with RNA-Seq data (Tables III and IV; Fig. 2; Supplemental Fig. S6).
Table IV. Confirmation of RNA-Seq expression profiles with qPCR.
Expression levels of PsR, PdCR, PsL, and PdL were calculated, and pairwise comparisons of roots (PdCR versus PsR) and leaves (PdL versus PsL) were calculated. Expression ratios, as fold change, for RNA-Seq data were calculated for roots [RNA-Seq (R)] and leaves [RNA-Seq (L)] with the DESeq program. Ratios for qPCR analysis were calculated for roots [qPCR (R)] and leaves [qPCR (L)] using the method described by Schmittgen and Livak (2008). All expression ratios are presented as Pi deficient/Pi sufficient. Negative values indicate that Pi sufficient has a higher expression level than Pi deficient, and expression ratios of 1 indicate no difference in expression between Pi sufficient and Pi deficient. ABC, ATP-binding cassette; ACC, 1-aminocyclopropane-1-carboxylate; AGTA, Ala glyoxlate transaminase; GPX-PDE, glycerophosphodiester phosphodiesterase; HAP2B, heme activator protein homolog 2B; MGDG, monogalactosyldiacylglycerol; SPX, SYG1/Pho8/XPR1; THFD, THF deformylase; ZIM, zinc finger protein expressed in inflorescence meristem.
| Sequence Identifier | RNA-Seq (R) | qPCR (R) | RNA-Seq (L) | qPCR (L) | Annotation |
|---|---|---|---|---|---|
| LAGI01_35427 | 451 | 546 | 1.5 | 25 | Purple acid phosphatase |
| LAGI01_58965 | 37 | 13 | 1 | 18 | Peroxidase |
| LAGI01_85917 | 27 | 30 | 1 | −7 | Ferric reductase |
| LAGI01_48402 | 23 | 12 | 2 | 1 | Formamidase |
| LAGI01_72004 | 22 | 5 | 1 | 3 | Ferric reductase3 |
| LAGI01_21605 | 19 | 33 | 2 | 2 | MATE |
| LAGI01_46294 | 17 | 14 | 1 | 5 | Low-phosphate root1 (LPR1) |
| LAGI01_74540 | 16 | 4 | 1 | 1 | Nodulin |
| LAGI01_77756 | 15 | 3 | 1 | 1 | Malate synthase |
| LAGI01_30950 | 12 | 16 | 1 | 7 | Cytochrome P450 (76C7) |
| LAGI01_51470 | 12 | 11 | 1 | 28 | bHLH TF |
| LAGI01_66840 | 9 | 1 | 1 | 6 | Unknown |
| LAGI01_48446 | 9 | 12 | 6 | 1 | GPX-PDE |
| LAGI01_42932 | 8 | 8 | 2 | 4 | Phosphatase |
| LAGI01_3168 | 7 | 9 | 14 | 1 | Phospholipase D |
| LAGI01_46560 | 6 | 22 | 4 | 7 | SPX3 (SPX domain3) |
| LAGI01_20132 | 4 | 1 | 3 | 3 | ABC TP |
| LAGI01_57111 | 4 | 3 | 2 | 1 | Sulfoqunovosyldiacylglycerol2 (SQD2) |
| LAGI01_51543 | 4 | 2 | 1 | 1 | HAP2B |
| LAGI01_28063 | 4 | 1 | 2 | 2 | Phosphatase |
| LAGI01_40436 | 4 | 4 | 3 | 2 | Unknown |
| LAGI01_50631 | 4 | 5 | 1 | 1 | Formate dehydrogenase |
| LAGI01_20605 | 4 | 12 | 2 | 4 | Phosphate transporter1 |
| LAGI01_52503 | 3 | 13 | 2 | 1 | SPX3 (SPX domain3) |
| LAGI01_23011 | 3 | 7 | 4 | 3 | Salt tolerance (SAT32) |
| LAGI01_23220 | 3 | 5 | 6 | 2 | MGDG synthase |
| LAGI01_29037 | 3 | 3 | 1 | 4 | CKX |
| LAGI01_32571 | 3 | 3 | 1 | 1 | S-Adenosyl-Met synthase |
| LAGI01_29617 | 3 | 2 | 1 | 1 | PHO1 (phosphate1) |
| LAGI01_14509 | 3 | 2 | 1 | 1 | Transferase |
| LAGI01_25695 | 3 | 1 | 1 | 1 | AGTA |
| LAGI01_45009 | 3 | 1 | −6 | 2 | Flavonoid |
| LAGI01_30478 | 2 | 5 | 1 | 2 | ACC synthase |
| LAGI01_48408 | 2 | 5 | 1 | 1 | THFD |
| LAGI01_59196 | 2 | 3 | 1 | 1 | Suc synthase |
| LAGI01_37805 | 1 | 9 | 2 | 8 | ACC oxidase |
| LAGI01_31303 | 1 | 3 | −3 | 1 | Pentatricopeptide repeat |
| LAGI01_14931 | 1 | 2 | 1 | 1 | Methylene tetrahydrofolate |
| LAGI01_18713 | 1 | 2 | −3 | 1 | Sugar transferase |
| LAGI01_26301 | 1 | 1 | 2 | 1 | TP |
| LAGI01_33647 | 1 | −4 | 2 | 1 | Short-chain dehydrogenase |
| LAGI01_8482 | −2 | 1 | 1 | 2 | Yellow stripe-like3 |
| LAGI01_6205 | −2 | 1 | 1 | 2 | DIS3-like exonuclease |
| LAGI01_8667 | −2 | 1 | 2 | 2 | Auxin response factor19 (ARF19) |
| LAGI01_69456 | −2 | 2 | 1 | 8 | Inositol polyphosphate 5-phosphatase |
| LAGI01_9188 | −2 | 2 | 1 | 1 | Time for coffee-like |
| LAGI01_57082 | −2 | −2 | 1 | 3 | Lysosomal α-mannosidase |
| LAGI01_73251 | −2 | −2 | −2 | 2 | Auxin induced in root12 |
| LAGI01_33259 | −2 | −2 | −4 | 1 | ZIM domain |
| LAGI01_63677 | −5 | 1 | 1 | 2 | GA3 stimulated |
| LAGI01_55506 | −5 | 1 | −5 | 1 | GA regulated |
| LAGI01_2053 | −9 | −4 | 1 | 1 | PHO2 (phosphate2) |
| LAGI01_51692 | −15 | −2 | 1 | 1 | Unknown |
MicroRNAs
Posttranscriptional regulation by microRNAs (miRNAs) is an important response to nutritional, biotic, and abiotic stresses (Sunkar and Zhu, 2004; Sunkar, 2010). Sequencing of miRNAs requires special library construction, so the identification of mature miRNAs is beyond the scope of this study. However, transcripts regulated by miRNAs should contain sequences with almost perfect complementarity to known miRNAs. We used the psRNAT program (http://plantgrn.noble.org/psRNATarget/) to screen the sequences of transcripts we identified as differentially expressed due to Pi deficiency for sequences potentially regulated by miRNAs using mature miRNAs from miRBase (all miRNAs from all plant species as of release September 16, 2010) as queries. We identified 261 transcripts potentially regulated by 127 miRNA families (Supplemental Tables S11 and S12). Two of the 127 miRNAs identified in this analysis (miR399 and miR395) have previously been shown to directly regulate sulfate and phosphate deficiency responses (Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Fujii et al., 2005; Bari et al., 2006; Chiou et al., 2006). Additionally, researchers have identified a number of miRNAs that are regulated by Pi deficiency stress (Hsieh et al., 2009; Zhu et al., 2009; Valdés-López et al., 2010); several of these were also identified in this analysis.
DISCUSSION
Because of the projected crisis in the availability of inexpensive Pi, plant acclimation to Pi stress has been a topic of considerable interest in plant biology. Several studies utilizing microarrays and limited EST sequencing have documented the responses of plants to Pi deficiency stress. In this study, we extended the fundamental understanding of plant acclimation to Pi status through an RNA-Seq whole-transcriptome analysis of Pi deficiency in white lupin. Although our analysis primarily focuses on PdCR, we also explore the effect of Pi on leaves. Our data provide the foundation for what is to our knowledge the first white lupin gene index (www.comparative-legumes.org). We report 125,821 unique sequences, of which more than 2,000 respond to the Pi status of the plant. These transcripts have been mapped to recently reported L. angustifolius BAC end sequences. We also report previously unrecognized transcriptional responses to Pi deficiency.
Comparison of the LAGI 1.0 Assembly with Other Plant Transcriptomes
Genomic sequencing and identification of predicted genes within a genome have exploded in recent years. The Arabidopsis genome is one of the smallest plant genomes at 135 Mb. Fully 24% of the Arabidopsis genome is predicted to be transcriptionally active. Soybean, with a genome size of 1,115 Mb, was fully sequenced in 2010 (Schmutz et al., 2010). Approximately 5% of the 1,115-Mb genome is predicted to encode traditional genes. Considering all fully sequenced legumes, approximately 6% of the genome is predicted to encode proteins (M. truncatula, 8.3%; Lotus japonicus, 4.4%; pigeon pea, 5.6%). Although not sequenced, the genome of the closely related L. angustifolius is approximately 924 Mb. Thus, the 145-Mb LAGI 1.0 assembly accounts for approximately 15% of the genome, while the 50,734 transcriptionally active transcripts represent 7.8% of the genome, closely resembling active transcriptional units seen in other species.
The closely related L. angustifolius is an important crop in Australia, where genetic resources are being developed to facilitate a whole-genome-sequencing project. Recently, a BAC library was developed, and BES were made publicly available (Gao et al., 2011). The L. angustifolius BES represents 8.89 Mb, while the LAGI 1.0 represents 145 Mb, a 16-fold difference. Both projects identified a GC content of 39% (data not shown), which is similar to other dicot species (Severin et al., 2010). Protein-coding regions were identified on 2,667 BES. A BLASTN comparison of the LAGI 1.0 assembly with the BES identified 13,453 sequence matches on 2,494 unique BES with an e-value of 1e-6 or less (Supplemental File S2), suggesting that the LAGI 1.0 assembly contains most of the protein-encoding regions identified on the BES. Based on the 1% of the genome covered by the BES, Gao et al. (2011) predict the L. angustifolius genome to contain between 42,656 and 52,204 traditional protein-coding regions. This is approximately one-half the number of sequences in the LAGI 1.0 assembly. We hypothesize that multiple transcripts in the LAGI 1.0 assembly may encode a single gene. This will be readily resolved with the release of the L. angustifolius genome but is currently beyond the scope of this project. We predict that the sequences assembled in LAGI 1.0 will facilitate gene and splice variant identification upon the completion of the L. angustifolius genome sequencing. Additionally, the LAGI 1.0 sequences will provide evidence of expression for the predicted genes in the L. angustifolius genome.
Genes Commonly Expressed Under Pi Deficiency
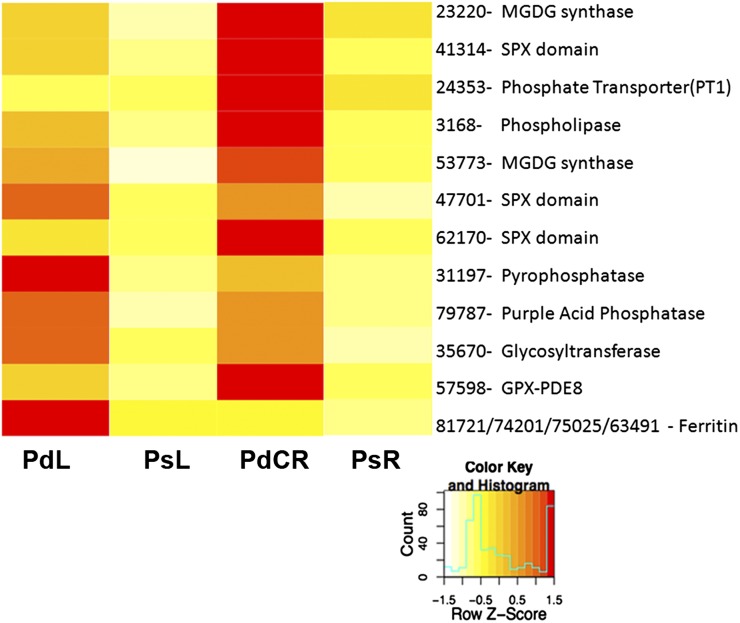
Pi deficiency has been the subject of a number of studies aimed to better understand plant responses to Pi-deficient growth conditions. Recently, Hammond et al. (2011) proposed a suite of 200 genes in potato (corresponding to 136 genes in Arabidopsis) as indicators of the Pi status of field-grown plants to limit phosphorus fertilizer usage. We compared the 2,128 white lupin transcript sequences differentially expressed in response to Pi deficiency with those identified in previous Pi deficiency microarray studies in potato (Hammond et al., 2011) and Arabidopsis (Misson et al., 2005; Morcuende et al., 2007; Thibaud et al., 2010). We identified six transcripts commonly differentially expressed in response to Pi deficiency in our experiment and all four microarray studies (Fig. 6). We also identified six transcripts differentially expressed in response to Pi deficiency in our study and three of the four microarray studies (Fig. 6). Genes commonly expressed across three species and in five laboratories are excellent candidates to be used as Pi deficiency indicators, as proposed by Hammond et al. (2011). All the transcripts identified were differentially expressed between PsR and PdCR. Of particular interest are six genes that also exhibit greater than 2-fold change in expression between PsL and PdL, with expression increasing under Pi-deficient growth conditions. These genes could be used to measure the Pi status of plants with a leaf punch rather than by destroying the plant to sample root tissues.
Figure 6.
Candidate genes for use as Pi deficiency indicators. The heat map illustrates the expression pattern, represented by Z scores, in our lupin RNA-Seq data of sequences identified as differentially expressed in multiple microarray experiments and lupin RNA-Seq data due to Pi deficiency. Red indicates high expression, yellow indicates intermediate expression, and white indicates low expression. All transcripts, except the ferritin sequences, are up-regulated in PdCR. Transcripts 47701, 31197, 79787, 35670, and multiple ferritin sequences are up-regulated in PdL.
Three genes with expression increasing in both Pi-deficient roots and leaves encode SPX domain-containing proteins (Fig. 6). SPX domain-containing proteins are essential for maintaining Pi homeostasis (Secco et al., 2012) and have been suggested to be involved in Pi signaling (Duan et al., 2008; Chiou and Lin, 2011). These proteins have been classified into four families: SPX, SPX-EXS, SPX-MFS, and SPX-RING. Twenty SPX domain-containing proteins have been identified in Arabidopsis and 15 in rice (Oryza sativa). Of these, six Arabidopsis and seven rice SPX domain-containing proteins are induced by Pi deficiency. In lupin, SPX domain-containing proteins differentially expressed due to Pi deficiency all belong to the SPX class. In Arabidopsis, this class is directly controlled by the AtPHR1 TF, a master regulator of Pi deficiency response (Secco et al., 2012).
TP and TF Expression Is Altered by Pi Availability
PdCR display enhanced Pi uptake compared with Pi-deficient normal roots (Liu et al., 2001). We identified eight phosphate TPs up-regulated in PdCR (Table I), including Lupin Phosphate Transporter1 (LaPT1), a high-affinity phosphate TP identified in lupin cluster roots under Pi-deficient conditions (Liu et al., 2001). In our RNA-Seq data, LaPT1 (LAGI01_24353) is up-regulated 5-fold in PdCR compared with PsR. In addition to increased phosphate TPs, we also identified five potassium TPs up-regulated in PdCR, suggesting that the transport of phosphorus may be coordinated and/or coregulated with potassium, as suggested previously by Wang et al. (2002). Pi deficiency also enhanced the expression patterns of ABC TPs, which are induced under a variety of abiotic stress responses (Huang et al., 2010; Kim et al., 2010), and divalent cation TPs, which are known to be involved in heavy metal transport (Mäser et al., 2001). In addition to increasing the transport of phosphorus into the plant, PdCR shows an increased transport of protons and organic acids (especially citrate) from roots to acidify the rhizosphere, releasing sparingly soluble Pi (and Fe2+) from various complexes (Zhu et al., 2005; Tomasi et al., 2009). In mature cluster roots, citrate exudation is coupled to changes in H+-ATPase activity (Tomasi et al., 2009). Two H+-ATPases, LHA1 and LHA2, were identified by Tomasi et al. (2009) as exhibiting Pi-dependent responses. In our RNA-Seq data, six H+-ATPase transcripts exhibited Pi-dependent expression changes in roots (Supplemental Table S2).
Interestingly, all the major TF families induced by Pi deficiency are directly related to either a general stress response (bHLH, MYB, and WRKY) or a specific hormone pathway (AP2-EREBP, AUX-IAA, PHOR1, TCP, and Pseudo-ARR). Four Arabidopsis TFs have been implicated in signaling during the Pi stress response: PHR1, WRKY75, ZAT6, and bHLH32. In Arabidopsis, the expression profiles of WRKY75, ZAT6, and bHLH32 are dependent on Pi availability. WRKY75 is up-regulated under Pi-deficient conditions to positively regulate phosphate stress-inducible genes and regulate root development. In the RNA-Seq data, one WRKY TF is up-regulated in PdCR. In addition, three WRKY TFs were up-regulated in PdL (Table II; Supplemental Table S7). ZAT6 is a C2H2 zinc finger TF implicated in Pi homeostasis by controlling root architecture, specifically repressing primary root growth (Devaiah et al., 2007). We evaluated the expression pattern of C2H2 zinc finger TFs differentially expressed due to Pi deficiency. None appear to be homologs of ZAT6. In PdCR, 14 bHLH TFs show increased expression (2-fold or greater change) compared with PsR (Table II; Supplemental Table S7). This is the largest class of TFs to be induced in roots under Pi-deficient conditions, suggesting that this TF family plays a major role in the Pi deficiency root response. Yi et al. (2005) showed that plants overexpressing the rice homolog of bHLH32 (OsPTF1) exhibit increased tolerance to Pi-deficient conditions, likely due to the increased root length and root area. In Arabidopsis, this TF negatively regulates Pi-deficient induced processes including root hair formation and anthocyanin formation (Chen et al., 2007). It also modulates the expression of phosphoenolpyruvate carboxylase kinase in Pi-sufficient conditions but does not induce phosphoenolpyruvate carboxylase kinase expression under Pi-deficient conditions (Chen et al., 2007). The up-regulation of another Arabidopsis bHLH TF, a homolog of FIT1 (LAGI_31032), in PdCR suggests the coordinated response of Pi and iron (Fe) deficiency responses and allows the possibility that FIT1 may play a role in the Pi deficiency response.
The largest class of TFs to be induced by Pi deficiency in leaves is the MYB TF family. A total of 33 MYBs were up-regulated due to Pi deficiency in lupin leaves (Table II), suggesting that this TF family plays a major role in the Pi deficiency response. Phosphate Starvation Response1 (PHR1), a MYB TF, has been identified as a master regulator of the Pi deficiency response. However, PHR1 is posttranslationally regulated by SIZ1 (Miura et al., 2005) and thus appears constitutively expressed in the LAGI 1.0 RNA-Seq data (RPKM = 3.4 ± 0.2 for all samples). In turn, PHR1 directly or indirectly regulates the expression of SPX-domain proteins to regulate Pi homeostasis, specifically Pi transport and signaling (Secco et al., 2012). In Arabidopsis, the promoters of all candidate genes for use as Pi deficiency indicators, except the ferritin sequences (Fig. 6), contain at least one PHR1-binding site (GNATATNC; data not shown), suggesting that these sequences are all regulated by PHR1. A recent study in common bean (Phaseolus vulgaris; Hernández et al., 2007) identified four TFs up-regulated in Pi-deficient roots, three of which belonged to the MYB family. Three MYBs homologous to those induced by Pi deficiency in common bean roots were also induced in Pi-deficient lupin roots (Table II; Supplemental Table S7). The conserved expression profile of these TFs across species and within tissues serves as an example of the impact that Pi deficiency has on the entire plant, not just within the roots.
Of particular interest to our laboratory is the TF Photoperiod Responsive1 (PHOR1), which has been implicated in the GA pathway (Amador et al., 2001). Inhibition of PHOR1 expression in potato resulted in earlier tuberization and increased tuber yield (Amador et al., 2001). These mutant plants resembled GA 20-oxidase-deficient mutant plants, and PHOR1 was shown to be involved in the GA signal transduction pathway of potato (Amador et al., 2001). More recently, PHOR1 has been shown to be a positive regulator of GA signaling, possibly regulating GA-mediated degradation of DELLA proteins (Thomas and Sun, 2004). LAGI 1.0 PHOR1 homologs (LAGI01_33328, LAGI01_27558, LAGI01_64796, and LAGI01_75918) show increased expression (average of 2.3-fold) in PsR compared with Pi-deficient roots (Supplemental Table S7). This Pi-dependent response highlights the importance of GA signaling in the lupin Pi deficiency response. Interestingly, PHOR1 was not expressed in either leaf sample, suggesting that the TF may be involved in a localized root process.
Oxidative Stress Response
Transcript accumulation for genes associated with the oxidative stress response is a hallmark of Pi deficiency gene expression experiments (Misson et al., 2005; Hernández et al., 2007; Torabi et al., 2009). Metabolic pathways continuously produce ROS as by-products. These ROS are usually scavenged by antioxidants, but abiotic/biotic and nutrient stresses can disrupt the equilibrium of ROS formation and detoxification, resulting in oxidative stress. ROS are known to both induce oxidative damage to cellular components and serve as signaling molecules in signaling cascades (Shin et al., 2005; Torabi et al., 2009). Those authors (Shin et al., 2005; Torabi et al., 2009) found the severity of Pi deficiency directly correlated with the severity of the oxidative stress. Increased oxidative stress and ROS accumulation are known photosynthesis inhibitors (Mackerness et al., 1999). The majority of transcripts down-regulated in leaves due to Pi deficiency (Fig. 1; Supplemental Table S1) are involved in photosynthetic processes. GSTs are dimeric proteins involved in cellular detoxification by catalyzing the conjugation of glutathione with free radicals (Torabi et al., 2009) and are up-regulated specifically in response to Pi deficiency (Misson et al., 2005; Yao et al., 2011). GST also binds anthocyanins, which may be up-regulated under Pi deficiency, and transports them to the vacuole for sequestration. Ascorbate peroxidase, which is required for anthocyanin production, is rapidly lost during severe oxidative stress (Malusa et al., 2002). Ascorbate peroxidase transcripts in our RNA-Seq data exhibit high expression in PsR and PsL but negligible expression in Pi-deficient tissues (Fig. 3; Supplemental Table S8), providing additional evidence that Pi deficiency induces severe oxidative stress.
Altering the Pi status of plants strongly affects Fe homeostasis. Phosphorus deficiency often results in increased Fe concentrations (Misson et al., 2005; Ward et al., 2008; Zheng et al., 2009). In PdCR, a bHLH TF homologous to FIT in Arabidopsis is up-regulated 6-fold compared with PsR. In Arabidopsis, FIT expression is induced by Fe deficiency and induces the expression of genes to promote Fe2+ uptake from soils (Yuan et al., 2008). Conversely, reducing available Fe under Pi-deficient growth conditions can improve plant growth by improving the utilization of cellular Pi (Ward et al., 2008; Zheng et al., 2009). Under normal conditions, vacuoles have a high concentration of Fe-Pi complexes, while under Pi-deficient conditions, vacuoles are relatively Fe free (Hirsch et al., 2006). Increased expression of transcripts encoding ferritin, an Fe storage protein, in PdL of our RNA-Seq study (Figs. 3 and 6; Supplemental Table S8) suggests that either increased root exudate activity facilitates greater Fe2+ uptake under Pi-deficient conditions or, more likely, Fe-Pi complexes normally occurring in the vacuole are being decoupled by one of the up-regulated phosphatases. The plant utilizes the Pi, and the Fe ion is transported to the ferritin molecule for storage. Ferritin proteins sequester excess Fe, oxygen, and hydrogen peroxide (Ravet et al., 2009; Briat et al., 2010), preventing ROS damage and the oxidative stress response. Further supporting the connection between Pi and Fe homeostasis is the up-regulation of MATE transport proteins in white lupin cluster roots (Uhde-Stone et al., 2003, 2005). In our RNA-Seq data, two of these MATEs show high similarity to FRD3, which is involved in Fe transport and localization throughout both roots and leaves (Green and Rogers, 2004; Roschzttardtz et al., 2011). These results support a strong correlation of Pi and Fe deficiency responses to prevent ROS damage and oxidative stress.
Pi Deficiency Modifies Root Metabolism
Previous studies have demonstrated that Pi deficiency has a striking effect on root carbon metabolism (Plaxton and Tran, 2011). Transcript and metabolomic analyses have demonstrated increased glycolysis and organic acid metabolism in Arabidopsis (Misson et al., 2005; Morcuende et al., 2007), common bean (Hernández et al., 2007), white lupin (Uhde-Stone et al., 2003), maize (Calderon-Vazquez et al., 2008), and rice (Li et al., 2010) during Pi deficiency. We not only reconfirm these initial findings but also extend the understanding of modified carbon metabolism in Pi-deficient roots. Our data show enhanced transcript abundance for steps in a modified glyoxylate-like cycle and one-carbon folate-Met pathways (Fig. 2). Enhanced expression of glyoxylate-like pathway genes in Pi-deficient roots may be important for at least two processes: (1) metabolism and recycling of acetyl-CoA generated from Pi deficiency-induced degradation of phospholipids; and (2) anaplerotic replenishment of organic acids lost through root exudation during Pi deficiency. It is well known that phospholipids are degraded during Pi deficiency, thus increasing the acetyl-CoA pool (Yu et al., 2002; Andersson et al., 2003; Hammond et al., 2003; Misson et al., 2005; Morcuende et al., 2007; Calderon-Vazquez et al., 2008). Our RNA-Seq expression analysis further validates and extends the suite of genes involved in phospholipid degradation, which increases in both roots and leaves under Pi-deficient conditions (Table IV). The acetyl-CoA formed from lipid degradation may then combine with glyoxylate generated through a modified glyoxylate cycle, formate, and amino acid transferases to anaplerotically regenerate malate and citrate lost through organic acid exudation (Fig. 2).
Our discovery of enhanced one-carbon metabolism also sheds new light on Pi deficiency-induced metabolic responses. We found evidence for pathways that lead to enhanced formate production and ethylene biosynthesis (Fig. 2). Previously, we reported enhanced expression of formamidase and formate dehydrogenase in PdCR (Uhde-Stone et al., 2003). Recently, Rath et al. (2010) expressed a recombinant form of white lupin Pi deficiency-induced formamidase and demonstrated activity with formamide. However, formamide is an unusual compound to be found in plants, and the Km of formamidase for formamide is high, suggesting that it may act on other substrates. A formate-glyoxylate synthase activity that carboxylates formate was reported to occur in potato tubers by Janave et al. (1993), but follow-up work has not been pursued. More recently, an alternative glyoxylate-formate pathway has been proposed to function during plant stress (Eisenhut et al., 2008; Allan et al., 2009). We detected increased abundance of transcripts encoding enzymes of THF metabolism, including a THF deformylase, which could give rise to formate. In addition, we identified transcripts for enzymes in the THF pathway leading to Met and subsequently ethylene (Fig. 2). Our finding that three primary transcripts in the Yang ethylene cycle have increased abundance in the PdCR, coupled with earlier findings that Pi-stressed lupin roots produce increased ethylene (Gilbert et al., 2000), support an interpretation that Pi deficiency increases ethylene production in roots. Ethylene is important in modifying root architecture, restricting primary root elongation, and promoting lateral root formation (Nagarajan and Smith, 2012). Our RNA-Seq data show that 17 members of the APETALA2/Ethylene Response Ethylene Binding (AP2_EREB) TF family are down-regulated in PdCR (Table III). Reduced AP2_EREB expression in PdCR may functionally mimic Arabidopsis ethylene-insensitive mutants, which show increased lateral roots during Pi deficiency (López-Bucio et al., 2002). Ethylene has also been shown to play a role in the Pi deficiency response of Arabidopsis, tomato (Solanum lycopersicum), and maize (López-Bucio et al., 2002; Ma et al., 2003; Kim et al., 2008). Interestingly, cyanide is a by-product of ethylene biosynthesis and is detoxified by β-cyanoalanine synthase (García et al., 2010). We found high expression of β-cyanoalanine synthase in both PdCR and PsR, providing further support for ethylene involvement in root acclimation to Pi deficiency. The ethylene biosynthesis pathway also serves as a branch point to polyamine biosynthesis, which is up-regulated in PdL (and to a lesser extent in roots; data not shown). RNA-Seq data and qPCR analysis confirm SAMDC, and spermine and spermidine biosynthesis increase 2- and 4-fold, respectively, in PdL. By contrast, we noted a down-regulation of transcripts associated with ethylene production in PdL. The presence of ethylene negatively regulates anthocyanin production (Lei et al., 2011). Down-regulating the expression of sequences in PdL associated with ethylene biosynthesis allows increased anthocyanin production in leaves, a well-characterized Pi deficiency response.
The Role of GA and CK in the Pi Deficiency Response
It has been long known that the interaction between and the balance of plant growth hormones regulate root development and architecture (Torrey, 1976; Werner et al., 2001; Tanimoto, 2005). Plant hormones have also been implicated in nutrient sensing and signaling (Lynch and Brown, 1997; López-Bucio et al., 2002; Zhang et al., 2007; Rubio et al., 2009; Krouk et al., 2011). Pi deficiency can alter hormone production, sensitivity, and transport (Chiou and Lin, 2011). Previously published data from our laboratory as well as others (Gilbert et al., 2000; Neumann et al., 2000; Cheng et al., 2011b) have provided evidence for the involvement of the plant growth hormones auxin and CK in the development of Pi deficiency-induced cluster roots.
The ratio of auxin and CK is known to control major developmental events, including the modification of root system architecture. Auxin has been implicated in regulating Pi deficiency-induced lateral root architectural changes in Arabidopsis (Jain et al., 2007; Pérez-Torres et al., 2008). Although root auxin concentration does not appear to increase under Pi deficiency, roots seem to have increased sensitivity to the auxin that is present. Early on, we showed that auxin was critical for Pi deficiency-induced cluster root development (Gilbert et al., 2000). More recently, our laboratory has shown that IAA7, which is important in lateral root formation (Okushima et al., 2007), has increased transcriptional activity in Pi-deficient roots (Cheng et al., 2011b); this was again confirmed in our RNA-Seq data (Fig. 3; Supplemental Table S8). The importance of auxin is exemplified by Low Phosphate Root1 (LPR1), originally identified as a quantitative trait locus in Arabidopsis involved in sensing external Pi (Reymond et al., 2006; Svistoonoff et al., 2007). Consistent with this, the homolog in lupin (LAGI01_46294) is up-regulated 20-fold in PdCR (Table IV). Recent experiments show that LPR1 alters Pi signaling and impairs sensitivity to auxin (Svistoonoff et al., 2007; Wang et al., 2010). Flavonoids and flavones, which are up-regulated in PdCR, inhibit polar auxin transport to increase localized auxin accumulation and accumulate in regions of lateral root emergence (Peer and Murphy, 2007). Although the mechanisms controlling LPR, flavonoid, and flavone interactions with auxin have yet to be uncovered, results from our experiments and others firmly establish auxin as an important component of the Pi deficiency stress response.
The Pi deficiency-induced root transcripts reported here were derived from PdCR and thus reflect the expression of genes not only involved in a generalized root response to Pi starvation but also transcripts directly related to cluster rootlet development and function (Supplemental Fig. S5). As initially shown by Neumann et al. (2000), application of exogenous CK reduced cluster root formation in Pi-deficient plants (Fig. 4). In addition, we found the CK application also reduced the number of cluster rootlet meristems, both emerged and preemergent (Z. Bozsoki, A. Rydeen, and C. Vance, unpublished data), suggesting that CK affects both meristem development and emergence. Similar effects have been noted for Arabidopsis, where CK addition inhibits root growth and reduces lateral root formation (Li et al., 2006; Ruzicka et al., 2009; Werner et al., 2010; Cui et al., 2011). Pi deficiency is known to reduce CK activity in roots (Lo et al., 2008; Werner et al., 2010) and root exudates (Menary and Staden, 1976; Salama and Waering, 1979; Martín et al., 2000; Franco-Zorrilla et al., 2004). This suggests that Pi deficiency-induced cluster root formation is dependent upon a reduction in CK availability. Interestingly, the expression profiles of PdCR transcripts suggest that CK activation appears to shift to a modified one-step pathway involving a homolog of the LONELY GUY gene in rice, as proposed by Kurakawa et al. (2007; Supplemental Fig. S5). We postulate that the increased CKX activity observed in PdCR reduces active CK content in Pi-deficient roots, thereby relieving the negative growth effect of CKs. Supporting this interpretation are experiments in Arabidopsis, tobacco (Nicotiana tabacum), and rice where a reduction of CKs, through overexpression of CKXs, results in increased lateral root formation (Laplaze et al., 2007; Lo et al., 2008; Werner et al., 2010). It is apparent that CK homeostasis plays a crucial role in Pi deficiency-induced root architecture changes.
The alteration of GA availability produced striking effects on PdCR formation and rootlet density. Most noticeable was the induction of cluster roots and increased rootlet density on Pi-sufficient plants when GA biosynthesis was blocked. Additionally, the Pi-independent responses of AP, MATE, and GAR (Table III) suggest that GA may, in part, regulate their expression. Similar to our results, Jiang et al. (2007) reported, in Arabidopsis, that exogenous GA did not reduce lateral root numbers but did reduce lateral root density. Jiang et al. (2007) also found that Pi deficiency reduced the level of bioactive GA by reducing transcript expression of GA biosynthesis and increasing the expression of transcripts involved in GA degradation, namely GA 2-oxidase. Silencing GA 2-oxidase in Populus spp. roots via RNAi increased root GA levels while decreasing the number of lateral roots (Gou et al., 2010). Overall, Gou et al. (2010) suggest that GA mediates the repression of lateral root density by negatively regulating lateral root primordia. We found GA 2-oxidase transcripts up-regulated in PdCR and GA receptor GID1b transcripts up-regulated in PdL and PdCR (Supplemental Table S8). These expression data, plus the effects of Paclo and GA applications to lupin roots (Table III), suggest that Pi deficiency-induced cluster root formation is modulated by either increased GA production under Pi deficiency or increased sensitivity of roots to GA. The TF MYB62 may regulate the Pi deficiency stress response by modulating GA availability (Devaiah et al., 2009; Chiou and Lin, 2011). We also noted increased expression of PIF1-like (for phytochrome-interacting factor) transcripts in PdL. Recent studies have implicated PIF-like genes in regulating GA metabolism through DELLA signaling in adaptation to cold stress, leaf oxidative stress, and ethylene-GA cross talk (Oh et al., 2007; Kidokoro et al., 2009; Zhong et al., 2009). To our knowledge, PIF-like genes responding to Pi status have not been reported previously.
CONCLUSION
Here, we present, to our knowledge, the first whole-transcriptome analysis of Pi-deficient white lupin plants and the first in-depth (three biological replicates) RNA-Seq analysis of leaf and root transcriptomes under optimal growth conditions. We identified 2,128 genes with expression patterns directly affected by the Pi status of the plant. Overall, this suite of experiments and results confirms previously reported responses to Pi deficiency while filling in previously unknown details and fundamentally advancing the understanding of how modified gene expression patterns facilitate acclimation to Pi deficiency.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
White lupin (Lupinus albus ‘Ultra’) seeds were surface sterilized, rinsed in sterile water, and allowed to imbibe overnight in the dark in a tray containing sterile water. Sterilized seeds were planted five seedlings per pot in 10-inch pots containing quartz sand the following day. Pots were placed in growth chambers at 20°C/15°C with a 16-h photoperiod (300 μmol photons m−2 s−1). Pots were watered once per day until seedlings emerged from the sand. From the time of emergence (0 DAE; approximately 7 d after planting), Pi-sufficient plants were given nutrient solutions of 3.0 mm KNO3, 2.5 mm Ca(NO3)2, 1.0 mm Ca(H2PO4)2, 1.0 mm MgSO4, 12 µm Fe (as FeEDTA), 4.0 µm MnCl2, 22.0 µm H3BO3, 0.4 µm ZnSO4, 0.5 µm NaMoO4, and 1.6 µm CuSO4. The Pi-deficient plants were treated identically, except that nutrient solutions contained 0.2 mm CaSO4 instead of 1 mm Ca(H2PO4)2 to maintain proper calcium concentrations. Total cluster roots and leaves were harvested from Pi-deficient plants, and total normal roots and leaves were harvested from Pi-sufficient plants at 16 DAE. Three biological replicates of each tissue sample per condition were used for further analysis.
Plants grown for the CK growth experiment were germinated and treated as described above, except that there were only three seedlings per pot. CK treatments were applied as a foliar spray of 1.6 m BA (Sigma; B9395), 0.01% dimethyl sulfoxide, and two drops of Triton X-100 on 3, 6, 9, and 11 DAE on both Pi-sufficient and Pi-deficient plants. Plants were sprayed until leaves were saturated. Three biological replicates (two pots per replicate) of normal and cluster roots were weighed and harvested from both CK-treated and control plants for RNA extraction and subsequent qPCR analysis. A paired Student’s t test was used to identify statistically significant (P = 0.05) changes in the percentage of root weight composed of cluster roots due to CK treatments (Fig. 4).
Plants grown for the GA growth experiment were also germinated as described above. All pots were provided appropriate (Pi-sufficient or Pi-deficient) nutrient solutions on 0, 1, and 2 DAE. GA (Sigma; G7645) treatments were given on 3, 6, 9, and 12 DAE on both Pi-sufficient and Pi-deficient plants. A 50× stock of GA was made fresh (8.65 mg of GA, 10 mL of ethanol, and 490 mL of water) and then diluted to 1× for GA treatments (300 mL, 1× GA). Plants were not given Pi-sufficient or Pi-deficient nutrient solutions on the days following GA treatments (4, 7, 10, and 12 DAE) but were given 100 to 150 mL of appropriate nutrient solutions on the remainder of the days. GA control plants were given 300 mL of an ethanol-water solution to mimic the ethanol contained in the GA treatment. Both Pi-sufficient and Pi-deficient pots were treated at 5 DAE with 1 mg of Paclo (Fluka; 46046) per 6-L pot. All treatments were replicated in three pots (three biological replicates per treatment). Plants were harvested 14 DAE and immediately placed on ice. Leaf, Pi-deficient cluster, and Pi-sufficient normal root samples were weighed, counted, and frozen for future RNA extraction and qPCR analysis. A paired Student’s t test was used to identify statistically significant differences (P = 0.05) in the number of cluster roots between treatments (Fig. 5).
RNA Extraction, cDNA Library Preparation, and Sequencing for RNA-Seq
Total RNA was purified from 12 samples {(Pi sufficient and Pi deficient) × (roots and leaves) × (three biological replicates)} using the RNeasy Plant Mini Kit (Qiagen). Contaminating genomic DNA was removed from each RNA sample using DNase I. RNA samples were quantified using Quant-iT RiboGreen RNA Reagent (http://www.invitrogen.com), and RNA integrity was checked with the RNA6000 Nano Assay using the Agilent 2100 Bioanalyzer (Agilent Technologies). cDNA library preparation and sequencing reactions were conducted in the Biomedical Genomics Center, University of Minnesota. Illumina library preparation, clustering, and sequencing reagents were used throughout the process following the manufacturer’s recommendations (http://www.illumina.com). Briefly, mRNAs were purified using poly-T oligonucleotide-attached magnetic beads and then fragmented. The first- and second-strand cDNAs were synthesized and end repaired. Adaptors were ligated after adenylation at the 3′ ends. After gel purification, cDNA templates were enriched by PCR. cDNA libraries were validated using a High Sensitivity Chip on the Agilent 2100 Bioanalyzer (Agilent Technologies). The cDNA library was quantified using the PicoGreen Assay and by qPCR. The samples were clustered on a flow cell using the cBOT, an Illumina automated clonal cluster generator. After clustering, the samples were loaded on the Illumina GA-II machine. The replicate1 cDNA libraries were run on one lane per library. For the replicate2 and replicate3 libraries, barcodes were given to each library during the cDNA library construction and two libraries were run together. The samples were sequenced using a single read with 76 cycles. Initial base calling and quality filtering of the Illumina GA-IIx image data were performed using the default parameters of the Illumina GA Pipeline GERALD stage (http://www.illumina.com). Additional filtering for homopolymers and read size (less than 76 bp) was performed using custom-written code.
De Novo Transcriptome Assembly
We first performed a test assembly using 109,555,059 Illumina reads derived from four rep1 libraries (Pi-sufficient and Pi-deficient roots and leaves) with a series of k-mers (19–65) comparing two programs designed for short read sequence assembly: ABySS (Simpson et al., 2009) and Velvet followed by Oases (Zerbino and Birney, 2008; Schulz et al., 2012). To select an optimal k-mer parameter and algorithm, we compared the summary statistics including N50s, total contig number, total number of hits, and average alignment length after a BLASTN search against soybean (Glycine max) coding sequences (less than 1e−10) and total number of Ns and N-containing sequences of each assembly generated by each k-mer and program (Supplemental Fig. S1). After careful consideration, we chose the Velvet/Oases program suite with a k-mer of 29. This generated the optimum transcript contiguity while preserving transcript diversity. Using these identified parameters, a total of 277,224,180 Illumina reads (76 bp) generated from all 12 cDNA libraries {(Pi sufficient and Pi deficient) × (roots and leaves) × (three replicates)} were de novo assembled into contigs. Additionally, 8,441 Sanger ESTs were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov) and combined with de novo assembled Illumina sequences using the CAP3 program with default parameters (Huang and Madan, 1999). Finally, the redundant sequences were collapsed using the CD-HIT-EST algorithm (Li and Godzik, 2006), producing a total of 125,821 LAGI 1.0 sequences. These sequences span 145,286,614 bp with average length of 1,155 bp (Supplemental File S1).
Gene Annotation and Functional Classification
We assigned putative functions for the unique sequences in LAGI 1.0 by conducting BLASTX queries against both the Arabidopsis (Arabidopsis thaliana) protein database (TAIR10_peptide; http://www.arabidopsis.org/) and the soybean protein database (Gmax_109_peptide; http://www.phytozome.net/soybean.php) using an e-value cutoff of 1e−10. Top protein matches were assigned to each of the LAGI 1.0 sequences. The Gene Ontology functional classes and pathways for each sequence in LAGI 1.0 were assigned based on Arabidopsis GO SLIM and pathway annotations (ftp://ftp.arabidopsis.org/home/tair/Ontologies/). In addition, LAGI 1.0 sequences were assigned to the MapMan gene functional classification system (Thimm et al., 2004) following the method described previously (Yang et al., 2009; Supplemental File S2). Functional class overrepresentation analysis was performed using the PageMan (Usadel et al., 2006) application within MapMan as described previously (Yang et al., 2009; Yang and Finnegan, 2010). The annotations of transcripts discussed throughout the text were manually curated to ensure accuracy.
Gene Expression Analysis
For gene expression analysis, the expression level of each gene in each library was calculated by quantifying the number of Illumina reads that mapped to each of the LAGI 1.0 sequences using the Bowtie program with default parameters (Langmead et al., 2009). The raw gene expression counts were normalized using the RPKM method (Mortazavi et al., 2008; Nagalakshmi et al., 2008) with custom R scripts. A single Pi-deficient leaf library was found to have spurious gene expression patterns, so this library was removed and gene expression counts were renormalized without it (Supplemental File S2). Genes exhibiting differential expression were identified using the DESeq program in R (Anders and Huber, 2010) to perform pairwise differential expression analysis. Differentially expressed genes identified by DESeq were required to have a 2-fold change and P ≤ 0.05. Additionally, one of the two tissues was required to have RPKM ≥ 3 (Fig. 1). Heat maps illustrating expression patterns of various subgroups of differentially expressed genes were generated in R as described by Severin et al. (2010).
qPCR
Total RNA was isolated with TRIzol reagent (Invitrogen) as described by Yoo et al. (2004). After DNase I (Ambion) treatment and further cleanup using an RNeasy column (Qiagen), the first-strand cDNA for each sample was made from 2 µg of total RNA using SuperScript II reverse transcriptase (Invitrogen) following the manufacturer’s recommendations and diluted 10 times before use in PCR. Gene-specific primers based on LAGI 1.0 sequences were subsequently designed using the BatchPrimer3 (version 1.0) program (http://probes.pw.usda.gov/batchprimer3/index.html; Supplemental Table S10). Samples and standards were run in triplicate on each plate using the QuantiFast SYBR Green PCR kit (Qiagen) on a StepOnePlus Real-Time PCR System (Applied Biosystems) following the manufacturer’s recommendations. qPCR was performed in a 12.5-µL reaction containing 3.11 µL of distilled, deionized water, 6.25 µL of 2× QuantiFastSYBR Green mix, 0.32 µL of forward primer (40 pmol µL−1), 0.32 µL of reverse primer (40 pmol µL−1), and 2.5 µL of template cDNA. The PCR conditions were as follows: 5 min of predenaturation at 95°C, 40 cycles of 10 s at 95°C and 30 s at 60°C, followed by steps for dissociation curve generation (15 s at 95°C, 60 s at 60°C, and 15 s at 95°C). The StepOnePlus software (Applied Biosystems) was used for data collection. Dissociation curves for each amplicon were carefully examined to confirm the lack of multiple amplicons at different melting temperatures. Relative transcript levels for each sample were obtained using the comparative cycle threshold method (Schmittgen and Livak, 2008) using the cycle threshold value of the tubulin gene for each sample as a standard.
Sequence data from this article can be accessed at http://lupal.comparative-legumes.org/#tabs-3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Total phosphorus content in developing white lupin shoot tissue.
Supplemental Figure S2. Optimization of the de novo assembly.
Supplemental Figure S3. Phenotype of the CKX RNAi (CKXi) white lupin PdCR.
Supplemental Figure S4. GUS staining of CRE expression.
Supplemental Figure S5. GUS staining of CKX expression.
Supplemental Figure S6. Gene expression patterns for CK activation and degradation.
Supplemental Figure S7. The effect of GA3 and Paclo on rootlet density of cluster roots.
Supplemental Table S1. Transcripts differentially expressed between Pi-sufficient and PdL.
Supplemental Table S2. Transcripts differentially expressed between PsR and PdCR.
Supplemental Table S3. Transcripts exhibiting tissue-specific expression patterns.
Supplemental Table S4. The most highly expressed transcripts in root and leaf tissues (either Pi sufficient or Pi deficient).
Supplemental Table S5. Housekeeping genes; these transcripts were identified as the most stably expressed across all tissues and conditions.
Supplemental Table S6. Differentially expressed TPs, expanded.
Supplemental Table S7. Differentially expressed TFs, expanded.
Supplemental Table S8. Transcripts and expression patterns corresponding to Figure 3.
Supplemental Table S9. All identified SPX proteins in the LAGI 1.0 assembly and their expression patterns.
Supplemental Table S10. Primer sequences used for real-time PCR.
Supplemental Table S11. Differentially expressed transcripts in leaves potentially regulated by miRNAs.
Supplemental Table S12. Differentially expressed transcripts in roots potentially regulated by miRNAs.
Glossary
- Pi
orthophosphate
- BAC
bacterial artificial chromosome
- BES
BAC end sequences
- NCBI
National Center for Biotechnology Information
- DAE
days after emergence
- PsR
orthophosphate-sufficient roots
- PdCR
orthophosphate-deficient cluster roots
- PsL
orthophosphate-sufficient leaves
- PdL
orthophosphate-deficient leaves
- RPKM
reads per kilobase per million reads
- qPCR
quantitative PCR
- TP
transporter
- TF
transcription factor
- MATE
multidrug and toxin efflux
- ROS
reactive oxygen species
- GST
glutathione S-transferase
- THF
tetrahydrofolate
- RNAi
RNA interference
- BA
benzyladenine
- CK
cytokinin
- CKX
cytokinin oxidase
- cDNA
complementary DNA
- CRE
cytokinin receptor
- Paclo
paclobutrazol
- KAO
ent-kaurenoic acid oxidase
- miRNA
microRNA
- Fe
iron
References
- Allan WL, Clark SM, Hoover GJ, Shelp BJ. (2009) Role of plant glyoxalate reductases during stress: a hypotheses. Biochem J 42: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador V, Monte E, García-Martínez J-L, Prat S. (2001) Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106: 343–354 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS. (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537: 128–132 [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible W-R. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, Morin RD, Zhao Y, Hirst M, Schein JE, et al. (2009) De novo transcriptome assembly with ABySS. Bioinformatics 25: 2872–2877 [DOI] [PubMed] [Google Scholar]
- Blanca J, Cañizares J, Roig C, Ziarsolo P, Nuez F, Picó B. (2011) Transcriptome characterization and high throughput SSRs and SNPs discovery in Cucurbita pepo (Cucurbitaceae). BMC Genomics 12: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat J-F, Ravet K, Arnaud N, Duc C, Boucherez J, Touraine B, Cellier F, Gaymard F. (2010) New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann Bot (Lond) 105: 811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Strand MR. (2012) Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor Bracovirus. J Virol 86: 3293–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh SH, Cavagnaro T, Jakobsen I. (2002) Functional diversity of arbuscular mycorrhizas extends to the expression of plant genes involved in P nutrition. J Exp Bot 53: 1593–1601 [DOI] [PubMed] [Google Scholar]
- Cakmak I, Hengeler C, Marschner H. (1994) Partitioning of shoot and root dry matter and carbohydrates in bean plants suffering from phosphorus, potassium, and magnesium deficiency. J Exp Bot 45: 1245–1250 [Google Scholar]
- Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L. (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59: 2479–2497 [DOI] [PubMed] [Google Scholar]
- Chen Z-H, Nimmo GA, Jenkins GI, Nimmo HG. (2007) BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem J 405: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Bucciarelli B, Liu J, Zinn K, Miller S, Patton-Vogt J, Allan D, Shen J, Vance CP. (2011a) White lupin cluster root acclimation to phosphorus deficiency and root hair development involve unique glycerophosphodiester phosphodiesterases. Plant Physiol 156: 1131–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Bucciarelli B, Shen J, Allan D, Vance CP. (2011b) Update on lupin cluster roots. Update on white lupin cluster root acclimation to phosphorus deficiency. Plant Physiol 156: 1025–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Aung K, Lin S-I, Wu C-C, Chiang S-F, Su C-L. (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Lin S-I. (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Cordell D, Drangert J, White S. (2009) The story of phosphorus: global food security and food for thought. Glob Environ Change 19: 292–305 [Google Scholar]
- Cui H, Hao Y, Kovtun M, Stolc V, Deng XW, Sakakibara H, Kojima M. (2011) Genome-wide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiol 157: 1221–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. (2007) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Dubey A, Farmer A, Schlueter J, Cannon SB, Abernathy B, Tuteja R, Woodward J, Shah T, Mulasmanovic B, Kudapa H, et al. (2011) Defining the transcriptome assembly and its use for genome dynamics and transcriptome profiling studies in pigeonpea (Cajanus cajan L.). DNA Res 18: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. (2008) The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiotically to plants. Proc Natl Acad Sci USA 105: 17199–17204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J. (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55: 285–293 [DOI] [PubMed] [Google Scholar]
- Franssen SU, Shrestha RP, Bräutigam A, Bornberg-Bauer E, Weber APM. (2011) Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chiou T-J, Lin S-I, Aung K, Zhu J-K. (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Gao L-L, Hane JK, Kamphuis LG, Foley R, Shi B-J, Atkins CA, Singh KB. (2011) Development of genomic resources for the narrow-leafed lupin (Lupinus angustifolius): construction of a bacterial artificial chromosome (BAC) library and BAC-end sequencing. BMC Genomics 12: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García I, Castellano JM, Vioque B, Solano R, Gotor C, Romero LC. (2010) Mitochondrial beta-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana. Plant Cell 22: 3268–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Patel RK, Tyagi AK, Jain M. (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Edwards M, Elser JJ. (2011) A transgenic approach to enhance phosphorus use efficiency in crops as part of a comprehensive strategy for sustainable agriculture. Chemosphere 84: 840–845 [DOI] [PubMed] [Google Scholar]
- Gilbert G, Knight J, Vance CP, Allan DL. (1999) Acid phosphatase activity in phosphorus-deficient white lupin roots. Plant Cell Environ 22: 801–810 [Google Scholar]
- Gilbert GA, Knight JD, Vance CP, Allan DL. (2000) Proteoid root development of phosphorus deficient lupin is mimicked by auxin and phosphonate. Ann Bot (Lond) 85: 921–928 [Google Scholar]
- Gou J, Strauss SH, Tsai CJ, Fang K, Chen Y, Jiang X, Busov VB. (2010) Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell 22: 623–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Rogers EE. (2004) FRD3 controls iron localization in Arabidopsis. Plant Physiol 136: 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Liu J, Zheng Y, Huang M, Zhang H, Gong G, He H, Ren Y, Zhong S, Fei Z, et al. (2011) Characterization of transcriptome dynamics during watermelon fruit development: sequencing, assembly, annotation and gene expression profiles. BMC Genomics 12: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, Bowen HC, Spracklen WP, Hayden RM, White PJ. (2011) Gene expression changes in phosphorus deficient potato (Solanum tuberosum L.) leaves and the potential for diagnostic gene expression markers. PLoS ONE 6: e24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, White PJ. (2011) Sugar signaling in root responses to low phosphorus availability. Plant Physiol 156: 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N. (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Hernández G, Ramírez M, Valdés-López O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, et al. (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC. (2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Hsieh L-C, Lin S-I, Shih ACC, Chen J-W, Lin W-Y, Tseng C-Y, Li W-H, Chiou T-J. (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-F, Yamaji N, Ma JF. (2010) Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol 153: 1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorizzo M, Senalik DA, Grzebelus D, Bowman M, Cavagnaro PF, Matvienko M, Ashrafi H, Van Deynze A, Simon PW. (2011) De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genomics 12: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG. (2007) Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol 144: 232–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janave MT, Ramaswamy NK, Nair PM. (1993) Purification and characterization of glyoxylate synthetase from greening potato-tuber chloroplasts. Eur J Biochem 214: 889–896 [DOI] [PubMed] [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X. (2007) Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol 145: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JF, Allan DL, Vance CP. (1994) Phosphorus stress-induced proteoid roots show altered metabolism in Lupinus albus. Plant Physiol 104: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JF, Allan DL, Vance CP, Weiblen G. (1996) Root carbon dioxide fixation by phosphorus-deficient Lupinus albus (contribution to organic acid exudation by proteoid roots). Plant Physiol 112: 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kaur S, Cogan NO, Pembleton LW, Shinozuka M, Savin KW, Materne M, Forster JW. (2011) Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genomics 12: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, et al. (2009) The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Jin JY, Alejandro S, Martinoia E, Lee Y. (2010) Overexpression of AtABCG36 improves drought and salt stress resistance in Arabidopsis. Physiol Plant 139: 170–180 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Lynch JP, Brown KM. (2008) Ethylene insensitivity impedes a subset of responses to phosphorus deficiency in tomato and petunia. Plant Cell Environ 31: 1744–1755 [DOI] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B. (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Kuo HF, Chiou TJ. (2011) The role of microRNAs in phosphorus deficiency signaling. Plant Physiol 156: 1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Lambers H, Finnegan PM, Laliberté E, Pearse SJ, Ryan MH, Shane MW, Veneklaas EJ. (2011) Update on phosphorus nutrition in Proteaceae. Phosphorus nutrition of Proteaceae in severely phosphorus-impoverished soils: are there lessons to be learned for future crops? Plant Physiol 156: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al. (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zhu C, Liu Y, Karthikeyan AS, Bressan RA, Raghothama KG, Liu D. (2011) Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol 189: 1084–1095 [DOI] [PubMed] [Google Scholar]
- Li L, Liu C, Lian X. (2010) Gene expression profiles in rice roots under low phosphorus stress. Plant Mol Biol 72: 423–432 [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 [DOI] [PubMed] [Google Scholar]
- Li X, Mo X, Shou H, Wu P. (2006) Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol 47: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Liu J, Uhde-Stone C, Li A, Vance CP, Allan DJP. (2001) A phosphate transporter with enhanced expression in proteoid roots of white lupin (Lupinus albus L.). Plant Soil 237: 257–266 [Google Scholar]
- Lo S-F, Yang S-Y, Chen K-T, Hsing Y-I, Zeevaart JAD, Chen L-J, Yu S-M. (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. (2011) Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol 156: 1041–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. (1997) Ethylene and plant responses to nutritional stress. Physiol Plant 100: 613–619 [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP. (2003) Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol 131: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerness SA-H, Surpls SL, Blake P, John CF, Buchanan-Wollaston V, Jordan BR, Thomas B. (1999) Ultraviolet-B-induced stresses and changes in gene expression in Arabidopsis thaliana: role of signalling pathways controlled by jasmonic acid, ethylene and reactive oxygen species. Plant Cell Environ 22: 1413–1423 [Google Scholar]
- Malusa E, Laurenti E, Juszczuk I, Ferrari RP, Rychter AM. (2002) Free radical production in roots of Phaseolus vulgaris subjected to phosphate deficiency stress. Plant Physiol Biochem 40: 963–967 [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, San Diego
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J. (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24: 559–567 [DOI] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonneau A, Langlade N, Léon S, Smutny J, Vogt E, Neumann G, Martinoia E. (2001) Metabolic changes associated with cluster root development in white lupin (Lupinus albus L.): relationship between organic acid excretion, sucrose metabolism and energy status. Planta 213: 534–542 [DOI] [PubMed] [Google Scholar]
- Menary RC, Staden JV. (1976) Effect of phosphorus nutrition and cytokinins on flowering in the tomato, Lycopersicon esculentum Mill. Aust J Plant Physiol 3: 201–205 [Google Scholar]
- Miller SS, Liu J, Allan DL, Menzhuber CJ, Fedorova M, Vance CP. (2001) Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiol 127: 594–606 [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrachi E, Hefer CA, Ranik M, Joubert F, Myburg AA. (2010) De novo assembled expressed gene catalog of a fast-growing Eucalyptus tree produced by Illumina mRNA-Seq. BMC Genomics 11: 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan VK, Smith AP. (2012) Ethylene’s role in phosphate starvation signaling: more than just a root growth regulator. Plant Cell Physiol 53: 277–286 [DOI] [PubMed] [Google Scholar]
- Nelson MN, Phan HTT, Ellwood SR, Moolhuijzen PM, Hane J, Williams A, O’Lone CE, Fosu-Nyarko J, Scobie M, Cakir M, et al. (2006) The first gene-based map of Lupinus angustifolius L.: location of domestication genes and conserved synteny with Medicago truncatula. Theor Appl Genet 113: 225–238 [DOI] [PubMed] [Google Scholar]
- Neumann G. (2010) Mining for nutrients: regulatory aspects of cluster root function and development. New Phytol 187: 879–882 [DOI] [PubMed] [Google Scholar]
- Neumann G, Martinoia E. (2002) Cluster roots: an underground adaptation for survival in extreme environments. Trends Plant Sci 7: 162–167 [DOI] [PubMed] [Google Scholar]
- Neumann G, Massonneau A, Langlade N, Dinkelaker B, Hengeler C, Romheld V, Martinoia E. (2000) Physiological aspects of cluster root function and development in phosphorus-deficient white lupin (Lupinus albus L.). Ann Bot (Lond) 85: 909–919 [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2010) Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol Plant 139: 129–143 [DOI] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G. (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS. (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Pérez-Torres C-A, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HT, Ellwood SR, Adhikari K, Nelson MN, Oliver RP. (2007) The first genetic and comparative map of white lupin (Lupinus albus L.): identification of QTLs for anthracnose resistance and flowering time, and a locus for alkaloid content. DNA Res 14: 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT. (2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rath M, Salas J, Parhy B, Norton R, Menakuru H, Sommerhalter M, Hatlstad G, Kwon J, Allan DL, Vance CP. (2010) Identification of genes induced in proteoid roots of white lupin under nitrogen and phosphorus deprivation, with functional characterization of a formamidase. Plant Soil 334: 137–150 [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat J-F, Gaymard F, Cellier F. (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57: 400–412 [DOI] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Medina C, Atkins CA, Mann AJ, Jordan ME, Smith PM. (2011) Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.). BMC Plant Biol 11: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Séguéla-Arnaud M, Briat J-F, Vert G, Curie C. (2011) The FRD3 citrate effluxer promotes iron nutrition between symplastically disconnected tissues throughout Arabidopsis development. Plant Cell 23: 2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Bustos R, Irigoyen ML, Cardona-López X, Rojas-Triana M, Paz-Ares J. (2009) Plant hormones and nutrient signaling. Plant Mol Biol 69: 361–373 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MC, Benková E. (2009) Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama AMSAE-D, Waering PF. (1979) Effects of mineral nutrition on endogenous cytokinins in plants of sunflower (Helianthus annuus L.). J Exp Bot 30: 971–981 [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Gutiérrez-Ortega A, Hernández-Abreu E, Herrera-Estrella L. (2006) Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol 140: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas L, Rengel Z, Tang C. (2001) Excess cation uptake, and extrusion of protons and organic acid anions by Lupinus albus under phosphorus deficiency. Plant Sci 160: 1191–1198 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J, Temple G, Temple SJ, Beschow H, Vance CP. (2006) Nitrogen fixation by white lupin under phosphorus deficiency. Ann Bot (Lond) 98: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J. (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon Y-T, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane M, Lambers HJP. (2005) Cluster roots: a curiosity in context. Plant Soil 274: 101–125 [Google Scholar]
- Shen J, Yuan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F. (2011) Phosphorus dynamics: from soil to plant. Plant Physiol 156: 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman DP. (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46: 1350–1357 [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19: 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AP, Jain A, Deal RB, Nagarajan VK, Poling MD, Raghothama KG, Meagher RB. (2010) Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol 152: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Rogers WL, Breton CM, Cai L, Malmberg RL. (2011) Transcriptome analysis of Sarracenia, an insectivorous plant. DNA Res 18: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R. (2010) MicroRNAs with macro-effects on plant stress responses. Semin Cell Dev Biol 21: 805–811 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Zhu J-K. (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Tanimoto E. (2005) Regulation of root growth by plant hormones: roles for auxin and gibberellin. Crit Rev Plant Sci 24: 249–265 [Google Scholar]
- Thibaud M-C, Arrighi J-F, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Sun TP. (2004) Update on gibberellin signaling: a tale of the tall and the short. Plant Physiol 135: 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Peel GJ, Lei Z, Aziz N, Dai X, He J, Watson B, Zhao PX, Sumner LW, Dixon RA. (2009) Transcript and proteomic analysis of developing white lupin (Lupinus albus L.) roots. BMC Plant Biol 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi N, Kretzschmar T, Espen L, Weisskopf L, Fuglsang AT, Palmgren MG, Neumann G, Varanini Z, Pinton R, Martinoia E, et al. (2009) Plasma membrane H-ATPase-dependent citrate exudation from cluster roots of phosphate-deficient white lupin. Plant Cell Environ 32: 465–475 [DOI] [PubMed] [Google Scholar]
- Torabi S, Wissuwa M, Heidari M, Naghavi M-R, Gilany K, Hajirezaei M-R, Omidi M, Yazdi-Samadi B, Ismail AM, Salekdeh GH. (2009) A comparative proteome approach to decipher the mechanism of rice adaptation to phosphorous deficiency. Proteomics 9: 159–170 [DOI] [PubMed] [Google Scholar]
- Torrey JG. (1976) Root hormones and plant growth. Annu Rev Plant Physiol 27: 435–459 [Google Scholar]
- Uhde-Stone C, Liu J, Zinn KE, Allan DL, Vance CP. (2005) Transgenic proteoid roots of white lupin: a vehicle for characterizing and silencing root genes involved in adaptation to P stress. Plant J 44: 840–853 [DOI] [PubMed] [Google Scholar]
- Uhde-Stone C, Zinn KE, Ramirez-Yáñez M, Li A, Vance CP, Allan DL. (2003) Nylon filter arrays reveal differential gene expression in proteoid roots of white lupin in response to phosphorus deficiency. Plant Physiol 131: 1064–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Yang SS, Aparicio-Fabre R, Graham PH, Reyes JL, Vance CP, Hernández G. (2010) MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol 187: 805–818 [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DLP. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Wang BL, Shen JB, Zhang WH, Zhang FS, Neumann G. (2007) Citrate exudation from white lupin induced by phosphorus deficiency differs from that induced by aluminum. New Phytol 176: 581–589 [DOI] [PubMed] [Google Scholar]
- Wang X, Du G, Wang X, Meng Y, Li Y, Wu P, Yi K. (2010a) The function of LPR1 is controlled by an element in the promoter and is independent of SUMO E3 ligase SIZ1 in response to low Pi stress in Arabidopsis thaliana. Plant Cell Physiol 51: 380–394 [DOI] [PubMed] [Google Scholar]
- Wang Y-H, Garvin DF, Kochian LV. (2002) Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots: evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol 130: 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fang B, Chen J, Zhang X, Luo Z, Huang L, Chen X, Li Y. (2010b) De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas). BMC Genomics 11: 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T. (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. (2010) Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22: 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Xu H, Zhai J, Li D, Luo H, He C, Huang X. (2011) RNA-Seq analysis and de novo transcriptome assembly of Hevea brasiliensis. Plant Mol Biol 77: 299–308 [DOI] [PubMed] [Google Scholar]
- Yang SS, Tu ZJ, Cheung F, Xu WW, Lamb JFS, Jung HJG, Vance CP, Gronwald JW. (2011) Using RNA-Seq for gene identification, polymorphism detection and transcript profiling in two alfalfa genotypes with divergent cell wall composition in stems. BMC Genomics 12: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SS, Xu WW, Tesfaye M, Lamb JFS, Jung H-JG, Samac DA, Vance CP, Gronwald JW. (2009) Single-feature polymorphism discovery in the transcriptome of tetraploid alfalfa. Plant Genome 2: 224–232 [Google Scholar]
- Yang XJ, Finnegan PM. (2010) Regulation of phosphate starvation responses in higher plants. Ann Bot (Lond) 105: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Sun H, Xu F, Zhang X, Liu S. (2011) Comparative proteome analysis of metabolic changes by low phosphorus stress in two Brassica napus genotypes. Planta 233: 523–537 [DOI] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138: 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. (2004) A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C. (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling H-Q. (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rong H, Pilbeam D. (2007) Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J Exp Bot 58: 2329–2338 [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Huang F, Narsai R, Wu J, Giraud E, He F, Cheng L, Wang F, Wu P, Whelan J, et al. (2009) Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol 151: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H. (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Yamagishi M, Osaki M, Masuda K. (2008) Sugar signalling mediates cluster root formation and phosphorus starvation-induced gene expression in white lupin. J Exp Bot 59: 2749–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yan F, Zörb C, Schubert S. (2005) A link between citrate and proton release by proteoid roots of white lupin (Lupinus albus L.) grown under phosphorus-deficient conditions? Plant Cell Physiol 46: 892–901 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Zeng H, Dong C, Yin X, Shen Q, Yang Z. (2009) MicroRNA expression profiles associated with phosphorus deficiency in white lupin (Lupinus albus L.). Plant Sci 178: 23–29 [Google Scholar]