Summary: Sulfite oxidase and the key elements of the sulfite network enzymes that include sulfite reductase, UDP-sulfoquinovose synthase, β-mercaptopyruvate sulfurtransferases, and adenosine-5′-phosphosulfate reductase has a important role in maintaining sulfite homeostasis, where sulfite appears to act as an orchestrating signal molecule.
Abstract
Plant sulfite reductase (SiR; Enzyme Commission 1.8.7.1) catalyzes the reduction of sulfite to sulfide in the reductive sulfate assimilation pathway. Comparison of SiR expression in tomato (Solanum lycopersicum ‘Rheinlands Ruhm’) and Arabidopsis (Arabidopsis thaliana) plants revealed that SiR is expressed in a different tissue-dependent manner that likely reflects dissimilarity in sulfur metabolism between the plant species. Using Arabidopsis and tomato SiR mutants with modified SiR expression, we show here that resistance to ectopically applied sulfur dioxide/sulfite is a function of SiR expression levels and that plants with reduced SiR expression exhibit higher sensitivity than the wild type, as manifested in pronounced leaf necrosis and chlorophyll bleaching. The sulfite-sensitive mutants accumulate applied sulfite and show a decline in glutathione levels. In contrast, mutants that overexpress SiR are more tolerant to sulfite toxicity, exhibiting little or no damage. Resistance to high sulfite application is manifested by fast sulfite disappearance and an increase in glutathione levels. The notion that SiR plays a role in the protection of plants against sulfite is supported by the rapid up-regulation of SiR transcript and activity within 30 min of sulfite injection into Arabidopsis and tomato leaves. Peroxisomal sulfite oxidase transcripts and activity levels are likewise promoted by sulfite application as compared with water injection controls. These results indicate that, in addition to participating in the sulfate assimilation reductive pathway, SiR also plays a role in protecting leaves against the toxicity of sulfite accumulation.
Sulfur, being a constituent of amino acids, iron-sulfur clusters, cofactors, polysaccharides, and lipids, is essential for all living organisms. Plants possess the ability to convert inorganic sulfate into reduced sulfur via the reductive sulfate assimilation pathway. Sulfate is first adenylated by ATP sulfurylase (ATP:sulfate adenylyltransferase; EC 2.7.7.4) to adenosine 5-phosphosulfate (APS). Next, APS is reduced by the plastidic enzyme 5′-phosphosulfate reductase (APR; EC 1.8.4.9). The toxic intermediate sulfite generated by APR enzymes is further reduced by sulfite reductase (SiR; EC 1.8.7.1) to sulfide, which is then incorporated into Cys by O-acetylserine(thiol)lyase (EC 2.5.1.47; Leustek and Saito, 1999; Nakayama et al., 2000).
SiR is encoded by a single-copy gene in the Arabidopsis (Arabidopsis thaliana) genome, and its crucial role in the normal growth of Arabidopsis plants has earned it the designation “bottleneck” in the reductive sulfate assimilation pathway (Khan et al., 2010). Plant SiR is a 70-kD enzyme that contains one iron-sulfur cluster and one siroheme as prosthetic groups and uses reduced ferredoxin as a physiological donor of electrons (Nakayama et al., 2000; Yonekura-Sakakibara et al., 2000). In addition to participating in the sulfate reduction pathway, SiR protein, which is localized to the plastids in both photosynthetic and nonphotosynthetic plant organs (Armengaud et al., 1995; Bork et al., 1998), acts as a binding protein for plastidic DNA organization (Cannon et al., 1999; Sato et al., 2001; Chi-Ham et al., 2002; Sekine et al., 2002, 2007; Kang et al., 2010).
Sulfite, the substrate for SiR activity, in addition to being generated in the chloroplasts by APR activity, can also penetrate from exogenous sources into plant tissues as gaseous sulfur dioxide (SO2), mainly via the stomata; SO2 is transformed into sulfite and bisulfite ions on the hydrated surface of guard cells and in the cytoplasmic fluid (Hänsch and Mendel, 2008). Being a strong nucleophile, sulfite/SO2 can harm the plant tissue via sulfitolysis, which leads to chlorophyll destruction, suppression of photosynthesis, necrotic damage, and growth retardation (Pfanz et al., 1987; Nandi et al., 1990; Noji et al., 2001).
In the detoxification process that takes place in the plant cells, SO2/sulfite is either reoxidized to sulfate or further assimilated in the sulfur reductive pathway (Hänsch and Mendel, 2008). The prevalent process, reoxidation of sulfite to sulfate, is known to be catalyzed by sulfite oxidase (SO; EC 1.8.3.1), a molybdenum cofactor-containing enzyme (Eilers et al., 2001). SO activity has recently been shown to play an essential part in protecting plants against SO2/sulfite toxicity (Brychkova et al., 2007; Lang et al., 2007).
While not much is known about the role of SiR in sulfite detoxification, up-regulation of SiR transcripts in response to SO2 application was recently demonstrated in Arabidopsis wild-type and SO-impaired plants (Brychkova et al., 2007). Similarly, a 1.5- to 2-fold increase in SiR activity was recorded in Arabidopsis plants after relatively long-term fumigation with SO2 for 60 h (Randewig et al., 2012).
Using Arabidopsis and tomato (Solanum lycopersicum) wild-type and SiR-modified plants, we describe here the fate of sulfite in the different transgenic plants and show, to our knowledge for the first time, that SiR plays an important role in protecting leaves against sulfite toxicity in at least two different species that show differences in SiR expression pattern. Additionally, we demonstrate that sulfite application can cause a rapid rise in SiR expression that, under these conditions, is essential for defense against sulfite toxicity.
RESULTS
SiR Expression in Arabidopsis and Tomato Plants
Microarray data have shown that Arabidopsis SiR transcript is expressed in a tissue-dependent manner (Hruz et al., 2008). To achieve a high-resolution picture of SiR expression in Arabidopsis plants, a 2.2-kb genomic DNA fragment containing SiR promoter was cloned to direct the expression of the reporter proteins Dronpa (a GFP-like protein; Habuchi et al., 2005) and GUS (EC 3.2.1.31).
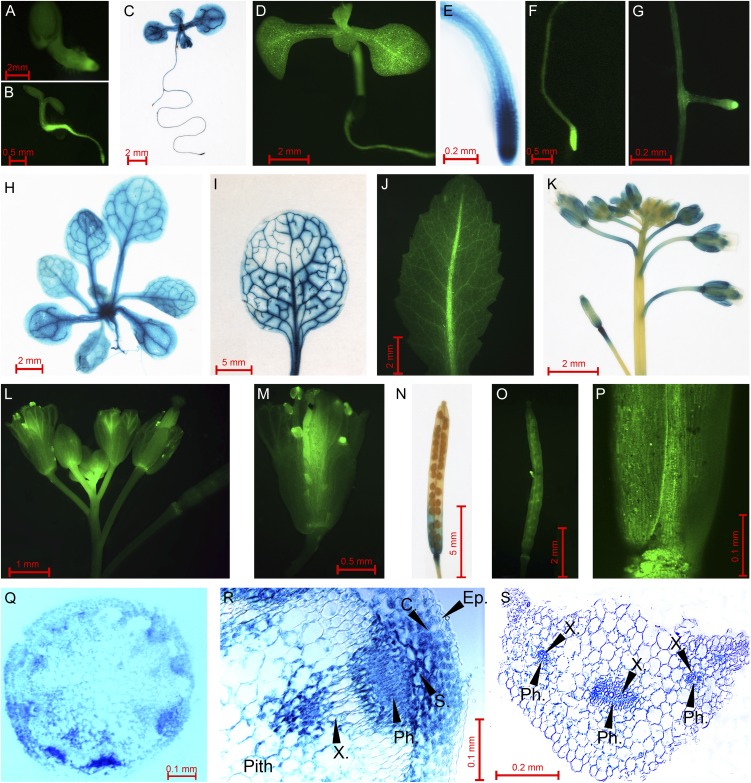
SiR promoter activity was detected in all organs of homozygous transgenic plants starting from the earliest stages of seedling development (Fig. 1, A–D). Relatively high levels of Dronpa and GUS proteins were observed in root tips of both primary and lateral roots (Fig. 1, E–G) and in flowers and developing siliques, especially their basal parts (Fig. 1, K–P). This indicates a possible role for SiR in the assimilation of sulfite both at entry into roots and during subsequent sulfur transportation or remobilization to the reproductive organs for seed formation. Additionally, the reporter proteins were expressed in the leaves, with clear-cut enhancement in the vascular system (Fig. 1, I and J). Cross sections of leaf petioles and flowering stems showed that GUS expression was associated with the vascular bundles. Relatively high levels of GUS expression were observed in xylem and phloem of flowering stalks and leaf petioles (Fig. 1, Q–S). Interestingly, high GUS activity was identified in flower stalk S-cells (cells with high sulfur content) identified in the area between the endodermis and cells belonging to the vascular bundle. Importantly, S-cells were shown to accumulate high levels of sulfur, mainly in the form of sulfur-rich glucosinolates (Koroleva et al., 2000, 2010). Elevated GUS activity was also observed in the cortex chlorenchyma, in the vicinity of the vascular bundle and S-cells in the flower stalk (Fig. 1R).
Figure 1.
SiR promoter activity in different tissues of Arabidopsis plants. A, B, and D, Seedlings expressing Dronpa fluorescence protein under the control of the SiR promoter at the ages of 1, 3, and 10 d. C and E, GUS activity in a 10-d-old seedling (C) and its root tip (E). F and G, Dronpa activity in the main root tip (F) and in an adventive root (G) of a 10-d-old seedling. H, GUS staining in a whole plant at the age of 3 weeks (the root part was removed). I and J, GUS and Dronpa expression in rosette leaves. K and L, Inflorescences of plants expressing GUS (K) and Dronpa (L). M, A flower expressing Dronpa activity. N and O, Siliques of plants expressing GUS (N) and Dronpa (O). P, Dronpa expression in the basal part of a silique. Q and S, GUS staining in a flowering stalk (Q) and in a leaf petiole (S). R, GUS activity in a bundle of a flowering stalk. C., Cortex chlorenchyma; Ep., epidermis; Ph., phloem zone; S., zone of S-cells; X., xylem zone.
Immunoblot analysis with specific antibody raised against SiR proteins was employed to determine whether SiR protein levels paralleled reporter gene activities (Fig. 2A, top). Indeed, Arabidopsis SiR protein was observed in all studied tissues where Dronpa and GUS were detected. In correspondence with the promoter results, the highest level of SiR protein was detected in parts rich in vascular tissue, including the lower part of flowering stems and leaf petioles, and less in leaf margins. Elevated expression of SiR was also observed in roots and flowers.
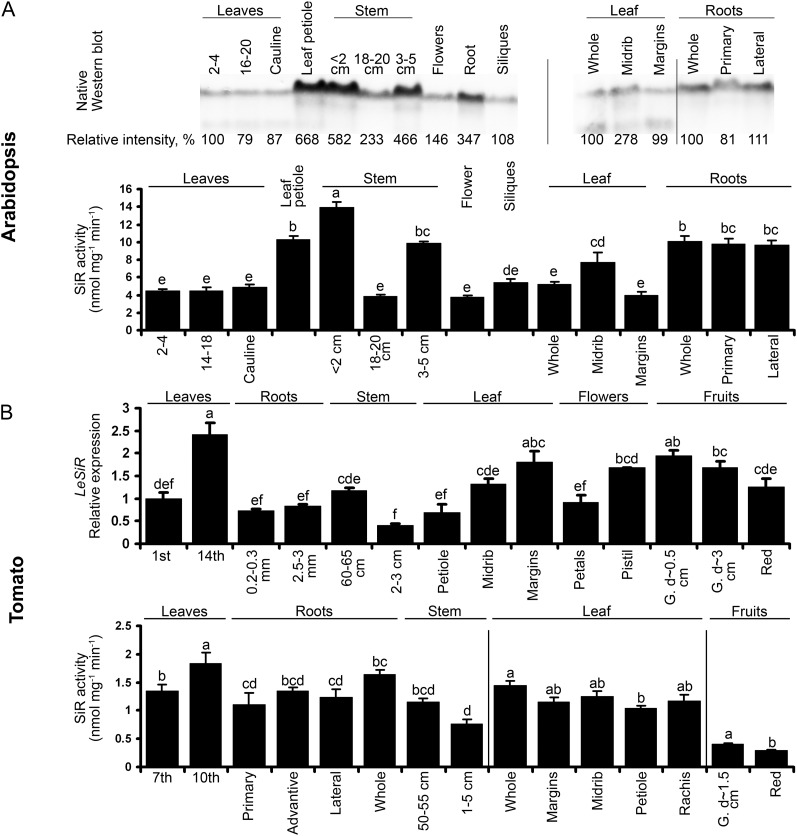
Figure 2.
SiR expression in Arabidopsis and tomato plants. A, The levels of SiR protein (top) and activity (bottom) in Arabidopsis plant organs. SiR proteins were extracted from 6-week-old Arabidopsis plants and detected by specific antibody (kindly supplied by Prof. S. Heinhorst, University of Southern Mississippi). The gel lanes were loaded with 5 µg of soluble protein. The positions of rosette leaves were counted from the bottom to the top (1 and 2 were the oldest leaves in the rosette, while 16–20 and 14–18 were the youngest). Stem length was defined from the bottom as well. The length of the sampled siliques was approximately 1 cm. Values are means ± se (n = 3). B, SiR expression in tomato plants. Quantitative reverse transcription-PCR of SiR transcript (SGN-U577417) was normalized using TFIID (SGN-U571616) as a housekeeping gene and presented relative to the first fully developed leaf from the ground (top). Primary and lateral roots were collected based on root shape and diameter. Except for the red fruits collected from older plants, all samples were collected from 2-month-old plants. Values are means ± se (n = 3). SiR activity was detected in organs of 5-month-old tomato plants (bottom). Leaves and stem positions were counted from the ground. Root samples were collected according to the distance from the stem. Old and young leaves (seventh and 10th from the ground, respectively) and root and stem samples were collected from the same plant, while leaf parts and fruits were taken from other plants. Values are means ± se (n = 4). G., Green fruit; d-, fruit diameter. Values with different letters are significantly different (Tukey-Kramer HSD test, P < 0.05).
It was of interest to compare the distribution of SiR in Arabidopsis with that found in tomato (Fig. 2). Important differences were detected. The overall level of SiR activity in tomato was significantly lower than in Arabidopsis plants, and tomato lines did not show preferable expression in the vascular system as seen in Arabidopsis. The pattern of SiR expression in tomato plants was characterized by a relatively high level of SiR transcript and activity in the young developing top leaves as compared with older leaves or stems and roots. In contrast to Arabidopsis, leaf petioles lacked an elevated level of SiR activity compared with leaflet vessels and margins. SiR expression decreased during fruit ripening, showing a possible role for SiR at early stages of fruit development (Fig. 2B).
Characterization of Plants with Modulated SiR Activity
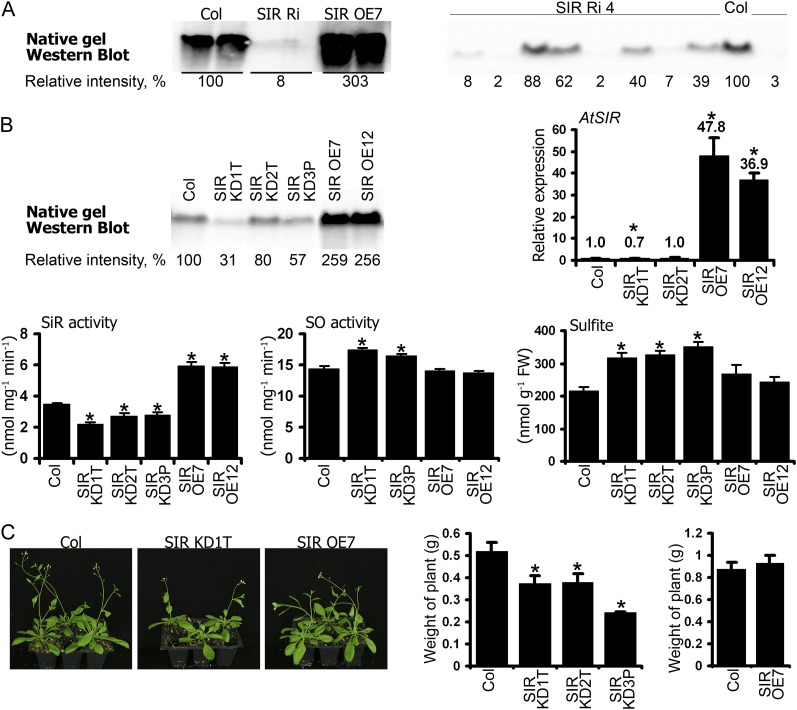
Plants with modulated SiR expression were generated to enable us to explore the role of SiR (Brychkova et al., 2012b). The homozygous Arabidopsis SIR OE7 and SIR OE12 lines (for overexpression), both of them exhibiting 37-, 2.5-, and 1.7-fold higher SiR transcripts, proteins, and activity levels, respectively, than the wild type (ecotype Columbia [Col]), were selected for further analysis (Fig. 3, A and B). In addition, five independent Arabidopsis plants with SiR impairment by RNA interference (SIR Ri; see “Materials and Methods”) exhibited strong suppression of SiR expression in leaves. Plants of the T1 generation demonstrated significantly reduced levels of SiR protein in the leaves (10%–33% of the wild-type level; see SIR Ri3 as a representative SIR Ri line in Fig. 3A). Since no homozygous plants could be selected from their progeny (see segregation of line SIR Ri4 select progeny in Fig. 3A, right panel), three homozygous transfer-DNA (T-DNA) insertion lines were selected (Nottingham Arabidopsis Stock Centre; http://arabidopsis.info/; Supplemental Fig. S1). In SAIL1223C03 (SIR KD3P; for knockdown), the T-DNA insertion is localized 521 bp upstream of the start codon, exhibiting diminished SiR protein and activity levels compared with the wild type (57% and 81%, respectively). Similarly, in SAIL867D09 (SIR KD2T) and SALK075776 (SIR KD1T), where T-DNA is inserted 119 and 168 bp downstream of the stop codon, respectively, protein levels were found to be 80% and 31%, and SiR activities 78% and 63%, of those shown by the wild type, respectively. SiR transcripts were reduced in SIR KD1T plants but were not affected in SIR KD2T (Fig. 3B). Reduction in protein levels and enzyme activities without parallel down-regulation at the transcriptional level has been described before for T-DNA-modified plants (Wang, 2008).
Figure 3.
Characterization of Arabidopsis plants with modulated SiR expression. A, Native western blot of SiR protein in SIR Ri3 and SIR OE plants. T1 heterozygous plants of SIR Ri and homozygous plants of SIR OE7 were used. Analysis of SIR Ri4 plants belonging to the T3 generation revealed segregation by SiR protein level among the Basta-resistant plants (right). B, Characterization of Arabidopsis SIR KD and SIR OE plants. A native western blot of SiR protein in leaf extracts of the modified and Col plants is shown at top left. Transcript levels of SiR were detected in the Arabidopsis plants by quantitative reverse transcription-PCR using ACTIN2 as housekeeping transcript for normalization, and the relative expression in the various plants was analyzed (top right). The SiR kinetic activity assay demonstrating the modified levels of SiR activity in the SIR KD and SIR OE plants is shown in the bottom left panel (n = 5). The elevated levels of SO activity (n = 6) and sulfite (n = 6) in SIR KD plants are shown in the bottom middle and right panels. Sulfite was detected by a colorimetric fuchsin-based method (Brychkova et al., 2012a). The values are means ± se. FW, Fresh weight. C, Growth phenotype of Arabidopsis modified SiR plants (left). Reduced biomass production is demonstrated in 5-week-old Arabidopsis SIR KD plants compared with the wild type (left panel) as well as for SIR OE7 plants at the age of 6 weeks (right panel). The values are means ± se (n = 4). Asterisks indicate significant differences between wild-type and transgenic plants (Student’s two-tailed t test, P < 0.05).
The Arabidopsis SIR KD mutants exhibited decreased biomass accumulation and an approximately 7-d delay in flower appearance relative to wild-type plants grown under a 12/12-h light regime. By contrast, the SIR OE lines did not differ from Col (Fig. 3C). Importantly, the suppression of SiR activity relative to the wild type was followed by a slight rise of 1.14- to 1.2-fold in SO activity and 1.5-fold higher sulfite level as compared with wild-type leaves (Fig. 3B).
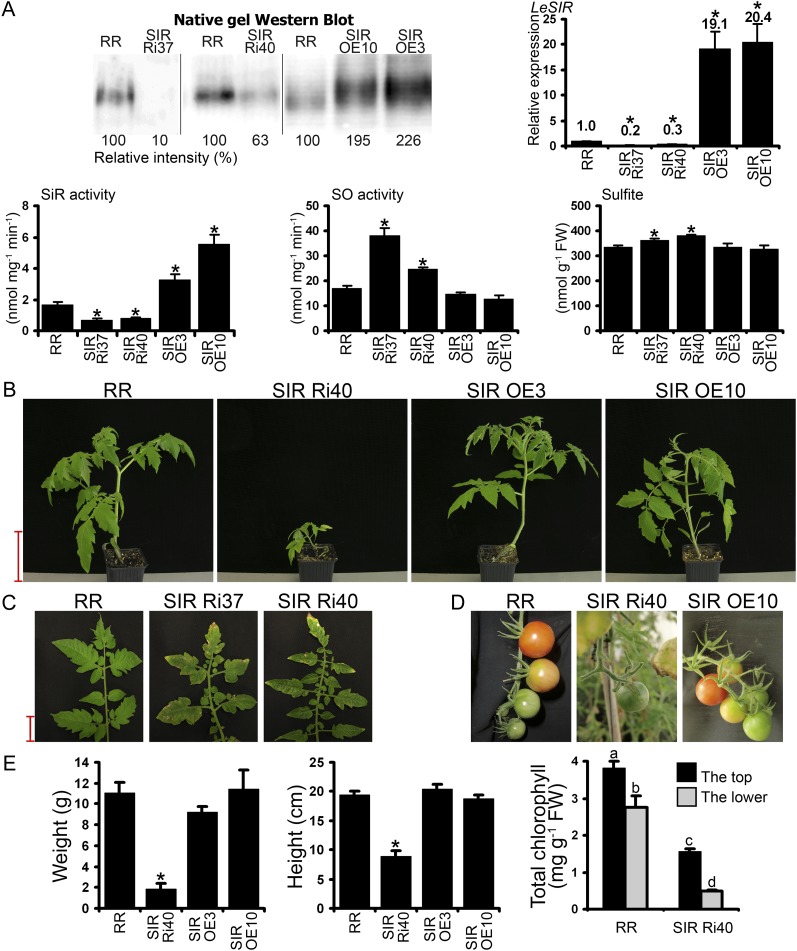
Analysis of the recently released tomato genome (Tomato Genome Consortium, 2012) revealed only one copy of the SiR gene. The full-length coding region of tomato SiR was cloned by us (SlSiR; GenBank accession no. JQ341913) and was employed to generate tomato (cv Rheinlands Ruhm) SiR mutants (see “Materials and Methods”). Homozygous tomato lines SIR OE3 and SIR OE10, which exhibited approximately 19- to 20-fold higher transcript levels than cv Rheinlands Ruhm and approximately 2-fold enhanced protein and activity levels (Fig. 4A), were selected for further experiments. Silencing SiR in tomato using the RNA interference technique was achieved in two independent SIR Ri lines: homozygous SIR Ri40 and heterozygous SIR Ri37, amenable to propagation only by vegetative means. The expression of SiR transcript was suppressed to 20% of the wild type in the heterozygous line and to 30% in the homozygous line, while protein levels were 10% and 63%, respectively, of levels in wild-type leaves (Fig. 4A). Correspondingly, SiR activity in SIR Ri37 and SIR Ri40 leaves was 43% and 50%, respectively, of that in wild-type leaves. In both lines, the decrease in SiR activity was accompanied by significant enhancement of SO activity and sulfite accumulation; in the SIR OE lines, by contrast, SO activity and sulfite did not differ from the wild type (Fig. 4A).
Figure 4.
Characterization of tomato plants with modulated SiR expression. A, SiR protein in the transgenic plants was detected by SiR antibody (a gift of Prof. S. Heinhorst, University of Southern Mississippi) after native PAGE fractionation of 10 µg of plant crude extract soluble proteins (top left). SiR transcript was quantified employing TFIID (SGN-U571616) for normalization (top right). The values are means ± se (n = 8 for cv Rheinlands Ruhm [RR], SIR OE3, and SIR Ri40, n = 4 for SIR OE10, n = 2 for SIR Ri37). Kinetic activity of SiR is shown in the bottom left lower panel (mean ± se, n = 3). SO activity (n = 6) and sulfite content (n = 4) in the transgenic plants are shown in the bottom middle and right panels. Sulfite was detected by a colorimetric fuchsin-based method (Brychkova et al., 2012a). The values are means ± se. B, Growth phenotypes of tomato plants with modulated SiR expression. Bar = 10 cm. C, Necrosis and yellowing on T1 heterozygous SIR Ri lower leaves compared with wild-type leaves. Bar = 2 cm. D, Abortion of fruits on SIR Ri40 plants compared with cv Rheinlands Ruhm and SIR OE10 plants. E, Weight and height of plants with modulated SiR activity (left and middle panels). The values are means ± se (n = 8). Chlorophyll content (right panel) was measured in individual cv Rheinlands Ruhm and SIR Ri40 plants using the top leaf (the first leaf from the top) and the bottom leaf (the first fully expanded leaf from the ground; n = 3). Values with different letters are statistically significant (Tukey-Kramer HSD test, P < 0.05). FW, Fresh weight. Asterisks indicate significant differences between wild-type and transgenic plants (Student’s two-tailed t test, P < 0.05).
The fruits of SIR Ri40 were aborted at the green stage and needed ripening in storage to form vital seeds (Fig. 4D). SIR Ri37 produced normal fruits containing seeds with extremely low germination rates (2%–4%). Some of the germinated SIR Ri37 seedlings stopped developing at an early stage, upon formation of the hypocotyl; the other SIR Ri37 seedlings stopped a few weeks later. The tomato SIR Ri40 plants showed reduced biomass accumulation and lower plant height (Fig. 4, B and E) and at the age of 1.5 months had fewer leaves than wild-type plants (5.2 ± 0.4 versus 7.5 ± 0.2 leaves per plant, respectively; P < 0.001 by two-tailed Student’s t test, n = 8). Importantly, the lower leaves of SIR Ri plants showed signs of early senescence (e.g. yellowing, partial necrosis, and leaf abortion); correspondingly, the chlorophyll content of these plants was considerably lower, with significant reduction in lower leaves as compared with top leaves (Fig. 4, C and E).
Response of Plants with Modulated SiR Activity to SO2/Sulfite Application
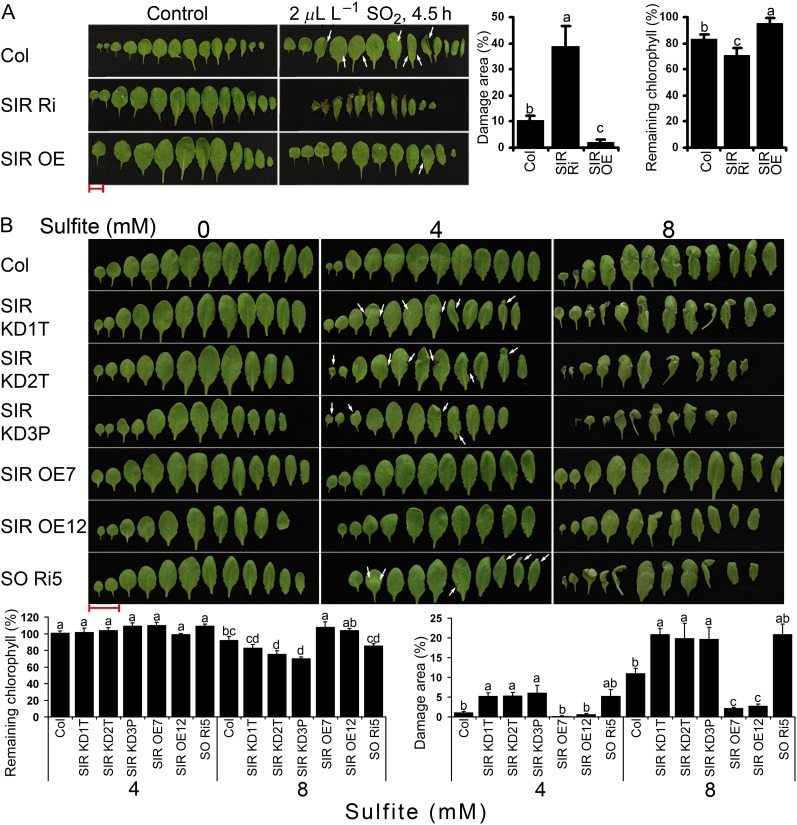
Chloroplast-localized SiR catalyzes the reduction of sulfite to sulfide in the reductive sulfate assimilation pathway. The SiR transcript enhancement detected in Arabidopsis and tomato plants in response to SO2 fumigation (Brychkova et al., 2007) suggests that SiR may participate in protecting plants against excessive sulfite levels. In order to confirm this hypothesis, SiR mutants and the wild-type Col were exposed to 2 μL L−1 SO2 fumigation for 4.5 h, a regime employed by us previously (Brychkova et al., 2007). The heterozygous SIR Ri plants exhibited more highly damaged leaf area (necrosis and shrinking of leaf area) and less remaining chlorophyll than Col and SIR OE plants, while SIR OE plants were only slightly damaged (Fig. 5A; Supplemental Fig. S2).
Figure 5.
Response of SiR-modified Arabidopsis plants to application of sulfite/SO2. A, Damaged appearance of leaves 24 h after plants were fumigated with 2 μL L−1 SO2 for 4.5 h. The photographs show leaves of Col, SIR Ri, and SIR OE7 plants that were fumigated with air (left) or SO2 (right). Damage area estimation is presented as means ± se (n = 3; middle panel). Remaining chlorophyll was calculated as the ratio of chlorophyll content in the treated plants to the control plants and expressed as a percentage (mean ± se, n = 5; right panel). The data are from one of three independent experiments that yielded identical results. B, Responses of Col, SiR-modulated plants, and SO Ri plants to sulfite injections. Leaves were photographed 72 h after injection with sodium sulfite at the indicated concentrations. Remaining chlorophyll is shown (bottom left panel; mean ± se, n = 4; the data are from one of two experiments that yielded essentially identical results). Sulfite-generated damage areas are presented as means of the values and are summarized from five independent experiments ± se (bottom right panel; n = 11 for Col and SIR KD1T, n = 7 for SIR KD2T, SIR KD3P, and SIR OE12, n = 5 for SIR OE7, n = 3 for SO Ri5). Slight damage from SO2/sulfite is indicated by arrows. Values followed by different letters are statistically different inside one treatment (Tukey-Kramer HSD test, P < 0.05). Bars = 2 cm.
Direct infiltration of sulfite by injection into the leaf makes it possible to control the amount applied and determine not only dose dependency but also the time course of the response to sulfite (Wu et al., 2011; Brychkova et al., 2012a). Monitoring the increment in leaf weight after the injection enabled us to estimate the quantity of sulfite added to leaves and to explore the fate of the sulfite (Brychkova et al., 2012a). The injection of 0.5, 4, and 8 mm sulfite into tomato and Arabidopsis leaves resulted in the following sulfite enhancements: 0.079, 0.632, and 1.264 µmol sulfite g−1 fresh weight, respectively, for Arabidopsis, and 0.174, 1.392, and 2.784 µmol sulfite g−1 fresh weight, respectively, for tomato. Arabidopsis and tomato wild types and SiR mutants, as well as Arabidopsis SO Ri5 (Brychkova et al., 2007), were used for sulfite and water control (mock) infiltration by injection. SO Ri5, described by Brychkova et al. (2007) as a line with impaired SO expression and high sensitivity to SO2/sulfite, was used as a “positive” control.
Injection of the lowest sulfite level (0.5 mm) produced no visible damage (Supplemental Figs. S3 and S4), while the higher sulfite levels (4 and 8 mm) resulted in damaged areas and chlorophyll bleaching, which were clearly related to SiR expression level (Fig. 5B; Supplemental Figs. S3 and S5). Importantly, the relatively small reduction in SiR expression in the Arabidopsis SIR KD mutants (SIR KD1T, SIR KD2T, and SIR KD3P) enhanced their sensitivity to 4 mm sulfite, a level that left wild-type and SIR OE plants undamaged. SIR OE plants injected with 8 mm sulfite exhibited significantly higher resistance to sulfite than the wild type, whereas the SIR KD mutants exhibited the highest sensitivity. As expected, the SO Ri5 plants, taken as sulfite-sensitive positive controls (Brychkova et al., 2007), exhibited a sulfite sensitivity similar to that of the SIR KD mutants injected with 4 and 8 mm sulfite (Fig. 5B).
Injection of sulfite into tomato plants revealed an identical pattern of dependence of damage intensity on SiR expression levels (Supplemental Fig. S5, A–C). SIR Ri40 plants with suppressed SiR expression were the most affected by 4 and 8 mm sulfite, exhibiting significantly greater damage than the SIR OE and wild-type plants. SIR OE3 and SIR OE10 were more tolerant to 8 mm sulfite than were the wild-type plants. A similar pattern of response was recorded when remaining chlorophyll content was monitored in leaf discs that had been sampled from tomato wild-type, SIR Ri37, and SIR Ri40 plants and then exposed to 7 mm Na2SO3 under constant light for 24 h. Both SIR Ri lines demonstrated significantly higher sensitivity to sulfite than wild-type plants, indicating that a normal SiR expression level is important for plant protection against sulfite toxicity (Supplemental Fig. S5D).
Conversion of Sulfite in SiR-Modified Plants
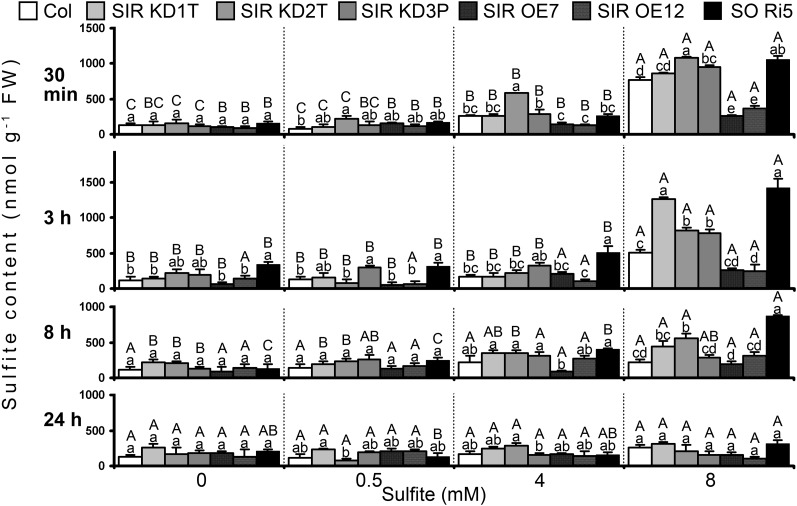
To investigate the capacity of SiR-modified mutants to convert sulfite, sulfite levels were monitored in the leaves of Arabidopsis and tomato wild-type and mutant plants injected with 0.5, 4, and 8 mm sulfite and compared with levels in plants injected with water (mock) as controls.
Sulfite levels in Arabidopsis and tomato leaves were detected as demonstrated recently (Brychkova et al., 2012a). The sulfite levels detected by a colorimetric fuchsin-based method (Figs. 3 and 4) were similar to those detected by employing commercial chicken SO (Fig. 6; Table I; Supplemental Fig. S6). Sulfite levels in SiR-modified Arabidopsis plants injected with 0.5 mm sulfite (nondamaging dose) were almost the same as in control mock-injected plants (Fig. 6A). However, in tomato SIR Ri40 mutants, an enhanced sulfite level was noticed 3 h after the injection of 0.5 mm sulfite that returned to the basal value 5 h later (Supplemental Fig. S6). These results demonstrate the capacity of the sulfite utilization mechanism(s) in tomato and Arabidopsis SiR-impaired plants, as well as in Arabidopsis SO-impaired plants, to efficiently utilize most of the 0.5 mm injected sulfite (0.079 and 0.174 µmol sulfite g−1 fresh weight in Arabidopsis and tomato, respectively; Fig. 6A; Table I; Supplemental Fig. S6).
Figure 6.
The effect of sulfite injections on sulfite in Col and SiR- and SO-modified Arabidopsis plants. The time course of sulfite content in sulfite-injected leaves is shown. Four levels of sulfite (mock [water], 0.5, 4, and 8 mm] were injected into the leaves. Sulfite levels were detected by a sulfite-specific detection assay using chicken SO (Brychkova et al., 2012a) and normalized to the fresh weight of the water-injected control leaves. The values are means ± se (n = 4). The data are from one of two independent experiments that yielded essentially identical results. Different uppercase letters indicate significant differences between plants of the same genotype at the indicated times after injection (Tukey-Kramer HSD test, P < 0.05). Different lowercase letters indicate significant differences between plants within a certain sulfite-injected level (Tukey-Kramer HSD test, P < 0.05). FW, Fresh weight.
Table I. Levels of sulfite, sulfate, Cys, glutathione, and other sulfur compounds detected in Arabidopsis 3 h after injection with sulfite solutions of different strengths.
ND, Not determined. Different uppercase letters indicate significant differences (P < 0.05) between the treatments within one line, while different lowercase letters show significant differences between the lines within one treatment, as calculated using Tukey’s HSD (JMP 8.0 software; http://www.jmp.com/).
| Genotypea | Sulfite (Increment)b | Detected Sulfitec | Detected Sulfated | Expected Sulfatee | Detected Cysf | Detected Glutathioneg | Other Sulfur Compoundsh | Total Sulfuri |
|---|---|---|---|---|---|---|---|---|
| mm (µmol g−1 fresh wt) | µmol g−1 fresh wt | |||||||
| Col | 0 (0) | 0.119 ± 0.044 B, b | 7.36 ± 0.54 B, ab | 7.36 ± 0.54 | 0.011 ± 0.002 C, a | 0.451 ± 0.011 A, a | 9.06 ± 0.37 A, b | 17.03 ± 0.43 B, b |
| SIR KD | 0.188 ± 0.026 B, b | 9.44 ± 0.69 A, a | 9.44 ± 0.69 | 0.008 ± 0.001 C, a | 0.414 ± 0.009 A, b | 11.99 ± 0.49 A, a | 22.07 ± 1.02 B, a | |
| SIR OE | 0.100 ± 0.023 B, b | 7.18 ± 0.27 A, b | 7.84 ± 0.76 | 0.009 ± 0.002 B, a | 0.268 ± 0.011 B, c | 10.73 ± 0.66 B, ab | 18.26 ± 1.02 A, b | |
| SO Ri5 | 0.342 ± 0.029 B, a | 7.68 ± 0.43 A, ab | 7.68 ± 0.43 | 0.009 ± 0.003 C, a | 0.378 ± 0.006 A, b | 9.83 ± 0.77 A, ab | 18.24 ± 0.78 A, b | |
| Col | 0.5 (0.079) | 0.129 ± 0.035 B, ab (87 ± 44%) | 7.23 ± 0.99 B, ab | 7.44 ± 0.54 | 0.008 ± 0.002 C, a | 0.327 ± 0.010 B, ab | ND | ND |
| SIR KD | 0.189 ± 0.039 B, ab (97 ± 50%) | 10.95 ± 1.11 A, a | 9.52 ± 0.69 | 0.011 ± 0.001 C, a | 0.371 ± 0.027 AB, a | ND | ND | |
| SIR OE | 0.064 ± 0.019 B, b (146 ± 24%) | 7.04 ± 0.55 A, b | 7.92 ± 0.76 | 0.012 ± 0.002 B, a | 0.302 ± 0.012 B, ab | ND | ND | |
| SO Ri5 | 0.315 ± 0.044 B, a (135 ± 56%) | ND | 7.76 ± 0.43 | 0.011 ± 0.001 C, a | 0.230 ± 0.006 C, b | ND | ND | |
| Col | 4 (0.632) | 0.160 ± 0.025 B, b (94 ± 4%) | 7.95 ± 0.31 B, ab | 7.99 ± 0.54 | 0.063 ± 0.008 B, ab | 0.359 ± 0.009 B, ab | ND | ND |
| SIR KD | 0.237 ± 0.030 B, b (92 ± 5%) | 10.51 ± 0.74 A, a | 10.07 ± 0.69 | 0.068 ± 0.009 B, a | 0.337 ± 0.022 BC, b | ND | ND | |
| SIR OE | 0.155 ± 0.025 AB, b (91 ± 4%) | 7.18 ± 0.79 A, b | 8.47 ± 0.76 | 0.033 ± 0.006 B, b | 0.417 ± 0.013 A, a | ND | ND | |
| SO Ri5 | 0.512 ± 0.086 B, a (73 ± 14%) | 6.73 ± 0.43 A, b | 8.31 ± 0.43* | 0.075 ± 0.004 B, a | 0.220 ± 0.011 C, c | ND | ND | |
| Col | 8 (1.264) | 0.505 ± 0.038 A, c (70 ± 3%) | 12.10 ± 0.02 A, ab | 8.62 ± 0.54* | 0.199 ± 0.003 A, a | 0.340 ± 0.008 B, b | 8.76 ± 0.94 A, b | 21.91 ± 1.48 A, b |
| SIR KD | 0.956 ± 0.071 A, b (39 ± 6%) | 12.72 ± 1.29 A, a | 10.71 ± 0.69 | 0.150 ± 0.010 A, b | 0.289 ± 0.006 C, c | 13.23 ± 0.89 A, a | 27.33 ± 1.24 A, a | |
| SIR OE | 0.252 ± 0.051 A, c (88 ± 4%) | 6.93 ± 0.37 A, b | 9.11 ± 0.76* | 0.088 ± 0.015 A, c | 0.403 ± 0.009 A, a | 12.90 ± 0.30 A, a | 20.59 ± 0.10 A, b | |
| SO Ri5 | 1.422 ± 0.122 A, a (15 ± 10%) | 7.61 ± 0.64 A, ab | 8.94 ± 0.43 | 0.167 ± 0.004 A, ab | 0.315 ± 0.005 B, bc | 10.95 ± 1.11 A, ab | 20.47 ± 1.24 A, b | |
Data on detected metabolite concentrations are means ± se for Col, SIR KD (SALK075776, SAIL867D09, and SAIL1223C03 lines), and SIR OE (SIR OE7 and SIR OE12) 4-week-old plants (n = 3 or 4). The values were normalized by dry weight content in the mock- and sulfite-injected plants, as described in “Materials and Methods.”
Freshly prepared sulfite solutions (pH 5.7). The increased concentration of sulfite was calculated based on weight increase after injection.
Sulfite was detected by the SO detection method (Brychkova et al., 2012a). The percentage of consumed exogenous sulfite is shown in parentheses.
Sulfate was detected in a formaldehyde-stabilized solution to prevent sulfite oxidation, as described (Brychkova et al., 2012a).
Expected sulfate was calculated by adding to the basal sulfate level the value of the injected sulfite at zero time, assuming that 100% of the converted sulfite is expected to be oxidized to sulfate. Asterisks show significant differences from the experimental data by two-tailed Student’s t test (P < 0.05).
Cys was determined as described (Brychkova et al., 2012a).
Glutathione was determined as described (Brychkova et al., 2012a).
Other sulfur compounds were calculated by subtracting sulfite, sulfate, Cys, and glutathione from total sulfur.
Total sulfur was measured in dried samples and calculated according to fresh weight content.
The response of plants to the application of higher sulfite concentrations pointed to a dependency of sulfite utilization on SiR and SO expression. Impressively, overexpression of Arabidopsis SiR enabled the utilization of approximately 83% of the 8 mm injected sulfite within 30 min, an additional 5% being converted within the next 2.5 h (Fig. 6; Table I). SIR KD and SO Ri mutants, on the other hand, exhibited only 39% ± 6% and 15% ± 10% conversion 3 h after sulfite injection. Wild-type Col plants converted 70% ± 3% of the 8 mm injected sulfite, resulting in sulfite enhancement as compared with the mock-injected plants for a period of 8 h after injection (Fig. 6; Table I). It should be noted that although SiR knockdown plants exhibited 92% ± 5% sulfite conversion within 3 h after their leaves were injected with 4 mm sulfite (Table I), leaves were significantly damaged, suggesting that sulfite utilization may not be a linear indicator of susceptibility to sulfite.
The pattern of sulfite conversion in tomato SiR-modified plants was similar to that observed in Arabidopsis. In SiR-impaired tomato plants, injection of the high doses of sulfite led to rapid and significant enhancement of sulfite concentrations to levels higher than recorded at any time point in cv Rheinlands Ruhm and SIR OE leaves (Supplemental Fig. S6). The fast and efficient conversion of sulfite by the SIR OE mutant and the impaired conversion by SIR KD and SIR Ri in Arabidopsis and tomato plants are in accord with the undamaged state of the SIR OE plants, on the one hand, and the sulfite-induced damage seen in the SiR-impaired plants, on the other, and support the notion of a role for SiR in protecting plant tissue against sulfite toxicity.
Effect of Applied Sulfite on Sulfate Level in Arabidopsis Leaves
Sulfite injected into plant leaves may be oxidized to sulfate (Brychkova et al., 2007; Lang et al., 2007) or else enter the reduction pathway, where it may be reduced to sulfide in a step on the Cys biosynthesis pathway (Leustek and Saito, 1999; Nakayama et al., 2000). Detection of sulfate levels in mock-injected Arabidopsis leaves revealed higher basal values in SIR KD mutants as compared with the wild type, SO Ri5, or SIR OE, the difference between the SIR KD mutants and SIR OE being significant (Table I). Surprisingly, while no significant sulfate enhancement was seen after injection of the low doses (0.5 and 4 mm), sulfate enhancement did occur in wild-type plants 3 h after injection of 8 mm sulfite; although only 1.264 µmol sulfite g−1 fresh weight had been injected, the wild-type plants exhibited an increment of 4.74 µmol sulfate g−1 fresh weight (Table I). None of the sulfite injection treatments induced significant changes in sulfate levels in SIR OE and SO Ri mutants (Table I). SIR KD mutant plants injected with 8 mm sulfite displayed sulfate levels higher than those detected in SIR OE and SO Ri plants but similar to the wild type, even though the initial sulfate content of the mutants (mock-injected plants) was higher (Table I). The lack of sulfite oxidation to sulfate in SO Ri is consistent with the 1.5- and 4.2-fold sulfite enhancement recorded for SO Ri mutants injected with 4 and 8 mm sulfite, respectively. At the same time, the sulfate level detected in the SIR OE plants was significantly lower than might be expected if all the sulfite had been completely oxidized to sulfate (Table I). The lack of sulfate enhancement in SIR OE and SO Ri leaves injected with the highest sulfite indicates that sulfate enhancement in SIR KD and Col leaves is due to the oxidation of sulfite (Table I) or the translocation to leaves from other plant organs rather than to enhanced uptake by the roots and translocation to the leaves.
Effect of Applied Sulfite on Major Thiols
The degree of preference for the reductive pathway in sulfite conversion may be deduced from the concentrations of the major thiols, Cys and glutathione, following sulfite application. While free Cys concentration 3 h after injection was not significantly affected in plants treated with 0.5 mm sulfite, it was significantly enhanced in all genotypes treated with 4 and 8 mm sulfite. Importantly, the undamaged SIR OE mutants exhibited significantly lower Cys concentrations than the damaged plants injected with the same high sulfite levels. These results suggest that Cys accumulation is a marker of cellular damage rather than an indication of reductive pathway activity in response to sulfite excess (Table I). Inspection of glutathione concentrations in response to sulfite revealed a significant decline in these metabolites in Col, SIR KD, and SO Ri plants in response to injected sulfite, which may indicate that the increase in free Cys described above in response to toxic sulfite is due to the degradation of sulfur-containing metabolites such as glutathione. In contrast to the Col, SIR KD, and SO Ri plants, the undamaged SIR OE plants exhibited a significant increase in glutathione levels under ectopically applied sulfite (Table I). These results suggest that the relatively low sulfite and sulfate levels detected in SIR OE leaves may be attributed to more efficient reduction of sulfite to sulfide in this genotype, leading to glutathione enhancement (Table I).
Effect of Applied Sulfite on Contents of Total Sulfur and Other Sulfur Compounds in Arabidopsis
As expected (Khan et al., 2010), analysis of the total sulfur in leaves of SIR KD plants revealed increments of up to 21% to 30% in the content of this element as compared with wild-type, SIR OE, and SO Ri plants (Table I). This increment is attributable to the accumulation of free sulfate as well as of sulfur compounds other than sulfite, Cys, or glutathione (Table I). Surprisingly, the increase in total sulfur detected 3 h after injection of high sulfite exceeded the amount of sulfur injected (1.264 µmol g−1 fresh weight; Table I). The injected wild-type and SIR KD plants demonstrated a significant increment in total sulfur as compared with the mock-injected plants (4.88 and 5.27 µmol g−1 fresh weight, respectively; Table I); the corresponding rise in total sulfur in SIR OE and SO Ri5 plants was less significant (2.3 and 2.23 µmol g−1 fresh weight, respectively; Table I). The difference in total sulfur between mock- and 8 mm sulfite-injected wild-type plants paralleled an increase in sulfate in the sulfite-treated plants (4.74 µmol g−1 fresh weight), which could be due to the oxidation of sulfite (Table I) or to the translocation to leaves from other plant organs. On the other hand, the increase in total sulfur in the SIR KD plants was mostly due to an increase in sulfate (3.28 µmol g−1 fresh weight; Table I) and nonassimilated sulfite (0.956 µmol g−1 fresh weight; Table I). In SIR OE and SO Ri5, the increase in total sulfur was of the same order as the amount of sulfite injected (1.264 µmol g−1 fresh weight).
The highest sulfite level also affected other sulfur compounds, whose content was estimated by subtraction of sulfate, sulfite, Cys, and glutathione from total sulfur. While the high treatment did not affect the concentration of these other compounds in the wild-type plants, it did enhance their concentrations in the SIR KD, SIR OE, and SO Ri5 plants. The largest increase in these compounds (2.17 µmol g−1 fresh weight) was registered in the leaves of SIR OE plants, which did not show an increase in sulfate following 8 mm sulfite injection (Table I).
Effect of Sulfite Injection on SiR and SO Expression
Since sulfite assimilation has been shown to be dependent on both SO (Brychkova et al., 2007, 2012a) and SiR plant modulation, we inspected the expression of both enzymes in the SiR-modified plants.
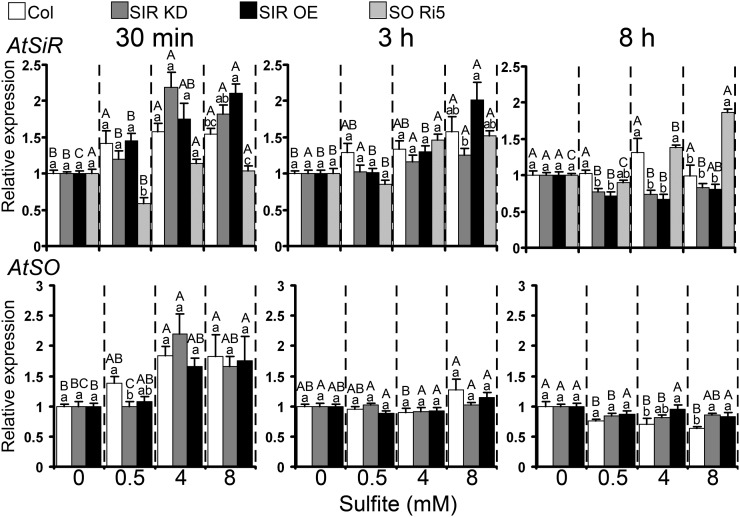
SiR transcript responded rapidly to sulfite infiltration by injection, enhancement being observed 30 min after injection in Col and Arabidopsis SiR-modified plants even at the lowest sulfite level (0.5 mm); enhancement persisted for 3 h in plants receiving 8 mm (Fig. 7). As in Arabidopsis, tomato SiR transcript showed a fast response to sulfite injection, up-regulation being recorded 30 min after sulfite treatment in tomato wild-type leaves. SiR transcript was still ongoing 3 h post injection in cv Rheinlands Ruhm, SIR Ri, and SIR OE plants injected with the highest sulfite level (Supplemental Fig. S7). In Arabidopsis SO Ri5 plants, no up-regulation of SiR transcript was detected at 30 min after injection of sulfite; however, up-regulation was observed at both 3 and 8 h post injection (Fig. 7).
Figure 7.
The effect of sulfite injection into leaves on SiR (top panel) and SO (bottom panel) transcripts in Arabidopsis Col, SIR KD, and SIR OE plants. The expression levels of AtSiR (AT5g04590) and AtSO (AT3g01910) in each sulfite-treated plant, sampled at the indicated times after sulfite injection (30 min, 3 h, and 8 h), were compared with the respective water-injected plant after normalization to the Arabidopsis ACTIN2 gene (At3g18780) and presented as relative expression. Values of SiR and SO expression are means ± se of SIR KD (SIR KD1T and SIR KD2T) and SIR OE (SIR OE7 and SIR OE12) plants from three independent experiments, each with three replicates. Different uppercase letters indicate significant differences between plants of the same genotype at the indicated times after injection (Tukey-Kramer HSD test, P < 0.05). Different lowercase letters indicate significant differences between plants within a certain sulfite-injected level (Tukey-Kramer HSD test, P < 0.05).
Rapid inductive response to sulfite injection was demonstrated for Arabidopsis SO transcript as well, up-regulation occurring 30 min after injection of 4 and 8 mm sulfite in Col, SIR KD, and SIR OE (Fig. 7). Unlike Arabidopsis, SO transcript in tomato was clearly induced only after 3 h in SIR Ri plants injected with 4 and 8 mm sulfite; up-regulation persisted for 8 h only in 8 mm-injected SIR Ri plants (Supplemental Fig. S7, bottom inset).
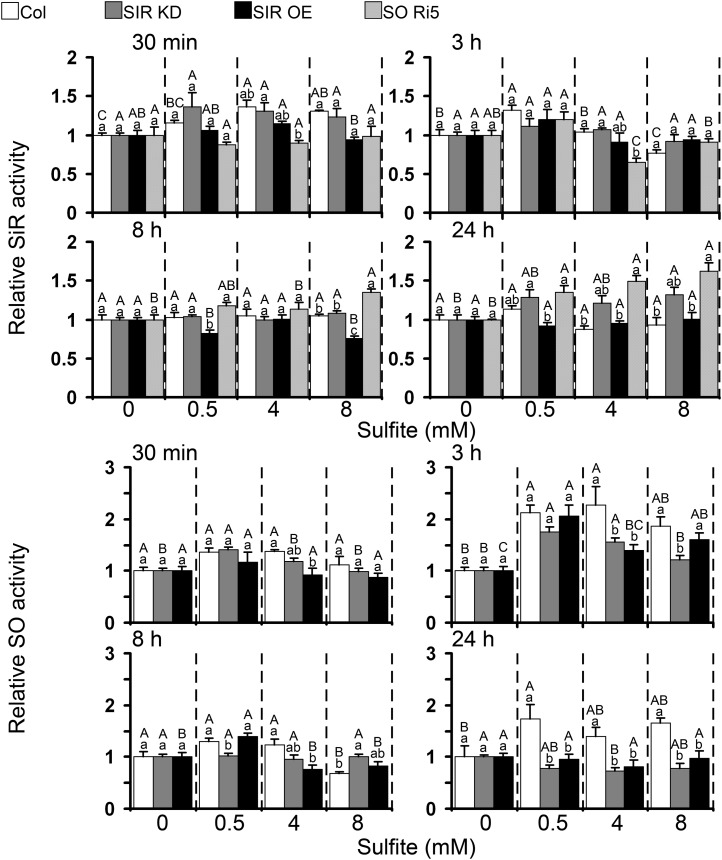
Being localized to the chloroplast, SiR activity is potentially a last line of defense protecting the organelle against toxic sulfite. Rapid enhancement of SiR activity, namely 30 min after sulfite injection, was observed in Col plants, while SIR OE plants, whose basal SiR activity, as noted above, is 1.7 times higher than that of Col, were not affected (Figs. 3B and 8; Supplemental Table S1). An increase in SiR activity was still evident 3 h post injection in Col plants injected with 0.5 mm sulfite (Fig. 8). Enhancement of SiR activity in SO Ri plants was delayed but persisted for longer relative to the other genotypes (Fig. 8; Supplemental Table S1). An increase in SiR activity was noticed in the tomato SiR OE10 plants 30 min after the injection with 8 mm sulfite as well as 3 h after 0.5 mm sulfite injection (Supplemental Fig. S8). The enhancement of SiR activity in response to sulfite injection in the wild-type tomato leaves was observed later than in Arabidopsis, being evident 3 h post injection with 8 mm sulfite and 8 h after the injection with 0.5 and 4 mm sulfite (Supplemental Fig. S8). These results show that SiR transcript and activity levels rose within minutes (30–180 min), and this rise extended over a period of a few hours in both plant species, demonstrating the inducible nature of SiR in response to sulfite application.
Figure 8.
Arabidopsis SiR (top panels) and SO (bottom panels) activities in leaves treated with sulfite. Samples were collected at fixed time times (30 min and 3, 8, and 24 h) after sulfite injections. Both enzyme activities were measured in soluble proteins extracted from one sample. The values are presented as relative levels of enzyme activity. Values of SiR and SO expression are means ± se for three SIR KD (SIR KD1T, SIR KD2T, and SIR KD3P) and two SIR OE (SIR OE7 and SIR OE12) plants (n = 3 or 4). SIR and SO activity values in water-injected plants are shown in Supplemental Tables S1 and S2. Different uppercase letters indicate significant difference between plants of the same genotype at the indicated times after injection (Tukey-Kramer HSD test, P < 0.05). Different lowercase letters indicate significant differences between plants within a certain sulfite-injected level (Tukey-Kramer HSD test, P < 0.05).
SO localized to the peroxisomes was recently deciphered as the key enzyme in the back oxidation and detoxification of sulfite by oxidation to sulfate (Brychkova et al., 2007; Randewig et al., 2012). Indeed, when SO Ri plants with negligible SO activity (Supplemental Table S2) were used as a positive control, they were found to be highly sensitive to 4 and 8 mm sulfite injections. As expected, and as demonstrated by others on SIR KD plants (Khan et al., 2010), we determined that the average basal level of SO activity in SiR-impaired mutants was higher than in the wild-type plants (Figs. 3 and 4; Supplemental Table S2). Importantly, the sulfite injection technique enabled us to reveal not only the rapid sulfite induction of SiR but also the induction of SO at the transcript and activity levels. SO was up-regulated in Col for 24 h beginning 30 min after sulfite injection, with a significant maximum at 3 h after the injections. SO activity in SIR KD mutants was enhanced 30 min and 3 h after 0.5 and 4 mm sulfite injection, respectively, while SO activity in SIR OE was enhanced only 3 h after sulfite injection and remained enhanced after 8 h only in 0.5 mm sulfite-injected plants (Fig. 8).
DISCUSSION
SiR Expression in Arabidopsis and Tomato
SiR was shown here to be expressed in all plant organs and tissues in both Arabidopsis and tomato plants (Figs. 1 and 2), but the precise patterns of SiR distribution showed fundamental differences. The most dramatic dissimilarity is that, in Arabidopsis, SO (Brychkova et al., 2007) and SiR were coexpressed in the vascular system. In tomato plants, SiR lacked enhanced levels in petioles, roots, and stems as compared with the activity in leaves. Additionally, unlike in Arabidopsis, where the level of SiR protein increased slightly with leaf age, in tomato, both SiR activity and SiR transcript were significantly higher in young top leaves than in older leaves (Fig. 2). Importantly, the overall level of SiR activity in different organs of tomato was significantly lower than in Arabidopsis (Fig. 2). These dissimilarities in SiR expression between the two plant species likely reflect important differences in sulfur metabolism. For example, high accumulation of the sulfur-containing glucosinolates is a unique attribute of the Cruciferae family (Hopkins et al., 2009; Björkman et al., 2011). Thus, in Arabidopsis, relatively high SiR promoter expression was observed in the vascular system in the vicinity of S-cells (Fig. 1), which are regarded as the storage site of sulfur metabolites, mainly glucosinolates (Koroleva et al., 2000, 2010). This suggests a pivotal role for SiR in sulfur metabolite biosynthesis and probably in the mobilization of these metabolites in Arabidopsis. A connection of SiR expression with species-specific defense systems poses an explanatory challenge, as it concerns links between primary and secondary sulfur metabolism. It is not clear why SiR level is elevated in the cells with active secondary sulfur metabolism if the branch point to glucosinolates in sulfur assimilation is at the level of APS, before sulfite reduction by SiR (Ravilious and Jez, 2012). The involvement of reduced glutathione and Met in glucosinolate synthesis may be one explanation (Sønderby et al., 2010; Ravilious and Jez, 2012). It should be noted that differences in tissue/organ SiR expression, in addition to reflecting participation in sulfur metabolism, may also indicate roles in chloroplast nucleoid metabolism, plastid gene expression, and chloroplast development (Cannon et al., 1999; Sato et al., 2001; Chi-Ham et al., 2002; Sekine et al., 2002, 2007; Kang et al., 2010). However, despite the dissimilarities in sulfur metabolism between tomato and Arabidopsis, both species responded in a similar way by employing both SO and SiR to protect plants against toxic levels of sulfite (Figs. 5–8; Supplemental Figs. S2–S7; Brychkova et al., 2007, 2012a; Lang et al., 2007).
SiR Is Essential in Protection against Excess Sulfite
SO has been characterized as the central player in plant protection against sulfite toxicity (Brychkova et al., 2007; Lang et al., 2007). However, in view of the similar Km values of SiR and SO for sulfite (10 μm [Krueger and Siegel, 1982] and 33.8 ± 3.2 μm [Eilers et al., 2001], respectively), it is reasonable to expect a possible role for SiR in the efficient detoxification of excess sulfite, at least in the chloroplast vicinity where SiR is localized. The role of SiR in protecting plant tissue against sulfite toxicity was explored in SiR-modified plants. In cases of SiR impairment, and more specifically, partial suppression of SiR activity in Arabidopsis and tomato (63%–81% and approximately 50% of wild-type SiR activity, respectively), we found that even limited reduction in SiR activity was sufficient to result in tissue damage upon exposure to toxic levels of sulfite/SO2 (Fig. 5; Supplemental Fig. S5). In contrast, Arabidopsis and tomato plants with overexpression of SiR (SiR activity approximately 2-fold higher than the wild type) exhibited no significant damage even when 8 mm sulfite was injected into the leaves, raising internal plant sulfite to levels at least 10 times higher than normal (Brychkova et al., 2012a; Fig. 5; Supplemental Fig. S5).
Thus, our results position SiR, one of the key enzymes in the sulfate assimilation reductive pathway (Khan et al., 2010), as an active component in sulfite detoxification in plant cells. Why does plant sulfur metabolism feature two alternative ways to rid itself of excess sulfite? There are fundamental metabolic cost differences between the two detoxification pathways. SO activity produces sulfate and hydrogen peroxide during sulfite oxidation. The sulfate accumulates in the vacuole, whereas hydrogen peroxide is scavenged in the peroxisome. In contrast, although the SiR pathway may be metabolically more robust than SO, its activity in detoxification would result in a futile pathway. Thus, reduced sulfite would be reinserted into the sulfur assimilation pathway from which sulfite might again be released as a result of sulfur-containing metabolite turnover (Kisker et al., 1997; Tsakraklides et al., 2002). The SiR pathway, therefore, is likely used only under strict metabolic control (i.e. in conditions where the chloroplast itself would suffer from excess sulfite). The inherent robustness of SiR activity and the metabolic cost of futile activity could explain the lack of a need for enhancement of SiR activity (2.74 ± 0.19 versus 2.78 ± 0.31 nmol mg−1 min−1 in Col and SO Ri, respectively) when compensating for SO impairment (Brychkova et al., 2007). In contrast, in the SiR KD mutants, exhibiting retarded growth and lower biomass accumulation (Figs. 3C and 4E; Khan et al., 2010), an increase in a less metabolically costly activity of SO (Figs. 3B and 4A) is essential to compensate for SiR reduction to protect the chloroplast from sulfite toxicity.
SiR Inducibility in Response to Sulfite
APR is known to be a major regulatory enzyme in the sulfate assimilation pathway in plants (Vauclare et al., 2002; Kopriva and Koprivova, 2004; Loudet et al., 2007). In contrast, the nonredundant SiR was thought to be an enzyme capable of having a mostly semiconstitutive expression pattern and, therefore, capable of only restricted fine-tuning (Bork et al., 1998; Hruz et al., 2008; Khan et al., 2010). However, it has been reported that partial down-regulation of the SiR enzyme in Arabidopsis SIR KD mutant plants results in growth retardation (Khan et al., 2010), demonstrating that SiR activity can be rate limiting and not in excess as assumed previously (Leustek, 2002; Kopriva, 2006). However, in addition to being a bottleneck along the biosynthetic pathway, SiR is shown here to protect plant tissue against sulfite toxicity, specifically in the chloroplast, where it is localized, as indicated by the more extensive chlorophyll degradation we observed in SiR-impaired plants exposed to toxic sulfite compared with wild-type plants (Fig. 5; Supplemental Fig. S5). Therefore, the chloroplastic localization of SiR may offer more effective protection of the photosynthetic apparatus, providing another explanation for the need for dual sulfite-scavenging systems.
The two plant enzymes that can detoxify sulfite, SiR and SO, are to a large extent constitutively expressed in plant tissue (Bork et al., 1998; Brychkova et al., 2007; Lang et al., 2007; Hruz et al., 2008; Khan et al., 2010). However, for prompt and efficient sulfite detoxification, the response to toxic levels of sulfite must be rapid. SiR transcript up-regulation in Arabidopsis wild-type and SO-impaired plants 2 h after fumigation with toxic levels of SO2 (2 μL L−1) may be indicative of an enhancement of SiR activity as well (Brychkova et al., 2007). An enhancement of SiR activity was recently demonstrated in response to relatively nontoxic SO2 (0.6 μL L−1) fumigation for 60 h (Randewig et al., 2012), possibly representing an adaptation to long-term SO2 exposure. Employing the technique of sulfite infiltration by injection into the leaves (Wu et al., 2011; Brychkova et al., 2012a), we were able to demonstrate a fast induction of SiR activity in response to sulfite. On the other hand, the 8-h delay observed in the up-regulation of SiR activity in response to the highest sulfite level in an SO Ri mutant indicates that SO activity is essential for the prompt induction of SiR activity (Fig. 8). Indeed, SiR transcript and activity were rapidly up-regulated, 30 min after sulfite application, in both wild-type and SIR KD plants (Figs. 7 and 8; Supplemental Fig. S7). Taking the chloroplastic localization of SiR into consideration, it is probable that under normal conditions, SiR is expressed in quantities that are adequate for reduction of the sulfite formed by the APRs. Faced with a sudden toxic rise in sulfite and the need to protect the chloroplast and its vicinity from damage, SiR has the capacity to respond by fast induction at both the transcript and activity levels.
Interplay between SiR and SO in Sulfite Detoxification
SiR and SO share sulfite as their substrate but are localized to different organelles, the chloroplast and peroxisome, respectively. SO, like SiR, has been shown to be constitutively expressed in Arabidopsis plant organs, with relative enrichment in root tips and the vascular system (Brychkova et al., 2007; Lang et al., 2007; Khan et al., 2010; Randewig et al., 2012). Infiltration of sulfite by injection into plant leaves allowed us to demonstrate the rapid (within 30 min) inducibility of both SiR and SO expression. The newly revealed inducibility of SO activity was further demonstrated when SO was induced by sulfite in SIR KD mutants (Fig. 8), despite being already up-regulated to compensate for the partial absence of normal SiR activity (Figs. 3 and 4; Supplemental Table S2).
Both SiR and SO enzymes play crucial roles in enabling Arabidopsis and tomato plants to cope with excess sulfite, as demonstrated by the finding that mutants impaired in these activities were damaged by toxic sulfite application while lines with overexpressed activities were more tolerant (Fig. 5; Supplemental Fig.S5; Brychkova et al., 2007; Lang et al., 2007). In Arabidopsis, SiR and SO activities in vitro are similar in their capacity to convert sulfite (in the range of nmol min−1 mg−1 protein; Randewig et al., 2012; Figs. 3 and 4; Supplemental Tables S1 and S2). However, it has been established that SO is the key enzyme in the back oxidation and detoxification of sulfite (Brychkova et al., 2007, 2012a; Lang et al., 2007; Randewig et al., 2012). The high sulfite-oxidizing capacity of SO in leaf tissue was demonstrated recently when the greater portion of doses of 0.799 and 1.74 µmol sulfite g−1 fresh weight (5 mm solution) injected into Arabidopsis and tomato wild type leaves, respectively, was oxidized to sulfate, thereby shielding plants from damage (Brychkova et al., 2012a). The resistance to damage (Supplemental Fig. S3) of SIR KD mutants injected with 0.5 mm sulfite can also be attributed mainly to sulfite oxidation by enhanced SO (Fig. 8), as may be deduced from the slight increase in sulfate levels in these mutants (Table I). However, injection of 4 and 8 mm sulfite into the same mutants led to severe damage, and while sulfite-induced SO activity rose rapidly, peaking at 3 h post injection, it was only partly able to compensate for the partially impaired SiR activity of the mutants (Figs. 5 and 8; Table I).
Although the reductive pathway has been shown to play a negligible role in sulfite assimilation (Brychkova et al., 2012a), a relatively small impairment of SiR activity in Arabidopsis SIR KD (63%–81% of the wild type) and tomato SiR Ri mutants (approximately 50%) was enough to result in chlorophyll degradation and leaf tissue damage in plants injected with 4 and 8 mm sulfite (Figs. 4, 5, and 8). The importance of SiR in protection against sulfite toxicity was further demonstrated when SIR OE mutants with approximately double the SiR activity of the wild type exhibited negligible tissue damage following 8 mm sulfite injection and SO2 fumigation, whereas wild-type plants subjected to the same treatments were significantly damaged (Figs. 5 and 8; Supplemental Table S2). Moreover, our finding that the enhancement of SO in SIR KD plants relative to wild-type plants was unable to compensate for a 19% to 37% reduction in SiR activity and thereby prevent tissue damage to these mutants (Figs. 5 and 8; Supplemental Table S2) reinforces the idea that normal SiR activity is essential to protect leaves from sulfite toxicity.
While SO is thought to function as a “safety valve” (Brychkova et al., 2007; Lang et al., 2007; Randewig et al., 2012) to detoxify excess sulfite, any sulfite outside the chloroplast that exceeds the oxidation capacity of SO activity (see SO Ri5 in Fig. 5; Table I) needs to be further detoxified; otherwise, the latter can penetrate and damage the chloroplasts, as occurred in the SIR Ri and SIR KD plants with their inadequate SiR activity (Fig. 5; Table I). The SIR OE plants, which have a high sulfite reduction capacity, escaped damage. Thus, the activities of SO and SiR are likely to be coordinated by an unknown mechanism to efficiently detoxify sulfite. The localization of SiR to the chloroplast, its lower Km for sulfite as compared with SO (10 μm [Krueger and Siegel, 1982] and 33.8 ± 3.2 μm [Eilers et al., 2001], respectively), and its semiconstitutive nature and rapid inducibility under high sulfite are the traits that most likely enable SiR to function as an essential chloroplastic “bodyguard” affording protection against excess levels of toxic sulfite.
The Fate of the Injected Sulfite
The Arabidopsis SIR KD mutant described by Khan et al. (2010) as exhibiting 28% of the SiR activity of the wild type showed a more than 3-fold higher total sulfur level than the latter, ultimately (approximately 75%) in the form of sulfate. In comparison, the SIR KD lines examined here were characterized by a significantly smaller degree of SiR repression (63%–83% of the wild-type level) and thus exhibited only a 30% increase in total sulfur compared with the wild type (Table I). Sulfate accounted for a significant part of the increased total sulfur in SIR KD (approximately 41%; Table I), although the concentration of sulfur compounds other than sulfate, sulfite, Cys, and glutathione was also enhanced (Table I).
We recently showed that endogenous sulfite levels were significantly higher than previous estimated. By employing two enzymatic sulfite-specific detection assays using SO and SiR, as well as the colorimetric fuchsin-based method, we demonstrated values for sulfite levels in the range of approximately 150 nmol g−1 fresh weight for Arabidopsis and approximately 200 to 500 nmol g−1 fresh weight for tomato (Brychkova et al., 2012a). However, despite the different sulfite detection methods and measured sulfite levels, plants with SiR impairment demonstrated the same alterations in sulfite content (i.e. its level was increased compared with wild-type plants in SiR-suppressed lines of Arabidopsis and tomato plants; Khan et al., 2010; Figs. 3 and 4). SiR is recognized to be a bottleneck in the reductive sulfate assimilation pathway (Khan et al., 2010), and apparently even a slight limitation of SiR activity is enough to produce significant perturbations in sulfur metabolism similar to those observed in plants with severe SiR impairment.
One would expect to see an increase in total sulfur in response to an injection of sulfite, as SO2/sulfite is assimilated by the plant (Van Der Kooij et al., 1997; Yang et al., 2006). Indeed, an increase in total sulfur was detected in all plant genotypes 3 h after infiltration with 8 mm sulfite. However, in both the wild-type and the SIR KD plants, the surpluses were significantly larger than the expected value of 1.264 µmol g−1 fresh weight (enhanced by 4.88 and 5.27 µmol g−1 fresh weight, respectively; Table I). The increase in total sulfur in these lines was mostly due to increased sulfate and sulfite; it is likely that the sulfite was converted to sulfate in the wild type but persisted unchanged in SIR KD for 3 h after the sulfite injections (Fig. 6; Table I). Total sulfur also increased slightly in SIR OE and SO Ri plants, which can be attributed to additional sulfur from the injected sulfite (Table I). Increases in the total soluble sulfur pool, significantly larger than expected (by 0.75 and 2.68 µmol g−1 fresh weight) upon assimilation of SO2 applied by fumigation, was evident in the Arabidopsis wild type and plants overexpressing SO activity. This surplus was mainly due to a rise in sulfate, which in turn was probably traceable to sulfate uptake from the soil, as speculated recently (Randewig et al., 2012). In support of this notion is the enhancement of sulfate uptake by roots in response to SO2 fumigation shown in Chinese cabbage (Brassica campestris ssp. pekinensis; Yang et al., 2006).
Sulfate enhancement appeared to be dependent on the level of SO and SiR activities. SO-impaired plants failed to accumulate more sulfate, while wild-type and SO OE plants accumulated more sulfate in response to sulfite injection or SO2 fumigation (Table I; Brychkova et al., 2007; Randewig et al., 2012). SIR OE plants exhibited an altered SiR/SO activity ratio recalling that of SO-impaired plants, which probably explains the absence of sulfate enhancement (Table I). The dependence of sulfate uptake on SiR expression is further supported by the 13-fold enhanced sulfate uptake by roots of plants with SiR repression to 28% of the wild-type level (Khan et al., 2010) and by the higher sulfate content in the unstressed SiR KD plant exhibiting a lower (81%–63%) SiR repression (Table I; Fig. 3B).
Importance of the Reductive Sulfate Assimilation Pathway in Sulfite Detoxification
The accumulation of Cys in response to sulfite injection into the leaves could be a consequence of sulfite reduction by SiR or could be due to toxic sulfite-induced protein degradation (Ranieri et al., 1995; Wen et al., 1996). The significantly lower incorporation rates of sulfate to generate Cys in the SIR KD mutant compared with the wild type (Khan et al., 2010) indicate a lower capacity of the mutant to generate Cys. Moreover, the significantly lower Cys enhancement observed in the SIR OE undamaged leaves as compared with the highly damaged Col, SIR KD, and SO Ri plants injected with the same sulfite levels (Table I) indicates that Cys accumulation is likely the result of tissue degradation rather than a higher rate of Cys biosynthesis.
A pivotal role of SiR in the assimilatory sulfate reduction pathway was recently demonstrated, where the total incorporation of sulfate into organic sulfur compounds, such as Cys and reduced glutathione, in the SIR KD mutant was 28-fold lower than in the wild type (Khan et al., 2010). Accordingly, the enhanced glutathione level in SIR OE mutants leaves after the injection of the highest sulfite level can be explained by the efficient reduction of the injected sulfite by the overexpressed SiR into Cys and glutathione (Fig. 8; Table I). The enhanced glutathione in the undamaged SIR OE and the lowered glutathione in damaged Col, SIR KD, and SO Ri sulfite-injected plants (Table I) may be linked to the capacity of plants to protect themselves against toxic sulfite not only through sulfite reduction via SiR toward enhanced Cys and glutathione levels, and/or through sulfite oxidation via SO to sulfate, but also by steady enhancement and regeneration of glutathione that can additionally protect plants against oxidative stress, as demonstrated in O-acetylserine(thiol)lyase overexpression mutants (Noji et al., 2001; Nakamura et al., 2009).
The high sulfite accumulation, lack of sulfate enhancement, and drop in glutathione content recorded in the highly damaged SO Ri mutant with deficient SO activity after injection of 4 and 8 mm sulfite (Table I; Fig. 5; Supplemental Table S2) confirm the crucial role of active SO in detoxifying excess sulfite by oxidation to sulfate (Brychkova et al., 2007, 2012a; Lang et al., 2007; Randewig et al., 2012). However, the tissue damage observed in plants with normal SO activity, such as SIR KD mutants upon injection of 4 and 8 mm sulfite and Col plants upon injection of 8 mm (but not 4 mm), points to the existence of a threshold above which the capacity of SO to detoxify excess sulfite declines (Figs. 5 and 8; Table I). The essential role of SiR in supporting SO-mediated sulfite detoxification is demonstrated by the finding that Col plants injected with 4 mm sulfite showed no sign of damage. However, SIR KD mutants with enhanced SO activity but a 19% to 37% reduction in SiR activity injected with the same level of sulfite did exhibit tissue damage (Table I; Figs. 3 and 5). Furthermore, the lack of sulfite and sulfate accumulation, low Cys accumulation, and enhanced glutathione level observed in the sulfite-tolerant SIR OE plants also support a role for SiR as a shield against sulfite toxicity. However, the enhanced sulfite resistance of the SIR OE plants was probably also due to the increase in SO activity seen 3 h after the 8 mm sulfite injections. It seems likely that SiR overexpression acted together with normal SO activities to prevent sulfite and sulfate accumulation in the SIR OE lines under sulfite stress (Figs. 5 and 8; Table I; Supplemental Tables S1 and S2). These results indicate that, in addition to being an important component in the sulfate assimilation reductive pathway, SiR plays a role in protecting leaves against sulfite toxicity.
Plants are unlikely to meet situations of acute rises in exogenous sulfite in nature; hence, the rapid metabolic perturbations recorded here as a result of such application are not the consequence of direct evolutionary pressure. It is more likely that shifting abiotic circumstances, such as those encountered during natural growth dynamics, provide the driving force for the development of sulfite control by SiR. The study of these conditions is part of our future work.
MATERIALS AND METHODS
Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col plants were germinated as described by Brychkova et al. (2012a). The seedlings were transferred to pots containing a low-nutrient soil at the age of 8 to 9 d. Tomato plants (Solanum lycopersicum ‘Rheinlands Ruhm’) were germinated on filter paper, soaked with water, and transferred at the stage of cotyledons to pots filled with a peat and vermiculite (4:1, v/v) mixture containing slow-release high-nitrogen Multicote 4 with microelements (0.3%, w/w; Haifa Chemicals; http://www.haifachem.com/). Arabidopsis and tomato plants were grown in a growth room under 12 h of light/12 h of dark, 22°C, 75% to 85% relative humidity, and 100 µmol m−2 s−1 light intensity. All plants in pots were supplemented with 20-20-20 (Haifa Chemicals) water-soluble fertilizer (1 g L−1) once per week.
Plants with Modulated SiR and SO Activities
The generation of AtSiR and SlSiR RNA interference lines with reduced SiR expression and AtSiR and SlSiR overexpression lines with enhanced SiR expression used here has been described before (Brychkova et al., 2012b). The copy number of the construct inserted into the genome of Arabidopsis and tomato SIR OE and SIR Ri lines (Brychkova et al., 2012b) was estimated first by resistance to Basta (glufosinate ammonium; Aventis CropScience; http://www.aventis.com) and kanamycin (kanamycin sulfate; Sigma-Aldrich), respectively. Finally, quantitative PCR was employed to verify the copy number of the inserted constructs in the genome of the transgenic plants. The primers 35S-Fw and 35S-Rw spanning the 35S promoter were used for Arabidopsis SIR OE and tomato SIR Ri and SIR OE lines. Arabidopsis SIR Ri lines were analyzed employing the sense primer 35SA-Fw and the antisense primer Rint-Rw spanning the 35S promoter and the intron regions of the pRNA69 plasmid (Supplemental Table S3). It was revealed that Arabidopsis SIR OE7 and SIR OE12 lines, as well as tomato SIR OE10 and SIR Ri40 lines, contained a single insertion, while tomato SIR OE3 and the Arabidopsis RNA interference lines contained two insertions. Additionally, the heterozygous tomato SIR Ri37 line, which was amenable to propagation only by vegetative means, contained at least two insertions of T-DNA.
Arabidopsis plants with T-DNA insertions in close proximity to the SiR gene were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/) and annotated as SIR KD1T (SALK075776), SIR KD2T (SAIL867D09), and SIR KD3P (SAIL1223C03). Homozygous forms of these lines were selected using PCR screening with the specific sets of primers (Supplemental Fig. S1). The following primers were used for the identification of homozygous SIR-KD1T lines: the T-DNA insertion (pBIN-pROK2 plasmid)-specific primer LBb1.3 and the primers SIR KD1T-LP and SIR KD1T-RP flanking the insertion site. The lines SIR-KD2T and SIR-KD3P, which belonged to the SAIL transgenic plant collection, were analyzed with the T-DNA (pCSA110-pDAP101 plasmid)-specific primer LB2 in combination with the gene-specific primer pairs SIR KD2T-LP and SIR KD2T-RP for SIR-KD2T and SIR KD3P-LP and SIR KD3P-RP for SIR-KD3P (Supplemental Table S3). The PCR products were sequenced to confirm the positions of T-DNA insertions in the mutant genomes.
Plants Harboring GUS and Dronpa Expression under the Control of the AtSiR Promoter and Their Analysis
The 2,187-bp fragment carrying the upstream region of the Arabidopsis SiR gene was amplified using genomic DNA as a template and the primers AtSIRpr-Fw and AtSIRpr-Rw (Supplemental Table S3). Following digestion with EcoRI and XhoI, the fragment was cloned into the pRITA and pART7:dronpa plasmids upstream of the coding regions of GUS and Dronpa, respectively. The NotI-excised expression cassettes were recloned into pMLBart to produce the final plasmids for Arabidopsis transformation. The cloned region of the SiR promoter was sequenced for verification.
The resulting constructs were transformed into Arabidopsis Col plants using the floral dip method (Clough and Bent, 1998). The presence of GUS and Dronpa genes in the genomes of the transgenic plants and their homozygosity were confirmed by performing Basta spray and PCR with gene-specific primers (GUS-Fw and GUS-Rw for GUS, Dronpa-Fw and Dronpa-Rw for Dronpa; Supplemental Table S3) with sequencing of the generated PCR fragments (191 and 194 bp, respectively). The lines containing one T-DNA insertion in the Arabidopsis genome were used for further analysis. Histochemical staining was performed to detect GUS activity employing 5-bromo-4-chloro-indolyl-galactopyranoside solutions during 1 to 2 h at 37°C (Jefferson et al., 1987). The stained tissues were analyzed using Stemi SV6 or Axio Imager A1 microscopes with an AxioCamMR5 digital camera (Carl Zeiss; http://www.zeiss.com/). Cross sections of flowering stem and leaf petioles were carried out with GUS-stained samples after their fixation in 4% paraformaldehyde and embedding in melted Paraplast. The photographs of Dronpa fluorescence in plants were taken under blue light illumination and with a green optical filter using stereomicroscope Stemi SV6 with attached AxioCamMR5 digital camera (Carl Zeiss). Dronpa expression under the control of the SiR promoter in the basal part of Arabidopsis siliques (Fig. 1P) was detected with the LSM 510 META Laser Scanning Microscope (Carl Zeiss) using a 488-nm laser beam and a BP505-520 filter.
Sulfite/SO2 Application
Four-week-old Arabidopsis and 5-week-old tomato plants were employed for the experiments with sulfite/SO2 applications. SO2 fumigation was performed as described (Brychkova et al., 2007). Solutions with 0.5, 4, and 8 mm sulfite for infiltration by injection into plant leaves were prepared by immersing sodium sulfite in doubly distilled water and adjusting the pH to 5.7 using concentrated HCl. The sulfite solutions were used promptly, within 30 min of preparation. The solutions were injected into all leaves in the case of Arabidopsis and into the third and fourth leaves from the bottom in tomato. Leaves injected with water without sulfite adjusted to pH 5.7 were used as controls (mock). The injected leaves (the third leaf of tomato plants and all leaves of Arabidopsis) were sampled 30 min, 3 h, 8 h, or 24 h after sulfite or water injections and stored at −80°C before RNA, metabolite, or protein extraction. To estimate the amount of sulfite supplemented by injection into leaves, 50 detached Arabidopsis and tomato leaves were weighed before and after injections, and the sulfite increments were expressed as µmol sulfite g−1 fresh weight.
Tomato leaf discs were treated as described (Brychkova et al., 2007). Leaves of plants subjected to sulfite/SO2 treatments were photographed 24 or 72 h after application, and the damaged area was evaluated by employing ImageJ software (http://rsbweb.nih.gov/ij/). The damaged area measured included the nonnecrotic leaf area that underwent shrinking as a result of leaf deformation by the toxic sulfite as well as the area of tissue with clear necrosis. The areas of damage were expressed as a percentage of the total initial leaf area. Chlorophyll content was measured in extracts of the leaves as described previously (Brychkova et al., 2007), and the remaining chlorophyll content was determined as the quantity of chlorophyll per leaf disc, divided by the same value in the control plant (mock), expressed as a percentage.
Preparation of RNA and Quantitative Real-Time Reverse Transcription-PCR
The Aurum total RNA mini kit and the iScript complementary DNA synthesis kit (Bio-Rad) were used to extract RNA and prepare complementary DNA according to the manufacturer’s recommendations. The quantitative analysis of transcripts in the treated and control plants was performed as described (Brychkova et al., 2007) employing suitable primers (for primers, their sequences, and expected PCR products, see Supplemental Table S4). Real-time PCR was normalized using ACTIN2 (At3g18780) and ELONGATION FACTOR1-α (At5g60390) for Arabidopsis and TRANSCRIPTION FACTORIID (TFIID, SGN-U571616) and ELONGATION FACTOR1-α (SGN-U196120) for tomato plants as housekeeping genes revealed similar results. The data are presented as relative expression (means ± se, n = 3 or 4).
The number of insertions in SIR OE and SIR Ri plants was determined by quantitative PCR on genomic DNA using primers to the 35S promoter (35S-Fw and 35S-Rw) or primers spanning the 35S promoter and the intron in pRNA69 plasmid (in the case of Arabidopsis SIR Ri, primers 35SA-Fw and Rint-Rw). The sequences of the primers are shown in Supplemental Table S3. The ACTIN2 (At3g18780) and ACTIN Tom41 (SGN-U60480) genes were used for normalization of DNA samples for Arabidopsis and tomato, respectively (Supplemental Table S4). All PCR fragments were sequenced for verification.
Soluble Protein, SiR and SO Immunoblot Analysis, Kinetic Activities and Metabolite Determination, and Total Sulfur Content Measurement
Protein extraction and immunoblot analysis of SiR protein, SiR activity, and SO activity were performed as described recently (Brychkova et al., 2012a, 2012b). SiR activity was expressed as nmol Cys mg−1 protein min−1, and SO activity was expressed as nmol sulfite mg−1 protein min−1. Determination of sulfite, Cys, glutathione, and sulfate was performed as described recently (Brychkova et al., 2012a) and normalized to the fresh weight of the water-injected control leaves (Table I). Soluble proteins were determined according to Bradford (1976).
Total sulfur content in the Arabidopsis plants was measured in dried samples (10 mg) that were digested in HNO3:HClO4 (83:17, v/v) in accordance with Zarcinas et al. (1987) employing an inductively coupled plasma spectrophotometer (SPECTRO model CIROS CCD; http://www.spectro.com) to detect sulfur in the samples.
Statistical Analysis
Two-tailed unpaired Student’s t test (two-sample unequal variance) was employed to show differences between pairs of samples. ANOVA (Tukey-Kramer honestly significant difference [HSD]) was used to compare multiple groups of samples (JMP 8.0 software; http://www.jmp.com/). The bands on SiR immunoblotting were analyzed with ImageJ software (http://rsbweb.nih.gov/ij/) in accordance with the developer’s recommendations.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JQ341913 (SlSiR).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genomic characterization of Arabidopsis SiR knockdown mutants.
Supplemental Figure S2. Appearance of SO2-related damage on the Arabidopsis wild type and SIR-modified plants.
Supplemental Figure S3. Response of Arabidopsis plants to injections with sulfite solutions.
Supplemental Figure S4. Tomato leaves injected with 0.5 mM sulfite show no damage.
Supplemental Figure S5. Response of tomato SiR mutants to sulfite.
Supplemental Figure S6. Time course of sulfite levels in tomato sulfite-injected leaves.
Supplemental Figure S7. Expression of SiR and SO in tomato plants in response to sulfite injections.
Supplemental Figure S8. SiR activity in tomato plants in response to sulfite injection.
Supplemental Table S1. SiR activity in water-injected plants.
Supplemental Table S2. SO activity in water-injected Arabidopsis plants.
Supplemental Table S3. List of primers used for transgenic plant production and verification.
Supplemental Table S4. List of primers used for quantitative real-time PCR.
Acknowledgments
We thank Dr. Olga Davidov (Department of Plant Sciences, Weizmann Institute) for generating the SiR-modified tomato plants.
Glossary
- EC
Enzyme Commission
- APS
adenosine 5-phosphosulfate
- APR
5′-phosphosulfate reductase
- SiR
sulfite reductase
- SO2
sulfur dioxide
- SO
sulfite oxidase
- Col
ecotype Columbia
References
- Armengaud J, Gaillard J, Forest E, Jouanneau Y. (1995) Characterization of a 2[4Fe-4S] ferredoxin obtained by chemical insertion of the Fe-S clusters into the apoferredoxin II from Rhodobacter capsulatus. Eur J Biochem 231: 396–404 [DOI] [PubMed] [Google Scholar]
- Björkman M, Klingen I, Birch AN, Bones AM, Bruce TJ, Johansen TJ, Meadow R, Mølmann J, Seljåsen R, Smart LE, et al. (2011) Phytochemicals of Brassicaceae in plant protection and human health: influences of climate, environment and agronomic practice. Phytochemistry 72: 538–556 [DOI] [PubMed] [Google Scholar]
- Bork C, Schwenn JD, Hell R. (1998) Isolation and characterization of a gene for assimilatory sulfite reductase from Arabidopsis thaliana. Gene 212: 147–153 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Xia Z, Yang G, Yesbergenova Z, Zhang Z, Davydov O, Fluhr R, Sagi M. (2007) Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J 50: 696–709 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Yarmolinsky D, Fluhr R, Sagi M. (2012a) The determination of sulfite levels and its oxidation in plant leaves. Plant Sci 190: 123–130 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Yarmolinsky D, Ventura Y, Sagi M. (2012b) A novel in-gel assay and an improved kinetic assay for determining in vitro sulfite reductase activity in plants. Plant Cell Physiol 53: 1507–1516 [DOI] [PubMed] [Google Scholar]
- Cannon GC, Ward LN, Case CI, Heinhorst S. (1999) The 68 kDa DNA compacting nucleoid protein from soybean chloroplasts inhibits DNA synthesis in vitro. Plant Mol Biol 39: 835–845 [DOI] [PubMed] [Google Scholar]
- Chi-Ham CL, Keaton MA, Cannon GC, Heinhorst S. (2002) The DNA-compacting protein DCP68 from soybean chloroplasts is ferredoxin:sulfite reductase and co-localizes with the organellar nucleoid. Plant Mol Biol 49: 621–631 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eilers T, Schwarz G, Brinkmann H, Witt C, Richter T, Nieder J, Koch B, Hille R, Hänsch R, Mendel RR. (2001) Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase: a new player in plant sulfur metabolism. J Biol Chem 276: 46989–46994 [DOI] [PubMed] [Google Scholar]
- Habuchi S, Ando R, Dedecker P, Verheijen W, Mizuno H, Miyawaki A, Hofkens J. (2005) Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci USA 102: 9511–9516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsch R, Mendel RR. (2008) Sulfite oxidation in plants. In Govindjee, J Amesz, J Barber, RE Blankenship, N Murata, WL Ogren, DR Ort, eds, Sulfur Metabolism in Phototrophic Organisms, Vol 27. Springer, Dordrecht, The Netherlands, pp 223–230
- Hopkins RJ, van Dam NM, van Loon JJ. (2009) Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu Rev Entomol 54: 57–83 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YW, Lee JY, Jeon Y, Cheong GW, Kim M, Pai HS. (2010) In vivo effects of NbSiR silencing on chloroplast development in Nicotiana benthamiana. Plant Mol Biol 72: 569–583 [DOI] [PubMed] [Google Scholar]
- Khan MS, Haas FH, Samami AA, Gholami AM, Bauer A, Fellenberg K, Reichelt M, Hänsch R, Mendel RR, Meyer AJ, et al. (2010) Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. Plant Cell 22: 1216–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisker C, Schindelin H, Rees DC. (1997) Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu Rev Biochem 66: 233–267 [DOI] [PubMed] [Google Scholar]
- Kopriva S. (2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot (Lond) 97: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Koprivova A. (2004) Plant adenosine 5′-phosphosulphate reductase: the past, the present, and the future. J Exp Bot 55: 1775–1783 [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Davies A, Deeken R, Thorpe MR, Tomos AD, Hedrich R. (2000) Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol 124: 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Gibson TM, Cramer R, Stain C. (2010) Glucosinolate-accumulating S-cells in Arabidopsis leaves and flower stalks undergo programmed cell death at early stages of differentiation. Plant J 64: 456–469 [DOI] [PubMed] [Google Scholar]
- Krueger RJ, Siegel LM. (1982) Spinach siroheme enzymes: isolation and characterization of ferredoxin-sulfite reductase and comparison of properties with ferredoxin-nitrite reductase. Biochemistry 21: 2892–2904 [DOI] [PubMed] [Google Scholar]
- Lang C, Popko J, Wirtz M, Hell R, Herschbach C, Kreuzwieser J, Rennenberg H, Mendel RR, Hänsch R. (2007) Sulphite oxidase as key enzyme for protecting plants against sulphur dioxide. Plant Cell Environ 30: 447–455 [DOI] [PubMed] [Google Scholar]
- Leustek T. (2002) Sulfate metabolism. The Arabidopsis Book 1: e0017, doi/10.1199/tab.0017 [DOI] [PMC free article] [PubMed]
- Leustek T, Saito K. (1999) Sulfate transport and assimilation in plants. Plant Physiol 120: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F. (2007) Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet 39: 896–900 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Kuramata M, Kasugai I, Abe M, Youssefian S. (2009) Increased thiol biosynthesis of transgenic poplar expressing a wheat O-acetylserine(thiol) lyase enhances resistance to hydrogen sulfide and sulfur dioxide toxicity. Plant Cell Rep 28: 313–323 [DOI] [PubMed] [Google Scholar]
- Nakayama M, Akashi T, Hase T. (2000) Plant sulfite reductase: molecular structure, catalytic function and interaction with ferredoxin. J Inorg Biochem 82: 27–32 [DOI] [PubMed] [Google Scholar]
- Nandi PK, Agrawal M, Agrawal SB, Rao DN. (1990) Physiological responses of Vicia faba plants to sulfur dioxide. Ecotoxicol Environ Saf 19: 64–71 [DOI] [PubMed] [Google Scholar]
- Noji M, Saito M, Nakamura M, Aono M, Saji H, Saito K. (2001) Cysteine synthase overexpression in tobacco confers tolerance to sulfur-containing environmental pollutants. Plant Physiol 126: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanz H, Martinoia E, Lange OL, Heber U. (1987) Flux of SO2 into leaf cells and cellular acidification by SO2. Plant Physiol 85: 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randewig D, Hamisch D, Herschbach C, Eiblmeier M, Gehl C, Jurgeleit J, Skerra J, Mendel RR, Rennenberg H, Hänsch R. (2012) Sulfite oxidase controls sulfur metabolism under SO2 exposure in Arabidopsis thaliana. Plant Cell Environ 35: 100–115 [DOI] [PubMed] [Google Scholar]
- Ranieri A, Castagna A, Bini L, Soldatini GF. (1995) Two-dimensional electrophoresis of thylakoid protein patterns in two wheat cultivars with different sensitivity to sulfur dioxide. Electrophoresis 16: 1301–1304 [DOI] [PubMed] [Google Scholar]
- Ravilious GE, Jez JM. (2012) Structural biology of plant sulfur metabolism: from assimilation to biosynthesis. Nat Prod Rep 29: 1138–1152 [DOI] [PubMed] [Google Scholar]
- Sato N, Nakayama M, Hase T. (2001) The 70-kDa major DNA-compacting protein of the chloroplast nucleoid is sulfite reductase. FEBS Lett 487: 347–350 [DOI] [PubMed] [Google Scholar]
- Sekine K, Fujiwara M, Nakayama M, Takao T, Hase T, Sato N. (2007) DNA binding and partial nucleoid localization of the chloroplast stromal enzyme ferredoxin:sulfite reductase. FEBS J 274: 2054–2069 [DOI] [PubMed] [Google Scholar]
- Sekine K, Hase T, Sato N. (2002) Reversible DNA compaction by sulfite reductase regulates transcriptional activity of chloroplast nucleoids. J Biol Chem 277: 24399–24404 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA. (2010) Biosynthesis of glucosinolates: gene discovery and beyond. Trends Plant Sci 15: 283–290 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides G, Martin M, Chalam R, Tarczynski MC, Schmidt A, Leustek T. (2002) Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J 32: 879–889 [DOI] [PubMed] [Google Scholar]
- Van Der Kooij TAW, De Kok LJ, Haneklaus S, Schnug E. (1997) Uptake and metabolism of sulphur dioxide by Arabidopsis thaliana. New Phytol 135: 101–107 [DOI] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, Krähenbühl U, den Camp RO, Brunold C. (2002) Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J 31: 729–740 [DOI] [PubMed] [Google Scholar]
- Wang YH. (2008) How effective is T-DNA insertional mutagenesis in Arabidopsis? Journal of Biochemical Technology 1: 11–20 [Google Scholar]
- Wen J-Q, Wen B-C, Wen H-G. (1996) Changes in protein and amino acid levels during growth and senescence of Nicotiana rustica callus. J Plant Physiol 148: 707–710 [Google Scholar]
- Wu Y, Zheng F, Ma W, Han Z, Gu Q, Shen Y, Mi H. (2011) Regulation of NAD(P)H dehydrogenase-dependent cyclic electron transport around PSI by NaHSO3 at low concentrations in tobacco chloroplasts. Plant Cell Physiol 52: 1734–1743 [DOI] [PubMed] [Google Scholar]
- Yang L, Stulen I, De Kok LJ. (2006) Impact of sulfate nutrition on the utilization of atmospheric SO2 as sulfur source for Chinese cabbage. J Plant Nutr Soil Sci 169: 529–534 [Google Scholar]
- Yonekura-Sakakibara K, Onda Y, Ashikari T, Tanaka Y, Kusumi T, Hase T. (2000) Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol 122: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarcinas BA, Cartwright B, Spouncer LR. (1987) Nitric acid digestion and multi-element analysis of plant material by inductively coupled plasma spectrometry. Commun Soil Sci Plant Anal 18: 131–146 [Google Scholar]