Summary: Under natural sunlight, this study demonstrates multiple and complex roles for the UV-B photoreceptor UV RESISTANCE LOCUS 8 in the acclimation of Arabidopsis plants to UV radiation.
Abstract
Photomorphogenic responses triggered by low fluence rates of ultraviolet B radiation (UV-B; 280–315 nm) are mediated by the UV-B photoreceptor UV RESISTANCE LOCUS8 (UVR8). Beyond our understanding of the molecular mechanisms of UV-B perception by UVR8, there is still limited information on how the UVR8 pathway functions under natural sunlight. Here, wild-type Arabidopsis (Arabidopsis thaliana) and the uvr8-2 mutant were used in an experiment outdoors where UV-A (315–400 nm) and UV-B irradiances were attenuated using plastic films. Gene expression, PYRIDOXINE BIOSYNTHESIS1 (PDX1) accumulation, and leaf metabolite signatures were analyzed. The results show that UVR8 is required for transcript accumulation of genes involved in UV protection, oxidative stress, hormone signal transduction, and defense against herbivores under solar UV. Under natural UV-A irradiance, UVR8 is likely to interact with UV-A/blue light signaling pathways to moderate UV-B-driven transcript and PDX1 accumulation. UVR8 both positively and negatively affects UV-A-regulated gene expression and metabolite accumulation but is required for the UV-B induction of phenolics. Moreover, UVR8-dependent UV-B acclimation during the early stages of plant development may enhance normal growth under long-term exposure to solar UV.
Plants use UV as an environmental cue to regulate a wide range of physiological processes. Low fluence rates of short-wavelength UV (280–315 nm; UV-B) induce photomorphogenic responses such as the inhibition of hypocotyl elongation, expression of UV-protective genes, and the accumulation of phenolic compounds, as well as regulating leaf growth and stomatal differentiation (Jenkins, 2009; Wargent et al., 2009b). These UV-B photomorphogenic responses are mediated by the UV-B photoreceptor, UV RESISTANCE LOCUS8 (UVR8; Rizzini et al., 2011). However, UV-B is very energetic, and high UV-B irradiance can induce the formation of reactive oxygen species, cause damage to plant cells, DNA, and proteins and the photosynthesis apparatus, and affect growth and development (Jenkins, 2009). These are considered stress responses and are thought to be regulated by other pathways that do not require UVR8 (Brown and Jenkins, 2008). While essentially all radiation in the shorter part of the UV-B (280–293 nm) is absorbed in the stratosphere by ozone, UV-A (315–400 nm) is the major UV component of the solar spectrum to which plants are exposed. UV-A and blue light are key factors in the photorepair of DNA damage caused by UV-B. In addition, UV-A and the high irradiance of photosynthetically active radiation (PAR) induce the expression of genes conferring UV protection and the accumulation of phenolics in plants (Ibdah et al., 2002; Götz et al., 2010; Morales et al., 2010).
The recent characterizations of UVR8 as a UV-B photoreceptor (Rizzini et al., 2011) and the mechanisms of UV-B absorption by UVR8 (Wu et al., 2011, 2012; Christie et al., 2012) have advanced our understanding of UV-B perception in plants. UVR8 is a seven-bladed β-propeller protein with sequence similarity to the human REGULATOR OF CHROMATIN CONDENSATION1 (RCC1; Kliebenstein et al., 2002). However, UVR8 and RCC1 differ in activity and function (Jenkins, 2009; Rizzini et al., 2011) and also in their monomeric topology (Wu et al., 2011; Christie et al., 2012). Under visible light (400–750 nm), UVR8 appears in plants as a dimer; however, after UV-B perception by Trp-285 and Trp-233, the salt bridges joining the dimer break, splitting UVR8 into monomers (Christie et al., 2012; Wu et al., 2012). UVR8 monomers interact with the E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1; Rizzini et al., 2011). The interaction between UVR8-COP1 occurs within minutes of UVR8 perception of UV-B and is crucial for relaying the signal that activates gene expression and UV-B acclimation in plants (Favory et al., 2009). Recent research with the uvr8-2 mutant has shown that 27 amino acids toward the C terminus of UVR8 are required for the interaction with COP1 and for the protein to be functional (Cloix et al., 2012). Accordingly, uvr8-2 fails to induce the UVR8-UV-B-regulated expression of CHALCONE SYNTHASE (CHS), the first enzyme committed in the flavonoid pathway (Brown et al., 2005; Cloix et al., 2012), and also shows phenotypic differences from its wild type under UV-B (Brown and Jenkins, 2008). Downstream of UVR8 and COP1, the transcription factors ELONGATED HYPOCOTYL5 (HY5) and the ELONGATED HYPOCOTYL5 HOMOLOG act redundantly to regulate the expression of most of the genes involved in the UVR8 photoregulatory pathway (Brown and Jenkins, 2008). Transcriptome analyses of uvr8 mutants exposed to low fluence rates of UV-B indoors have shown that UVR8 is required for the induction of genes with important functions in UV protection (flavonoid and alkaloid pathways), photorepair of DNA damage induced by UV-B, oxidative stress, chloroplast proteins, and several transcription factors (Brown et al., 2005; Favory et al., 2009). Other data have indicated that the proteins REPRESSOR OF PHOTOMORPHOGENESIS1 (RUP1) and RUP2 interact with UVR8 and are negative regulators of the UVR8 pathway (Gruber et al., 2010; Cloix et al., 2012). While the roles of UVR8 in orchestrating transcript accumulation following UV-B absorption are now clear, we have yet to elucidate how UVR8 regulates the accumulation of metabolites through downstream gene expression.
Plants are highly efficient in the synthesis and accumulation of phenolic compounds, especially flavonoids and hydroxycinnamic acids to provide protection from UV radiation. Several studies report that uvr8 mutants exposed to supplementary UV-B under controlled-environment conditions have lower amounts of phenolics than their respective wild types (Kliebenstein et al., 2002; Brown et al., 2005; González Besteiro et al., 2011; Demkura and Ballaré, 2012). However, the manner by which UVR8 regulates the accumulation of phenolics and other metabolites under natural sunlight has not yet been studied, and furthermore, the role of UVR8 and its possible interaction with UV-A/blue light signaling in mediating the UV-A induction of several metabolites is not well understood. This information is important for understanding the mechanisms, driven by UVR8 and/or other photoreceptors, that provide solar UV protection in plants. In natural environments, plants are usually exposed to high and variable fluxes of PAR and also much higher irradiances of UV-A than of UV-B radiation. Field studies have shown that besides UV-B, solar UV-A regulates gene expression and the accumulation of flavonoids in silver birch (Betula pendula) leaves (Morales et al., 2010). Moreover, it is usually specific compounds rather than the total phenolics that are differentially regulated by different doses of solar UV-A and UV-B radiation (Kotilainen et al., 2009; Morales et al., 2010). In addition to phenolics, plants synthesize antioxidants such as vitamins C and E, glutathione, and carotenoids to cope with oxidative stress (Chen and Xiong, 2005). Pyridoxine (vitamin B6) is another essential antioxidant that may provide protection from UV-B (Ristilä et al., 2011). Two proteins are involved in vitamin B6 biosynthesis, PYRIDOXINE BIOSYNTHESIS1 (PDX1) and PDX2 (Denslow et al., 2007). It has been reported that Arabidopsis (Arabidopsis thaliana) leaves exposed to supplementary UV-B quickly accumulate PDX1 and vitamin B6 (Ristilä et al., 2011). Furthermore, transcripts of PDX1.3 induced by low levels of UV-B radiation are thought to be regulated by UVR8 and COP1 (Ristilä et al., 2011). However, it remains unclear how PDX1 accumulates under natural fluxes of solar UV-A and UV-B radiation and if UVR8 is involved in this process.
Most studies characterizing the UVR8 signaling pathway in Arabidopsis have been conducted under controlled environmental conditions (Kliebenstein et al., 2002; Brown et al., 2005, 2009; Oravecz et al., 2006; Brown and Jenkins, 2008; Favory et al., 2009; Wargent et al., 2009a; González Besteiro et al., 2011; Demkura and Ballaré, 2012). These studies have substantially expanded our mechanistic understanding of plant responses to low fluence rates of UV-B by identifying major regulators in the UVR8 pathway and by elucidating gene functions. In addition, important functions of UVR8 have been determined in photomorphogenesis, UV-B acclimation, and defense against pathogens. However, a paucity of information on how the UVR8 pathway operates under solar radiation limits our understanding of the possible roles of UVR8 in mediating plant responses to environmental factors other than UV-B and how UVR8 interacts with other light signaling pathways under solar radiation. Spectral balances of PAR, UV-A, and UV-B radiation frequently used in experiments indoors differ from that of solar radiation in the natural environment and may influence plant responses to UV-B at different molecular levels (Casati et al., 2011).
Here, outdoor experiments were designed to investigate the roles of UVR8 in regulating gene expression and metabolite accumulation under natural sunlight. We hypothesized that (1) UVR8 plays key functions in plant acclimation and survival under solar UV, but, in addition, (2) high PAR and UV-A irradiance, as present in natural sunlight, substantially modify UVR8-mediated responses to UV-B.
RESULTS
The role of different bands of solar UV in UVR8-mediated responses was studied by subjecting wild-type Arabidopsis Landsberg erecta (Ler) and uvr8-2 (Brown et al., 2005) transferred from greenhouse to field conditions under filters used to manipulate UV-A and UV-B irradiance. Samples from the wild type and uvr8-2 were taken 12 and 36 h after transfer of the seedlings outdoors and used to analyze gene expression (microarrays and real-time quantitative PCR [qPCR]), protein accumulation, and metabolite signatures.
UVR8 Is a Key Regulator of Gene Expression in Arabidopsis Leaves under Solar UV
Microarray analysis identified patterns of gene expression in wild-type and uvr8-2 plants exposed for 12 h to solar UV. In the wild type, 96 genes were differentially expressed (with at least 1.4-fold change; P ≤ 0.05) under near-ambient solar UV (UV A+B treatment; Supplemental Table S1). Genes with increased transcript accumulation included biosynthesis of flavonoids and anthocyanins (PHENYLALANINE AMMONIA LYASA [PAL], CHS, CHALCONE ISOMERASE, TRANSPARENT TESTA7 [TT7], DIHYDROFLAVONOL REDUCTASE [DFR], FLAVONOL SYNTHASE1, FLAVANONE HYDROXYLASE, LEUCOANTHOCYANIDIN DIOXYGENASE [LDOX], and UDP-GLUCOSYL TRANSFERASE78D2), protection against oxidative stress (EARLY LIGHT INDUCIBLE PROTEINs), vitamin B6 (PDX2), RUP2, and several proteins with unknown function. In contrast, genes involved in cell expansion (At2g44080, ARL), defense and stress responses (PATHOGENESIS RELATED GENE1 [PR1], BASIC PATHOGENESIS RELATED PROTEIN1, transcription factor WRKY33, RESPONSIVE TO DESSICATION22, GLUTATHIONE TRANSFERASE2 [GSTF2], GSTF8, and ALD1 [At2g13810]), and MITOGEN ACTIVATED PROTEIN KINASE3 (MPK3) had lower transcript abundance in the wild type under solar UV-A plus UV-B than under exclusion of solar UV-A and UV-B (UV 0 treatment; Supplemental Table S1). The candidate UVR8-dependent UV-regulated genes were identified by comparing transcripts of the wild type and uvr8-2 under solar UV-A plus UV-B. This comparison showed differential accumulation for 89 transcripts (P ≤ 0.07; Supplemental Table S2); at this level of significance, we found statistical differences in the expression of several genes using qPCR. Transcripts of genes involved in the biosynthesis of flavonoids and anthocyanins (DFR, LDOX, AT5MAT [At3g29590], and PRODUCTION OF ANTHOCYANIN PIGMENT1), jasmonic acid (JA) biosynthesis (ALLENE OXIDE SYNTHASE [AOS], ALLENE OXIDE CYCLASE1 [AOC1], AOC3, and OXOPHYTODIENOATE REDUCTASE3), JA signaling (transcription factor WRKY70, JASMONATE ZIM DOMAIN1 [JAZ1], SYNTAXIN RELATED PROTEIN1, and GRX480 [At1g28480]), glucosinolate biosynthesis (ATR4 [At4g31500] and SULFOTRANSFERASE17 [SOT17]), and auxin signaling(At5g47370, HAT2) were accumulated at lower levels in uvr8-2 than in the wild type. In addition, genes involved in defense responses (At4g12490, LURP1 [At2g14560], JAZ1, SYNTAXIN RELATED PROTEIN1, WRKY70, and MITOGEN ACTIVATED PROTEIN KINASE KINASE4) and salt stress (JASMONATE RESPONSIVE1 [At1g16850], AOC3, and ANNAT1 [At1g35720]) had low transcript abundance in uvr8-2 compared with the wild type (Supplemental Table S2). In contrast, genes that are known to respond to UV-B (MEB5.2 [At3g17800]) and blue and red light (SIGMA FACTOR5 [SIG5]) had increased transcript accumulation in uvr8-2. The same response was observed for genes implicated in reactive oxygen species response (FERRETIN1 and FERRETIN3), reproduction (GLYCINE RICH PROTEIN19 and N-METHYLTRANSFERASE1), lipid biosynthetic processes (N-METHYLTRANSFERASE1 and At1g11880), l-ascorbic acid biosynthesis (PHOSPHOMANNOSE ISOMERASE2 [PMI2]), zinc ion binding (At5g01520), brassinosteroid signaling (At3g05800, AIF1), and ubiquinone biosynthetic processes (SOLANESYL DIPHOSPHATE SYNTHASE [SPS]). Transcripts of several transposable elements and proteins of unknown function were also highly accumulated under solar UV-A plus UV-B in uvr8-2 (Supplemental Table S2).
The microarray data were first compared with previous transcriptome analysis of uvr8 mutants exposed to fluence rates of 3 µmol m−2 s−1 UV-B (Q-Panel UVB-313, broadband; Brown et al., 2005) and 1.5 µmol m−2 s−1 UV-B (Philips TL20W/01RS narrowband UV-B tubes; Favory et al., 2009). The low irradiances used in the latter study help to distinguish UV-B-induced photomorphogenesis from UV-B stress responses, while in the former it is possible that both photomorphogenic and stress pathways may be activated (Favory et al., 2009). The list of 96 UV-regulated genes in the wild type overlapped with 52 UV-B-regulated genes in wild-type plants from these previous studies; excluding At3g16530, all of the genes share the same patterns of expression (Supplemental Fig. S1A). However, for the UVR8-regulated genes detected under our conditions, six overlapped with Brown et al. (2005) and 12 with Favory et al. (2009; Supplemental Fig. S1, B and C). Interestingly, and in contrast with these previous studies, the transcript accumulation of many genes, including SIG5, SPS, MEB5.2, AIF1, PMI2, and At5g01520, increased in uvr8-2 under solar UV-A plus UV-B (Supplemental Table S2; Supplemental Fig. S1, B and C). We compared two gene lists (Supplemental Tables S1 and S2) with transcriptome data from Oravecz et al. (2006) to search for COP1-regulated genes under UV-B and white light. COP1 plays a central role in the UVR8 pathway as a positive regulator of gene expression under UV-B (Favory et al., 2009) but is also a negative regulator of photomorphogenesis under visible light (Oravecz et al., 2006). The Venn diagrams show that several of the UV-regulated genes under our conditions are COP1 dependent under UV-B and white light (Supplemental Fig. S1D). The comparison with data from Kleine et al. (2007) shows overlap with blue-light-regulated genes (30 in the wild type and 17 with the UVR8 list; Supplemental Fig. S1E). Genes responding to high-light treatment via CRYPTOCHROME1 (CRY1) and HY5 (Kleine et al., 2007) also overlapped in our data (Supplemental Fig. S1F).
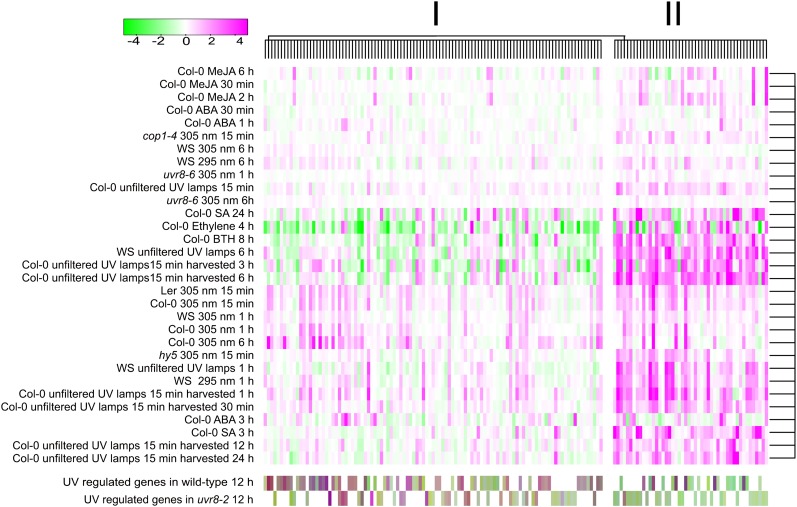
In addition, a total of 153 UV-regulated genes (96 from the wild type plus 57 from uvr8-2) were clustered with previous UV-B and plant hormone experiments performed on Affymetrix ATH1 chips using a Bayesian hierarchical clustering approach (Fig. 1). We included UV-B experiments using both filtered and unfiltered radiation (i.e. the latter includes a small, but very active, component of UV-C) in order to separate UV-B-regulated changes in gene expression from those changes arising from stress caused by high-energy radiation (Supplemental Table S3). The Bayesian clustering identified two main groups of genes (Fig. 1). Genes encoding proteins with antioxidant activity as well as flavonoid and anthocyanin biosynthesis, which are involved in UV-B acclimation, were mainly grouped in cluster I. Several genes induced by biotic and abiotic stress also appeared in cluster I; however, these types of genes and others related to secondary metabolism were more abundant in cluster II (for a complete description of the genes in each cluster, see Supplemental Table S4). Furthermore, genes in cluster II, including salicylic acid (SA) signaling (PR-1), regulation of SA signaling (WRKY70), and JA biosynthesis and signaling (JAZ1 and AOC3), had increased expression in plants treated with SA and the SA analog benzothiadiazole S-methylester (BTH). The expression of genes grouped in cluster II was consistently increased by unfiltered UV-B, which, together with their SA regulation, suggested that this group of genes responds to UV-B conditions that induce stress rather than UV-B acclimation. In contrast, many genes with increased expression in response to UV in cluster I had decreased expression in plants treated with ethylene and in some cases with SA or BTH (Fig. 1). In addition, several genes whose expression increased with filtered UV-B were present in both clusters I and II, although in cluster II unfiltered UV gave stronger induction.
Figure 1.
Bayesian hierarchical clustering of selected UV-regulated genes in UV-B and plant hormone experiments. Magenta and green indicate increased and decreased expression, respectively, in treated plants compared with untreated controls. Detailed information on the experimental data sets, gene identification, fold change induction, and cluster association is provided in Supplemental Tables S3 and S4. ABA, Abscisic acid; Col-0, Columbia; MeJA, methyl jasmonate; WS, Wassilewskija.
Effects of Solar UV-A and UV-B Radiation on Gene Expression as Measured with qPCR
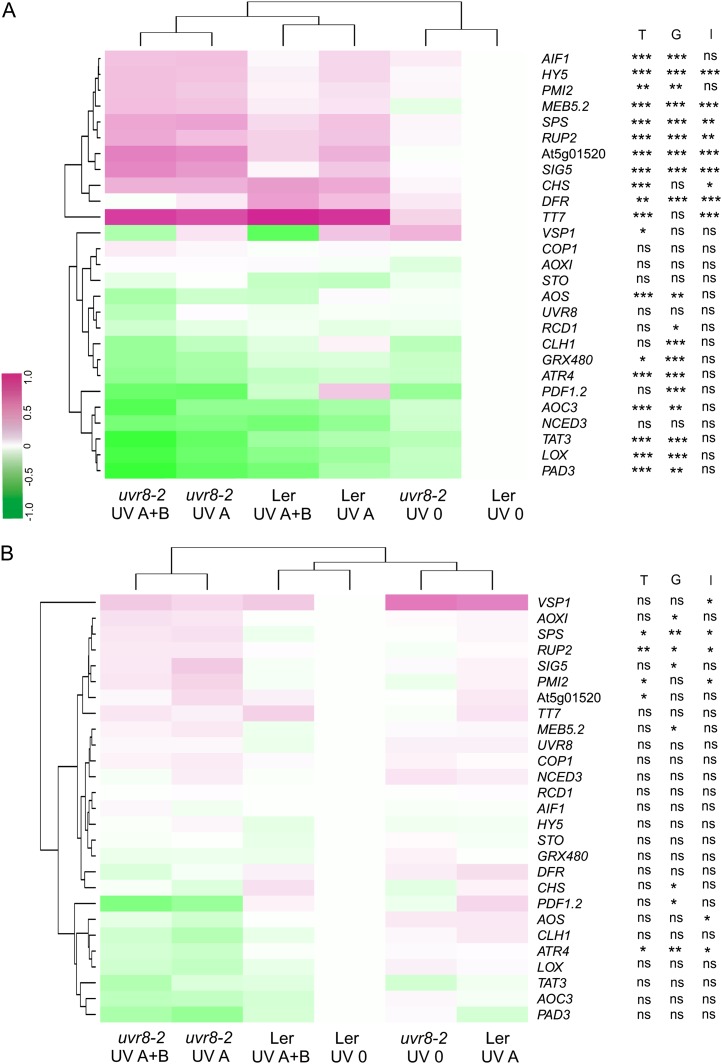
qPCR was used to confirm the microarray results and also to determine the effects of solar UV-A and UV-B radiation on gene expression after 12 and 36 h of exposure outdoors. Transcript accumulation of 15 genes showing altered expression as measured with microarrays was further studied with qPCR. In addition, several known UV-B and defense-related markers were included in the analysis (Fig. 2). After 12 h, more genes were differentially regulated than after 36 h (Fig. 2). Statistical analysis of the qPCR data using two-way ANOVA demonstrated significant main effects of the UV treatments and genotypes on the expression of 19 genes after 12 h. The UV treatment × genotype interaction was also significant (P ≤ 0.05) for nine genes (Fig. 2A; Supplemental Table S5). For those genes exhibiting a significant interaction, contrasts were fitted between the UV treatments within each genotype, and the effects of solar UV-A and UV-B on gene expression were determined from the contrasts UV 0 versus exclusion of solar UV-B (UV A treatment) and UV A versus UV A+B, respectively. In both genotypes, UV-A enhanced the transcript accumulation of UV-B-regulated genes (HY5, MEB5.2, RUP2, CHS, and TT7), SPS, At5g01520, and SIG5. Another gene responding to UV-B (DFR) was only induced by UV-A and UV-A plus UV-B in wild-type plants. Transcript accumulation of RUP2 was further increased by UV-B in uvr8-2 plants. In contrast, the wild type had less HY5, SPS, At5g01520, and SIG5 transcripts in plants that received UV-A and UV-B than UV-A alone (Fig. 2A; Supplemental Table S6). Because the interaction UV treatment × genotype was marginally nonsignificant for AIF1 (P = 0.08), PMI2 (P = 0.15), and AOS (P = 0.10), we could not detect significant differences in the expression of these three genes between genotypes using qPCR. However, the expression values obtained for these genes with qPCR were consistent with patterns of expression identified with microarrays (Fig. 2A; Supplemental Table S2).
Figure 2.
Heat maps showing patterns of gene expression measured by qPCR in wild-type (Ler) and uvr8-2 plants after 12 h (A) and 36 h (B) outdoors. Means of log-transformed expression values relative to wild-type UV 0 are presented (n = 3). Main significant effects of the UV treatments (T), genotype (G), and the interaction UV treatment × genotype (I) on the expression of each gene are denoted with asterisks (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001), and ns indicates nonsignificant effects.
For genes where ANOVA only showed significant main effects of the UV treatments and genotypes but no significant interaction, the effects of solar UV-A and UV-B radiation on gene expression were determined by fitting contrasts between the UV treatments pooling the genotypes. It was found that UV-A lowered the expression of genes with activity in JA biosynthesis (AOC3 and LIPOXYGENASE4 [LOX4]), JA response (TYROSINE AMINOTRANSFERASE3 [TAT3]), biosynthesis of glucosinolates (ATR4), and camalexin (PHYTOALEXIN DEFICIENT3 [PAD3]) but increased AIF1 and PMI2. Transcripts of defense-related genes (VEGETATIVE STORAGE PROTEIN1 [VSP1] and PLANT DEFENSIN1.2 [PDF1.2]) and AOS were decreased by UV-B (Fig. 2A; Supplemental Table S6). The marginally nonsignificant interaction might be related to a larger effect of UV-B on AIF1 expression in the wild type compared with uvr8-2 (Fig. 2A; Supplemental Table S6). After 36 h outdoors, the interaction UV treatment × genotype was significant for transcripts of VSP1, SPS, RUP2, PMI2, AOS, and ATR4 (Fig. 2B; Supplemental Table S5). After fitting contrasts, we did not find significant effects of UV-A or UV-B on the expression of these genes in the wild type. In uvr8-2, UV-A increased the levels of SPS, RUP2, and PMI2 and reduced the transcript accumulation of AOS and ATR4. At this time point, UV-B had no effects on gene expression in uvr8-2 (Fig. 2B; Supplemental Table S6). For VSP1 and other genes with a nonsignificant interaction (AOXI, SIG5, At5g01520, MEB5.2, CHS, and PDF1.2), the contrasts between UV treatments were not significant (Fig. 2B; Supplemental Table S6). We also fitted linear mixed-effect models within the UV treatments for genes having a significant UV treatment × genotype interaction after 12 and 36 h. The ANOVAs revealed a significant effect of the genotype under the treatments UV A and UV A+B but not under UV 0 (Supplemental Table S6), indicating that the differences between genotypes detected for these genes in the full statistical model are due to the effect of UVR8 in the presence of UV radiation (see also Supplemental Fig. S4).
UVR8 Modulates PDX1 Accumulation in Arabidopsis Leaves under Solar UV
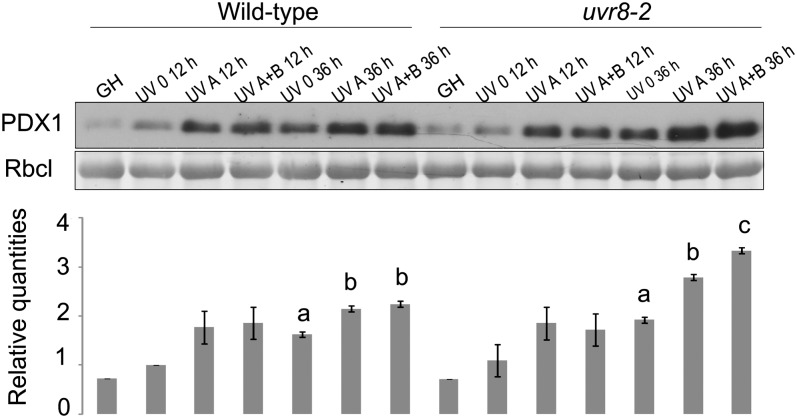
PDX1 is a protein involved in the biosynthesis of vitamin B6 that accumulates greatly in tissues exposed to UV-B (Ristilä et al., 2011). To test the role of UVR8 on PDX1 accumulation under natural sunlight, the induction of PDX1 in leaves of the wild type and uvr8-2 exposed for 12 and 36 h to solar UV-A and UV-B was monitored by western blot. Fold changes in PDX1 expression scaled to wild-type UV 0 after 12 h were used for statistical analysis (Fig. 3). The ANOVA revealed a significant main effect of the UV treatments (P ≤ 0.001) on PDX1 expression after 12 h outdoors. The main effect of genotype (P = 0.90) and the interaction UV treatment × genotype (P = 0.63) were not significant. The contrasts between the treatments revealed that levels of PDX1 were increased by solar UV-A in both genotypes (Fig. 3). After 36 h, ANOVA showed significant main effects of the UV treatments and genotypes on PDX1 accumulation, and the interaction UV treatment × genotype was also significant (P ≤ 0.01; Fig. 3). Solar UV-A enhanced PDX1 in leaves of both genotypes (P ≤ 0.01), whereas UV-B increased PDX1 to higher levels than UV-A alone only in uvr8-2 (P ≤ 0.01; Fig. 3). The expression of PDX1 was also measured in the wild type and uvr8-2 grown in the greenhouse to compare changes in PDX1 induction between greenhouse and field conditions. Interestingly, when both genotypes were transferred from the greenhouse to the treatment UV 0 outdoors, PDX1 increased in leaves from 12 to 36 h (Fig. 3).
Figure 3.
Accumulation of PDX1 protein in wild-type (Ler) and uvr8-2 leaves after 12 and 36 h of solar UV exposure as determined by western blot. Levels of RbcL were used to indicate equal protein loading among samples. Means of relative quantities of PDX1 scaled to wild-type UV 0 at 12 h are plotted, with se estimated for models fitted in each time point (n = 3). Significant differences (P ≤ 0.05) among the UV treatments for the expression of PDX1 after 36 h are represented with different letters. Greenhouse samples (GH) were not included in the statistical analysis; they were used to compare PDX1 induction in greenhouse and field conditions.
Solar UV-A and UV-B Radiation Induce Metabolite Accumulation in Arabidopsis Leaves via UVR8
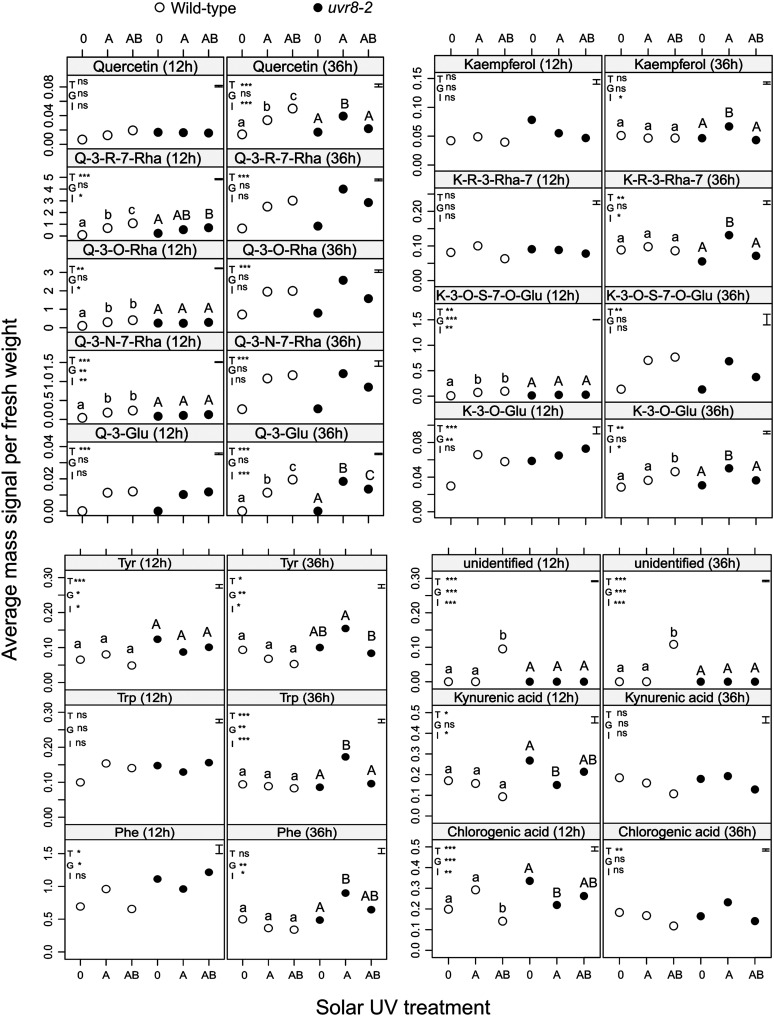
Ultra-performance liquid chromatography (UPLC) with tandem mass spectrometry (MS/MS) analysis identified 24 metabolites in Arabidopsis leaves (Supplemental Table S7). In addition to flavonoids and phenolic acids, mass signals for amino acids, Trp derivatives, and other phenylpropanoids were detected (Supplemental Table S7). After 12 h of exposure outdoors, ANOVA revealed significant main effects of the UV treatments and genotypes on the accumulation of 16 and 14 metabolites, respectively (Supplemental Table S5). A significant UV treatment × genotype interaction was detected for eight metabolites (Fig. 4). For compounds exhibiting a significant interaction, the contrasts between UV treatments within genotypes indicated that solar UV-A decreased the amount of kynurenic and chlorogenic acids in uvr8-2. UV-A increased quercetin-3-O-rutinoside-7-O-rhamnoside, quercetin-3-O-rhamnoside, quercetin-3-O-neohesperidoside-7-rhamnoside, and kaempferol-3-O-sophoroside-7-O-glucoside in the wild type (Fig. 4; Supplemental Table S6). There was no effect of solar UV-B on the accumulation of these eight metabolites in uvr8-2. However, wild-type plants responded in two different ways to solar UV-B. On the one hand, UV-B enhanced quercetin-3-O-rutinoside-7-O-rhamnoside to higher levels than UV-A alone and also induced an unidentified metabolite. On the other hand, UV-B significantly reduced the amount of chlorogenic acid in the wild type (Fig. 4; Supplemental Table S6). For metabolites where the UV treatments and genotypes had significant main effects, quercetin-3-glucoside, kaempferol-3-glucoside, and 12-hydroxy jasmonic acid 12-hexose were induced by UV-A (Fig. 4). The concentration of 12-hydroxy jasmonic acid 12-hexose was reduced by UV-B (Supplemental Table S6).
Figure 4.
Mass signals per mg of fresh weight of metabolites where ANOVA detected significant UV treatment × genotype interaction after 12 or 36 h outdoors. Main significant effects of the UV treatments (T), genotype (G), and the interaction UV treatment × genotype (I) on the accumulation of particular metabolites are denoted by asterisks (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001), and ns stands for nonsignificant effects. Average mass signals for each metabolite are presented (n = 5). The se estimated for each model is also plotted at the left top corner of every panel. Significant differences among treatments (P ≤ 0.05) are represented with different lowercase letters for the wild type and uppercase letters for uvr8-2.
After 36 h outdoors, ANOVA showed significant main effects of the UV treatments and genotypes for 18 and six metabolites, respectively (Supplemental Table S5). The interaction UV treatment × genotype was also significant for nine compounds (Fig. 4). Contrasts fitted within genotypes showed that in uvr8-2, the levels of kaempferol, kaempferol-3-O-rutinoside-7-O-rhamnoside, kaempferol-3-glucoside, quercetin, quercetin-3-glucoside, Trp, and Phe were increased by UV-A compared with UV 0, while UV-B significantly reduced this increase (Fig. 4). In addition, the amount of Tyr was reduced by UV-B in uvr8-2. In wild-type plants, the amounts of quercetin and quercetin-3-glucoside were enhanced to a greater extent by UV-A plus UV-B than by UV-A alone (Fig. 4). Kaempferol-3-glucoside and an unidentified compound were only induced by UV-B in the wild type (Fig. 4). Of the metabolites exhibiting a significant main effect of UV treatments but no significant interaction, quercetin-3-O-rutinoside-7-O-rhamnoside, quercetin-3-O-rhamnoside, quercetin-3-O-neohesperidoside-7-rhamnoside, kaempferol-3-O-sophoroside-7-O-glucoside, 12-hydroxy jasmonic acid 12-hexose, and isorhamnetin-3-O-glucoside-7-O-rhamnoside were increased by UV-A (Fig. 4; Supplemental Table S6). The amount of chlorogenic acid was reduced by UV-B (Fig. 4; Supplemental Table S6).
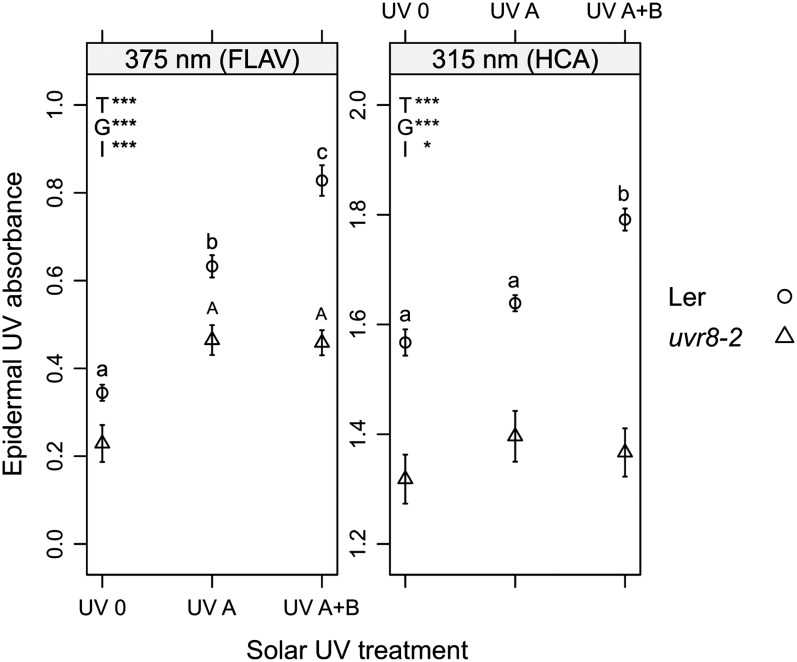
The leaf epidermal UV absorbance measured after 50 h outdoors was affected by the UV treatments, genotypes, and the interaction UV treatment × genotype as indicated by ANOVA (Fig. 5; Supplemental Table S6). Solar UV-A increased the contents of flavonoids in the epidermis measured in vivo with Dualex FLAV in leaves of both genotypes, whereas UV-B further increased flavonoid content only in wild-type plants (Fig. 5; Supplemental Table S6). The contents of hydroxycinnamic acids in the epidermis, estimated with Dualex HCA, were enhanced by UV-B in the wild type, while no effects of UV-A and UV-B were detected in uvr8-2 (Fig. 5). The ANOVA revealed a significant effect of the genotype (P ≤ 0.01) for leaf chlorophyll contents estimated with SPAD, while the UV treatments (P = 0.32) and the interaction UV treatment × genotype (P = 0.41) had no significant effect. The wild type had higher chlorophyll content than uvr8-2 under all UV treatments (approximately 13% under UV A and UV A+B and 17% under UV 0; Supplemental Table S6).
Figure 5.
Epidermal UV absorbance at the adaxial epidermis measured with Dualex FLAV and Dualex HCA in wild-type (Ler) and uvr8-2 leaves after 50 h of exposure outdoors. Means ± se (n = 30) are presented. Main significant effects of the UV treatments (T), genotype (G), and the interaction UV treatment × genotype (I) on the epidermal UV absorbance are denoted with asterisks (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001), and ns stands for nonsignificant effects. Significant differences among treatments (P ≤ 0.05) are represented with different lowercase letters for the wild type and uppercase letters for uvr8-2.
UVR8 Controls Growth and Gene Expression under Long-Term Exposure to Solar UV
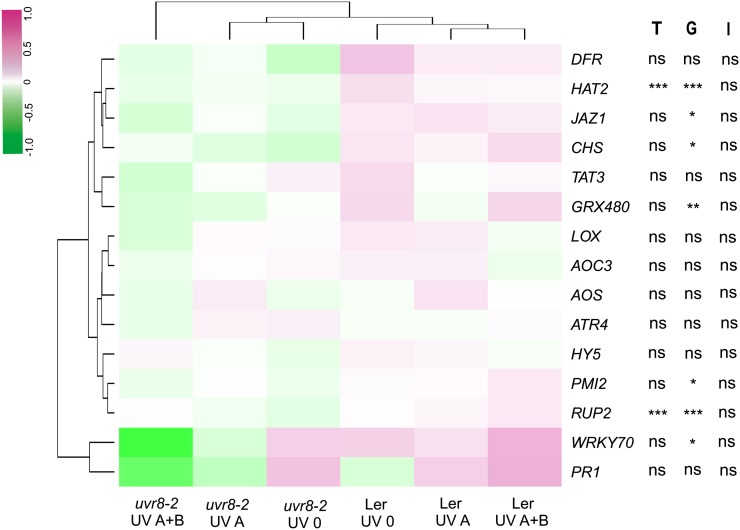
In a different experiment, we evaluated the performance of wild-type and uvr8-2 plants grown for 3 weeks under solar UV-A and UV-B radiation during summer, using the same filter types for UV treatments as in the first experiment. Both genotypes successfully germinated, developed without visible symptoms of damage, and produced flowers under the UV treatments outdoors. However, after 3 weeks, uvr8-2 plants were visibly smaller than wild-type plants and also showed a delay in flowering in all UV treatments (Supplemental Fig. S2). The ANOVA showed a significant effect of the genotype (P ≤ 0.01) and the UV treatment (P = 0.03) on the number of flowering plants, whereas the interaction UV treatment × genotype (P = 0.06) had no significant effect. Fitting contrasts between the UV treatments showed that UV-B increased the number of flowering plants in both genotypes (Supplemental Table S6). We also studied the role of UVR8 in regulating gene expression in these plants germinated and grown for 3 weeks under solar UV-A and UV-B (Fig. 6). ANOVA showed significant main effects (P ≤ 0.01) of the UV treatments on the expression of RUP2 and HAT2 and also effects of the genotype (P ≤ 0.05) for HAT2, JAZ1, CHS, GRX480, PMI2, RUP2, and WRKY70 (Fig. 6). The interaction UV treatment × genotype was not significant for any of the genes studied (Fig. 6; Supplemental Table S5), most probably due to high variation. However, there were clear differences in the expression between genotypes (Fig. 6), where the wild type and uvr8-2 under solar UV-A plus UV-B were the most distant clusters. Fitting contrasts between the UV treatments revealed that RUP2 expression was affected by UV-A and UV-B, while GRX480 was only affected by UV-B (Fig. 6; Supplemental Table S6).
Figure 6.
Heat map showing patterns of gene expression measured by qPCR in wild-type (Ler) and uvr8-2 plants germinated and grown for 3 weeks outdoors under the UV treatments. Means of log-transformed expression values are presented (n = 3). Main significant effects of the UV treatments (T), genotype (G), and the interaction UV treatment × genotype (I) on the expression of particular genes are denoted by asterisks (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001), and ns stands for nonsignificant effects.
DISCUSSION
Our results demonstrate that UVR8 is a key regulator of fundamental plant responses to solar UV. At the transcript level, UVR8 regulates the expression of genes involved in UV protection, defense responses, and the biosynthesis and signaling of JA, SA, and glucosinolates. In our experiment, under an environment of near-ambient solar radiation, solar UV-A represented approximately 98% of the ambient UV photon irradiance, and as a consequence, large effects of solar UV-A on gene expression, the accumulation of metabolites, and the expression of PDX1 were detected in Arabidopsis leaves. Moreover, our findings indicate that UVR8 may interact with UV-A/blue light signaling pathways to modulate UV-A responses. UVR8 had a UV-B-specific role in the accumulation of several phenolics. Finally, functional UVR8 may be required for normal patterns of growth and gene expression during long-term exposure to solar UV radiation but not for survival.
UVR8 Has Multiple Functions in Regulating Gene Expression in Arabidopsis Leaves Exposed to Solar UV-A and UV-B Radiation
Here, we used microarrays to detect changes in gene expression in wild-type and uvr8-2 plants exposed for 12 h to average UV-B and UV-A irradiances of 1.25 and 72.01 µmol m−2 s−1, respectively. These irradiation conditions were appropriate to study UV-B signaling under natural sunlight, as indicated by the large overlap with UV-regulated genes in wild-type plants (Brown et al., 2005; Favory et al., 2009) and the high number of genes involved in UV acclimation grouped in cluster I (Fig. 1). In agreement with Brown et al. (2005), we show that UVR8 is required for the induction of phenylpropanoid and other defense genes under solar UV. However, contrary to what might have been expected from a mutant lacking a functional UV-B photoreceptor, the expression of many UV-regulated genes was enhanced in uvr8-2. Thus, our findings indicate that transcript regulation by UVR8 in natural sunlight may be more complex than was previously anticipated from the results of indoor studies under controlled environmental conditions. This finding is supported by the low number of UVR8-regulated genes in common with Brown et al. (2005) and Favory et al. (2009) and also by the opposite trends for the expression of some of these genes (Supplemental Fig. S1, B and C). Differences in the experimental setups among studies could explain these contradictions. For example, those previous studies harvested tissue a few hours after UV-B treatment, whereas substantially longer exposures were used in our study. Hence, transcripts that increased transiently in response to UV-B may not have been detected over longer time periods, possibly explaining the smaller number of genes induced by UV-B compared with Brown et al. (2005) and Favory et al. (2009). Although these previous studies have provided valuable information on the mechanisms by which UVR8 regulates gene expression under low fluence rates of UV-B, they have focused less on the effects of UV-A irradiance, which is much higher than UV-B irradiance in natural sunlight.
Here, the effects of UV-B were determined in the presence of UV-A by comparing responses between the treatments UV A versus UV A+B. Our results indicate that in the presence of high UV-A irradiance, UVR8 may have dual functions in regulating the UV-B gene expression response. We show that UVR8 had a negative role in the transcript accumulation of genes involved in the UVR8-dependent pathway (HY5 and SIG5; Brown and Jenkins, 2008), genes regulated by UVR8 independently from HY5 (SPS; Brown et al., 2005), and genes induced in response to UV-B, such as MEB5.2 (Brosché et al., 2002; Kalbina et al., 2008), RUP2 (Gruber et al., 2010), and At5g01520 (Oravecz et al., 2006). Interestingly, most of these genes are also regulated at the transcription level by either UV-A or blue light (Supplemental Fig. S1E). Other studies report that high fluence rates of UV-A increase transcripts of HY5 and CHS independently from the UV-B-specific UVR8 pathway (Wade et al., 2001; Brown et al., 2005). In addition, blue light via CRY1 regulates the expression of CHS (Fuglevand et al., 1996), HY5 (Kleine et al., 2007), and SIG5 (Kleine et al., 2007; Onda et al., 2008). Under our conditions, it is likely that these genes are responding to UV-A and not to blue light, because of the differences in expression detected between the treatments UV 0 and UV A. Interestingly, COP1 regulates the expression of HY5, CHS, SIG5, SPS, MEB5.2, PMI2, RUP2, and At5g01520 under UV-B (Oravecz et al., 2006), and it is also a positive regulator of SIG5, MEB5.2, and At5g01520 under white light (Supplemental Fig. S1C). Taken together with these previous reports, our results indicate that under natural sunlight, the UV-A/blue light signaling pathway, most probably mediated by cryptochromes, interacts with UVR8 to modulate UV-A responses in the presence of UV-B. Furthermore, COP1 could play a significant role in this interaction. COP1 is a multifunctional protein involved in light responses mediated by cryptochromes, phytochromes, and UVR8, and its WD40 domain is a common point of interaction with these three photoreceptors (Heijde and Ulm, 2012). Evidence exists that the interaction of UVR8 with COP1 under extended UV-B exposure might include removing COP1 from the phytochrome and cryptochrome signaling pathways (Favory et al., 2009). Thus, it is possible that under solar radiation, cryptochromes compete with UVR8 to interact with COP1, and high UV-A irradiances could mask the UV-B-driven gene expression mediated by the UVR8-COP1 pathway.
The positive role of UVR8 in the expression of DFR, which is not regulated by COP1, under UV-B (Oravecz et al., 2006) supports the hypothesis that an interaction between photoreceptors is mediated by COP1. The interaction/competition between photoreceptors to drive light responses in plants has been characterized in previous studies. For example, COP1, despite mediating opposing effects on light responses through phytochromes A and B, determines the most effective light signal between these two photoreceptors to promote photomorphogenesis under red light (Boccalandro et al., 2004). It is now clear that Trp-285 and Trp-233 play major roles as the active chromophore in the UVR8 protein, allowing dimer dissociation after UV-B absorption (Christie et al., 2012; Wu et al., 2012) and the subsequent interaction with other proteins regulating gene expression (Rizzini et al., 2011). Although there is no indication that UVR8 can directly mediate UV-A responses, here we show that UVR8 influences UV-A-regulated gene expression under natural sunlight (Fig. 2). Weighting our UV doses with the absorbance spectrum of UVR8 (Table I) suggests that it is possible that the amount of radiation under the treatment UV A may have been sufficient to drive UVR8-mediated gene expression. The possibility that UVR8 interacts with UV-A photoreceptors/UV-A signaling to regulate transcript accumulation under solar UV-A should not be excluded; however, further research is required to validate both hypotheses.
Table I. UVBE (kJ m−2) and PAR (MJ m−2) estimated for every UV treatment and the ambient sunlight.
The UVBE every hour was calculated with four BSWFs: two formulations of Caldwell’s generalized plant action spectrum, GEN(G) given by Green et al. (1974) and GEN(T) given by Thimijan et al. (1978); FLAV (Ibdah et al., 2002) for the accumulation of mesembryanthin in Mesembryanthemum crystallinum; and PG, the new plant growth action spectrum formulated by Flint and Caldwell (2003). Doses weighted with the absorbance spectrum of UVR8 (see Supplemental Fig. S3A in Christie et al., 2012) are also given. All BSWFs were normalized to 1 at a wavelength of 300 nm. Accumulated UVBE and PAR after 12, 36, and 50 h were calculated by summing the corresponding hourly values. In addition, the accumulated nonweighted UV-A and UV-B doses (mmol m−2) in every treatment are reported to allow comparisons with previous research. Ambient (no filter) is not a treatment; these doses were included for comparative purposes.
| UV Treatment | Time | GEN(G) | GEN(T) | FLAV | PG | UVR8 | UV-A | UV-B | PAR |
|---|---|---|---|---|---|---|---|---|---|
| h | |||||||||
| UV 0 | 12 | 0.001 | 0.005 | 0.01 | 0.04 | 0.01 | 122.3 | 0.06 | 6.2 |
| 36 | 0.002 | 0.012 | 0.03 | 0.10 | 0.02 | 328 | 0.15 | 17.2 | |
| 50 | 0.004 | 0.014 | 0.04 | 0.12 | 0.03 | 374.4 | 0.16 | 19.8 | |
| UV A | 12 | 0.01 | 0.46 | 1.8 | 12.7 | 0.56 | 2,395 | 1.1 | 6.6 |
| 36 | 0.03 | 0.91 | 4.6 | 33.2 | 1.40 | 6,323.8 | 2.8 | 18.2 | |
| 50 | 0.04 | 1.02 | 5.1 | 37.7 | 2.12 | 7,195.9 | 3.1 | 21 | |
| UV A+B | 12 | 2.3 | 3.6 | 7.4 | 17.1 | 4.92 | 2,634.2 | 45.2 | 6.4 |
| 36 | 5.6 | 8.8 | 18.3 | 44.3 | 12.05 | 6,934.8 | 109.8 | 17.8 | |
| 50 | 6 | 9.6 | 20.2 | 50 | 18.14 | 7,883.8 | 120.1 | 20.4 | |
| Ambient | 12 | 2.8 | 4.3 | 8.8 | 20.3 | 5.89 | 3,111.1 | 54.1 | 7.4 |
| 36 | 6.7 | 10.5 | 21.8 | 52.5 | 14.42 | 8,189.7 | 131.7 | 20.6 | |
| 50 | 7.2 | 11.5 | 24.1 | 59.2 | 21.72 | 9,310.1 | 143.9 | 23.7 |
The number of experiments measuring gene expression in response to solar UV-B in Arabidopsis remains small. This limits comparisons between these types of studies and makes it difficult to extrapolate results from indoor experiments to natural environments. For other plants such as maize (Zea mays), field studies dealing with plant responses to solar UV-B at several molecular levels are more common (Casati and Walbot, 2003; Casati et al., 2011). Furthermore, evidence exists at the transcript, metabolite, and protein levels that there are more differences than similarities shared between greenhouse and field experiments into maize responses to UV-B (Casati et al., 2011). Here, in agreement with Casati et al. (2011), we show that transcript regulation induced by UV-B irradiance indoors, even when equivalent to ambient levels of solar UV-B, does not necessarily predict changes in gene expression occurring in natural sunlight (Fig. 1). Furthermore, misinterpretation of gene expression data are more likely when less realistic irradiation treatments, such as unfiltered UV lamps, are used. Thus, our comparison clearly shows the importance of using appropriate irradiation conditions in experiments indoors if they are to predict gene expression regulated by UV-B in natural sunlight. The comparison of our data with previous hormone studies in agreement with Mackerness et al. (1999) reveals that under natural sunlight, ethylene and to some extent SA may be involved in responses to solar UV at the transcript level (Fig. 1). We also show that transcript accumulation of several JA biosynthesis and signaling genes was dependent on UVR8 after 12 h of solar UV exposure. Thus, our findings indicate that under natural sunlight, the UVR8 pathway interacts with the JA signaling pathway and could induce resistance against pathogens and herbivores (Izaguirre et al., 2003; Demkura et al., 2010; Demkura and Ballaré, 2012).
UVR8 Moderates PDX1 Accumulation under Solar UV
There is clear evidence for the critical role of vitamin B6 and the enzyme PDX1 in conferring plant protection against abiotic stresses (Chen and Xiong, 2005; Denslow et al., 2007) and supplementary UV-B radiation (Ristilä et al., 2011). Here, a significant induction of PDX1 was observed in Arabidopsis leaves subjected to ambient solar UV-A and UV-B, indicating the importance of PDX1 for plants exposed to natural sunlight. Furthermore, our results show that UVR8 regulates PDX1 levels under solar UV. In our experiment, PDX1 accumulation within a short period of exposure (12 h) appeared to be regulated by solar UV-A independently of UVR8 (Fig. 3). However, after a longer exposure period (36 h), our findings reveal that UVR8 in the presence of UV-B is a negative modulator of the UV-A response. Under our conditions, the transcript levels of PDX1.3 remained unchanged, whereas other studies report that UV-B exposure increases the expression of PDX1.3 (Brosché et al., 2002; Ulm et al., 2004; Kalbina et al., 2008). It is possible that the high levels of UV-A present in our conditions could have masked the UV-B induction of PDX1 transcripts as for the other UV-B-specific markers (Fig. 2). Although PDX1 content was always higher under UV A and UV A+B than under UV 0, plants grown in the greenhouse also had increased PDX1 after being transferred to the treatment UV 0 outdoors. This result suggests that other environmental factors, such as blue light or high PAR, may induce PDX1 accumulation under natural sunlight. The average PAR values detected during the first 12 h under the treatment UV 0 (654 µmol m−2 s−1) were higher than those from the greenhouse (450 µmol m−2 s−1) and may have enhanced the transcript accumulation of genes involved in vitamin B6 biosynthesis (Denslow et al., 2007; Kleine et al., 2007). Overall, analysis of PDX1 protein contents supports the gene expression data in showing that when plants receive UV-A and UV-B, UVR8 can modulate UV-A responses having a negative role on PDX1 accumulation.
UVR8 Is Required for the UV-B Induction of Phenolics and Also Affects the UV-A-Regulated Accumulation of Metabolites in Arabidopsis Leaves
Having determined patterns of gene expression and protein accumulation in the wild type and uvr8-2 under natural sunlight, we also analyzed the accumulation of leaf phenolic compounds and other metabolites. The phenolic profiles of wild-type plants exposed to solar UV are positively correlated with the increased expression of genes involved in the flavonoid pathway: PAL1 and PAL2, CHS, CHALCONE ISOMERASE, TT7, DFR, FLAVONOL SYNTHASE1, and FLAVANONE HYDROXYLASE detected with microarrays. In general, solar UV-A had larger effects than UV-B on the accumulation of metabolites after 12 and 36 h outdoors, most probably due to the relatively high levels of UV-A in our experiment compared with indoor studies. In agreement with previous research in Arabidopsis (Götz et al., 2010) and in silver birch (Kotilainen et al., 2009; Morales et al., 2010), individual phenolic compounds were differentially regulated by solar UV-A and UV-B, indicating that their contribution to solar UV protection may vary. It has been argued that quercetin derivatives play a more efficient role in UV-B protection than kaempferols (Götz et al., 2010). Accordingly, the number of UV-B-induced quercetins was larger than that of kaempferol derivatives (Fig. 4). Furthermore, our findings provide evidence that UVR8 may be involved in the UV-A regulation of individual metabolites. UVR8 was required for the UV-A induction of kynurenic and chlorogenic acids, but UVR8 also had a negative role in the accumulation of Trp, Phe, kaempferol, and kaempferol-3-rhamnoside. These results for metabolite responses confirm the role of UVR8 as a negative modulator of UV-A responses detected at the transcript and protein levels. In addition, and in better agreement with the previous UV-B-specific roles reported for UVR8 (Kliebenstein et al., 2002; Brown et al., 2005; Favory et al., 2009; Demkura and Ballaré, 2012), we show that UVR8 is a positive regulator of the UV-B induction of kaempferol-3-glucoside, quercetin, quercetin-3-glucoside, and one unidentified metabolite in plants that receive both solar UV-A and UV-B. The uvr8-2 mutant retained the capacity for UV-A induction of epidermal flavonoids, but its UV-B induction of both flavonoids and hydroxycinnamic acids was impaired (Fig. 5). Thus, consistent with the UPLC-MS/MS analysis, our findings reveal that UVR8 is required for the UV-B induction of phenolics in the leaf epidermis and to enhance the content of epidermal flavonoids in those environments that expose plants to solar UV-B radiation.
UVR8 Affects Plant Performance under Natural Sunlight
The importance of the UVR8 signaling pathway in promoting plant survival in natural environments was suggested by Brown et al. (2005). More recently, Favory et al. (2009) argued that UV-B acclimation mediated by UVR8 is required for plants to survive in sunlight, and used a sun simulator to show that uvr8 mutants that received UV-B for 27 d displayed leaf curling and cell death and were smaller than uvr8 grown under filtered UV-B. In an attempt to corroborate these previous findings under natural sunlight, the wild type and uvr8-2 were germinated and grown outdoors under near-ambient solar UV-A and UV-B to study plant survival and gene expression. We show that uvr8-2 germinated, developed, and produced flowers under the UV treatments outdoors; however, this mutant had reduced growth compared with the wild type, especially in plants exposed to solar UV-A plus UV-B. In contrast, such a difference in growth between genotypes was not evident during the previous experiment in plants grown in the greenhouse without UV-B. Thus, it cannot be ruled out that the very small amount of scattered UV-B radiation entering through the sides of the filters had an effect on the growth of uvr8-2 that was visible after 3 weeks outdoors under the treatments UV 0 and UV A. Further evidence that UVR8 is needed to maintain leaf expansion and organ size in the presence of supplementary UV-B comes from a study where weighted UV-B doses with the generalized plant action spectrum (Caldwell, 1971) in the range of 6.7 to 10 kJ m−2 d−1 reduced the size of uvr8-2 leaves compared with the wild type (Wargent et al., 2009a). Our results agree with Favory et al. (2009) and Wargent et al. (2009a) in attributing a functional role for UVR8 in promoting growth under natural sunlight. The UV-B acclimation mediated by UVR8 (Favory et al., 2009; González Besteiro et al., 2011) and its positive role in regulating the expression of genes involved in auxin signaling (HAT2) may help plants to grow normally when they start to perceive solar UV. The fact that uvr8-2 survived under our conditions indicates that other pathways that are not dependent on UVR8 (Brown and Jenkins, 2008) may become active and enhance survival in the absence of UVR8. In addition to the short-term UV-B exposures, we also measured the expression of several genes 3 weeks after germination and growth outdoors. In this case, the lack of a significant UV treatment × genotype interaction, most probably due to high variation, did not allow us to identify specific roles for UVR8 regulating gene expression after long-term exposure to solar UV-A and UV-B. However, genes for which significant effects of the UV treatments and genotype were detected exhibited the same trends in expression as determined with microarrays and qPCR after 12 h outdoors. In addition, the results of the two very different experiments (12 h versus 3 weeks) were also consistent for most of the genes analyzed (Figs. 2 and 6). Therefore, the trends in gene expression observed in this study may reveal that functional UVR8 is needed for the stable expression of genes involved in JA signaling (GRX480 and JAZ1), plant defense (WRKY70), and auxin signaling (HAT2) under natural sunlight. Furthermore, our findings suggest that the expression of these genes may be needed in the plant for long-term acclimation to solar UV radiation.
CONCLUSION
The findings of this outdoor study under natural sunlight are consistent with those published experiments using artificial light to demonstrate that UVR8 has a key function in promoting the acclimation of Arabidopsis plants to solar UV radiation. Our findings show that UVR8 is required for the transcript accumulation of genes involved in UV protection, oxidative stress, defense against herbivores, and the signaling and biosynthesis of several plant hormones. These results support our first hypothesis that UVR8 enables plants to acclimate under solar UV. The results also support our second hypothesis, which predicted an altered response through UVR8 when plants are exposed to the full spectrum of solar radiation, compared with exposure to UV-B alone, or to UV-B with a low background irradiances of UV-A and PAR. This finding suggests that under ambient PAR and high UV-A irradiance, UVR8 may interact with other photoreceptors to modulate UV-A responses in the presence of UV-B. UVR8 impacts on solar UV-A responses, acting as a positive and negative regulator of gene expression and of the accumulation of individual metabolites. We propose that such an interaction could be caused by competition among different photoreceptors for the same pool of COP1. However, further research is needed to test this.
Our results not only provide a novel insight into how UVR8 regulates gene expression and metabolite accumulation under solar radiation but also show that solar UV-A elicits some of the same responses as solar UV-B. Under natural sunlight, UVR8-independent pathways may enhance plant survival; however, UV acclimation, which is especially dependent on UVR8 at early stages of plant development, is important for the normal growth of Arabidopsis plants and is required for the expression of several genes involved in plant defense against herbivores and other stresses. To our knowledge, this study is the first attempt to elucidate the complex and multiple roles of UVR8 under natural sunlight. The evidence presented here suggests that future research covering the interactions between UVR8 and other photoreceptors is required to understand the acclimation of plants to their natural environment.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and UV Treatments
The experiment was conducted in the greenhouse and field area of the Viikki campus of Helsinki University (60°13′N, 25°1′E). Arabidopsis (Arabidopsis thaliana) seeds of uvr8-2 backcrossed twice to the wild type (Brown et al., 2005) and the wild-type Ler were sown on May 28, 2010, in plastic pots (5 × 5 cm) containing a 1:1 mixture of peat and vermiculite. The seeds were kept in darkness at 4°C for 3 d and then transferred to environmentally controlled growth rooms at 23°C/19°C (day/night) and 70%/90% relative humidity under 12-h photoperiod with 280 µmol m−2 s−1 white light irradiance. Seedlings were transplanted to individual plastic pots (4 × 4 cm) using the same substrate on June 8, 2010. After transplanting, plants were kept for 14 d in a greenhouse compartment where they received sunlight passing through the 4-mm glass of the roof of the greenhouse. This roof transmits approximately 7% of the ambient UV-A but no UV-B radiation, as detected with a spectroradiometer (Maya2000 Pro; Ocean Optics). For this period, the average values are (minimum, mean, and maximum) temperature of 19.2°C, 22°C, and 25.4°C and relative humidity of 50.4%, 62.3%, and 74.6%, respectively. PAR was measured with a light meter (LI-250A; LI-COR) during sunny days, and the average clear-sky value at noon was 450 µmol m−2 s−1.
Outdoors, three UV treatments were created by using three types of plastic film that selectively exclude different bands of the UV spectrum. Exclusion of solar UV-A and UV-B (UV 0) was provided by a theatrical “gel” (Rosco E+ 226), exclusion of solar UV-B (UV A) was provided with polyester film (0.125 mm thick; Autostat CT5; Thermoplast), and near-ambient solar UV (UV A+B) was provided by polythene film (0.05 mm thick; 04 PE-LD; Etola). The treatments were randomly assigned to one filter frame within five complete blocks or biological replicates. The films were placed on top of 1-m × 0.80-m wooden frames. The frames faced south at a slight inclination, higher toward the north edge, to avoid the accumulation of rain water on top of the films. The frames were adjusted to keep the top of the plants at about 8 cm below the films. The transmittance of the films was measured with a spectrophotometer equipped with an integrating sphere (Shimadzu UV-2501 PC UV-VIS) at the beginning and the end of the experiment. During a sunny day at noon, spectral irradiances (W m−2) of UV-B (280–315 nm) and UV-A (315–400 nm) were measured under the filters with a Maya2000 Pro spectroradiometer (Supplemental Fig. S3), and the readings were corrected according to the procedure described by Ylianttila et al. (2005) with further correction for stray light based on a cutoff filter adapted from Kreuter and Blumthaler (2009). The unweighted photon irradiances from every UV treatment were then compared with the ambient irradiance. The UV-B and UV-A irradiances under the treatment UV 0 represented 0.2% and 2.7% of the ambient conditions, respectively. The treatment UV A had about 0.4% of UV-B and 72% of UV-A compared with the ambient conditions, while treatment UV A+B received approximately 73% of the ambient UV-B and UV-A.
In the greenhouse, 20 plants from each genotype were randomly assigned to the same plastic tray, and two trays were allocated at random to each frame outdoors on June 22, 2010, between 7:00 and 7:30 am. Samples from the wild type and uvr8-2 grown in the greenhouse were collected to compare patterns of gene and protein expression between greenhouse and field conditions. Each biological sample collected consisted of five pooled rosettes frozen in liquid nitrogen and stored at –80°C. After 12 and 36 h outdoors, samples from each filter frame were collected by block with treatments and genotypes in random order within each block. For both time points, samples were harvested during daylight at 7:00 pm, Eastern European Summer Time (5:38 pm local solar time). Plants received sunlight throughout the first 12 h outdoors, while the 36-h time point included 5 h of night. The collected leaf material was ground in liquid nitrogen, and the powder was divided into subsamples for gene and protein expression analysis and metabolite profiling analysis. The temperature and relative humidity during the experiment were recorded with iButtons (Dallas Semiconductors) placed under the filters. The calculated average values for minimum, mean, and maximum were temperature of 11.2°C, 16.4°C, and 23.3°C and relative humidity of 37.5%, 72.1%, and 98.9%, respectively. Outdoors, water was supplied to plants through subirrigation.
Optical Measurements of Epidermal UV Absorbance and Chlorophyll in the Leaves
Epidermal UV absorbance of the leaves was measured in vivo with Dualex FLAV 3.3 and Dualex HCA 3 (FORCE-A). These fluorometers make noninvasive in vivo estimations of leaf phenolic contents. Dualex FLAV measures the UV absorbance of the leaf epidermis by double excitation of chlorophyll fluorescence using UV-A (375 nm) and red light (650 nm) and has been used to reliably estimate epidermal flavonoid contents in several plant species (Goulas et al., 2004). Dualex HCA has the same measurement principles as Dualex FLAV but uses UV-B (315 nm) and red light (650 nm) to estimate the concentration of hydroxycinnamic acids. The chlorophyll content of the leaves was measured with a SPAD-502 chlorophyll meter (Minolta Camera). The adaxial epidermis of the two youngest leaves in the rosettes of 30 plants per genotype/UV treatment was measured with Dualex FLAV, Dualex HCA, and SPAD-502 after 50 h of UV exposure outdoors.
Microarray Analysis
Two-color microarrays were used to identify genes regulated by solar UV radiation in the wild type and uvr8-2 after 12 h of exposure outdoors. Two biological repeats (blocks) out of five in this experiment were randomly selected for the analysis. RNA was extracted by using a Spectrum Plant Total RNA kit (Sigma-Aldrich), and the RNA concentration was measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Details on complementary DNA (cDNA) synthesis, probe labeling, microarray hybridization, and data analysis are available from the Array-Express database (http://www.ebi.ac.uk/microarray-as/ae/; accession no. E-MTAB-1093). To avoid bias, dyes were swapped in each of the two biological repeats. Genes regulated by solar UV in the wild type were identified by fitting a linear model (Wettenhall and Smyth, 2004) into the normalized array data, taking into account the dye-related differences in labeling, and comparing wild-type UV 0 versus wild-type UV A+B. An empirical Bayesian shrinkage was then performed on the t statistics obtained from the linear model fit for each of the genes. The resulting moderated t statistics for each of the genes were then subjected to a Benjamini and Hochberg false discovery rate adjustment. Significantly expressed genes were then obtained by using a threshold false discovery rate of less than 0.05. Comparisons were also made between the wild type and uvr8-2 under UV A+B to find UVR8-regulated genes under solar UV.
Genes with significantly altered expression, as identified above, were first compared with other studies of UVR8 activity (Brown et al., 2005; Favory et al., 2009) and with light responses in Arabidopsis (Kleine et al., 2007) using Venn diagrams. Comparisons were also made with other UV-B experiments and with expression data from plant hormone assays with SA, abscisic acid, methyl jasmonate, ethylene, and the SA analog BTH (Supplemental Table S3). The raw .cel files were obtained from public databases and normalized with robust multiarray average normalization, and for each experiment, the log2-base fold changes of treatment versus control were computed. The preprocessed data were clustered using Bayesian hierarchical clustering as described by Wrzaczek et al. (2010). Publicly available experiments using the Affymetrix ATH1-121501 platform were obtained from several data sources: NASC Arrays, http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl (UV-B, NASCARRAYS-137 and NASCARRAYS144; abscisic acid, NASCARRAYS-176; SA, NASCARRAYS-192; BTH, NASCARRAYS-392); Array-Express, http://www.ebi.ac.uk/microarrayas/ae/ (methyl jasmonate, EATMX-13; UV-B, E-MEXP-550, E-MEXP-557, E-MEXP-1957); Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/ (SA, GSE14961; ethylene, GSE14247).
Expression Analysis by qPCR
Gene expression after 12 and 36 h was further analyzed with qPCR using three independent biological repeats (blocks not used in microarray analysis). Total RNA was extracted according to Jaspers et al. (2009) and treated with DNase I (Fermentas; now part of ThermoScientific). Five micrograms of DNA-free RNA was reverse transcribed with RevertAid Premium (ThermoScientific) in a 20-μL reaction volume. The cDNA reactions were diluted to a final volume of 120 μL, and 1 μL was used as a template for PCR using iQ SYBR Green Super mix (Bio-Rad) on a CFX 384 Real-Time PCR detection system (Bio-Rad) in triplicate. Primers were designed by using the Primer-Blast tool from the National Center for Biotechnology Information. Primer sequences and amplification efficiencies are available in Supplemental Table S8. A dilution series of pooled cDNA was used to calculate the amplification efficiency in Bio-Rad CFX Manager (version 1.6). PCR was initiated at 95°C for 5 min followed by 44 cycles at 95°C for 10 s, 60°C for 10 s, and 72°C for 30 s. The cycle thresholds were determined using Bio-Rad CFX Manager and were imported in qbasePLUS 2.0 (Biogazelle). In qbasePLUS 2.0, the expression values were normalized with three reference genes (At4g34270, At4g33380, and At4g35510). The reference genes were stably expressed throughout the experiment (average geNorm expression value M = 0.18, coefficient of variation = 0.07). All samples belonging to the same block and time point were run on the same plate. In every run, the normalized expression values were scaled to wild-type UV 0, log10 transformed, and exported from qbasePLUS as Excel files for statistical analyses in R (R Development Core Team, 2010) version (2.12.0).
Analysis of Metabolites by UPLC-MS/MS
The powdered samples used for metabolite profiling were first weighed, and metabolite extraction was done according to Morales et al. (2010). A Waters Acquity UPLC system equipped with a sample manager and binary solvent manager was used for the analyses. In addition, a Waters Synapt GS HDMS mass spectrometer was interfaced with the UPLC system via an electrospray ionization source. Mass range was set from 50 to 1,200. Samples were analyzed in positive ion mode, with capillary voltage at 3.0 kV. The source temperature was 120°C and desolvation temperature was set to 350°C; the cone gas flow rate was 20 L h−1 and desolvation gas flow rate was 1,000 L h−1. The compounds were separated on an Acquity UPLC BEH C18 column (1.7 µm, 50 × 2.1 mm; Waters) at 40°C. The mobile phase consisted of (A) water and (B) acetonitrile (Chromasolv grade; Sigma-Aldrich), both containing 0.1% HCOOH (Sigma-Aldrich). A gradient of eluents was used as follows: linear gradient of 95% A to 10% in 9 min, then back to 95% in 9.1 min, and left to equilibrate for 1 min. The injection volume was 2 µL, and the flow rate of the mobile phase was 0.6 mL min−1. Mass spectra and peak analysis were performed using MarkerLynx software (Waters), and the array of peak areas was exported as an Excel file. The identification of metabolites was done according to Malitsky et al. (2008). Five biological replicates representing all blocks outdoors were analyzed. Mass signals per fresh weight for each metabolite within each genotype and UV treatment were used as raw data for statistical analysis.
Expression of PDX1 in the Leaves
Induction of the protein PDX1 in leaves after 12 and 36 h of exposure outdoors was monitored by western blot according to Ristilä et al. (2011). Total proteins were extracted from approximately 100 mg of frozen powdered leaf material, and 5 µg of total protein was used for immunoblot analyses. All samples belonging to the same block outdoors were blotted to the same membrane; one greenhouse sample per genotype was also included for comparative purposes (Fig. 3). Three biological replicates with two technical repeats each were analyzed. The fold changes in PDX1 expression scaled to wild-type UV 0 at 12 h were calculated using ImageJ software (http://rsb.info.nih.gov/ij/) and used for statistical analysis (n = 6).
Performance of Wild-Type and uvr8-2 Plants Grown under Solar UV
We studied the performance of wild-type and uvr8-2 plants germinated and grown outdoors under natural sunlight. Seeds of uvr8-2 and the wild type were sown on July 16, 2010, in plastic pots (4 × 4 cm) containing a 1:1 mixture of peat and vermiculite. The seeds were vernalized at 4°C in darkness for 72 h and transferred outdoors under three UV treatments (UV 0, UV A, and UV-A+B) created by plastic films as described above. Plants were grown under the UV treatments for 3 weeks after germination; thereafter, the number of flowering plants per pot was counted, and samples from three independent biological replicates were collected as indicated above and stored at −80°C. Long-term effects of solar UV-A and UV-B radiation on the expression of selected genes were also measured with qPCR. Temperature and relative humidity during the experiment were recorded with iButtons placed under the filters. The calculated average values for minimum, mean, and maximum were temperature of 17.3°C, 24.4°C, and 33.7°C and relative humidity of 41.5%, 76.1%, and 99.7%, respectively.
Statistical Analysis
All statistical analyses were done in R. Linear mixed-effects models with blocks as random-grouping factors were fitted with the NLME package (Pinheiro and Bates, 2000). The models were fitted with and without a power variance covariate weighting to test whether this term improved the fit. Factorial ANOVA was used to assess the significance of the main effects (UV treatments and genotypes) and of the interaction UV treatment × genotype for all variables measured throughout the experiments. The ANOVA results should be interpreted as follows: main significant effects of the UV treatments or genotypes means that these factors systematically affect a response variable, while a significant UV treatment × genotype interaction indicates that genotypes respond in different ways to the UV treatments. In models where ANOVA indicated a significant interaction, treatment effects within genotypes (P ≤ 0.05) were assessed by fitting contrasts using the package gregmisc (Warnes, 2005). The effect of solar UV was determined from contrasts between the treatments (UV 0 versus UV A+B), while the contrasts (UV 0 versus UV A) and (UV A versus UV A+B) allowed us to identify specific UV-A and UV-B effects, respectively. From model fits where the main effects were significant, but not the interaction, contrasts were fitted to assess differences between UV treatments. P values from multiple contrasts were adjusted using Holm’s procedure. Figures 4 and 5 were drawn in the lattice package (Sarkar, 2008).
Calculation of Biologically Effective UV Doses Outdoors and PAR
The biologically effective UV dose (UVBE) and PAR during the two experiments for each UV treatment and the ambient sunlight were calculated as described by Morales et al. (2010). The ambient solar UV radiation spectral irradiance for the experimental site was first estimated at hourly intervals as described by Lindfors et al. (2009). The estimates were then convoluted with measured filter transmittance spectra and action spectra (normalized to action = 1 at 300 nm). Four of the most commonly used biological spectral weighting functions (BSWFs) in plant research were used to compare the UVBE between the UV treatments and ambient sunlight. Two formulations of Caldwell’s generalized plant action spectrum were used, GEN(G) given by Green et al. (1974) and GEN(T) given by Thimijan et al. (1978). FLAV is the spectrum for the accumulation of mesembryanthin in Mesembryanthemum crystallinum (Ibdah et al., 2002) and PG is the “plant growth” spectrum Flint and Caldwell (2003). For more details on the action spectra and dose calculations, see Flint and Caldwell (2003) and Kotilainen et al. (2009). Doses weighted with the absorbance spectrum of UVR8 (see Supplemental Fig. S3A in Christie et al., 2012) were calculated using the package UVcalc (P.J. Aphalo, unpublished data) in R. For the first experiment, the UV doses (kJ m−2) and PAR (MJ m−2) were calculated for every hour that plants were exposed outdoors, and the accumulated UV and PAR after 12, 36, and 50 h were determined by summing the corresponding hourly values (Table I). For the purposes of comparison with previous research, we also calculated nonweighted UV-A and UV-B using UVcalc. During the experiment, UV-A and UV-B irradiances had maximum ambient values of 142.4 and 2.9 µmol m−2 s−1, respectively; the accumulated values are shown in Table I. For the performance experiment, the daily UVBE and PAR were calculated as described above. The minimum, average, and maximum values for the whole duration of the experiment are indicated in Supplemental Table S9. For both experiments, with all the BSWFs, the UV doses differed between treatments, while PAR differed by less than 5.6% (Table I; Supplemental Table S9).
Microarray data from this article can be found in the Array-Express database (http://www.ebi.ac.uk/microarray-as/ae) under accession number E-MTAB-1093.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of our microarray data with previous studies using Venn diagrams.
Supplemental Figure S2. Images of wild-type (Ler) and uvr8-2 plants germinated and grown for 3 weeks outdoors under the UV treatments.
Supplemental Figure S3. Spectral irradiance (W m−2 nm−1) under the UV treatments measured with a Maya2000 Pro spectroradiometer during a sunny day at noon.
Supplemental Figure S4. Patterns of gene expression measured by qPCR in the wild type (Ler) and uvr8-2 grown in the greenhouse.
Supplemental Table S1. Genes differentially expressed in the wild type after 12 h under solar UV A+B as detected by microarrays.
Supplemental Table S2. Genes differentially expressed in uvr8-2 after 12 h under solar UV A+B as detected by microarrays.
Supplemental Table S3. Detailed information on experiments represented in the Bayesian cluster (Fig. 1).
Supplemental Table S4. Detailed information on genes represented in the Bayesian cluster (Fig. 1).
Supplemental Table S5. Summary of the ANOVA models used to estimate significant effects of the UV treatments, genotype, and the interaction UV treatment × genotype on the expression of selected genes measured by qPCR, accumulation of PDX1 and other metabolites, and optical measurements.
Supplemental Table S6. Tests of the effects of the UV treatments on gene expression, accumulation of PDX1 and other metabolites, and optical measurements.
Supplemental Table S7. Metabolites identified in wild-type and uvr8-2 leaves by UPLC-MS/MS.
Supplemental Table S8. Detailed information of primers used in qPCR.
Supplemental Table S9. UVBE and PAR calculated for the whole duration of the experiment.
Acknowledgments
We acknowledge the Finnish Microarray Center for manufacturing the microarrays and FORCE-A for lending us a Dualex HCA device. We are grateful to the European Union COST action FA0906 (UV4growth) for giving us the opportunity to interact with coauthors of this paper. We thank Nigel Paul, Carlos Ballaré, and Mathew Robson for several relevant discussions of our research and two anonymous reviewers for constructive criticisms on the manuscript. Yohama Puentes, Liz Carolina Eklund, Pekka Lönnqvist, and Leena Grönholm are also acknowledged for technical assistance. Lasse Ylianttila calibrated the spectrometer used.
Glossary
- PAR
photosynthetically active radiation
- qPCR
quantitative PCR
- Ler
Landsberg erecta
- JA
jasmonic acid
- SA
salicylic acid
- BTH
benzothiadiazole S-methylester
- UPLC
ultra-performance liquid chromatography
- MS/MS
tandem mass spectrometry
- UVBE
biologically effective UV dose
- BSWFs
biological spectral weighting functions
- UV 0
exclusion of solar UV-A and UV-B
- UV A
exclusion of solar UV-B
- UV-A+B
near-ambient solar UV
References
- Boccalandro HE, Rossi MC, Saijo Y, Deng XW, Casal JJ. (2004) Promotion of photomorphogenesis by COP1. Plant Mol Biol 56: 905–915 [DOI] [PubMed] [Google Scholar]
- Brosché M, Schuler MA, Kalbina I, Connor L, Strid Å. (2002) Gene regulation by low level UV-B radiation: identification by DNA array analysis. Photochem Photobiol Sci 1: 656–664 [DOI] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI. (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Headland LR, Jenkins GI. (2009) UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochem Photobiol 85: 1147–1155 [DOI] [PubMed] [Google Scholar]
- Brown BA, Jenkins GI. (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MM. (1971) Solar UV irradiation and the growth and development of higher plants. In AC Giese, ed, Photophysiology. Vol VI. Current Topics in Photobiology and Photochemistry. Academic Press, New York, pp 131–177 [Google Scholar]
- Casati P, Campi M, Morrow DJ, Fernandes J, Walbot V. (2011) Transcriptomic, proteomic and metabolomic analysis of maize responses to UV-B: comparison of greenhouse and field growth conditions. Plant Signal Behav 6: 1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V. (2003) Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol 132: 1739–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xiong L. (2005) Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J 44: 396–408 [DOI] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, O’Hara A, Smith BO, Christie JM, Jenkins GI. (2012) C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci USA 109: 16366–16370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demkura PV, Abdala G, Baldwin IT, Ballaré CL. (2010) Jasmonate-dependent and -independent pathways mediate specific effects of solar ultraviolet B radiation on leaf phenolics and antiherbivore defense. Plant Physiol 152: 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demkura PV, Ballaré CL. (2012) UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol Plant 5: 642–652 [DOI] [PubMed] [Google Scholar]
- Denslow SA, Rueschhoff EE, Daub ME. (2007) Regulation of the Arabidopsis thaliana vitamin B6 biosynthesis genes by abiotic stress. Plant Physiol Biochem 45: 152–161 [DOI] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint SD, Caldwell MM. (2003) A biological spectral weighting function for ozone depletion research with higher plants. Physiol Plant 117: 137–144 [Google Scholar]
- Fuglevand G, Jackson JA, Jenkins GI. (1996) UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell 8: 2347–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Besteiro MA, Bartels S, Albert A, Ulm R. (2011) Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J 68: 727–737 [DOI] [PubMed] [Google Scholar]
- Götz M, Albert A, Stich S, Heller W, Scherb H, Krins A, Langebartels C, Seidlitz HK, Ernst D. (2010) PAR modulation of the UV-dependent levels of flavonoid metabolites in Arabidopsis thaliana (L.) Heynh. leaf rosettes: cumulative effects after a whole vegetative growth period. Protoplasma 243: 95–103 [DOI] [PubMed] [Google Scholar]
- Goulas Y, Cerovic ZG, Cartelat A, Moya I. (2004) Dualex: a new instrument for field measurements of epidermal ultraviolet absorbance by chlorophyll fluorescence. Appl Opt 43: 4488–4496 [DOI] [PubMed] [Google Scholar]
- Green AES, Sawada T, Shettle EP. (1974) The middle ultraviolet reaching the ground. Photochem Photobiol 19: 251–259 [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R. (2010) Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Ulm R. (2012) UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Ibdah M, Krins A, Seidlitz HK, Heller W, Strack D, Vogt T. (2002) Spectral dependence of flavonol and betacyanin accumulation in Mesembryanthemum crystallinum under enhanced ultraviolet radiation. Plant Cell Environ 25: 1145–1154 [Google Scholar]
- Izaguirre MM, Scopel AL, Baldwin IT, Ballaré CL. (2003) Convergent responses to stress: solar ultraviolet-B radiation and Manduca sexta herbivory elicit overlapping transcriptional responses in field-grown plants of Nicotiana longiflora. Plant Physiol 132: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P, Blomster T, Brosché M, Salojärvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F, et al. (2009) Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 60: 268–279 [DOI] [PubMed] [Google Scholar]
- Jenkins GI. (2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Kalbina I, Li S, Kalbin G, Björn LO, Strid Å. (2008) Two separate UV-B radiation wavelength regions control expression of different molecular markers in Arabidopsis thaliana. Funct Plant Biol 35: 222–227 [DOI] [PubMed] [Google Scholar]
- Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand Å. (2007) Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL. (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotilainen T, Venäläinen T, Tegelberg R, Lindfors A, Julkunen-Tiitto R, Sutinen S, O’Hara RB, Aphalo PJ. (2009) Assessment of UV biological spectral weighting functions for phenolic metabolites and growth responses in silver birch seedlings. Photochem Photobiol 85: 1346–1355 [DOI] [PubMed] [Google Scholar]
- Kreuter A, Blumthaler M. (2009) Stray light correction for solar measurements using array spectrometers. Rev Sci Instrum 80: 096108. [DOI] [PubMed] [Google Scholar]
- Lindfors A, Heikkilä A, Kaurola J, Koskela T, Lakkala K. (2009) Reconstruction of solar spectral surface UV irradiances using radiative transfer simulations. Photochem Photobiol 85: 1233–1239 [DOI] [PubMed] [Google Scholar]
- Mackerness SAH, Surplus SL, Blake P, John CF, Buchanan-Wollaston V, Jordan BR, Thomas B. (1999) Ultraviolet-B-induced stress and changes in gene expression in Arabidopsis thaliana: role of signalling pathways controlled by jasmonic acid, ethylene and reactive oxygen species. Plant Cell Environ 22: 1413–1423 [Google Scholar]
- Malitsky S, Blum E, Less H, Venger I, Elbaz M, Morin S, Eshed Y, Aharoni A. (2008) The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol 148: 2021–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales LO, Tegelberg R, Brosché M, Keinänen M, Lindfors A, Aphalo PJ. (2010) Effects of solar UV-A and UV-B radiation on gene expression and phenolic accumulation in Betula pendula leaves. Tree Physiol 30: 923–934 [DOI] [PubMed] [Google Scholar]
- Onda Y, Yagi Y, Saito Y, Takenaka N, Toyoshima Y. (2008) Light induction of Arabidopsis SIG1 and SIG5 transcripts in mature leaves: differential roles of cryptochrome 1 and cryptochrome 2 and dual function of SIG5 in the recognition of plastid promoters. Plant J 55: 968–978 [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R. (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. (2000) Mixed-Effects Models in S and S-Plus. Springer, New York [Google Scholar]
- R Development Core Team (2010) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org (October 15, 2010)
- Ristilä M, Strid H, Eriksson LA, Strid A, Sävenstrand H. (2011) The role of the pyridoxine (vitamin B6) biosynthesis enzyme PDX1 in ultraviolet-B radiation responses in plants. Plant Physiol Biochem 49: 284–292 [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Sarkar D. (2008) Lattice: Multivariate Data Visualization with R. Springer, New York [Google Scholar]
- Thimijan RW, Carns HR, Campbell LE. (1978) Radiation Sources and Related Environmental Control for Biological and Climatic Effects UV Research (BACER). Final report (EPA-IAG-DG-0168). Environmental Protection Agency, Washington, DC [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F. (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade HK, Bibikova TN, Valentine WJ, Jenkins GI. (2001) Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. Plant J 25: 675–685 [DOI] [PubMed] [Google Scholar]
- Wargent JJ, Gegas VC, Jenkins GI, Doonan JH, Paul ND. (2009a) UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol 183: 315–326 [DOI] [PubMed] [Google Scholar]
- Wargent JJ, Moore JP, Roland Ennos A, Paul ND. (2009b) Ultraviolet radiation as a limiting factor in leaf expansion and development. Photochem Photobiol 85: 279–286 [DOI] [PubMed] [Google Scholar]
- Warnes GR. (2005) gregmisc: Greg’s Miscellaneous Functions. R package version 2.1.1. http://cran.r-project.org/package=gregmisc (October 15, 2010)
- Wettenhall JM, Smyth GK. (2004) limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20: 3705–3706 [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J. (2010) Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, et al. (2012) Structural basis of ultraviolet-B perception by UVR8. Nature 484: 214–219 [DOI] [PubMed] [Google Scholar]
- Wu M, Grahn E, Eriksson LA, Strid Å. (2011) Computational evidence for the role of Arabidopsis thaliana UVR8 as UV-B photoreceptor and identification of its chromophore amino acids. J Chem Inf Model 51: 1287–1295 [DOI] [PubMed] [Google Scholar]
- Ylianttila L, Visuri R, Huurto L, Jokela K. (2005) Evaluation of a single-monochromator diode array spectroradiometer for sunbed UV-radiation measurements. Photochem Photobiol 81: 333–341 [DOI] [PubMed] [Google Scholar]