Summary: Through regulation by light, a mobile mRNA of potato is involved in controlling processes of growth and in mediating the activation of its own gene.
Abstract
BEL1- and KNOTTED1-type proteins are transcription factors from the three-amino-loop-extension superclass that interact in a tandem complex to regulate the expression of target genes. In potato (Solanum tuberosum), StBEL5 and its Knox protein partner regulate tuberization by targeting genes that control growth. RNA movement assays demonstrated that StBEL5 transcripts move through the phloem to stolon tips, the site of tuber induction. StBEL5 messenger RNA originates in the leaf, and its movement to stolons is induced by a short-day photoperiod. Here, we report the movement of StBEL5 RNA to roots correlated with increased growth, changes in morphology, and accumulation of GA2-oxidase1, YUCCA1a, and ISOPENTENYL TRANSFERASE transcripts. Transcription of StBEL5 in leaves is induced by light but insensitive to photoperiod, whereas in stolon tips growing in the dark, promoter activity is enhanced by short days. The heterodimer of StBEL5 and POTH1, a KNOTTED1-type transcription factor, binds to a tandem TTGAC-TTGAC motif that is essential for regulating transcription. The discovery of an inverted tandem motif in the StBEL5 promoter with TTGAC motifs on opposite strands may explain the induction of StBEL5 promoter activity in stolon tips under short days. Using transgenic potato lines, deletion of one of the TTGAC motifs from the StBEL5 promoter results in the reduction of GUS activity in new tubers and roots. Gel-shift assays demonstrate BEL5/POTH1 binding specificity to the motifs present in the StBEL5 promoter and a double tandem motif present in the StGA2-oxidase1 promoter. These results suggest that, in addition to tuberization, the movement of StBEL5 messenger RNA regulates other aspects of vegetative development.
As part of an elaborate long-distance communication system, plants have evolved a unique signaling pathway that takes advantage of connections in the vascular tissue, predominantly the phloem. This information superhighway has been implicated in regulating development, responding to biotic stress, delivering nutrients, and as a vehicle commandeered by viruses for spreading infections (Lough and Lucas, 2006). Numerous full-length transcripts have been identified in the sieve elements of several plant species (Asano et al., 2002; Vilaine et al., 2003; Omid et al., 2007; Deeken et al., 2008; Gaupels et al., 2008; Kehr and Buhtz, 2008). One of these mobile RNAs is StBEL5, a BEL1-like transcription factor that is expressed in potato (Solanum tuberosum; Banerjee et al., 2006a). BEL1-like transcription factors are members of the three-amino-loop-extension superclass that interact with KNOTTED1 (KN1)-like partners to regulate numerous aspects of development. Both types are ubiquitous among plants, and the BEL1 types function in the floral pathway (Kanrar et al., 2008; Rutjens et al., 2009), inflorescence stem growth (Smith and Hake, 2003; Bhatt et al., 2004; Ragni et al., 2008), stem cell fate (Byrne et al., 2003), leaf architecture (Kumar et al., 2007), ovule formation (Ray et al., 1994), and the establishment of egg cell fate in the mature embryo sac (Pagnussat et al., 2007).
In potato, the BEL1-like transcription factor, StBEL5, and its Knox protein partner regulate tuber formation by targeting genes that control growth. Overexpression of StBEL5 consistently produced plants with enhanced tuber yields. RNA detection methods and heterografting experiments demonstrated that StBEL5 transcripts are present in phloem cells and move across a graft union to localize in stolon tips, the site of tuber induction (Banerjee et al., 2006a). This movement of RNA originates in leaf veins and petioles and is induced by a short-day photoperiod, regulated by the untranslated regions, and correlated with enhanced tuber production (Banerjee et al., 2006a, 2009). In general, these results suggest that the movement of StBEL5 is not solely regulated by source/sink relations but instead is controlled by sequence-specific motifs and daylength-mediated gating. The promoter of StBEL5 is light activated in leaves and in stolon tips induced by a short-day photoperiod (Chatterjee et al., 2007). The positive correlation of short day-activated movement of StBEL5 transcripts and promoter activity in stolon tips, an underground organ, suggests the possibility of an active mechanism for the transductive enhancement of a photoperiod signal to organs growing in the dark.
Here, the movement of a mobile RNA and its correlation with root growth are established in potato ssp. andigena. The mechanism for photoperiod regulation of the promoter of StBEL5 is demonstrated in underground organs that perceive no light signals. These results suggest that regulation of the StBEL5 promoter in stolons and roots is mediated by phloem-associated movement of the mRNA of StBEL5 from its transcriptional source in leaves. This remarkable whole-plant communication system involves light induction of transcription in the leaf, photoperiod-activated mobilization of the StBEL5 mRNA through the phloem, and short-day regulation of StBEL5 promoter activity in target organs growing underground in the dark.
RESULTS
Movement of StBEL5 RNA into Roots Is Correlated with Increased Root Growth
Using two different promoters in transgenic potato lines, the movement of StBEL5 RNA from leaves to stolons in response to a short-day photoperiod was previously demonstrated (Banerjee et al., 2006a). One of the promoters, for galactinol synthase (GAS), is leaf specific, with its activity restricted to the minor veins of the leaf mesophyll, and has been used before in phloem-mobility studies (Banerjee et al., 2006a, 2009; Srivastava et al., 2008). In theory, any RNA driven by the GAS promoter that is detected in organs other than the leaf is the result of long-distance transport.
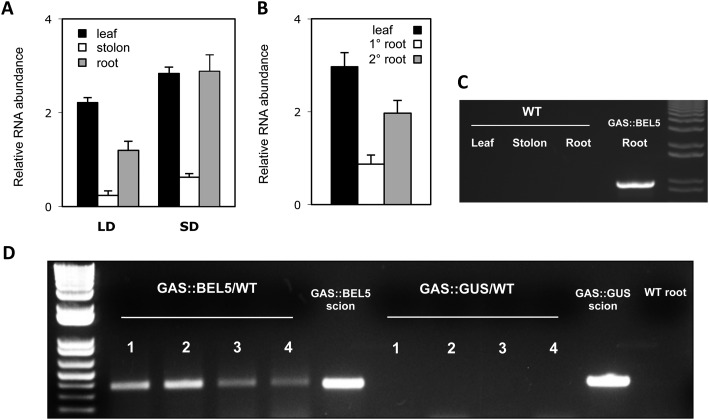
As expected, the transport of full-length transgenic StBEL5 RNA into stolons occurred under both long- and short-day conditions, with enhanced movement under short days (Fig. 1A; Banerjee et al., 2006a). The movement of transgenic StBEL5 was also observed into roots of soil-grown plants (Fig. 1A). The relative abundance of this transported mRNA was greater than that observed for stolons under both photoperiod conditions. The relative abundance of transported StBEL5 RNA in roots under short days was approximately 4-fold greater than the levels in short-day stolons (Fig. 1A). The movement data of Figure 1A were based on RNA extracted from total root harvests, making it impossible to ascertain whether the transport of StBEL5 RNA was to primary roots (also called crown roots) and/or secondary roots. To determine the accumulation pattern of mobile transcripts of StBEL5 in roots, RNA was extracted separately from primary and secondary roots of GAS:BEL5 plants grown under short-day conditions and quantitative reverse transcription (qRT)-PCR was performed. Under these conditions, more than twice the relative amount of transgenic StBEL5 mRNA was transported to secondary roots than to primary roots (Fig. 1B). The specificity of the transgenic primers used in Figure 1, A and B, was verified on RNA from wild-type andigena leaf, stolons, and roots using the same PCR conditions (Fig. 1C). To verify the movement of StBEL5 transcripts to roots, heterografts of GAS:BEL5 scions and wild-type stocks were performed with reverse transcription (RT)-PCR assays of RNA from the roots of wild-type stock material (Fig. 1D). As a negative control, GAS:GUS transgenics were grafted as scions onto wild-type stocks. Transgenic StBEL5 RNA was detected in lateral roots of wild-type stock from four separate GAS:BEL5/wild-type heterografts, whereas no GUS RNA was detected in lateral roots from wild-type stock from four separate GAS:GUS/wild-type heterografts (Fig. 1D).
Figure 1.
Movement of transgenic StBEL5 mRNA from leaf to stolon or root. A, Quantification of movement was performed on transgenic lines expressing full-length StBEL5 RNA driven by the GAS promoter of melon (Cucumis melo). This promoter is predominantly expressed in the minor veins of leaf mesophyll (Ayre et al., 2003; Banerjee et al., 2009). Relative levels of transgenic StBEL5 RNA were quantified from total RNA extracted from new leaves (black bars), 0.5-cm samples from the tip of the stolon (white bars), and root samples (gray bars). B, In a separate experiment, relative levels of transgenic StBEL5 RNA were quantified from total RNA extracted from new leaves (black bars) and from either primary (white bars) or secondary (gray bars) root samples of a short-day (SD)-grown GAS:BEL5 transgenic plant. One-step RT-PCR was performed using 200 to 250 ng of total RNA, a primer for the NOS terminator sequence specific to all transgenic RNAs, and a gene-specific primer for the full-length StBEL5 transcript. These primers specifically amplify only transgenic BEL5 RNA. All PCRs were standardized and optimized to yield product in the linear range. Homogenous PCR products were quantified by using ImageJ software (Abramoff et al., 2004) and normalized by using 18S rRNA values. se values of three replicate samples are shown. LD, Long days. C, The specificity of the transgenic primers used in A and B was verified on RNA from wild-type potato ssp. andigena leaf, stolons, and roots using the same PCR conditions. D, For heterografts, micrografts were performed with replicates of either GAS:BEL5 scions on wild-type andigena stocks or GAS:GUS scions on wild-type andigena stocks. After 2 weeks in culture, grafts were moved to soil and grown under long days for 3 weeks and then under short days for 2 weeks before harvest of roots and leaves. After RNA extraction, RT-PCR with gene-specific primers was performed on RNA from wild-type lateral roots of both heterografts. A second PCR was performed with nested primers for both types. RNA from scion leaf samples was used as a positive control (scion samples). Two different gene-specific primers were used with a nonplant sequence tag specific for the transgenic StBEL5 RNA to discriminate from the native RNA. Four plants were assayed for both heterografts and are designated 1 to 4. Wild-type RNA from lateral roots of whole plants (andigena) was used in the RT-PCR, with StBEL5 transgenic gene-specific primers as a negative control (WT root). Similar negative results were obtained with RNA from wild-type leaves.
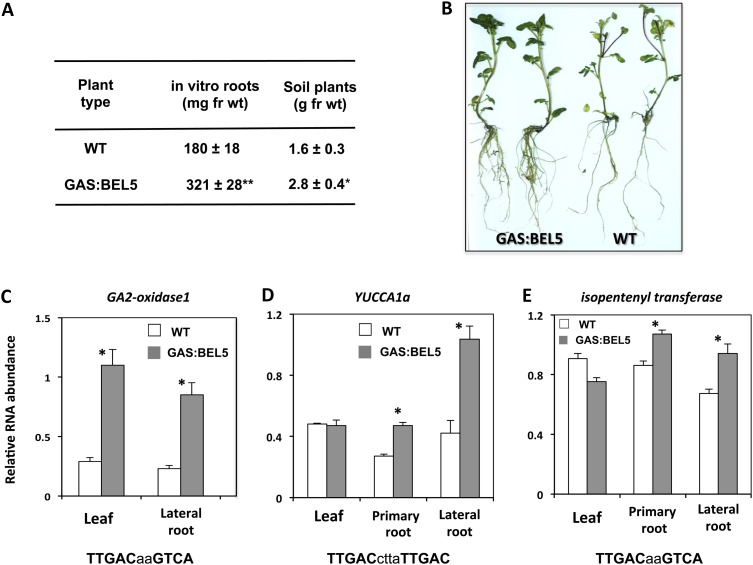
Because StBEL5 is a transcription factor that works in tandem with KN1 types to regulate hormone levels that control plant growth (Chen et al., 2003), root growth was measured in these GAS:BEL5 transgenic lines. In both in vitro-grown plantlets and soil-grown plants, root growth of these transgenic lines was increased by approximately 75% (Fig. 2A). Roots from in vitro-grown plants were generally longer and more robust than control roots (Fig. 2B). Because increased activity of GA 2-oxidase1 (GA2ox1) is known to enhance lateral root growth (Gou et al., 2010), transcript levels for StGA2ox1 were assayed in lateral roots from wild-type and transgenic GAS:BEL5 plants. In the transgenic line, StGA2ox1 (PGSC0003DMT400054348) mRNA levels increased by greater than 3-fold in both leaf and root RNA samples relative to controls (Fig. 2C). A routine screening of upstream sequence from StGA2ox1 and selected hormone synthesis genes revealed the presence of core tandem TGAC motifs (Fig. 2, C–E) all within 1.8 kb of the start codon for StGA2ox1, YUCCA1a (auxin synthesis; PGSC0003DMT400067103), and ISOPENTENYL TRANSFERASE (IPT; cytokinin synthesis; PGSC0003DMT400068271). RNA levels for these latter two genes increased significantly in both primary and lateral roots of the GAS:BEL5 transgenic line (Fig. 2, D and E).
Figure 2.
Root development of transgenic lines of potato ssp. andigena grown in vitro and in soil. A, For root fresh weight (fr wt) harvests, in vitro plantlets were grown for 4 weeks at 27°C under 16 h of light/8 h of dark. B, Roots from in vitro transgenic lines were generally longer and more robust than wild-type controls (WT). Soil plants were grown in pots in a growth chamber under long days (16 h of light/8 h of dark) at 24°C days and 18°C nights and harvested after 7 weeks. The se of several plants is shown in A. C to E, Accumulation of StGA2ox1 mRNA in leaves and lateral roots (C) and YUCCA1a (D) and IPT (E) mRNA in leaves and primary and lateral roots of wild-type andigena or the transgenic andigena line expressing full-length StBEL5 with a leaf-specific promoter, designated GAS. The transgenic leaf and root samples assayed in C to E are the same ones used in Figure 1B. RT-PCR was performed with gene-specific primers and standardized to yield product in the linear range, normalized using rRNA primers, and quantified by using ImageJ software (Abramoff et al., 2004). Values represent means ± se for three biological replicates. Asterisks indicate significant differences (*P < 0.05, **P < 0.01) using Student’s t test.
To separate the effect of the shoot on root growth, 1.0- to 2.0-cm root tip explants were cultured on medium in vitro. Overall, lateral root production more than doubled in root tips from GAS:BEL5 plants (Table I). Lateral roots from these transgenic plants grew almost twice as long as control roots (20.4 versus 11.5 mm). These results are consistent with previous work showing that overexpression of GA2ox1 lowers GA levels and enhances the production of lateral root primordium (Gou et al., 2010).
Table I. Lateral root growth from root tip explants of GAS:BEL5 plants after 10 d of in vitro culture.
Root tip explants approximately 1.8 cm in length and without any lateral roots were excised from 12 4-week-old plantlets and cultured on Murashige and Skoog medium plus 2.0% Suc under long-day conditions. The asterisk indicates a significant difference (P < 0.01) using Student’s t test.
| Sample | No. of Explants | No. of Lateral Roots per Explant | Lateral Root Length |
|---|---|---|---|
| mm | |||
| GAS:BEL5 | 47 | 1.19 | 20.4 ± 1.8* |
| Wild type | 54 | 0.46 | 11.5 ± 1.2 |
Gel-Shift Assay for the GA2ox1 Double Motif
Similar to the motif identified in the StGA20ox1 promoter (Chen et al., 2004), a TTGACXXXTTGAC motif that bound strongly to KN1 was identified in an intron of the GA2ox1 gene of maize (Zea mays; Bolduc and Hake, 2009). This KN1-binding site is conserved in the GA2ox1 genes of several grasses (Bolduc and Hake, 2009). Upstream sequence of the potato gene that encodes GA2ox1 contains two tandem motifs 85 nucleotides apart, both containing TGAC elements on opposite strands two nucleotides apart (Figs. 2C, boldface, and 3A, boldface and underlined). Other than the two nonconserved nucleotide linkers, the opposite strands of the separated tandem motifs (in a 5′ to 3′ direction) form a palindrome:
 |
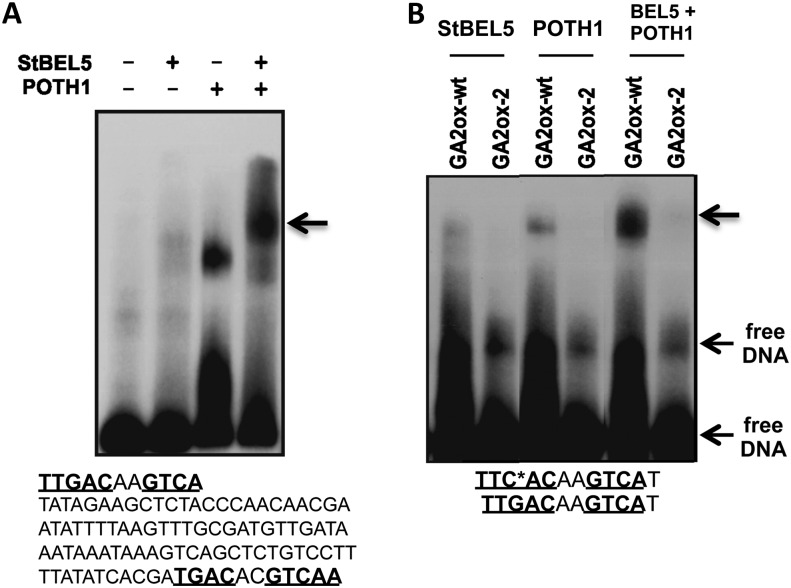
Figure 3.
Binding of StBEL5 and POTH1 to upstream regulatory sequences of GA2ox1. The core bait DNA is listed below each panel, and TTGAC and TGAC motifs are designated in boldface and underlined. The asterisk in the mutated sequence of the single GA2ox1 motif (B) represents the G-to-C point mutation and is designated GA2ox-2. The wild-type sequence is designated GA2ox-wt (B). Each DNA sequence was incubated with POTH1 or StBEL5 protein alone or together in a binding reaction mix. The GA2ox1 sequence in A contains two tandem motifs separated by 85 nucleotides. The two TGAC motifs of GA2ox1 are arranged on opposite strands in a tail-to-tail orientation (A). The arrow in A and the top arrow in B represent supershifted bands likely retarded by the tandem protein complex. The strongest interactions were observed with both proteins in the reaction mix.
Unlike the StGA20ox1 TTGAC motifs, the TGAC motifs in the StGA2ox1 sequence are aligned in a tail-to-tail orientation. To assess the binding affinity of the double tandem motifs present in the StGA2ox1 promoter, gel-shift analyses were undertaken with the StBEL5 and POTH1 proteins on this extended sequence (Fig. 3A). Some binding was observed with either protein alone, but the strongest interaction occurred with both proteins, with clear evidence of a supershifted band (Fig. 3A, arrow). Overall, these results suggest a very strong interaction of the double motifs of StGA2ox1 with the BEL5/POTH1 complex. The gel-shift results on the modified single GA2ox1 motif demonstrate that a single point mutation (G→C) in this 11-nucleotide sequence reduces any StBEL5 or POTH1 protein interaction and essentially eliminates tandem binding (Fig. 3B, top arrow). These results are consistent with those of Chen et al. (2004), where a similar G-to-C point mutation completely abolished the transcriptional regulation of StGA20ox1.
Morphology of Roots from GAS:BEL5 Plants
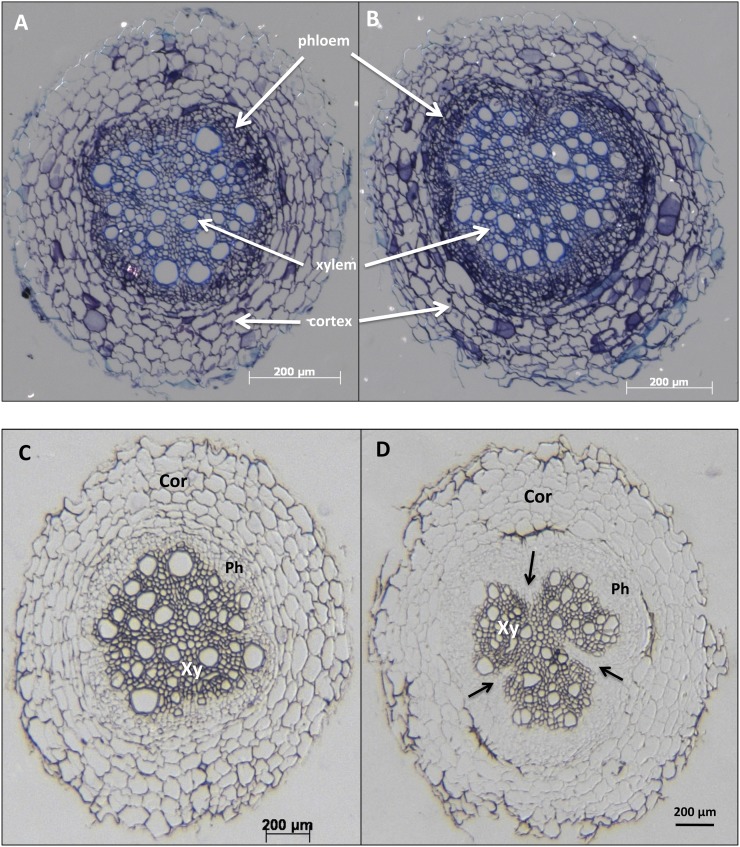
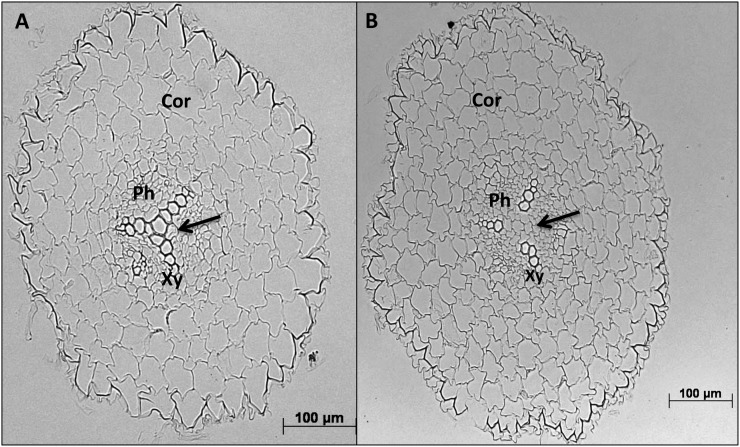
To determine if an enhanced accumulation of StBEL5, StGA2ox1, YUCCA1, and IPT mRNAs in roots had any correlation with changes in root morphology, transverse sections of numerous distinct root pieces from several wild-type and GAS:BEL5 plants were examined, and the root diameters were measured. In primary roots of GAS:BEL5 plants, the stele, the central region of the root containing cells arising from the vascular cambium, makes up a larger proportion of the overall root area (66.4% compared with 58.4%; Table II). One example of this increased area of the stele is shown in representative stained transverse sections (Fig. 4, A and B). Both the xylem and phloem regions of GAS:BEL5 roots are larger than in wild-type roots (Fig. 4, A and B, arrows). There was no difference, however, in the overall diameter of primary roots between the two types (Table II). GAS:BEL5 roots also exhibited a structural anomaly in the xylem core. Fifteen out of 20 random sections that were examined exhibited cleavage in the xylem core at a level that was generally not observed in wild-type roots (Fig. 4, C and D, arrows). Three out of 16 wild-type primary roots exhibited a very shallow form of this xylem cleavage (data not shown). It is possible that this xylem cleavage is the result of an excessive growth of phloem cells leaking into the xylem core (Fig. 4D, arrows). Overall, primary roots from GAS:BEL5 plants appeared to contain more phloem cells than wild-type roots (compare phloem of Fig. 4A to that of Fig. 4B and the phloem of Fig. 4C to that of Fig. 4D). In most wild-type secondary roots, the triarch of mature xylem cells is tightly linked (Fig. 5A, arrow). These cells are adjacent or very close together. This morphology was observed in 13 out of the 17 random sections for wild type secondary roots. All 17 exhibited a well-organized, distinguishable triarch structure. Secondary roots from GAS:BEL5 plants, however, either exhibited an open structure for the triarch of xylem cells that were not adjacent (Fig. 5B, arrow) or had no observable triarch organization. Thirteen out of the 16 random sections for GAS:BEL5 roots exhibited this deviant organization: either an open triarch structure or no triarch of xylem cells. The openness of the xylem cells may reflect induced cell division within the stele region. In several secondary roots from GAS:BEL5 plants, excessive numbers of cortical cells were also observed (Fig. 5B, Cor).
Table II. Phenotypes of primary roots of wild-type and transgenic GAS:BEL5 andigena plants.
The mean of the ratio of the diameter of the stele (consisting of endodermis, xylem, and phloem) to the total diameter of the primary root section was calculated from sections of roots from soil-grown plants. Diameters were measured using the Image Analysis Program by averaging two widths as described in “Materials and Methods.” These sections were randomly selected from three to four lines from embedded tissue harvested near the middle length of the primary root sample. A significant difference (P < 0.01; Student’s t test) is indicated with an asterisk.
| Root Sample | Mean Ratio | sd of the Mean Ratio | Mean Total Diameter | Mean Core Diameter | n |
|---|---|---|---|---|---|
| μm | |||||
| GAS:BEL5 | 0.664* | 0.0546 | 1,687 | 1,120 | 20 |
| Wild type | 0.584 | 0.0400 | 1,685 | 984 | 16 |
Figure 4.
Transverse sections of primary roots of wild type (A and C) and GAS:BEL5 (B and D) plants. Plants were grown under short-day conditions, and roots were harvested at the 12- to 13-leaf stage. Sixteen to 20 sections were examined for each line. These sections came from random root pieces from three to four plants for each line. These micrographs are representative of each type. Micrographs A and B were stained with toluidine blue to enhance cells in the vascular cylinder. The arrows in D designate xylem cleavage. Sections were viewed and photodocumented with an Olympus BX40 microscope and a Carl Zeiss AxioCam MRc 5 digital camera. Cor, Cortex; Ph, phloem; Xy, xylem.
Figure 5.
Transverse sections of secondary roots of wild-type (A) and GAS:BEL5 (B) plants. Plants were grown under short-day conditions, and roots were harvested at the 12- to 13-leaf stage. Sixteen or 17 sections were examined for each line. These sections came from random root pieces from three to four plants of each line. These micrographs are representative of each type. Sections were viewed and photodocumented with an Olympus BX40 microscope and a Carl Zeiss AxioCam MRc 5 digital camera. Cor, Cortex; Ph, phloem; Xy, xylem. The arrows show the separation of the xylem triarch that occurs in secondary roots of GAS:BEL5 plants (B) but not the wild type (A).
Transcriptional Regulation of StBEL5 in Stolon Tips and New Tubers
Under normal conditions, the promoter activity of StBEL5 in leaves is insensitive to photoperiod and induced by low irradiance levels (Chatterjee et al., 2007). Most of this foliar activity is observed in primary veins and petioles (Banerjee et al., 2006a). Promoter activity was observed in underground stolon tips from plants grown under both long and short days (Banerjee et al., 2006a). Enhanced activity was observed, however, in correlation with short days (Chatterjee et al., 2007). This photoperiod-regulated activity occurs in short-day stolon tips despite the fact that in emerging stolons that grow above the soil line in response to light, StBEL5 promoter activity is repressed (Supplemental Fig. S1).
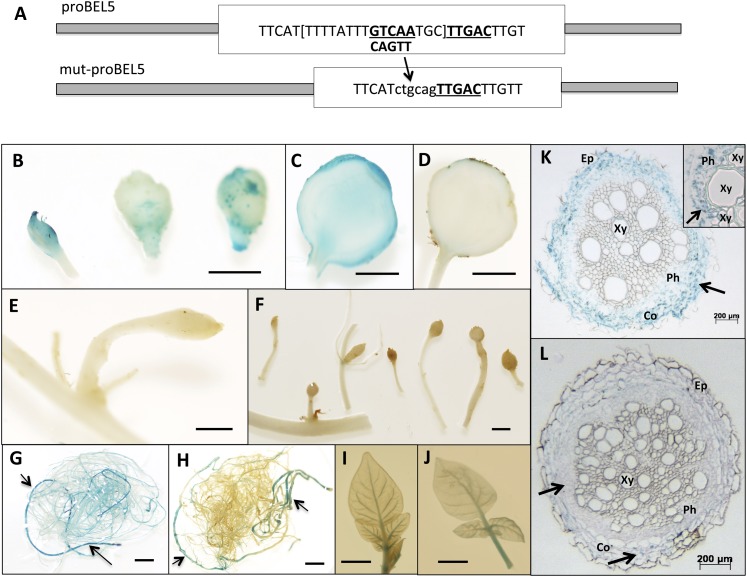
The correlation of short day-activated movement of StBEL5 transcripts to stolon tips with promoter activity in these same underground organs suggested the possibility that StBEL5 is involved in regulating its own transcription underground. A double TTGAC motif specific for the StBEL5/POTH1 tandem complex (Chen et al., 2004) is present on the StBEL5 promoter 820 nucleotides upstream from the transcription start site. In this particular example, the two TTGAC motifs were located on opposite strands three nucleotides apart (Fig. 6A, proBEL5, underline). Because two complete TTGAC motifs in close proximity were necessary for the tandem complex of transcription factors to bind and affect transcription (Chen et al., 2004), a mutated form of the wild-type promoter was constructed by deleting one TTGAC motif and replacing it with the PstI restriction site, CTGCAG (Fig. 6A, mut-proBEL5). Based on this design, only one complete conserved motif remained intact in this mutated form of the promoter (Fig. 6A, mut-proBEL5, underline).
Figure 6.
A, Schematic of the modification of the wild-type StBEL5 promoter sequence. To create the mutated StBEL5 promoter used in the transgenic lines reported here, one of the tandem TTGAC cis-elements (underlined and boldface) that make up the binding motif for StBEL5 and its KN1-like partner, POTH1 (Chen et al., 2004), was deleted. To facilitate cloning, this five-base motif plus the TGC linker and eight other bases (all in brackets) were removed and replaced by the ctgcag sequence. The intact wild-type double motif sequence begins 820 nucleotides upstream from the start of the StBEL5 5′ UTR (Chatterjee et al., 2007). The wild-type StBEL5 promoter sequence analyzed here was 2,002 nucleotides in length. B to L, StBEL5 promoter activity in newly formed tubers (B–F), roots (G, H, K, and L), and leaves (I and J) of andigena plants grown under short-day conditions (8 h of light/16 h of dark). Transgenic lines contained constructs of the wild-type StBEL5 promoter (B, C, G, I, and K) or a mutated form lacking one of the tandem TTGAC motifs (D–F, H, J, and L), both driving a GUS marker gene. Arrows in G and H indicate GUS activity in primary roots. GUS activity of the wild-type StBEL5 promoter (K) and the mutated form (L) in transverse sections of primary roots can be observed in phloem cells (inset in K, arrow), in the cortex of lines with the wild-type promoter (K, arrow), and in the cortex of roots from the mutated promoter lines (L, bottom arrow). Very little GUS activity was observed in phloem cells of the mutated promoter lines (L, top arrow). In primary roots, the cortex layer may become compressed in response to the expanding vascular cylinder. Co, Cortex; Ep, epidermis; Ph, phloem; Xy, xylem. These samples are representative of several independent lines. The P-StBEL5 line from Chatterjee et al. (2007) is shown here for the wild-type proBEL5 construct, whereas the mut-proBEL5 line is CI-12-8. For G through L, similar results were obtained from plants grown under long-day conditions. Bars = 5.0 mm (B–F) and 10 mm (G–J).
In transgenic lines expressing GUS with the mutated promoter of StBEL5, GUS activity was suppressed in stolons and newly formed tubers (Fig. 6, compare B and C with D–F) and reduced in secondary roots (Fig. 6H). Despite this repression of activity in stolon tips and secondary roots, the mutated promoter lines exhibited wild-type-like promoter activity in some primary (crown) roots (Fig. 6, G and H, arrows) and in leaves (Fig. 6, I and J). In transverse sections of primary roots of potato, wild-type StBEL5 promoter activity was observed in cortical cells, phloem parenchyma, and phloem cells (Fig. 6K, inset, arrow). Very little, if any, activity was observed in epidermal cells (Fig. 6K, Ep) or mature xylem cells (Fig. 6K, Xy). GUS expression was observed in cortical cells of primary roots of transgenic lines with the mutated StBEL5 promoter (Fig. 6L, bottom arrow), but very little GUS activity was observed in phloem cells (Fig. 6L, top arrow).
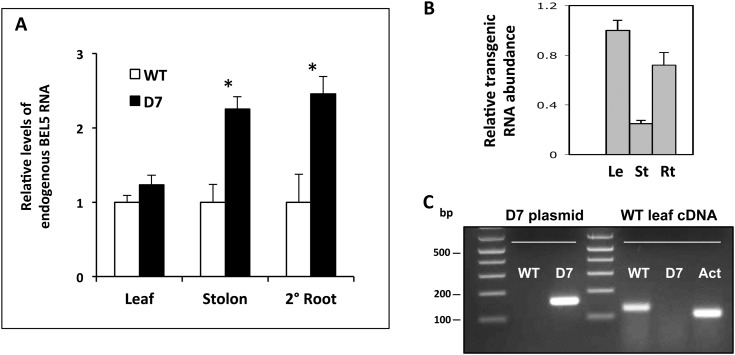
If mobile StBEL5 transcripts are involved in autoregulation, then one would expect to observe an increase in the endogenous level of StBEL5 RNA in those organs where accumulation of the transgenic RNA occurs. To address this question, endogenous RNA levels were quantified in leaves, tuberizing stolons, and lateral roots of an StBEL5 transgenic line that expresses the coding sequence plus the 500-nucleotide 3′ untranslated region (UTR) of StBEL5 driven by the GAS promoter (Fig. 7). This construct (minus the 5′ UTR) was used to readily distinguish endogenous and transgenic StBEL5 RNAs. This GAS:BEL5 line, designated D7, was previously confirmed to support significant transport of StBEL5 to stolons (Banerjee et al., 2009). Using qRT-PCR, in this line, endogenous levels of StBEL5 RNA increased 2.2- and 2.4-fold above levels in the wild type in tuberizing stolons and lateral roots, respectively (Fig. 7A). No significant difference in endogenous levels in leaves was observed. These results are consistent with the reduction in promoter activity in both new tubers and lateral roots observed in the mut-proBEL5 transgenic line (Fig. 6, B–J).
Figure 7.
Effect on endogenous levels of StBEL5 in transgenic plants that accumulate transgenic StBEL5 RNA in leaves, stolon tips, and lateral roots. A, Relative levels of endogenous StBEL5 transcript were quantified using total RNA extracted from new leaves (Leaf), 0.5-cm samples from the tip of tuberizing stolons (Stolon), and lateral roots (2° Root) of transgenic lines of andigena expressing the D7 construct (lacking the 5′ UTR sequence) of StBEL5 RNA driven by the GAS promoter of melon (black bars) or wild-type plants (WT; white bars). Asterisks indicate significant differences (P < 0.05) using Student’s t test. B, To assess the degree of mobility for transgenic StBEL5 RNA, relative levels of transgenic RNA were quantified in leaves (Le), stolon tips (St), and lateral roots (Rt) of the D7 line. All samples were harvested from plants growing under short days for 20 d. Using real-time qRT-PCR, an StBEL5 gene-specific primer plus either a primer for the NOS terminator sequence specific to all transgenic RNAs or an endogenous RNA-specific primer for the 5′ UTR of StBEL5 were used. Deletion of the 5′ UTR in the D7 construct made it possible for the primers to specifically amplify either endogenous (+5′ UTR; A) or transgenic (−5′ UTR; B) StBEL5 RNA. The expression of each target gene was normalized to endogenous reference genes StACT8 (A) or StUBQ (B). The fold change in expression of endogenous and transgenic StBEL5 transcripts in D7 tissue samples was calculated as the comparative threshold cycle method value relative to the mean values obtained in wild-type (A) or D7 (B) leaf control tissues. C, Specificity of the endogenous StBEL5 (wild type) and transgenic (D7) primers used was verified on a plasmid containing the D7 construct and wild-type leaf cDNA. Actin primers (Act) were used against the cDNA template as a PCR control. In RT-PCR using gene-specific primers for transgenic StBEL5 RNA, no PCR product was detected in any wild-type organs (Supplemental Fig. S2). se values of three biological replicate samples are shown.
Gel-Shift Assay for the StBEL5 Double Motif
To verify the interaction of the StBEL5/POTH1 protein complex with the double TTGAC motifs of the StBEL5 promoter, gel-shift assays were performed with both intact and mutated forms of the promoter (Fig. 8). In contrast to the BEL5/Knox target promoter of StGA20ox1 that contained two TTGAC motifs located tail to head on the same DNA strand (Chen et al., 2004), the TTGAC motifs present on the StBEL5 promoter were located on opposite strands in a head-to-head orientation (Fig. 8, BE5 wt). Similar to the StGA2ox1 gel-shift analysis, where some binding of both proteins alone was observed (Fig. 3), the strongest interaction and a supershifted band for the StBEL5 motif were observed only when both proteins were included (Fig. 8, arrow). A shift occurred with either StBEL5 tandem motif (wild type or mutated) with one or the other protein alone, but the mutated form of the StBEL5 motif (BE5 mut) affected a decrease in the supershifted band with both proteins (Fig. 8, arrow). In general, these results are consistent with the analyses of the motifs identified in the promoters of both StGA2ox1 (Fig. 3) and StGA20ox1 (Chen et al., 2004). Mutations in the central nucleotides of either of the TTGAC motifs of GA20ox1 abolished binding to the BEL5/POTH1 complex and blocked the repression activity of the heterodimer (Chen et al., 2004).
Figure 8.
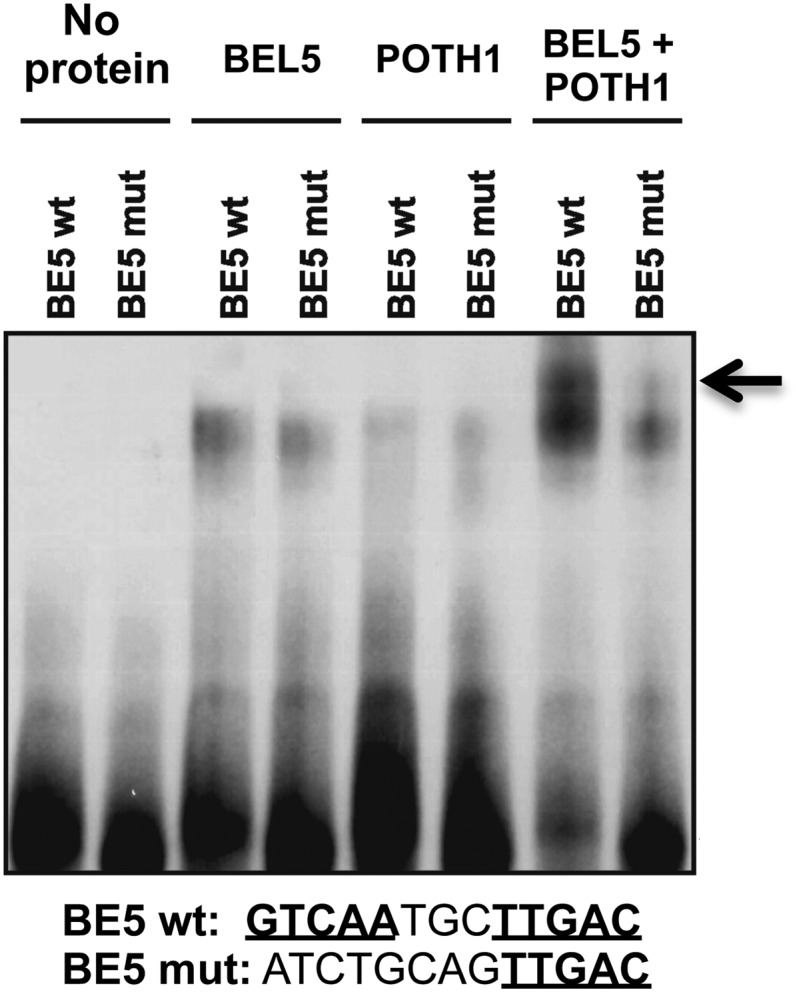
Binding of StBEL5 and POTH1 to upstream regulatory sequences of StBEL5. The core bait DNA is listed at bottom, and TTGAC motifs are designated in boldface and underlined. The wild-type (BE5 wt) and mutated (BE5 mut) forms of the proStBEL5 motifs are the same sequences shown in Figure 6A. Each DNA sequence was incubated with POTH1 or StBEL5 protein alone or together in a binding reaction mix. The two TTGAC motifs of StBEL5 are arranged on opposite strands in a head-to-head orientation. The arrow designates supershifted bands likely retarded by the tandem protein complex. The strongest interactions were consistently observed with both proteins in the reaction mix.
DISCUSSION
Short day-induced transport of StBEL5 transcripts from leaves to stolon tips has been previously demonstrated by using heterografts and two types of promoters (Banerjee et al., 2006a, 2009). Here, the long-distance transport of StBEL5 RNA to roots is also demonstrated. Similar to movement to stolon tips, some transport to roots may occur under a long-day photoperiod; however, maximum mobility of the RNA into roots was observed under short days. These results suggest that a common mechanism may be involved in transport to these underground organs. The morphology of these two, however, is quite different. Roots arise from the crown, contain no chlorophyll, and grow from their apex gravitropically. They generally have no capacity to revert to a shoot but, in some cases, may form adventitious buds that develop into aboveground shoots. Potato stolons are specialized stems that grow horizontally. Under favorable conditions, a tuber may form from the subapical region of the stolon tip. Stolons arise from the stem, contain no chlorophyll, but have internodes, axillary buds, rudimentary, scale-like leaves, and, if the apex perceives light, may grow phototropically to develop into a mature shoot. Movement of a molecule like RNA could certainly transverse the same vascular connections belowground and then separate at the stem/root junction.
The function of StBEL5 in activating tuber formation has been well established (Chen et al., 2003; Banerjee et al., 2006a, 2009). The promoter is induced by light in leaves, and the mRNA is then transported to stolons and roots under short-day conditions. But why is the StBEL5 promoter suppressed in light-grown stolons (Supplemental Fig. S1)? To explain this observation, consider that there are several examples linking the overexpression of BEL1-like or KN1-type transcription factors to the suppression of GA activity (Tanaka-Ueguchi et al., 1998; Hay et al., 2002; Chen et al., 2003; Rosin et al., 2003). This can occur through transcriptional repression of GA20ox1, a gene encoding a GA biosynthetic enzyme, or by activation of the GA catabolic gene GA2ox1 (Bolduc and Hake, 2009). Tuberization has long been associated with reduced levels of GA in stolons (Racca and Tizio, 1968; Xu et al., 1998; Chen et al., 2003). During stem and stolon elongation in the light, however, GA levels increase. Suppression of StBEL5 transcriptional activity in these newly emerging stolons would be consistent with a concomitant increase in GA accumulation. This pattern of StBEL5 gene activity could be controlled by an organ-specific activator or receptor in leaves but absent in stolons (like phytochrome or cryptochrome) or by a repressor active only in light-grown stolons. At least three light-repression motifs have been identified in the StBEL5 promoter, GGGCC, ATAAAACGT, and another involved in shade-avoidance responses (Steindler et al., 1999).
Whereas the tuberization function of StBEL5 has been documented (for review, see Hannapel, 2010), a putative role in root growth had not yet been demonstrated. Evidence of increased growth in the root correlated with RNA accumulation and active StBEL5 promoter activity in both phloem and cortical cells suggest a direct role for StBEL5 in activating cell growth in the root. Increases in RNA levels for three target genes involved in hormone synthesis (GA, auxin, and cyokinins) suggest that StBEL5’s role in regulating their expression could be modulating hormone activity. The increase of cytokinin levels mediated by StBEL5 expression (Chen et al., 2003) could explain the increased stele diameter of primary roots (Fig. 4, A and B) and the aberrant root morphology of GAS:BEL5 plants (Figs. 4, C and D, and 5, A and B). Cytokinins function in root meristem maintenance (Dolan et al., 1993) and act in restricted regions of the root meristem to mediate cell differentiation and determine the root meristem size (Dello Ioio et al., 2007). They also regulate the number of cell files within the vascular bundle and the pericycle (Dettmer et al., 2009; Perilli et al., 2010). Using a promoter:GUS fusion, expression of the cytokinin biosynthetic isopentenyl transferase gene from Arabidopsis, AtIPT3, was specific to phloem cells in the root (Miyawaki et al., 2004). Cytokinins play an important role in root growth, with many aspects of development coordinated by subtle spatial differences in the concentrations of auxin and cytokinin. Cross talk between these hormones can regulate the position of auxin transport proteins and signaling pathways (for review, see Bishopp et al., 2011). Hormone profiling in maize revealed a preferential accumulation of auxins in the stele and a predominant localization of several cytokinins in the cortical parenchyma (Saleem et al., 2010). As auxin biosynthetic enzymes, proteins encoded by the YUCCA gene family play an important role in the Trp-dependent indole-3-acetic acid pathway (Yamamoto et al., 2007). In rice, a YUCCA protein functions to regulate root-to-shoot ratios and, by this mechanism, contributes to maintaining water homeostasis (Woo et al., 2007). A YUCCA1 gene of potato is strongly induced in stolons after the switch to a short-day photoperiod (Roumeliotis et al., 2012). Beyond its role in regulating YUCCA gene expression, however, it is certainly plausible that StBEL5 is also targeting other auxin synthesis or signaling genes (Bolduc et al., 2012).
The high levels of StGA2ox1 mRNA in leaves correlates with the leaf-specific activity of the GAS:StBEL5 transgene (Fig. 2C). The high levels of StGA2ox1 transcripts in roots concomitant with enhanced levels of transported StBEL5 RNA (Fig. 1) further suggests a causal relationship between the two. Based on the DNA-binding assays, it is conceivable that the BEL/Knox complex activates StGA2ox1 transcription in roots. During the early stages of tuberization, transcript levels of StGA2ox1 increase more than 60-fold in the newly formed tuber (Kloosterman et al., 2007). The accumulation of StGA2ox1 RNA in lateral roots and the positive correlation with root growth in this study was unexpected. Recent work, however, has demonstrated the role of GA2ox1 in regulating lateral root development (Gou et al., 2010). Transgenic Populus spp. plants constitutively overexpressing PcGA2ox1 exhibited increased lateral root growth under both in vitro and greenhouse conditions. In light of the effect of StBEL5 mRNA on root morphology and growth, it is conceivable that StBEL5 in tandem with one of its Knox partners is regulating root growth through the transcriptional control of select genes involved in hormone synthesis.
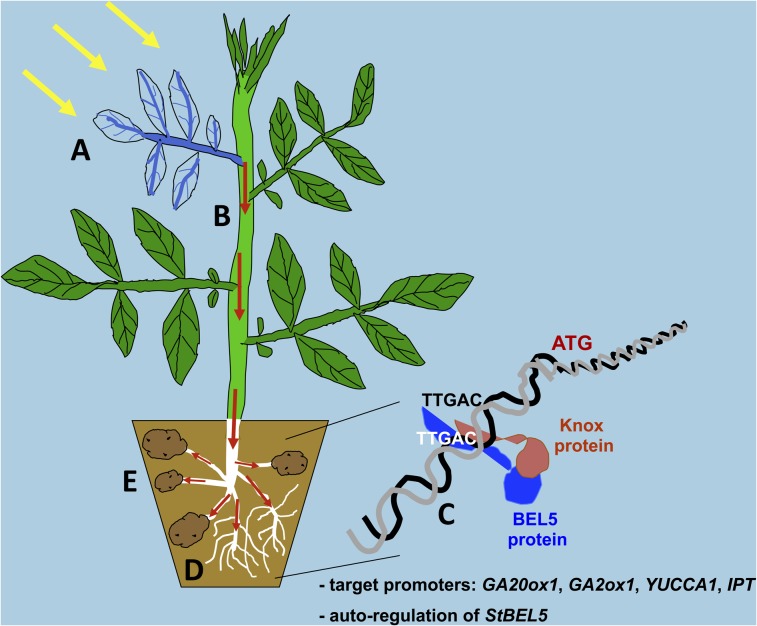
In our model, the leaf perceives a light signal that activates the transcription of StBEL5 in the veins and petiole (Fig. 9A). After accumulating in the leaf veins, a short-day photoperiod facilitates movement of the StBEL5 RNA through the petiole junction into the stem (Fig. 9B, red arrows) by instigating the activation or expression of appropriate RNA-binding proteins. Under these conditions, transcripts may be escorted to stolon tips or roots (Fig. 9, red arrows) via RNA/protein complexes (Ham et al., 2009). Translation of StBEL5 occurs on site, and the StBEL5 protein interacts with its Knox protein partner, creating the heterodimer that regulates the transcription of BEL/Knox target genes (Fig. 9C). Previous work has shown that StBEL5 interacts with a Knox-like protein, POTH1, to regulate the transcription of a specific target gene, GA20ox1 (Chen et al., 2004), and this study confirms the identity of three more target genes, GA2ox1, YUCCA1a, and IPT. The BEL5/POTH1 complex binds to a tandem motif, TTGAC, in the promoters of these genes, which has also been identified in the promoter of the StBEL5 gene. Our results also suggest the possibility of the autoregulation of StBEL5 transcription in stolons, newly formed tubers, and lateral roots to further augment the StBEL5 signal. Overall, this transcriptional complex leads to enhanced root and tuber growth (Fig. 9, D and E).
Figure 9.
Model showing the impact of mobile StBEL5 RNA on root and tuber development. Previously, the long-distance transport of StBEL5 RNA was shown to be correlated with the induction of tuber formation in potato (Banerjee et al., 2006a). This signaling pathway is based on the initial activation of transcription by light (A, yellow arrows) of the StBEL5 gene in the veins of leaves and petioles (A, blue). A short-day photoperiod facilitates movement of the StBEL5 RNA to stolon tips, whereas movement to roots occurs regardless of daylength (B, red arrows). Under these conditions, RNA may be escorted to site-specific targets, like stolon tips or roots, via protein chaperones (Ham et al., 2009). Enhanced translation then occurs in the stolon tip or root followed by binding to a Knox protein partner (C) and subsequent activation of the transcription and regulation of select genes (e.g. GA20ox1, GA2ox1, YUCCA1, IPT, and StBEL5) by binding to the tandem TTGAC core motif of the target promoter. In this model, transcriptional regulation then leads to enhanced growth of roots (D) and tubers (E) modulated by hormone levels. (Modified from figure 10.4 in Hannapel, 2012. This material is reproduced with permission of John Wiley & Sons.)
MATERIALS AND METHODS
Construct Designs
For the GAS promoter transgenic lines (Figs. 1 and 2), both constructs, GAS:GUS and GAS:BEL5, were PCR cloned into the XmaI/SacI site downstream from the GAS promoter that had been previously cloned into pBI101.2 (Banerjee et al., 2006a). For the GUS transgenic lines in Figure 6, both constructs, proBEL5:GUS and mut-proBEL5:GUS (Fig. 6A), were cloned by using wild-type BEL5 promoter sequence (Chatterjee et al., 2007) as a template in a PCR strategy. For the construct proBEL5, the promoter sequence was amplified by PCR with restricted digestion sites and inserted into pBI101.2 vector. For mut-proBEL5, the construct was amplified into two fragments, an upstream part and a downstream part, with SphI/PstI ends (primer set: forward, 5′-TTGCATGCGGAAAGTTGCAAGGATT-3′; and reverse, 5′-CGCCTGCAGATGAACAGAAAAATAT-3′) and PstI/SpeI ends (primer set: forward, 5′-AAACTGCAGTTGACTTGTTGTCACTCT-3′; and reverse, 5′-CGCACTAGTAGGGAAATATGAATAAA-3′). One of the TTGAC motifs was omitted between the two fragments. The two fragments were PCR subcloned into pGEM-T Easy vector separately. The sequence of the fragments was confirmed by sequencing, and only those clones in the correct orientation were used for further cloning for both fragments. The upstream promoter fragment was excised from the pGEM-T Easy vector by PstI digestion and ligated into the pGEM-T Easy vector in front of the downstream promoter fragment. This mutated promoter combination was excised from the pGEM-T Easy vector with SpeI single digestion at both the PCR-added SpeI site and another SpeI site present in the pGEM-T Easy vector and then inserted into the pBI101.2 vector at the SpeI cloning site. Correct orientation was again confirmed by sequencing. All sequencing was performed by the DNA Facility at Iowa State University.
Plant Material and Generation of Transgenic Lines
Transformation was implemented on the photoperiod-responsive potato (Solanum tuberosum ssp. andigena; Banerjee et al., 2006b). In photoperiod-adapted genotypes like andigena, short-day photoperiods (less than 12 h of light) are required for tuber formation, whereas under long-day conditions, no tubers are produced. Twenty to 25 independent transgenic lines that rooted on kanamycin were screened for GUS expression or RNA accumulation by using transgene-specific primers. Three to four high-expressing lines were selected from each construct and were used in evaluating growth or expression phenotypes. The results shown here are from one representative line of these latter groups. The wild-type proBEL5:GUS and GAS:BEL5 lines were screened during earlier studies (Banerjee et al., 2006b; Chatterjee et al., 2007). The in vitro transgenic potato plants were maintained in a growth chamber (Percival Scientific) at 27°C with a photoperiod of 16 h of light/8 h of dark and a fluence rate of 40 μmol m−2 s−1. Soil-grown plants were maintained in a growth chamber under either a long-day (16 h of light at 22°C, 8 h of dark at 18°C) or short-day (8 h of light at 22°C, 16 h of dark at 18°C) photoperiod with a fluence rate of 400 μmol m−2 s−1.
Sample Harvest
Leaves, stolons, and roots from wild-type, GAS:GUS, and GAS:BEL5 plants were harvested from soil-grown plants. They were grown in a growth chamber until the 12- to 13-leaf stage and randomly sorted into either short- or long-day conditions for 2 weeks. All environmental conditions except daylength were the same for these two groups. Young, healthy leaves were harvested and frozen in liquid nitrogen. Stolon and root samples for Figure 1 were harvested, washed with tap water, and dried by blotting. Primary and secondary root types could be distinguished by their morphology. Primary roots are thicker and often exhibit a light purple color, whereas secondary roots are bright white. Roots were harvested by washing the soil away gently in tap water, until the root was clean, and then blotted. Samples for RT-PCR were frozen in liquid nitrogen immediately after harvest and stored at −80°C prior to RNA extraction. Fresh root samples for the paraffin sections used in Figures 4 and 5 were cut into 5-cm lengths followed by fixation in 45% ethanol, 5% acetic acid, and 1.8% formaldehyde (FAA) and vacuum infiltration overnight. Paraffin-sectioning procedures are described below. Roots for paraffin sectioning were sampled from approximately the middle of several different roots, 2 to 4 cm from the tip, from different transgenic clones. Because root thickness is reasonably uniform along the middle root zone, this zone was randomly sampled, avoiding the proximal and distal ends of the root, where the diameter was more variable.
Movement Assay
Primary and secondary roots and new leaves were harvested from three to five plants and pooled. Three separate RNA extraction reactions were run as replicates with the RNeasy Plant Mini Kit (Qiagen). One-step RT-PCR (with the SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase) was performed using 200 ng of total RNA, a nonplant-sequence primer fused to all transgenic RNAs, and a gene-specific primer (5′-GGGAGATTTTGGAAGGTTTG-3′) from the StBEL5 coding sequence. Use of the nonplant-sequence primer, NT-142 (5′-CGGGACTCTAATCATAAAAAC-3′), corresponding to sequence from the nos terminator (Banerjee et al., 2006b), makes it possible to distinguish transgenic RNA from native StBEL5 RNA. The internal control for PCR was 18S ribosomal RNA (rRNA). The PCR cycle numbers were adjusted to 32 for StBEL5 and 17 for the 18S rRNA to be in the linear range. The cycling program for StBEL5 is 50°C for 30 min, 94°C for 2 min, 32 cycles of 94°C for 15 s, 55°C for 30s, and 68°C for 1 min, followed by one cycle of incubation at 68°C for 5 min. Seventeen cycles were used for 18S rRNA with the same conditions. Homogenous PCR products were run on a 1.0% agarose gel and quantified by using ImageJ software (Abramoff et al., 2004) and normalized by using 18S rRNA values.
Heterografts
Micrografts were made using GAS:BEL5 or GAS:GUS transgenic lines for scion material and wild-type andigena for stocks and were grown in vitro for 2 weeks before transfer to soil. Heterografts were then grown for 3 weeks under long-day conditions (16 h of light, 8 h of dark, 25°C) and then 2 weeks under short days (8 h of light, 16 h of dark, 25°C) before sample harvest, RNA extraction, and nested RT-PCR.
GUS Histochemical Analysis
Expression of the GUS reporter gene driven by the StBEL5 promoters (Fig. 6) was analyzed by incubating the samples 24 h at 37°C in GUS buffer containing 0.1 m phosphate buffer, pH 7.0, 10 mm EDTA, pH 8.0, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 0.1% Triton X-100, and 0.7 mg mL−1 5-bromo-4-chloro-3- indolyl-β-d-glucuronic acid. Samples were cleared with 100% ethanol and photodocumented with an Olympus E-500 digital camera. The proBEL5:GUS and mut-proBEL5:GUS root samples used for paraffin sectioning were bleached and stored in 70% ethanol. After 30 min in 50% ethanol, the samples were fixed in FAA solution overnight under a vacuum. Paraffin-sectioning procedures are described below.
Light Microscopy
After harvest, samples were fixed in FAA solution and cut into approximately 1.0-cm pieces. The samples were dehydrated at 4°C in 50% ethanol for 30 min, 70% ethanol for 30 min, 85% ethanol for 30 min, and 100% ethanol overnight. The following day, samples were incubated in 100% ethanol for 30 min and tertiary butyl alcohol (TBA):ethanol (1:1) for 30 min at room temperature. After changing to 100% TBA, samples were kept in a 60°C oven overnight. The following day, samples are incubated in fresh 100% TBA for 30 min and then in TBA plus liquid paraffin (1:1) at 60°C. Over the next 2 d, samples were incubated in TBA plus paraffin (1:3) and then in 100% paraffin at 60°C. Samples were then embedded into paraffin blocks and stored at 4°C to facilitate sectioning. Paraffin ribbons were placed in water on slides and heated at 40°C to position the ribbon evenly. Excess water was blotted with a tissue, and slides were left at room temperature until completely dry (12–18 h). To make permanent sections, select slides were heated in a 60°C oven for 30 min, soaked in xylene for 10 min three times, and then covered with a cover slide and Permount. In Figure 4, A and B, transverse sections of primary roots were stained with toluidine blue O. After heating in a 60°C oven for 30 min, slides were treated with xylene for 5 min three times, with 100% ethanol for 1 min two times, with 95% ethanol for 1 min and 70% ethanol for 1 min, with distilled water for 1 min, with 1% toluidine blue O stain in 1% borax for 45 s, followed by a tap water wash about 10 times until the water was clear. The stained slide was then dipped in 70% ethanol 10 times, 95% ethanol 10 times, 100% ethanol 10 times, 1:1 ethanol:xylene for 1 min, and xylene for 3 min two times. Permount was then added for any permanent sections. Sections were photodocumented with an Olympus BX40 microscope and a Carl Zeiss AxioCam MRc 5 digital camera operated by the systems program Zeiss AxioVision AC. Because many of the root cross sections were oval in shape, root and stele diameters were measured as the average of the longest and shortest distances across the center. Diameters were measured with the Image Analysis Program in the Soft Imaging System from Olympus, calibrated with the scale bar in each image. The central stele of the primary root includes the endodermis, xylem, and phloem, surrounded by cortical cells, which are much larger than the phloem and endodermal cells. Sectioning and image analysis were performed in the Microscopy and Nanoimaging Facility at Iowa State University.
Gel-Shift Assays
Recombinant protein expression, purification, and DNA-binding assays were performed as described previously (Viola and Gonzalez, 2006). For the double tandem target sequence of StGA20x-1, a 210-bp region including both tandem motifs was amplified by PCR with primer sets F (5′-cgggatccTAAACTTGGGGCATGATTGA-3′) and R (5′-cgggatcCAGTAGGAAACAAAATATAC-3′; BamHI sites are in lowercase throughout). The full sequence of this region is as follows (underline marks the primers, and motifs are highlighted in boldface): 5′-cgggatccTAAACTTGGGGCATGATTGATTTTCATTCGTTCATTTAAATTACCTTTTTATTTATTCGATTAAATTGACAAGTCATATAGAAGCTCTACCCAACAACGAATATTTTAAGTTTGCGATGTTGATAAATAAATAAAGTCAGCTCTGTCCTTTTATATCACGATGACACGTCAACAAGAACATTAAGCTTTAGTATATTTTGTTTCCTACTGgatcccg-3′. As a single tandem target, both strands of a 100-bp oligonucleotide that includes only one of the two tandem motifs were synthesized with a 5′ overhang at both ends. The sense sequence is 5′-gatccATTTTCATTCGTTCATTTAAATTACCTTTTTATTTATTCGATTAAATTGACAAGTCATATAGAAGCTCTACCCAACAACGAATATTTTAAGTTTGCGATGg-3′. The single-stranded oligonucleotides were annealed and end labeled with [32P]dATP by filling in the 3′ ends with the Klenow fragment of DNA polymerase. The G-to-C mutated sequence (Fig. 3B) was prepared and labeled in the same way. For the StBEL5 promoter bait, a 97-bp region was synthesized in the same way as the StGA2ox1 motifs. The sequence is 5′-gatccAAGAGTGAATAATAAAATATATTTTTCTGTTCATTTTTATTTGTCAATGCTTGACTTGTTGTCACTCTCTTTAGTACTAATATTAATAAACTTTTAAg-3′. For the mutated sequence, the underlined region was replaced with CTGCAG, eliminating one of the TTGAC motifs. A complete list of the primers used is included in Supplemental Table S1.
Real-Time qRT-PCR
Total RNA was extracted from all the plant tissues using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. To avoid genomic DNA contamination, total RNA was treated with RNase-free DNase Set (Qiagen). The quantity and quality of RNA samples were estimated using a Nano spectrophotometer (ND-1000; Thermo Scientific). RNA samples with a 260:280 ratio from 1.9 to 2.1 and a 260:230 ratio from 2.0 to 2.5 were used for qRT-PCR analysis. qRT-PCR analysis was performed with the qScript One-Step SYBR Green qRT-PCR Kit (Quanta Biosciences) following the manufacturer’s protocol. Briefly, 50-ng aliquots of total RNA template were subjected to each qRT-PCR in a final volume of 15 µL containing 7.5 µL of One-Step SYBR Green Master Mix and 0.3 µL of qScript One-Step Reverse Transcriptase along with target specific primers (200 nm). All reactions were performed in triplicate using an Illumina Eco qPCR machine with fast quantitative PCR cycling parameters (complementary DNA [cDNA] synthesis: 50°C, 5 min; Taq activation: 95°C, 2 min; PCR cycling [40 cycles]: 95°C, 3 s/60°C, 30 s). The StACT8 (accession no. GQ339765) and StUBC (accession no. DQ222513) genes were used as endogenous controls for normalization of the total RNA template in a reaction. The relative gene quantification (comparative threshold cycle) method (Livak and Schmittgen, 2001) was used to calculate the expression levels of different target genes. Primers ranged from 98 to 160 bp and were mostly designed spanning the introns in order to detect any genomic DNA contamination (Supplemental Table S1). The specificity of primers was determined by melting-curve analyses and agarose gel (3%) electrophoresis performed following the qRT-PCR experiments. A standard curve was generated based on six-point (10-fold) serial dilutions of cDNA to calculate the gene-specific PCR efficiency. PCR efficiencies of primers ranged from 97% to 110%.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Activity of the StBEL5 promoter in stolons grown in the light.
Supplemental Figure S2. Specificity of gene-specific primers for transgenic StBEL5 RNA.
Supplemental Table S1. Oligonucleotides and primers designed for the synthesis of promoter sequences used in gel-shift assays and primers used in qRT-PCR.
Acknowledgments
Thanks to Tracey Pepper and Jack Horner for their wonderful support with the microscopy, to Dan Stessman for his contributions to the transformation work, to Nathan Butler for help with grafting, and to Mithu Chatterjee for establishing the profile for StBEL5 promoter activity. Thanks also to Hao Chen for his important contributions to our understanding of StBEL5 biology.
Glossary
- GAS
galactinol synthase
- qRT
quantitative reverse transcription
- RT
reverse transcription
- UTR
untranslated region
- FAA
45% ethanol, 5% acetic acid, and 1.8% formaldehyde
- TBA
tertiary butyl alcohol
- cDNA
complementary DNA
- rRNA
ribosomal RNA
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophot Int 11: 36–42 [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K. (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32: 401–408 [DOI] [PubMed] [Google Scholar]
- Ayre BG, Blair JE, Turgeon R. (2003) Functional and phylogenetic analyses of a conserved regulatory program in the phloem of minor veins. Plant Physiol 133: 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Chatterjee M, Yu Y, Suh SG, Miller WA, Hannapel DJ. (2006a) Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18: 3443–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Prat S, Hannapel DJ. (2006b) Efficient production of transgenic potato (S. tuberosum L. ssp. andigena) plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci 170: 732–738 [Google Scholar]
- Banerjee AK, Lin T, Hannapel DJ. (2009) Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiol 151: 1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H. (2004) VAAMANA: a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328: 103–111 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Benková E, Helariutta Y. (2011) Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr Opin Plant Biol 14: 10–16 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Hake S. (2009) The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S. (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Groover AT, Fontana JR, Martienssen RA. (2003) Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130: 3941–3950 [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Banerjee AK, Hannapel DJ. (2007) A BELL1-like gene of potato is light activated and wound inducible. Plant Physiol 145: 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ. (2004) The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J 38: 276–284 [DOI] [PubMed] [Google Scholar]
- Chen H, Rosin FM, Prat S, Hannapel DJ. (2003) Interacting transcription factors from the three-amino acid loop superclass regulate tuber formation. Plant Physiol 132: 1391–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R. (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Dettmer J, Elo A, Helariutta Y. (2009) Hormone interactions during vascular development. Plant Mol Biol 69: 347–360 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Gaupels F, Buhtz A, Knauer T, Deshmukh S, Waller F, van Bel AJE, Kogel KH, Kehr J. (2008) Adaptation of aphid stylectomy for analyses of proteins and mRNAs in barley phloem sap. J Exp Bot 59: 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J, Strauss SH, Tsai CJ, Fang K, Chen Y, Jiang X, Busov VB. (2010) Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell 22: 623–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham BK, Brandom JL, Xoconostle-Cázares B, Ringgold V, Lough TJ, Lucas WJ. (2009) A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21: 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannapel DJ. (2010) A model system of development regulated by the long-distance transport of mRNA. J Integr Plant Biol 52: 40–52 [DOI] [PubMed] [Google Scholar]
- Hannapel DJ. (2012) The effect of long-distance signaling on development. In AJE van Bel, GA Thompson, eds, Phloem: Molecular Cell Biology, Systemic Communication, Biotic Interactions. John Wiley & Sons, New York, pp 209–226
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Kanrar S, Bhattacharya M, Arthur B, Courtier J, Smith HMS. (2008) Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J 54: 924–937 [DOI] [PubMed] [Google Scholar]
- Kehr J, Buhtz A. (2008) Long distance transport and movement of RNA through the phloem. J Exp Bot 59: 85–92 [DOI] [PubMed] [Google Scholar]
- Kloosterman B, Navarro C, Bijsterbosch G, Lange T, Prat S, Visser RG, Bachem CW. (2007) StGA2ox1 is induced prior to stolon swelling and controls GA levels during potato tuber development. Plant J 52: 362–373 [DOI] [PubMed] [Google Scholar]
- Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW. (2007) The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19: 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lough TJ, Lucas WJ. (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57: 203–232 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Omid A, Keilin T, Glass A, Leshkowitz D, Wolf S. (2007) Characterization of phloem-sap transcription profile in melon plants. J Exp Bot 58: 3645–3656 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H-J, Sundaresan V. (2007) Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19: 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Moubayidin L, Sabatini S. (2010) The molecular basis of cytokinin function. Curr Opin Plant Biol 13: 21–26 [DOI] [PubMed] [Google Scholar]
- Racca RW, Tizio R. (1968) A preliminary study of changes in the content of gibberellin-like substances in the potato plant in relation to the tuberization mechanism. Eur Potato J 11: 213–220 [Google Scholar]
- Ragni L, Belles-Boix E, Günl M, Pautot V. (2008) Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 20: 888–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Robinson-Beers K, Ray S, Baker SC, Lang JD, Preuss D, Milligan SB, Gasser CS. (1994) Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc Natl Acad Sci USA 91: 5761–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin FM, Hart JK, Horner HT, Davies PJ, Hannapel DJ. (2003) Overexpression of a knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol 132: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumeliotis E, Kloosterman B, Oortwijn M, Kohlen W, Bouwmeester HJ, Visser RG, Bachem CW. (2012) The effects of auxin and strigolactones on tuber initiation and stolon architecture in potato. J Exp Bot 63: 4539–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. (2009) Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J 58: 641–654 [DOI] [PubMed] [Google Scholar]
- Saleem M, Lamkemeyer T, Schützenmeister A, Madlung J, Sakai H, Piepho HP, Nordheim A, Hochholdinger F. (2010) Specification of cortical parenchyma and stele of maize primary roots by asymmetric levels of auxin, cytokinin, and cytokinin-regulated proteins. Plant Physiol 152: 4–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Hake S. (2003) The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. (2008) Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Tanaka-Ueguchi M, Itoh H, Oyama N, Koshioka M, Matsuoka M. (1998) Over-expression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J 15: 391–400 [DOI] [PubMed] [Google Scholar]
- Vilaine F, Palauqui JC, Amselem J, Kusiak C, Lemoine R, Dinant S. (2003) Towards deciphering phloem: a transcriptome analysis of the phloem of Apium graveolens. Plant J 36: 67–81 [DOI] [PubMed] [Google Scholar]
- Viola IL, Gonzalez DH. (2006) Interaction of the BELL-like protein ATH1 with DNA: role of homeodomain residue 54 in specifying the different binding properties of BELL and KNOX proteins. Biol Chem 387: 31–40 [DOI] [PubMed] [Google Scholar]
- Woo YM, Park HJ, Su’udi M, Yang JI, Park JJ, Back K, Park YM, An G. (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65: 125–136 [DOI] [PubMed] [Google Scholar]
- Xu X, van Lammeren AA, Vermeer E, Vreugdenhil D, (1998) The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol 117: 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]