Summary: Transcriptome analysis of an early cone-setting mutant identified candidate genes active during reproductive phase change in the conifer Picea abies.
Abstract
Conifers normally go through a long juvenile period, for Norway spruce (Picea abies) around 20 to 25 years, before developing male and female cones. We have grown plants from inbred crosses of a naturally occurring spruce mutant (acrocona). One-fourth of the segregating acrocona plants initiate cones already in their second growth cycle, suggesting control by a single locus. The early cone-setting properties of the acrocona mutant were utilized to identify candidate genes involved in vegetative-to-reproductive phase change in Norway spruce. Poly(A+) RNA samples from apical and basal shoots of cone-setting and non-cone-setting plants were subjected to high-throughput sequencing (RNA-seq). We assembled and investigated 33,383 expressed putative protein-coding acrocona transcripts. Eight transcripts were differentially expressed between selected sample pairs. One of these (Acr42124_1) was significantly up-regulated in apical shoot samples from cone-setting acrocona plants, and the encoded protein belongs to the MADS box gene family of transcription factors. Using quantitative real-time polymerase chain reaction with independently derived plant material, we confirmed that the MADS box gene is up-regulated in both needles and buds of cone-inducing shoots when reproductive identity is determined. Our results constitute important steps for the development of a rapid cycling model system that can be used to study gene function in conifers. In addition, our data suggest the involvement of a MADS box transcription factor in the vegetative-to-reproductive phase change in Norway spruce.
The two most commonly grown and economically important conifers in Sweden, Scots pine (Pinus sylvestris) and Norway spruce (Picea abies), both go through a long vegetative growth phase before they begin to produce cones; the vegetative growth period is 20 to 25 years and 8 to 20 years for Norway spruce and Scots pine, respectively. Mature Scots pine trees produce cones every year, while mature Norway spruce trees produce cones only every 3rd to 5th year. The timing of cone establishment is largely synchronized within the various spruce populations and is determined by a combination of genetic and environmental factors (Lindgren et al., 1977; Högberg and Eriksson, 1994). The long generation time and the irregular cone setting between different years pose major obstacles for breeding of Norway spruce both with respect to genetic gain and retained diversity. To guarantee a sufficient seed supply, forest companies have to plant and maintain large seed orchards and in addition store large quantities of seeds to cover the demand during years of little or no seed production. Both conifer breeding programs and the production of improved seed for forest regeneration would benefit from methods to control the length of the vegetative period before the trees start to produce cones as well as the possibilities to control cone setting and cone production itself. Despite that, our knowledge of the genetic mechanisms that regulate the transition from vegetative growth to reproductive development in conifers is limited. Here, we utilize the intrinsic properties of a naturally occurring homeotic mutant of spruce, acrocona, which produces cones every year, to identify candidate genes of importance for reproductive phase change in spruce. In the acrocona mutant, female cones are regularly set on terminal positions of shoots on both top shoots and scaffold branches, at positions where wild-type spruce never produces female cones, hence the Latin name acrocona for “top cone.” Reports of acrocona specimens from different localities in Uppland, Sweden, stem from the mid 19th century, and sporadic occurrence of the acrocona mutant has since then also been reported from other parts of Sweden (Fries, 1890; Joneborg, 1945). For horticultural purposes, the acrocona mutant has been widely propagated by grafting, and acrocona plants can now be found in gardens all over the temperate world. In a previous study including the acrocona mutant, early cone setting was mapped to spruce chromosome 6 (Acheré et al., 2004). However, the gene or genes responsible for the acrocona phenotype are not known.
Genetic and molecular analyses of flowering-time mutants in the angiosperm model species Arabidopsis (Arabidopsis thaliana) have demonstrated that the phase change from vegetative to reproductive development in flowering plants follows four distinct pathways: the autonomous pathway and those induced by photoperiod, vernalization, or gibberellin (for review, see Bäurle and Dean, 2006; Lee and Lee, 2010). In short, pathways influenced by intrinsic and environmental signals (e.g. photoperiod and temperature) regulate the onset of flowering by increasing the expression of the FLOWERING LOCUS T (FT) gene, whose encoded protein has been proposed to be the elusive mobile flower-inducing factor “florigen” (Wigge et al., 2005; Lifschitz et al., 2006). The FT protein regulates the transition of a vegetative meristem to an inflorescence meristem by activating the expression of genes belonging to the MADS box gene family of transcription factors, APETALA1 and SUPPRESSOR OF CONSTANS1 (SOC1). Hormonal induction of flowering also occurs independently of FT by the GA3-dependent activation of SOC1, which, in turn, activates the floral meristem identity gene LEAFY (LFY; Moon et al., 2003). Based on advances in the understanding of the genetic programs controlling flowering in Arabidopsis, various biotechnological approaches have been undertaken to shorten the time to flowering in angiosperm trees. For instance, expression of the FT gene induces early flowering by shortening the juvenile phase in Populus trichocarpa (Böhlenius et al., 2006). Similarly, overexpression of the Citrus unishu FT homolog (CiFT) can accelerate flowering in Poncirus trifoliata (Endo et al., 2005). Interestingly, recent evolutionary studies suggest that the angiosperm FT gene is the result of a duplication event that occurred in the angiosperm lineage after the split between angiosperms and gymnosperms approximately 300 million years ago (Smith et al., 2010; Karlgren et al., 2011). This indicates that the phase change from vegetative to reproductive development in gymnosperms and angiosperms is regulated by distinct genetic mechanisms that involve both orthologous and evolutionarily unrelated genes that may or may not exert similar functions.
Studies of transcriptional regulators belonging to the MADS box gene family in Norway spruce suggest that members of this gene family have putative functions in determining the length of the juvenile period and the vegetative or reproductive identity of a shoot (Carlsbecker et al., 2003, 2004). Expression of the gene DEFICIENS AGAMOUS-LIKE1 (DAL1) increases with age and conforms to a spatial pattern that marks physiological and morphological features associated with the transition to the reproductive phase. In addition, the expression of DAL1 in transgenic Arabidopsis shortens the vegetative phase, and the plants begin to flower extremely early, sometimes forming embryonic flowers (Carlsbecker et al., 2004). Similarly, the spatial expression patterns of the Norway spruce gene DAL10 together with phenotypic studies of Arabidopsis plants, harboring DAL10 under the control of a constitutive promoter, suggest that this MADS box gene may be involved in specifying reproductive identity in the male and female shoots (cones) of Norway spruce (Carlsbecker et al., 2003).
Recent advances in high-throughput sequencing technologies allow for global transcriptome profiling of wild-type and mutant plants and can be used to study the differential expression of all genes involved in a particular developmental process (Wang et al., 2009). This holds true also for spruce, although analyses are hampered by the current lack of a reference genome and the fact that spruce, like other conifers, displays a high genetic diversity (Tollefsrud et al., 2008), which makes it difficult to choose a relevant wild-type comparator. Here, we have developed a bioinformatics pipeline that allows us to accurately assemble Norway spruce transcripts de novo without prior knowledge of the genomic sequence. To avoid the identification of false positives due to genetic diversity in Norway spruce, we have analyzed differentially expressed genes in a segregating sibling population of the acrocona mutant using massively parallel sequencing methods and also assessed selected candidate genes involved in reproductive phase change using targeted quantitative real-time (qRT)-PCR from another segregating sibling population.
RESULTS
Offspring from Controlled Crosses of the acrocona Mutant Show an Early Cone-Setting Phenotype
To examine the segregation pattern of the acrocona phenotype and to produce a population of acrocona siblings, two ramets of the acrocona clone were crossed, and the resulting progeny were grown under accelerated growth conditions in a phytotron. Nineteen out of 75 inbred acrocona plants (derived from crossings made in 2006) initiated cones already during the second growth cycle (Fig. 1; Supplemental Table S1). Open-pollinated acrocona progeny (n = 150) and control plants (n = 75) were grown together with the inbred acrocona plants. Only two of the open-pollinated acrocona progeny and none of the control plants initiated cones during the first four growth cycles. A similar segregation pattern was obtained in a subsequent growth experiment of inbred acrocona plants (derived from crossings made in 2009), in which three out of 16 plants initiated cones during the second growth cycle and a fourth plant initiated cones during the third growth cycle, while none of the control plants initiated cones. Together, the segregation of early cone setting in acrocona plants [19/75 and (3+1)/16] constitutes an excellent fit (χ2 value, P = 0.003) with the Mendelian segregation expected assuming that the parents were heterozygous for the acrocona mutation and that only homozygous plants displayed the early cone-setting phenotype. During the fourth growth cycle, additional intermediate shoot phenotypes were initiated in inbred acrocona plants but not in open-pollinated acrocona progeny or in control plants: apical shoots with a large number of lateral short shoots and vegetative shoots with broad and bract-like needles (Supplemental Fig. S1, E–H; Supplemental Table S1).
Figure 1.
Cone formation in spruce. A, Apical transition cone developed on an early cone-setting inbred acrocona plant grown under accelerated growth conditions after three growth cycles. B, Close up of the apical transition cone in A. Note the transition from needles in the lower part to ovuliferous scales in the upper part of the apical shoot. C, Vegetative shoot phenotype on an inbred acrocona plant. D, Female cone on an open-pollinated wild-type tree. Bars = 2 cm (A and D) and 0.5 cm (B and C).
Cone-setting acrocona plants regularly initiated cones on the apical shoot, although individual plants produced additional cones on lateral branches (Fig. 1; Supplemental Fig. S1). Both apical and lateral shoots that initiated cones produced a transition cone phenotype, with vegetative needles in the basal part and reproductive ovuliferous scales subtended by bracts in the apical part (Fig. 1, A and B). The number of needles initiated before the transition to reproductive development differed among individual plants and among cones on plants that initiated multiple cones. Hence, in the acrocona plants, the transition from vegetative to reproductive identity often occurred at different stages during bud development.
Apart from the early cone-setting phenotype, the acrocona plants often displayed a bushy appearance, suggesting a reduced apical dominance, and reduced height as compared with open-pollinated acrocona progeny and control plants. The seasonal growth response to increased light and temperature was similar in both cone-setting acrocona plants and control plants (Supplemental Fig. S2), which indicates that the reduced height was an effect neither of daylight sensing nor of differences in the length of the growth cycle.
Transcriptome Sequencing and Analysis
In order to identify candidate genes involved in the early cone-setting acrocona phenotype, we performed a massively parallel sequencing (RNA-seq) of 14 poly(A+)-enriched (i.e. mRNA) samples extracted from needles taken from inbred cone-setting acrocona plants, inbred non-cone-setting acrocona plants, and control plants. For each individual, a sample pair was extracted: one sample from the apical, potentially cone-setting shoot, and one sample from a shoot at the base of the plant, where no cone-setting occurs (Fig. 2A). The sequencing effort yielded 136 Gb of RNA sequence (between 58 and 270 Mb per sample), for an estimated average coverage on the order of 100× for exonic regions.
Figure 2.
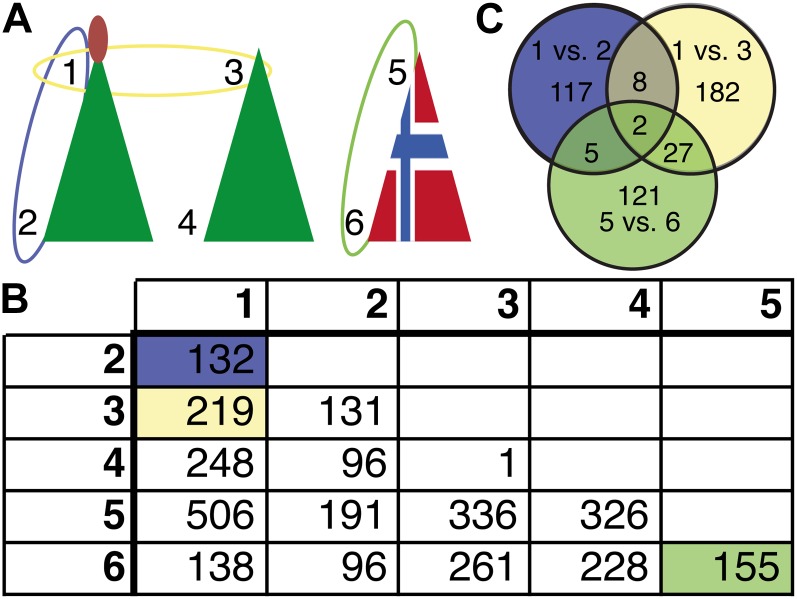
Unbiased whole-transcriptome differential expression analysis. A, Schematic view of different needle samples that were tested for differential expression: apical shoot (1) and basal shoot (2) from two acrocona plants that initiated cones in the second growth cycle; apical shoot (3) and basal shoot (4) from three acrocona plants that did not initiate cones in the second growth cycle; and apical shoot (5) and basal shoot (6) from two control plants that did not initiate cones in the second growth cycle. B, Number of differentially expressed genes between all six needle samples (multiple testing corrected P < 0.005). C, Venn diagram with the number of shared and unique differentially expressed genes between sample 1 compared with 2, 1 compared with 3, and 5 compared with 6.
Since there is no Norway spruce genome sequence available, the acrocona transcripts from four different samples from one acrocona plant were reconstructed both de novo (i.e. without aligning to any reference sequence) and ab initio, using a comprehensive set of 27,720 Picea glauca transcripts as reference (Rigault et al., 2011). With the ab initio approach, we detected 83% (22,924 transcripts) of the P. glauca transcripts, indicating that homologs of these transcripts were expressed in our acrocona RNA-seq data. With the de novo approach, we assembled 67,857 different acrocona transcripts (including isoforms), of which 54,775 transcripts contained at least one identified open reading frame (ORF), amounting to a total of 83,650 ORFs in total (Supplemental Fig. S3A).
Putative orthologs were detected using OrthoMCL (Li et al., 2003), which constructs groups of orthologous proteins across species. The input data sets were, in addition to our translated acrocona ORFs, comprehensive protein sets from P. glauca and Arabidopsis. The acrocona ORFs were represented in 19,865 orthologous groups, and we note that 14,109 (71%) of these groups also contain P. glauca and/or Arabidopsis proteins (Supplemental Fig. S3, B and C), indicating that at least the corresponding set of 19,439 (35%) reconstructed ORF-containing acrocona transcripts are real.
Conversely, 93% of the groups of orthologous proteins that contained P. glauca proteins also contained acrocona proteins. On the transcript level, 14,520 (52%) of the P. glauca transcripts had an ortholog in our de novo-assembled acrocona set. Further details about the reconstruction and analysis of the acrocona and wild-type spruce transcriptomes are available (J. Reimegård and O. Emanuelsson, unpublished data), and the expression levels of the identified ORFs are presented in Supplemental Data Set S1.
Unbiased Differential Gene Expression Analysis Indicates Up-Regulation of a MADS Box Gene in Early Cone-Setting acrocona Apical Shoots
To identify differentially expressed genes between cone-setting and non-cone-setting acrocona plants in an unbiased way, relative expression levels between relevant sample pairs were established from our RNA-seq data (specifically, the 83,650 identified ORFs) using Bowtie2, Cufflinks, Cuffmerge, and Cuffdiff (Trapnell et al., 2012; Fig. 2). This process yielded expression estimates for 33,383 ORFs, and we define these as the set of potential protein-coding transcripts present in our acrocona samples. By comparing transcript abundance in samples from apical cone-setting shoots with samples from non-cone-setting basal shoots of the same cone-setting acrocona plants, 132 (four up and 128 down) significantly differentially expressed transcripts were identified (Fig. 2B). Similarly, 219 (86 up and 133 down) transcripts were significantly differentially expressed in apical shoots from cone-setting acrocona plants compared with non-cone-setting acrocona plants. Expression levels and annotation of the differentially expressed genes are listed in Supplemental Data Set S2. By combining the two data sets, 10 transcripts were identified as being significantly up- or down-regulated in cone-setting shoots of acrocona (adjusted for multiple testing; P < 0.005) as compared with their prevalence in both (1) non-cone-setting regions (basal shoots) of the same plant individual and (2) potential cone-setting regions (apical shoots) with cone-setting ability but where no cones were set in other acrocona individuals (1 versus 2 and 1 versus 3, respectively, in Fig. 2C). Sequence comparison showed that all 10 transcripts had sequence similarity to distinct genes in the model species Arabidopsis, and we thus conclude that they likely originated from 10 different genes in acrocona. Two of these genes were also differentially expressed (apical versus basal) in control plants. We believe that the remaining eight genes constitute a reasonable set of first-pass candidate genes involved in the early cone-setting in acrocona (Table I). Seven of these genes were down-regulated in cone-setting individuals. Functional annotation and sequence similarity to known model plant proteins suggest that they have enzymatic functions of importance for cell wall composition during meristem or organ development, cell signaling, and plant stress response, and one gene encodes a protein with unknown function.
Table I. Eight differentially expressed genes in cone-setting regions compared with non-cone-setting regions.
See also the Venn diagram in Figure 2C.
| Name | Differential Expression | Arabidopsis | Panther | Function |
|---|---|---|---|---|
| Acr34511_3 | − | PRX52 | PTHR31388 | Peroxidase |
| Acr38476_2 | − | AT3G49190 | PTHR31650 | O-Acyltransferase |
| Acr38884_3 | − | AT5G36800 | PTHR31999 | Unknown function |
| Acr42124_1 | + | SOC1 | PTHR11945:SF19 | MADS box protein |
| Acr55424_3 | − | GH9B13 | PTHR22298:SF3 | Endo-1,4-β-glucanase |
| Acr1417_5 | − | ATCAT3 | PTHR11465 | Catalase |
| Acr15193_3 | − | XTH9 | PTHR31062 | Xyloglucan endotransglucosylase |
| Acr31311_1 | − | AT1G72110 | PTHR31650 | O-Acyltransferase |
One candidate gene, Acr42124_1, was up-regulated in apical cone-setting shoots compared with both basal shoots of cone-setting acrocona plants and apical shoots of non-cone-setting acrocona plants. The transcript corresponding to Acr42124_1 was 100% identical (over its entire protein-coding region as generated by our transcript reconstruction approach; BLAST E value of 0.0) to a wild-type spruce transcript identified and named DAL19 in a previous study (P. Engström, personal communication). DAL19 encodes a putative transcription factor belonging to the MADS box gene family. Apart from the conserved MADS domain, Acr42124_1 and DAL19 harbor a less conserved intervening region, a K domain, and a C-terminal extension; hence, both transcripts belong to a subclass of the MADS box gene family called the MIKC type (Ma et al., 1991). Based on the sequence identity and the occurrence of all MIKC sequence elements, we concluded that the transcript Acr42124_1 originated from the gene DAL19.
The MADS Box Gene DAL19 Shows High Similarity to a Class of Angiosperm Genes That Play Important Roles in the Transition from Vegetative to Inflorescence Meristem Identity
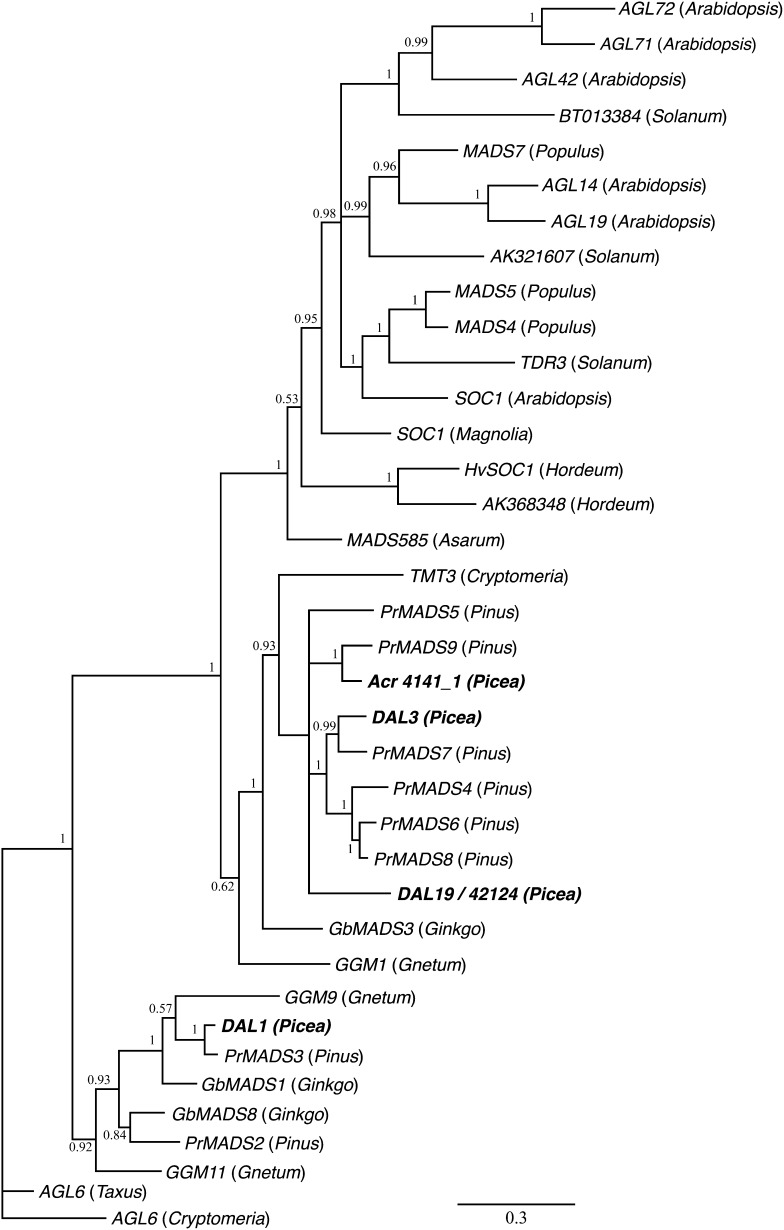
BLAST searches indicated that Acr42124_1/DAL19 (henceforth DAL19) is most similar to the previously cloned spruce DAL3 gene, which, in phylogenetic analyses covering all MIKC-type genes from the model species Arabidopsis, groups close to a clade of angiosperm genes that includes, but is not limited to, the Arabidopsis genes SOC1 and AGAMOUS LIKE42 (AGL42; Tandre et al., 1995; Carlsbecker et al., 2004). In order to establish the evolutionary relationship between DAL19, other similar conifer genes, and angiosperm genes previously annotated to the SOC1 clade, we performed a phylogenetic analysis using conifer and angiosperm representatives of the distantly related AGL6 clade as an outgroup. The phylogenetic analysis showed that DAL19 together with the spruce gene DAL3 and other gymnosperm MADS box genes form an orthologous sister clade to the entire angiosperm SOC1 clade (Fig. 3). This is in agreement with previously published results and suggests that both the angiosperm SOC1 clade and the gymnosperm clade harboring DAL19 have gone through several rounds of gene duplication after the split between gymnosperms and angiosperms.
Figure 3.
Phylogenetic relationship of MADS box genes from angiosperms and gymnosperms that show orthology to the DAL19 gene. Shown is a 50% majority rule tree derived using Bayesian phylogenetics. Numbers beside each node indicate posterior probabilities. The taxon of origin is shown in parentheses after each gene name, and spruce gene names are highlighted in boldface. Names and accession numbers are found in Supplemental Table S2.
Directed qRT-PCR Expression Analyses of Candidate Genes for the Early Cone-Setting Response Indicate MADS Box Gene Involvement in the Initiation of Cone Set and Reproductive Maturity
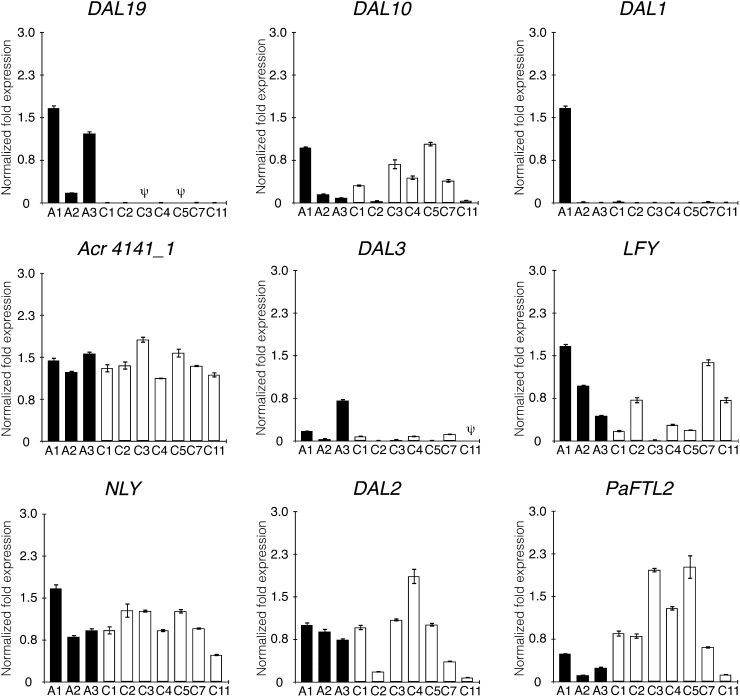
To provide independent acrocona gene expression data for DAL19 and an additional set of eight genes previously implicated during reproductive phase change in conifers, relative transcript levels were assayed using qRT-PCR (Fig. 4). Expression levels of these nine genes were measured in needles from lateral shoots directly subtending the apical shoot. The inbred acrocona plants were grouped into two main categories: A, inbred acrocona plants initiating at least four cones; and C, inbred acrocona plants that did not initiate cones. Since cones were either initiated on apical shoots or, in cases of more than one cone, on lateral shoots directly subtending the apical shoot, we expected needle samples from shoots in category A (four or more cones) to be comparable to needle samples from apical shoots used in our transcriptome analysis. In samples from needles collected during the second growth cycle, the expression of DAL19 was significantly higher in acrocona plants that initiated more than four cones (i.e. category A) compared with non-cone-setting acrocona plants of category C, where low or no expression could be detected (Student’s t test, P = 0.0046). None of the other genes tested differed significantly between cone-setting plants and non-cone-setting plants. These results are also in agreement with the transcriptome analysis, which identified full-length transcripts from a majority of the genes assayed in the qRT-PCR experiments but differential expression only of transcripts corresponding to DAL19 (Supplemental Fig. S4).
Figure 4.
qRT-PCR analyses of nine genes in needles from the second growth cycle. The samples were collected from inbred cone-setting acrocona plants initiating four or more cones (A) and inbred vegetative, non-cone-setting acrocona plants (C). Numbers beside the categories A and C indicate individual acrocona plants. The expression of DAL19, DAL10, DAL1, Acr4141_1, DAL3, LFY, NLY, DAL2, and PaFTL2 was assayed. Needles were sampled from one lateral shoot subtending the apical shoot and collected during the vegetative growth of the shoot before winter dormancy. Expression levels are presented as unscaled expression values based on mean values of three technical replicates. ψ represents no detectable expression. Error bars denote se of technical replicates. Expression values of each gene are normalized against the expression of three reference genes, POLYUBIQUITIN, ACTIN, and HISTONE2A.
To further examine gene expression in our acrocona plants, samples from apical buds collected from the same shoots as the needle samples presented in Figure 4 were also assayed (Supplemental Fig. S5). In the bud sample experiments, we included a third plant category: B, inbred cone-setting acrocona plants initiating fewer than four cones. Expression of DAL19 was detected in buds from acrocona plants that initiated more than four cones (category A), whereas no expression could be detected in buds from acrocona plants that initiated fewer than four cones (category B) or no cones (category C), indicating that DAL19 was specifically up-regulated in putative cone-setting shoots. Interestingly, Norway spruce genes paralogous to DAL19, such as the gene corresponding to transcript Acr4141_1 and DAL3, did not show any differential expression between cone-setting and non-cone-setting acrocona plants in any of our qRT-PCR experiments (Fig. 4; Supplemental Fig. S5).
The MADS box genes DAL10 and DAL1, which both have suggested functions in different aspects of reproductive phase change, were analyzed in our qRT-PCR experiments (Fig. 4; Supplemental Fig. S5). DAL10 was expressed in needles from both cone-setting and non-cone-setting acrocona plants during the second growth cycle. No expression could be detected in buds from the same growth cycle. However, during the third growth cycle, significantly higher DAL10 levels were detected in plants that already had initiated at least one cone (i.e. in shoots belonging to categories A and B) compared with acrocona plants lacking cones (category C), which expressed DAL10 only at very low levels (Student’s t test, P = 0.0426; Fig. 4; Supplemental Fig. S5). Interestingly, expression of DAL1 was consistently high during the second growth cycle in both needles and buds of one individual plant that initiated as many as 10 cones (individual A1 in Fig. 4; Supplemental Fig. S5).
Other genes that have been implicated in reproductive development and phase change are the angiosperm LFY orthologs NEEDLY (NLY) and PaLFY. Both NLY and PaLFY were expressed at varying levels in all examined samples, and no distinct changes between cone-setting and non-cone-setting plants could be detected for NLY (Fig. 4; Supplemental Fig. S5). Expression levels of PaLFY, however, were significantly higher in buds from the third growth cycle of cone-setting acrocona plants as compared with non-cone-setting plants (Student’s t test, P = 0.007; Fig. 4; Supplemental Fig. S5). The putative reproductive organ identity gene DAL2 or the FT/TFL1-like gene PaFTL2 did not show expression patterns that correlated with the cone-setting phenotype (Supplemental Fig. S4). The expression of candidate genes was also assayed using qRT-PCR in bud and needle samples of control plants (Supplemental Fig. S6). In these experiments, we could not detect any significant differences in expression levels between tissue samples, although individual plants expressed different genes at different levels.
Altogether, the qRT-PCR experiments corroborate the differential expression of DAL19 in early cone-setting acrocona plants before or at the onset of cone initiation and indicate that DAL10 is activated in apical buds of acrocona plants that have made the phase change from vegetative to reproductive development.
DISCUSSION
Theodor Fries and colleagues first recognized the acrocona mutant in the mid-19th century from the aberrant shape and position of cones and vegetative shoots. Fries (1890) described the aberrant structures as monstrous cones. Here, we interpret these phenotypes as more or less covering the transition between vegetative and reproductive shoot identity. Hence, the female cones formed on the parental ramets can be categorized either as complete cones or as partial transition cones, which is in agreement with the structures reported both by Fries (1890) and Joneborg (1945). Both types of cones have the possibility to produce seeds. This is also true for inbred acrocona plants, which produced seeds after they had been planted outdoors and received normal growth cycles. One additional interesting aspect of the adult plants used to produce the segregating sibling population of inbred acrocona plants is that the adult plants initiate cones frequently (i.e. every growth season), even though cone setting in spruce, by and large, is coordinated and occurs only once every 3 to 5 years. One molecular interpretation of the combined phenotypes observed in adult acrocona plants is that one or more of the signals that act as reproductive integrators is ectopically active. By making inbred crosses of two acrocona ramets, we hoped to enhance the cone-inducing properties of the acrocona mutant and produce a population of segregating acrocona plants that could be used for further molecular studies. In addition to the frequent cone setting observed in the adult acrocona ramets, one-fourth of the plants in our inbred crosses made the phase change from vegetative growth to reproductive growth extremely early and initiated cones on apical shoots already during their second growth cycle. Occurrences of additional acrocona phenotypes, such as the intermediate shoot phenotypes that develop in the segregating plants during the fifth growth cycle, suggest that the acrocona trait is semidominant. In contrast, linkage analysis between genetic markers and the early cone-setting phenotype has previously suggested that early cone setting is a dominant monogenic trait (Acheré et al., 2004), while early cone setting in inbred acrocona mutants has been reported to occur with a much lower frequency (3%) than reported here (Flachowsky et al., 2009). This indicates that the accelerated growth conditions are of importance for the penetrance of the early cone-setting phenotype or that different variants of the acrocona mutant have been collected over the past 150 years.
The finding of an early cone-setting mutant in the gymnosperm Norway spruce is interesting in two respects. First, it may be developed into a technical tool for studying gene function in a conifer. Second, the mutant phenotype itself provides an opportunity to study vegetative-to-reproductive phase change in a conifer system. Functional studies of individual genes in conifers largely rely on cell-based systems such as somatic cell lines or comparative studies using the heterologous angiosperm model system (Sundström and Engström, 2002; Sundström et al., 2009). Adult characteristics are difficult and time consuming to study, since most conifers enter the reproductive phase after 15 to 25 years of juvenile growth. The short generation time of inbred acrocona plants makes this an interesting system that, in combination with embryogenic cell lines (to allow for mass propagation of acrocona clones) and efficient transformation protocols, can be developed into a rapid-cycling conifer model system. One key aspect of this is to characterize the genetic mechanism underlying the early cone-setting phenotype in the acrocona mutant. To accomplish this, we used large-scale sequencing methods and bioinformatics analyses to compile and present the transcriptome of cone-setting and non-cone-setting acrocona plants as well as of control plants of Norway spruce.
We performed massively parallel sequencing (RNA-seq) on needle samples from inbred acrocona plants and control plants and obtained an estimated coverage of the exonic regions of, on average, 100× for different samples. We used state-of-the-art transcript reconstruction methods, trying both ab initio and de novo approaches. With our final transcript reconstruction process, we found support through orthology in P. glauca and/or Arabidopsis for 35% of our acrocona transcripts. This is rather low; including more species in the orthology analysis would probably increase this percentage, as would relaxing the requirements for orthology. Also, many detected acrocona ORFs are probably fragments, as the length distribution of the ORFs with no orthologs has a large peak in the lower end of the length range (Supplemental Fig. S3C, “No orthologs”).
Almost all groups of orthologous proteins containing P. glauca proteins also contained acrocona proteins (de novo reconstructed set), which on the transcript level translates to a detection rate of 52% of the to-date most comprehensive published P. glauca transcriptome (Rigault et al., 2011). The ab initio approach to transcript reconstruction provided, at 83%, a greater coverage of the P. glauca transcripts, further supporting our suggestion that the ortholog detection process is rather conservative. Given that we only have a single tissue type (needles from apical and basal shoots) and a limited number of conditions or developmental stages, this is a reasonable fraction, indicating that we have reached a sufficient sequencing depth and that our sequence data analysis is reasonable, albeit not perfect. Also, of the 7,501 orthologous groups shared between Arabidopsis and P. glauca, 95% are present in acrocona, indicating that most of the well-conserved proteins are present in the P. glauca transcriptome. Turning the argument around, out of the 8,296 orthologous groups shared between acrocona and Arabidopsis, 86% are also found in P. glauca, indicating that our de novo-assembled transcriptome is at least as representative of the total spruce transcriptome as the published P. glauca transcriptome. Altogether, since we are interested in the mRNAs expressed in needles just before acrocona sets cones, and since the quality of the de novo-assembled transcriptome is on par with the P. glauca transcriptome, we decided to use our de novo-assembled transcriptome as a reference for the differential expression analysis, thus minimizing the risk of completely missing any tissue- or development-specific mRNAs.
We were interested in identifying transcripts that were significantly differentially expressed in apical cone-setting shoots compared with non-cone-setting shoots in acrocona plants and that at the same time were not differentially expressed (apical versus basal shoots) in control plants. As a result, we could drastically reduce the number of candidate genes associated with early cone setting from 33,383 down to eight, and among these, the MADS box gene DAL19 stood out as the most interesting candidate. Our unbiased approach (RNA-seq coupled with de novo transcript assembly) enabled us to suggest DAL19 as a candidate gene, which, since it was not previously published or deposited in any database, would have been practically impossible using a directed search (e.g. microarrays or any PCR-based method) or our ab initio-assembled transcripts. The process of extracting candidate genes through differential expression was rather conservative, and by easing the requirements, we would have ended up with a larger number of candidates. One possible candidate that could have been annotated as being up-regulated in cone-setting shoots is the MADS box gene DAL1, which shows a similar expression pattern to that of DAL19 in our transcriptome analysis, albeit with a higher variance between sample pairs, precluding it from being detected as statistically significant. This notion is supported by the qRT-PCR experiments that revealed high expression of DAL1 in buds and needle samples of one individual plant that initiated high numbers (more than 10) of cones during the second growth cycle.
In our phylogenetic analysis, DAL19 together with three other Norway spruce paralogs form a sister clade to the angiosperm SOC1/AGL42 clade. Several of the angiosperm genes in this clade have experimentally been shown to act as floral integrators during the transition from vegetative to reproductive phase (Dorca-Fornell et al., 2011), although other genes (e.g. AGL14 and AGL19) also have functions during root development (Gan et al., 2005). This is in agreement with the prevailing notion that the MADS box gene family has evolved through a series of gene duplications followed by subfunctionalization or neofunctionalization and that, although phylogenetic position may be indicative of a common ancestral function, orthologs can have acquired distinct and different activities (Irish and Litt, 2005). Here, we present evidence to suggest that the gymnosperm lineage orthologous to the SOC1/AGL42 clade has gone through several independent gene duplications. Furthermore, based on the expression profiles in our unbiased gene expression analysis (RNA-seq) and qRT-PCR experiments, we show that DAL19 is the only spruce paralog in the clade that shows a clear up-regulation in cone-setting shoots as compared with non-cone-setting shoots.
Recent experimental evidence suggests that SOC1 is a floral integrator that regulates the activity of both floral meristem identity genes as well as floral organ identity genes (Immink et al., 2012). In our qRT-PCR experiments, we assayed the expression of previously described Norway spruce orthologs to the floral meristem identity gene LFY: PaLFY and NLY. Both PaLFY and NLY are expressed in all tissues examined; however, the level of PaLFY expression increased slightly but significantly in buds of cone-setting acrocona plants during the third growth cycle. Known Norway spruce orthologs to the angiosperm floral organ identity genes did not show any differential expression in our experiments. Nor did we, in our unbiased approach, identify any differential expression of putative Norway spruce orthologs to microRNA156-targeted SQUAMOSA PROMOTOR BINDING-LIKE transcription factors, which recently have been shown to regulate SOC1 activity and vegetative-to-reproductive phase change in angiosperms (Huijser and Schmid, 2011). As previously noted, gymnosperms also lack an apparent ortholog to the angiosperm FT gene (Karlgren et al., 2011), and further studies of gymnosperm-specific genes are crucial for the understanding of reproductive phase change in conifer trees. Interestingly, we observed differential expression of the DAL10 gene, which belongs to a gymnosperm-specific clade of the MADS box gene family. DAL10 has been suggested to be a marker for reproductive shoot identity (Carlsbecker et al., 2003) and is expressed in buds after, but not before, the phase change from vegetative to reproductive growth. Hence, the data presented here suggest that DAL10 is activated as a result of reproductive competence, whereas up-regulation of DAL19 coincides with or precedes reproductive phase change. It is tempting to speculate that the ancestral function of the SOC1/AGL42 clade was to determine the length of the juvenile growth period and to regulate the phase change from vegetative to reproductive growth. Here, we present evidence to suggest that DAL19 may be involved in a similar process in gymnosperms, which would imply that at least part of this gene function was present already in the last common ancestor of angiosperms and gymnosperms.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Two parental ramets, derived from shoots collected from a homeotic mutant of Norway spruce (Picea abies var acrocona) that have been grafted onto rootstocks, were used in this study. Grafted plants were planted in the Genetic Garden in Uppsala (latitude 59°51′0″ N) and in the park of Alnarp (latitude 55°42′0″ N) around 1975. The genetic uniformity between one plant in the Genetic Garden and one plant in the Alnarp park was confirmed by microsatellite analysis, which revealed no discrepancy between studied loci (n = 7; data not shown). To establish a segregating sibling population of inbred acrocona plants, controlled crossings between the two selected ramets of the acrocona mutant were performed. Pollen collected from the ramet growing in the Alnarp park was used as pollen donor. Controlled crosses were made by isolation of female cones from surrounding pollen as they started to elongate and mature during early spring. The acrocona pollen was subsequently sprayed onto the female cones during the pollination phase. Open-pollinated progeny were derived from open-pollinated seeds from the acrocona plant in the Genetic Garden and in the Alnarp park. Seeds were collected from mature cones late in autumn. Seeds collected from open-pollinated trees in the seed orchard FP-65 Rörby (latitude 59°54′ N), which harbor 27 unrelated clones with an average latitudinal origin of 59°24′ N, were used for regenerating control plants.
Seeds were germinated for 1 month and then potted in stone wool (Grodiana) in the phytotron. The potted plants were initially grown under continuous light (240 µmol m−2 s−1) at 20°C and 75% relative humidity for 8 weeks. This was followed by a 16-h night treatment and with gradually lowered temperature essentially as described (Ununger et al., 1988; i.e. 1 week at 10°C, 1 week at 5°C, 1 week at 2°C, followed by 1 week at 10°C). During the subsequent growth cycles, the plants followed the following growth regime: 4 weeks with 8/16-h day/night conditions at 20°C/15°C; 7 weeks with progressive increase of the night period by 1 h per week at 20°C/15°C; 2 weeks with 8/16-h day/night conditions at 20°C/15°C; followed by a 16-h night treatment and gradually lowered temperature as described above. Under these accelerated growth conditions, the plants were cultured during four to five growth cycles.
All plant material for molecular analyses was collected when the shoots had reached approximately 90% of their final length, which corresponds to the time point when bud identity is determined in Norway spruce (Lindgren et al., 1977; Högberg and Eriksson, 1994). At the end of the growth cycle, it is only possible to determine the reproductive or vegetative identity of a shoot by detrimental microdissection of the developing bud. Therefore, the collected plant material was snap frozen in liquid nitrogen and stored at −70°C until the cone-setting status of the plant/shoot could be determined in the following growth cycle.
Phenological data (e.g. height, bud set, and branching pattern) were recorded each week under the growing periods during the first three growth cycles from 75 inbred acrocona plants (derived from crossings made in 2006), 150 open-pollinated acrocona progeny, and 75 control plants. During each growth cycle, one lateral shoot, directly subtending the apical shoot, was sampled from each plant. Isolated needles and buds from these shoots were used in subsequent qRT-PCR analyses.
Four inbred acrocona plants (two cone-setting plants and two non-cone-setting plants, derived from crossings made in 2009) together with two control plants were used in the transcriptome analysis. During the second and third growth cycles, needles (five to seven) directly subtending the forming bud on the top apical shoots (i.e. potentially cone-setting shoots) and from vegetative lateral shoots were sampled. Ten acrocona samples were sequenced: two samples from apical cone-setting shoots, three samples from apical non-cone-setting acrocona shoots, and five samples from basal acrocona shoots. Four samples were sequenced from control plants: two samples from apical non-cone-setting shoots and two samples from basal shots. Thus, in total, 14 samples were assayed in the transcriptome sequencing.
RNA Isolation
Total RNA for qRT-PCR analyses was isolated using the Spectrum Plant Total RNA kit (Sigma-Aldrich). Total RNA was subjected to on-column DNase digestion according to the manufacturer’s instructions (Sigma-Aldrich). The integrity of total RNA was assessed by Bioanalyzer (Agilent). Complementary DNA was prepared from 0.5 to 1 μg of total RNA using the Maxima First Strand Synthesis Kit for RT-qPCR (Thermo Scientific).
Total RNA for massively parallel sequencing was isolated using a slightly modified protocol described by Azevedo et al. (2003). RNA was subjected to DNase digestion according to the manufacturer’s instructions (DNA-free; Life Technologies).
Library Generation and Massively Parallel Sequencing
RNA libraries for sequencing were prepared using TruSeq RNA kits (Illumina) according to the manufacturer’s instructions with the following changes: the protocols were automated using an MBS 1200 pipetting station (Nordiag) as described previously (Stranneheim et al., 2011), and all purification steps and gel cuts were replaced by magnetic bead cleanup methods (Borgström et al., 2011).
The clustering was performed on a cBot cluster generation system using an Illumina HiSeq paired-end read cluster generation kit according to the manufacturer’s instructions (Illumina). The 14 samples were sequenced on an Illumina HiSEquation 2000 as paired-end reads to 100 bp, using all eight lanes of a flow cell. Each sample was divided into four equal aliquots and sequenced in four different lanes, thus with seven samples in each lane using multiplexing tags to separate samples logically. All lanes were spiked with a 1% to 2% phiX control library. One lane failed (low cluster density) and was resequenced 31 d later. The sequencing runs were performed according to the manufacturer’s instructions. Base conversion was done using Illumina’s OLB version 1.9.
De Novo Transcript Assembly and ORF Identification
The transcript reconstruction process has been described in detail (J. Reimegård and O. Emanuelsson, unpublished data). Briefly, sequence reads from Illumina HiSeq2000 were filtered with cutAdapt (Marcel, 2011) and an in-house script to remove whole reads or parts of reads with low quality. Only pairs where both reads passed the quality test were further analyzed. Trinity (Grabherr et al., 2011; build 11.05.27) was used to build the de novo-assembled transcripts. Potential ORFs were extracted from the transcripts with getOrf from the EMBOSS package (Rice et al., 2000). An ORF was accepted if it contained a start codon, an ORF longer than 150 nucleotides, and a stop codon. Multiple ORFs per transcript were allowed provided that they did not overlap on the transcript. ORFs on one transcript were selected in a hierarchal order based on their length, where the longest was selected first. If multiple ORFs from different transcripts had identical sequences, only one was kept. Each ORF was translated into peptide sequence using Transeq from the EMBOSS package.
Generating Groups of Orthologous Proteins
Groups of orthologous proteins from acrocona, Picea glauca, and Arabidopsis (Arabidopsis thaliana) were generated by OrthoMCL version 1.4 (Li et al., 2003). The Arabidopsis proteins (The Arabidopsis Information Resource 10) were downloaded from phytozome.net (Lamesch et al., 2012). P. glauca proteins were generated by subjecting its transcripts (Rigault et al., 2011) to the same ORF detection protocol as described for acrocona (above) and then translated into protein sequences.
Functional Annotation of acrocona Peptide Sequences
The identified acrocona peptide sequences were analyzed with Panther 7.2 (Mi et al., 2010). Genes that were identified as belonging to a Panther class were annotated with Gene Ontology terms according to Panther 7.2 Gene Ontology annotation. Genes with no Panther class were annotated based on orthologs in Arabidopsis and P. glauca (see above). If an acrocona protein belonged to an ortholog group that contained multiple Arabidopsis proteins, then the acrocona protein was assigned the functional annotation of the Arabidopsis protein with the highest BLAST (version 2.2.24) score.
Differential Gene Expression Analysis
The quality-filtered reads were mapped onto the ORFs generated from the transcriptome (see above) with Bowtie2 (Langmead and Salzberg, 2012), allowing only for concordant mapped pairs (–no-mixed–no-discordant). Cufflinks together with Cuffmerge (Trapnell et al., 2012) were used to identify the full-length transcripts, and Cuffdiff was used to identify differently expressed genes. Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to create the Venn diagrams, while expression patterns for genes were generated using the cummeRbund package for R (Trapnell et al., 2012).
Phylogenetic Analysis
Annotated MADS box genes with high similarity to DAL19 from selected angiosperms and gymnosperms were retrieved from GenBank using BLAST searches (Altschul et al., 1997; for accession numbers, see Supplemental Table S2). Selected sequences belonging to the closely related gymnosperm AGL6 clade were also included in the analysis to serve as outgroup sequences. The obtained sequences were translationally aligned using the MAFFT module within Geneious (Geneious Pro version 5.6 created by Biomatters; http://www.geneious.com). The data set was then reduced such that partial sequences and putative close orthologs and paralogs were omitted. The resulting data set comprised 37 taxa and included 666 characters, excluding the highly variable C-termini-encoding sequences.
Phylogenetic analysis of the resulting nucleotide alignment was carried out using MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003). The selected model of evolution was GTR + I + G, which assumes a general time reversibility (GTR), a certain proportion of invariable sites (I), and a gamma approximation (G) of the rate variation among sites. Four chains of the Markov Chain Monte Carlo were run in parallel, sampling one tree every 500 generations for 2.5 million generations starting with a random tree. The search reached stationarity after approximately 250,000 generations. The first 250,000 generations (the burn in) were omitted in generating the consensus phylogeny.
qRT-PCR
qRT-PCR was performed using the iQ5 Real-Time Detection System on iCycler iQ 96-well PCR plates with adhesive seals (Bio-Rad Laboratories). Primers used to quantify expression levels are presented in Supplemental Table S3. The expression data of each gene were normalized against the expression of three reference genes, ACTIN, POLYUBIQUITIN, and HISTONE2A (Supplemental Table S3). Amplifications were carried out using the DyNAmo Flash Sybr Green qPCR kit (Thermo Scientific). PCR cycling conditions were as advised by the manufacturer, with annealing and extension at 60°C for 30 s. The reactions were run for 40 cycles, and at the end of each run, melt curves were generated to ensure product uniformity. Samples were added to the plates in triplicate. In all studies, interrun connector samples were included to correct for the use of multiple plates. All calculations and normalizations were done using the iQ5 software based on the “Pfaffl methods” (Bio-Rad).
Sequence data from this article can be found in the Sequence Read Archive data libraries under accession number SRA064221.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of inbred acrocona plants grown under accelerated growth conditions.
Supplemental Figure S2. Seasonal growth responses of acrocona plants and control plants.
Supplemental Figure S3. Transcriptome sequence analysis.
Supplemental Figure S4. Expression pattern of protein-coding transcripts.
Supplemental Figure S5. qRT-PCR expression data of candidate genes from bud samples in cone-setting and non-cone-setting inbred acrocona plants.
Supplemental Figure S6. qRT-PCR expression data of candidate genes from needle and bud samples in non-cone-setting wild-type plants.
Supplemental Table S1. Phenotype frequencies of inbred acrocona plants.
Supplemental Table S2. Gene names and accession numbers used in the phylogeny.
Supplemental Table S3. Primer sequences used for qRT-PCR analyses.
Supplemental Data Set S1. Expression levels of all identified ORFs.
Supplemental Data Set S2. Expression levels and annotation of the differentially expressed genes.
Acknowledgments
We thank the Science for Life Laboratory (Stockholm), Swedish National Infrastructure for Large-Scale Sequencing, and Uppmax for providing assistance in massively parallel sequencing and computational infrastructure. The Troëdsson Research Foundation is acknowledged for providing support for cultivation facilities.
Glossary
- qRT
quantitative real-time
- ORF
open reading frame
References
- Acheré V, Faivre-Rampant P, Jeandroz S, Besnard G, Markussen T, Aragones A, Fladung M, Ritter E, Favre JM. (2004) A full saturated linkage map of Picea abies including AFLP, SSR, ESTP, 5S rDNA and morphological markers. Theor Appl Genet 108: 1602–1613 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo H, Lino-Neto T, Tavares RM. (2003) An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Mol Biol Rep 21: 333–338 [Google Scholar]
- Bäurle I, Dean C. (2006) The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Borgström E, Lundin S, Lundeberg J. (2011) Large scale library generation for high throughput sequencing. PLoS ONE 6: e19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Sundström J, Tandre K, Englund M, Kvarnheden A, Johanson U, Engström P. (2003) The DAL10 gene from Norway spruce (Picea abies) belongs to a potentially gymnosperm-specific subclass of MADS-box genes and is specifically active in seed cones and pollen cones. Evol Dev 5: 551–561 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Tandre K, Johanson U, Englund M, Engström P. (2004) The MADS-box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies). Plant J 40: 546–557 [DOI] [PubMed] [Google Scholar]
- Dorca-Fornell C, Gregis V, Grandi V, Coupland G, Colombo L, Kater MM. (2011) The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J 67: 1006–1017 [DOI] [PubMed] [Google Scholar]
- Endo T, Shimada T, Fujii H, Kobayashi Y, Araki T, Omura M. (2005) Ectopic expression of an FT homolog from citrus confers an early flowering phenotype on trifoliate orange (Poncirus trifoliata L. Raf.). Transgenic Res 14: 703–712 [DOI] [PubMed] [Google Scholar]
- Flachowsky H, Hanke MV, Peil A, Strauss SH, Fladung M. (2009) A review on transgenic approaches to accelerate breeding of woody plants. Plant Breed 128: 217–226 [Google Scholar]
- Fries TM. (1890) Strödda bidrag till kännedom om Skandinaviens barrträd. Bot Not 1: 250–260 [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG. (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222: 730–742 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högberg K-A, Eriksson U. (1994) Effects of root pruning and stem injections with gibberellin A 4/7 on flowering and cone harvest in three Picea abies seed orchards. Scand J For Res 9: 323–328 [Google Scholar]
- Huijser P, Schmid M. (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Immink R, Pose D, Ferrario S, Ott F, Kaufmann K, Leal Valentim F, De Folter S, Van der Wal F, van Dijk ADJ, Schmid M, et al. (2012) Characterisation of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Litt A. (2005) Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev 15: 454–460 [DOI] [PubMed] [Google Scholar]
- Joneborg S. (1945) Monstrous cone formation in spruce: vegetative shoot or cone? Sven Skogsvardsforen 43: 453–462 [Google Scholar]
- Karlgren A, Gyllenstrand N, Källman T, Sundström JF, Moore D, Lascoux M, Lagercrantz U. (2011) Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol 156: 1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I. (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren K, Ekberg I, Eriksson G. (1977) External factors influencing female flowering in Picea abies (L.) Karst. Studia Forestalia Suecia 142: 1–53 [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. (1991) AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev 5: 484–495 [DOI] [PubMed] [Google Scholar]
- Marcel M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12 [Google Scholar]
- Mi HY, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. (2010) PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res 38: D204–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rigault P, Boyle B, Lepage P, Cooke JEK, Bousquet J, MacKay JJ. (2011) A white spruce gene catalog for conifer genome analyses. Plant Physiol 157: 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Smith SA, Beaulieu JM, Donoghue MJ. (2010) An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proc Natl Acad Sci USA 107: 5897–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranneheim H, Werne B, Sherwood E, Lundeberg J. (2011) Scalable transcriptome preparation for massive parallel sequencing. PLoS ONE 6: e21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström J, Engström P. (2002) Conifer reproductive development involves B-type MADS-box genes with distinct and different activities in male organ primordia. Plant J 31: 161–169 [DOI] [PubMed] [Google Scholar]
- Sundström JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, Tiwari BS, Rodriguez-Nieto S, Zamyatnin AA, Jr, Välineva T, et al. (2009) Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol 11: 1347–1354 [DOI] [PubMed] [Google Scholar]
- Tandre K, Albert VA, Sundås A, Engström P. (1995) Conifer homologues to genes that control floral development in angiosperms. Plant Mol Biol 27: 69–78 [DOI] [PubMed] [Google Scholar]
- Tollefsrud MM, Kissling R, Gugerli F, Johnsen O, Skrøppa T, Cheddadi R, Van der Knaap WO, Latałowa M, Terhürne-Berson R, Litt T, et al. (2008) Genetic consequences of glacial survival and postglacial colonization in Norway spruce: combined analysis of mitochondrial DNA and fossil pollen. Mol Ecol 17: 4134–4150 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ununger J, Ekberg I, Kang H. (1988) Genetic control and age-related changes of juvenile growth characters in Picea abies. Scand J For Res 3: 55–66 [Google Scholar]
- Wang Z, Gerstein M, Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]