Summary: This research describes how a mobile genetic element recruits the TaMATE1B gene into a role associated with aluminum tolerance by altering its expression level and pattern.
Abstract
The TaMATE1B gene (for multidrug and toxic compound extrusion) from wheat (Triticum aestivum) was isolated and shown to encode a citrate transporter that is located on the plasma membrane. TaMATE1B expression in roots was induced by iron deficiency but not by phosphorus deficiency or aluminum treatment. The coding region of TaMATE1B was identical in a genotype showing citrate efflux from root apices (cv Carazinho) to one that lacked citrate efflux (cv Egret). However, sequence upstream of the coding region differed between these two genotypes in two ways. The first difference was a single-nucleotide polymorphism located approximately 2 kb upstream from the start codon in cv Egret. The second difference was an 11.1-kb transposon-like element located 25 bp upstream of the start codon in cv Carazinho that was absent from cv Egret. The influence of these polymorphisms on TaMATE1B expression was investigated using fusions to green fluorescent protein expressed in transgenic lines of rice (Oryza sativa). Fluorescence measurements in roots of rice indicated that 1.5- and 2.3-kb regions upstream of TaMATE1B in cv Carazinho (which incorporated 3′ regions of the transposon-like element) generated 20-fold greater expression in the apical 1 mm of root compared with the native promoter in cv Egret. By contrast, fluorescence in more mature tissues was similar in both cultivars. The presence of the single-nucleotide polymorphism alone consistently generated 2-fold greater fluorescence than the cv Egret promoter. We conclude that the transposon-like element in cv Carazinho extends TaMATE1B expression to the root apex, where it confers citrate efflux and enhanced aluminum tolerance.
Intraspecific variation in aluminum (Al3+) tolerance is evident in many crop species and has been shown to be under either simple or complex genetic control (Ma et al., 2004; Magalhaes et al., 2007; Krill et al., 2010; Famoso et al., 2011). In wheat (Triticum aestivum), Al3+ tolerance is primarily associated with efflux of organic anions from root tips, with at least two independent mechanisms being involved. The first and most widely distributed among genotypes is the Al3+-activated efflux of malate from root apices (Delhaize et al., 1993, Ryan et al., 1995). More recently, a second and less prevalent mechanism has been identified that relies on the constitutive efflux of citrate from root apices (Ryan et al., 2009).
Organic anions protect roots by chelating and detoxifying Al3+ in the apoplast and rhizosphere around sensitive root apices (Delhaize et al., 1993, 2012; Ryan et al., 2001). For example, Al3+-tolerant genotypes of wheat release significantly more malate than sensitive genotypes (Ryan et al., 1995). The efflux of malate from root apices is mediated by the TaALMT1 (for Al3+-activated malate transporter) gene, which resides on chromosome 4DL (Raman et al., 2005, 2008; Zhou et al., 2007). TaALMT1 encodes an Al3+-activated anion channel permeable to malate located on the plasma membrane of root cells (Sasaki et al., 2004; Yamaguchi et al., 2005; Piñeros et al., 2008; Zhang et al., 2008). Similarly, ALMT1 genes contribute to Al3+ tolerance in Arabidopsis (Arabidopsis thaliana), oilseed rape (Brassica napus), and rye (Secale cereale; Ryan et al., 2011). While all members of the ALMT family characterized to date encode transport proteins, only a minority are involved in Al3+ tolerance. The others contribute to physiological processes related to ionic relations and osmotic adjustment (Kovermann et al., 2007; Gruber et al., 2010; Meyer et al., 2010, 2011; Sasaki et al., 2010).
In many plant species, citrate efflux from root apices also confers Al3+ tolerance. The genes controlling citrate efflux were first identified in sorghum (Sorghum bicolor; SbMATE [for multidrug and toxic compound extrusion]; Magalhaes et al., 2007) and barley (Hordeum vulgare; HvAACT1; Furukawa et al., 2007), with additional genes subsequently identified in Arabidopsis (Liu et al., 2009), maize (Zea mays; Maron et al., 2010), rice (Yokosho et al., 2009), and rice bean (Vigna umbellate; Yang et al., 2011). These genes belong to a large and ubiquitous family of MATE genes that were first identified in prokaryotes (Hvorup et al., 2003). SbMATE and HvAACT1 are part of a subset of plant MATE genes that facilitate citrate efflux (Liu et al., 2009; Magalhaes, 2010) with roles in Al3+ tolerance and iron (Fe) nutrition. For example, FRD3 from Arabidopsis and OsFRDL1 from rice both encode MATE proteins located in vascular tissue, where they release citrate into the xylem to enable Fe transport to shoots (Durrett et al., 2007). Recent evidence suggests that the Al3+ tolerance gene in barley, HvAACT1, was coopted from an original function in Fe nutrition by a random mutation. Al3+-tolerant genotypes of barley have a 1-kb insertion in the 5′ untranslated region (UTR) of the HvAACT1 coding region that alters its expression pattern. The insertion extends HvAACT1 expression to the root apices, which, in the presence of Al3+, causes citrate efflux from the apices and enhanced Al3+ tolerance (Fujii et al., 2012).
Evidence that citrate efflux confers Al3+ tolerance in some genotypes of wheat was provided by Ryan et al. (2009). The trait was mapped to the long arm of chromosome 4B and cosegregated with an EST showing 94% sequence identity to HvAACT1. This EST was expressed only in the root apices of wheat lines that showed citrate efflux (e.g. cv Carazinho; Ryan et al., 2009), suggesting that a MATE gene encodes citrate efflux from root apices. Here, we describe the isolation and characterization of a MATE gene from wheat named TaMATE1B that encodes a citrate transporter located on the plasma membrane. A large transposable element-like sequence was found to be inserted near the start of the TaMATE1B coding region specifically in genotypes that release citrate. We demonstrate that the fragment functions as a promoter that extends TaMATE1B expression to root apices and discuss how this mutation recruited a gene to confer a new phenotype associated with Al3+ tolerance.
RESULTS
Isolation of TaMATE1B and Gene Structure
We isolated a wheat complementary DNA (cDNA) encoding a MATE protein with strong sequence homology to the citrate transporter protein HvAACT1 from barley. This was achieved by designing primers (Supplemental Table S1) that would amplify the predicted coding region of a contig assembled from multiple wheat ESTs. Although the ESTs were derived from different but closely related MATE genes, these primers amplified a single product from cDNA derived from root apices of cv Carazinho, whereas no product was obtained from root apices of cv Egret. The sequence of this product, named TaMATE1B, included the EST previously shown to be associated with citrate efflux on chromosome 4BL. The TaMATE1B product was used to design additional primers to amplify and sequence genomic DNA. Three related genes (TaMATE1B and two others) were identified with similar exon and intron structures and almost identical coding sequences (Supplemental Fig. S1). The genomic sequence of TaMATE1B spans 3,933 bp with 12 introns ranging from 84 to 1,029 bp (Fig. 1). The location of TaMATE1B on chromosome 4B was confirmed with PCR analysis by the presence of a product in a wheat line nullisomic for 4D (tetrasomic for 4A) and by the absence of a product in a durum wheat (Triticum turgidum) line where the 4B chromosome is substituted by the 4D chromosome (data not shown). Another homeolog was located to chromosome 4D (TaMATE1D), and the remaining homeolog was presumed to be located on chromosome 4A (TaMATE1A). The sequences of the TaMATE1B exons and introns were identical in cv Carazinho and Egret except for two additional Cs within a C-rich region (eight to 10 nucleotides) in the 11th intron of the cv Carazinho allele.
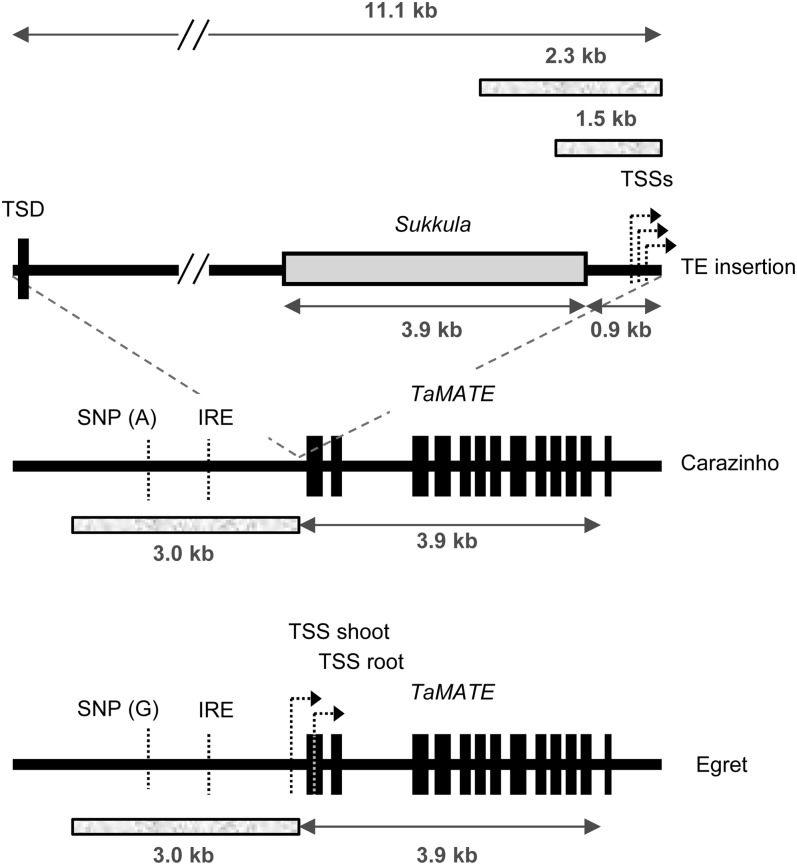
Figure 1.
TaMATE gene structure. The structures of the TaMATE1B gene of wheat ‘Carazinho’ and ‘Egret’ are shown highlighting differences between the two alleles. Exons are shown as 13 black boxes separated by 12 introns. The cv Carazinho allele differs from the cv Egret allele primarily by the presence of an 11.1-kb insert (TE) 25 bp upstream of the ATG start codon. The 3′ region of this insert possesses sequence homology (approximately 3.9 kb; gray box) to a Sukkula retrotransposon, with the remaining fragment similar to uncharacterized repeated regions within the wheat genome. The TE altered the TSSs from single positions for shoot and root in cv Egret to multiple TSSs in cv Carazinho for both roots and shoots. The TE also generated a TSD of 113 bp. The TaMATE1B alleles also differed by a SNP located approximately 2 kb upstream of the start codon and by a variable string of cytosines in the last intron (eight in cv Egret and 10 in cv Carazinho). A motif for an Fe-responsive element (IRE) is located in the upstream region. Four fragments of approximately 1.5, 2.3, and 3.0 kb (shaded boxes) were used to prepare promoter constructs driving a GFP reporter gene.
TaMATE1 sequences were also obtained from two bacterial artificial chromosome (BAC) libraries prepared from cv Renan and Chinese Spring. Three related TaMATE1 genes were identified on different BACs, which is consistent with the existence of three homeologs. Regions upstream and downstream of the TaMATE1B coding region were sequenced from a representative BAC clone generated from cv Renan and Chinese Spring. Sequence obtained from the BAC clones enabled equivalent genomic regions to be amplified and sequenced in cv Carazinho and Egret. The 2-kb region upstream of TaMATE1B in cv Egret was identical to the BAC clones, but initial attempts to amplify the equivalent region from cv Carazinho, the genotype with the citrate efflux phenotype, were unsuccessful. Products were only obtained from cv Carazinho by long-range PCR, which generated a greater than 10-kb product compared with the approximately 1-kb product from cv Egret with the same primer pair. Sequencing of these products showed that cv Carazinho had an 11.1-kb insertion located 25 bp upstream of the predicted ATG start codon (Fig. 1). The insertion (transposon-like element [TE]) was flanked by 113 bp of duplicated sequence (target site duplication [TSD]; Fig. 1), and approximately 4 kb was homologous to the Sukkula retrotransposons, which possess long terminal repeats (Kalendar et al., 2004). The TSD encompassed the 25 bp immediately upstream of the ATG start codon along with 88 bp of the first TaMATE1B exon. The TE present in cv Carazinho, therefore, appears to be a hybrid between a Sukkula retrotransposon and other uncharacterized repetitive elements present within the wheat genome.
The only other difference between the 2.5-kb sequence upstream of TaMATE1B in cv Egret and Carazinho was a single-nucleotide polymorphism (SNP; A/G substitution) located approximately 2 kb upstream of the start codon of cv Egret and approximately 12 kb upstream from the start codon in cv Carazinho (Fig. 1). Both promoter regions contained a putative Fe-responsive element, and no differences were apparent between the cultivars in the 790-bp region downstream of the stop codon. A cleaved-amplified polymorphic sequence (CAPS) marker was developed to target the upstream SNP and served to differentiate cv Carazinho from cv Egret at the TaMATE1B locus. The cv Carazinho allele, as defined by this CAPS marker, segregated with citrate efflux in an F2:3 population derived from an cv Egret × Carazinho cross. This same allele was detected in other Brazilian cultivars with the citrate efflux phenotype (cv Maringa, Toropi, and Trintecinco; Ryan et al., 2009) but not in cultivars and landraces that lacked this phenotype (data not shown; 75 genotypes screened). One European landrace, Seville 18, had the cv Egret allele for the CAPS marker yet showed citrate efflux (see below). Sequencing upstream of TaMATE1B in Seville 18 indicated that it contained the same large TE insertion found in cv Carazinho, which suggests that a recombination event occurred between a genotype that possessed the cv Egret SNP allele and one that possessed the cv Carazinho SNP and TE. Alternatively, the Seville 18 haplotype may have been the progenitor of the cv Carazinho haplotype, where a point mutation has generated the cv Carazinho SNP allele subsequent to the insertion of the TE.
Since the TE insertion was inferred to interrupt the 5′ UTR of TaMATE1B, we compared the transcription start site (TSS) of TaMATE1B in cv Carazinho with the TSS in cv Egret. cv Carazinho TaMATE1B transcripts in the roots and shoots contained several closely clustered TSSs 130 to 160 bp upstream of the ATG start codon (Fig. 1). By contrast, in cv Egret, one TSS was identified in shoots (269 bp upstream of the ATG) and a different TSS was identified in roots (24 bp downstream of the ATG). No differences were detected in the 3′ UTRs of TaMATE1B in the roots of cv Carazinho and Egret, with both varying from 266 to 324 bp after the stop codon.
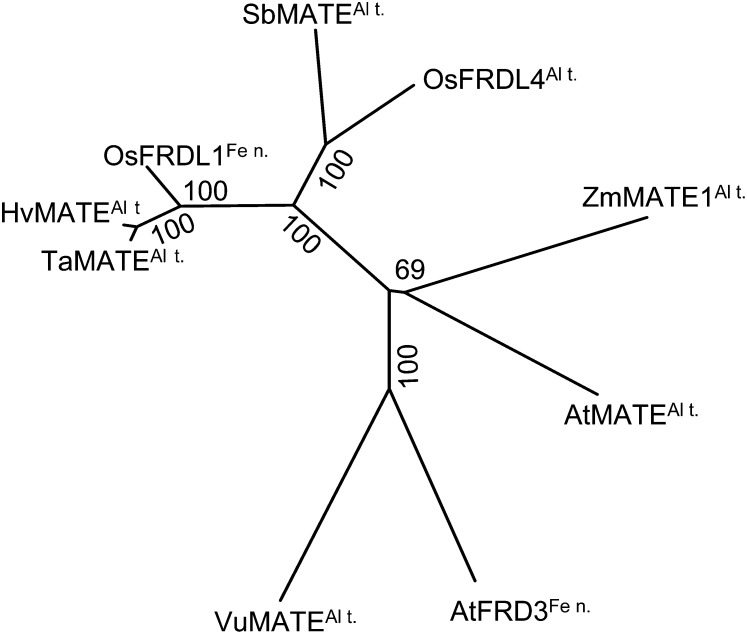
TaMATE1B encodes a 554-amino acid protein with seven to 11 predicted transmembrane domains depending on the algorithm used. The putative protein has 44% to 94% identity with other plant MATE proteins implicated in citrate transport, and phylogenetic analysis identifies HvAACT1 from barley as the homolog most similar to TaMATE1B (Fig. 2).
Figure 2.
Phylogenetic relationships of TaMATE1B and plant MATE proteins previously shown to transport citrate: AtFRD3 (Arabidopsis; Durrett et al., 2007), AtMATE (Arabidopsis; Liu et al., 2009), ZmMATE1 (maize; Maron et al., 2010), SbMATE (sorghum; Magalhaes et al., 2007), OsFRDL1 (rice; Yokosho et al., 2009), HvMATE (barley; Furukawa et al., 2007), and VuMATE (rice bean; Yang et al., 2011). The unrooted neighbor-joining tree was generated with MEGA5 (Tamura et al., 2011). Bootstrap values from 1,000 replicates are indicated at each branch. Proposed functions of the proteins are indicated (Al t., Al3+ tolerance; Fe n., Fe nutrition).
TaMATE1B Is Located on the Plasma Membrane
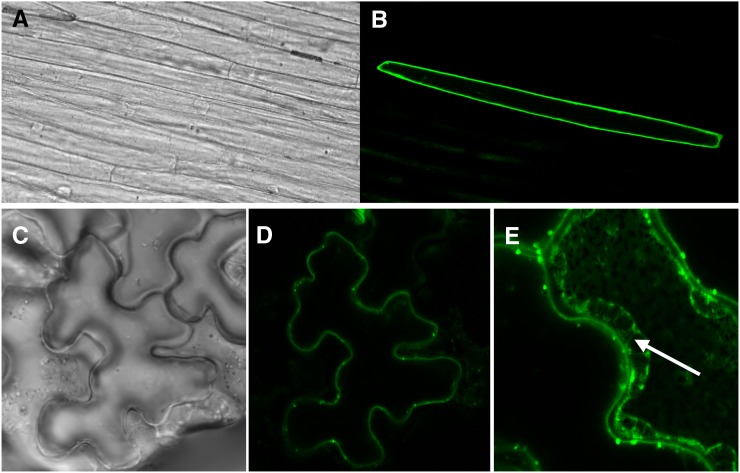
To examine the subcellular localization of the TaMATE1B protein, chimeric constructs were prepared by fusing GFP to the N- and C-terminal ends of the coding region. These constructs were expressed transiently in leek (Allium ampeloprasum) by particle bombardment and in Nicotiana benthamiana leaves using Agrobacterium tumefaciens infiltration. GFP fluorescence in leek with the N-terminal construct (GFP::TaMATE) was only observed at the cell periphery and not around any internal organelles (Fig. 3, A and B), which is consistent with the localization of TaMATE1B to the plasma membrane. The N- and C-terminal constructs (GFP::TaMATE and TaMATE::GFP, respectively) in N. benthamiana both produced similar patterns of fluorescence, with the signal restricted to the cell periphery (Fig. 3, C and D). Consistent with the localization of TaMATE1B to the plasma membrane was the appearance of GFP fluorescence in the Hechtian strands formed when the N. benthamiana leaf cells were plasmolyzed with 100 mm Suc (Fig. 3E). As the cytosol volume decreases, these cytoplasmic strands occur when the retracting plasma membrane remains attached to points on the cell wall (Lang-Pauluzzi and Gunning, 2000).
Figure 3.
TaMATE1B protein is located at the plasma membrane. GFP was fused to the N- and C-terminal ends of the TaMATE1B protein and transiently expressed in leek (A and B) and N. benthamiana (C–E) leaves. A, Bright-field image of leek tissue bombarded with a construct containing an N-terminal fusion of GFP to TaMATE1B. B, Fluorescence image of the same tissue showing GFP fluorescence around the periphery of a single cell. C, Bright-field image of N. benthamiana tissue transformed with a construct containing a C-terminal fusion of GFP to TaMATE1B. D, Fluorescence image of the same tissue showing GFP fluorescence around the periphery of a single cell. E, An N. benthamiana cell expressing the TaMATE:GFP fusion protein after plasmolysis with 100 mm Suc. The Hechtian strands that connect the cell wall with the retreating protoplast (white arrow) are consistent with TaMATE1B being located at the plasma membrane.
TaMATE Expression Generates Citrate Efflux to Transgenic Plants and Xenopus laevis Oocytes
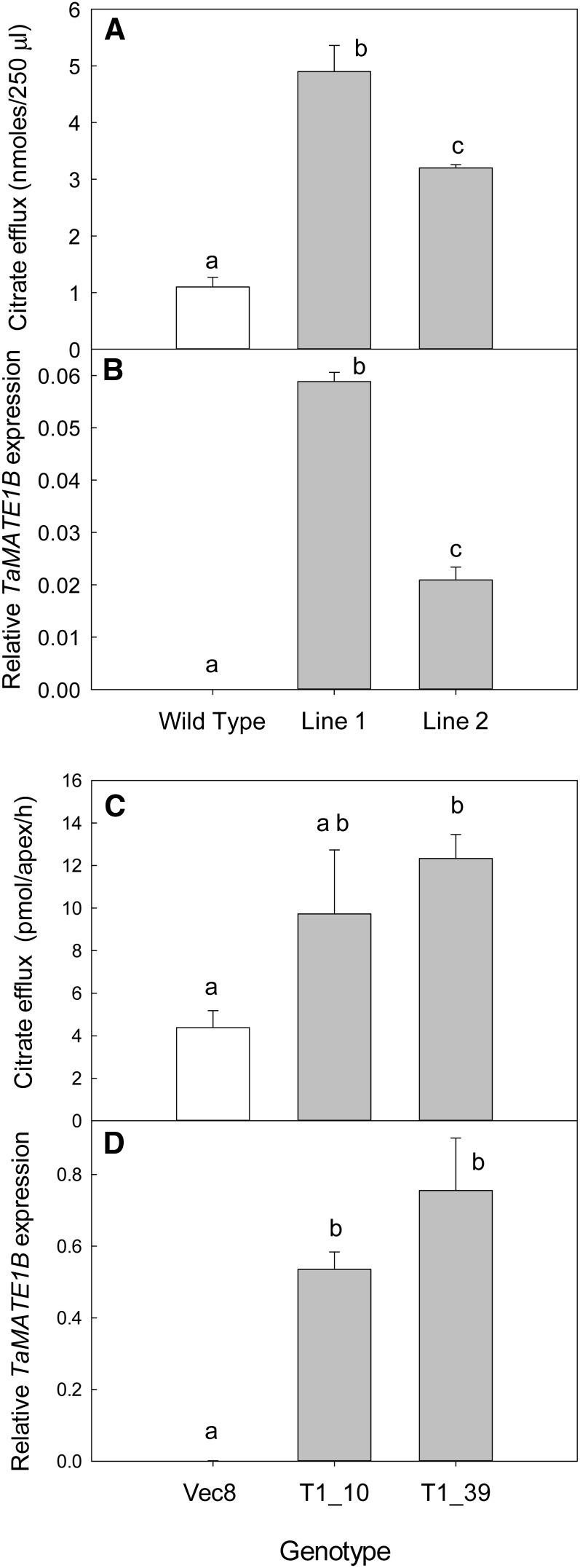
To verify that TaMATE1B encodes a citrate transporter, the gene was constitutively expressed in tobacco (Nicotiana tabacum) and rice. Citrate efflux from whole tobacco seedlings and from the excised root apices of rice was 2- to 4-fold greater from the transgenics than from the controls, which included untransformed plants (tobacco) and plants transformed with an empty plasmid (rice; Fig. 4, A and C). Relative levels of TaMATE1B expression in these transgenic plants, measured by quantitative real-time (qRT)-PCR, matched the relative citrate efflux from these T1 families, and no expression was detected in the controls (Fig. 4, B and D).
Figure 4.
Expression of TaMATE1B in tobacco and rice increases constitutive citrate efflux. TaMATE1B cDNA was constitutively expressed in tobacco and rice plants using A. tumefaciens-mediated transformation. Independent T0 plants were used to generate T1 seed for measurements of citrate efflux and TaMATE1B expression in tobacco (A and B) and rice (C and D). Citrate efflux data (A and C) show means ± se (n = 3 or 5). Efflux from tobacco was measured on three replicate batches of 40 seedlings for each T1 line. Citrate efflux from rice was measured on excised root apices collected from five plants in each T1 family. TaMATE1B expression is expressed relative to the Malate Dehydrogenase gene in tobacco (B) and to GAPDH in rice (D). Expression data are means ± se from three (tobacco) or four (rice) biological replicates. Different letters indicate genotypes that are significantly different from one another (P < 0.05) as identified by one-way ANOVA.
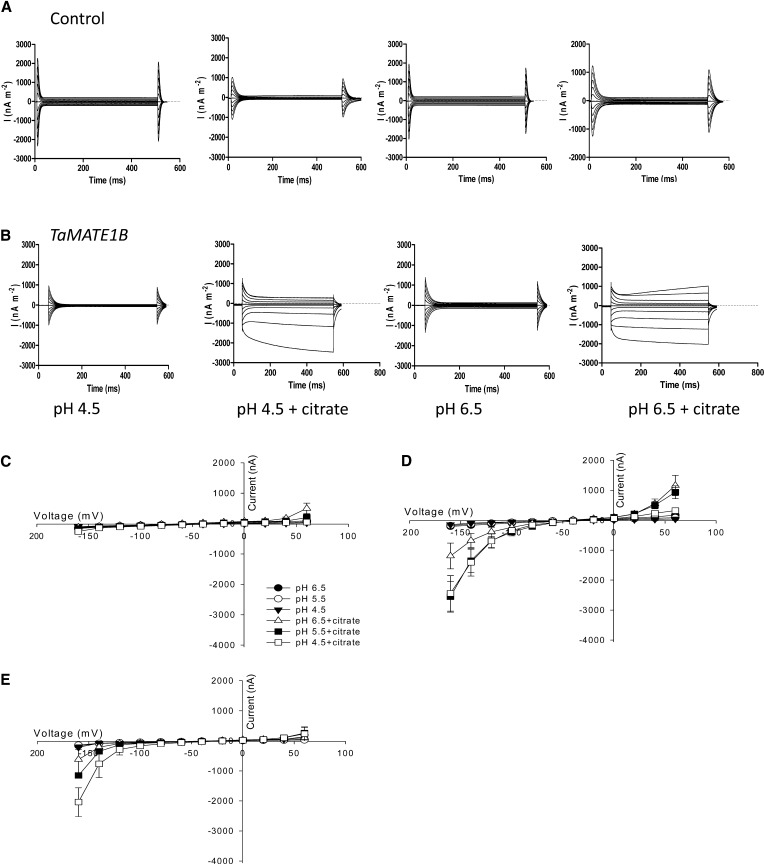
TaMATE1B function was also examined by expressing the copy RNA (cRNA) in X. laevis oocytes and using two-electrode voltage clamping (TEVC) to measure net currents across the membrane. Figure 5 shows representative sets of currents in control oocytes and oocytes injected with TaMATE1B cRNA at voltages between −160 and +60 mV with bathing solutions adjusted to pH 4.5 or 6.5. Control oocytes showed small currents regardless of pH and the presence of citrate in the bathing solution (Fig. 5A). Oocytes expressing TaMATE1B also showed small currents when citrate was absent from the bathing solution. However, when 10 mm citrate was added to the bathing solution, novel inward (negative) and outward (positive) currents were detected in oocytes expressing TaMATE1B that were absent in water-injected controls (Fig. 5B). These inward and outward currents activated rapidly over most of the voltage range but showed a time dependency at the extreme voltages of −160 and +60 mV. Figure 5, C to E, summarizes the current-voltage responses collected from all oocytes. Oocytes expressing TaMATE1B and loaded with citrate (Fig. 5D) showed inward and outward currents not present in the water-injected oocytes (also loaded with citrate; Fig. 5C). Again, these currents were only detected when 10 mm citrate was added to the bathing solution. The magnitude of the inward currents increased as the pH of the bathing solution decreased, whereas the outward currents were greater at pH 6.5 than at pH 4.5. For most voltages, the magnitudes of currents in oocytes injected with TaMATE1B cRNA and loaded with sodium citrate prior to measurements (Fig. 5D) were larger than those from oocytes not loaded with citrate (Fig. 5E). For example, the magnitudes of the average currents at −160 mV for oocytes injected with citrate were −1.2 ± 0.4, −2.5 ± 0.5, and −2.4 ± 0.6 µA at pH 6.5, 5.5, and 4.5 respectively, compared with −0.6 ± 0.5, −1.1 ± 0.0, and −2.0 ± 0.5 µA for oocytes not injected with citrate.
Figure 5.
TaMATE1B expression in X. laevis oocytes generates novel currents. A and B, Representative currents measured in oocytes injected with water (A) or TaMATE1B cRNA (B) with the bathing solution at pH 4.5 or 6.5 with or without 10 mm citrate. Voltages were pulsed to between +60 and −160 mV for 0.5 s. C to E, Current-voltage curves were generated from data collected at the end of the voltage pulses of oocytes injected with water (C) or TaMATE1B cRNA (D and E). Symbols indicate bathing solution at pH 4.5, 5.5, or 6.5 with and without 10 mm citrate. All oocytes were injected with citrate prior to measurement except those in E. pH was adjusted with MES/Tris, and osmolality was 200 mosmol. Data show means and se (n = 3–4).
TaMATE1B Is Highly Expressed in Root Apices of cv Carazinho But Not cv Egret
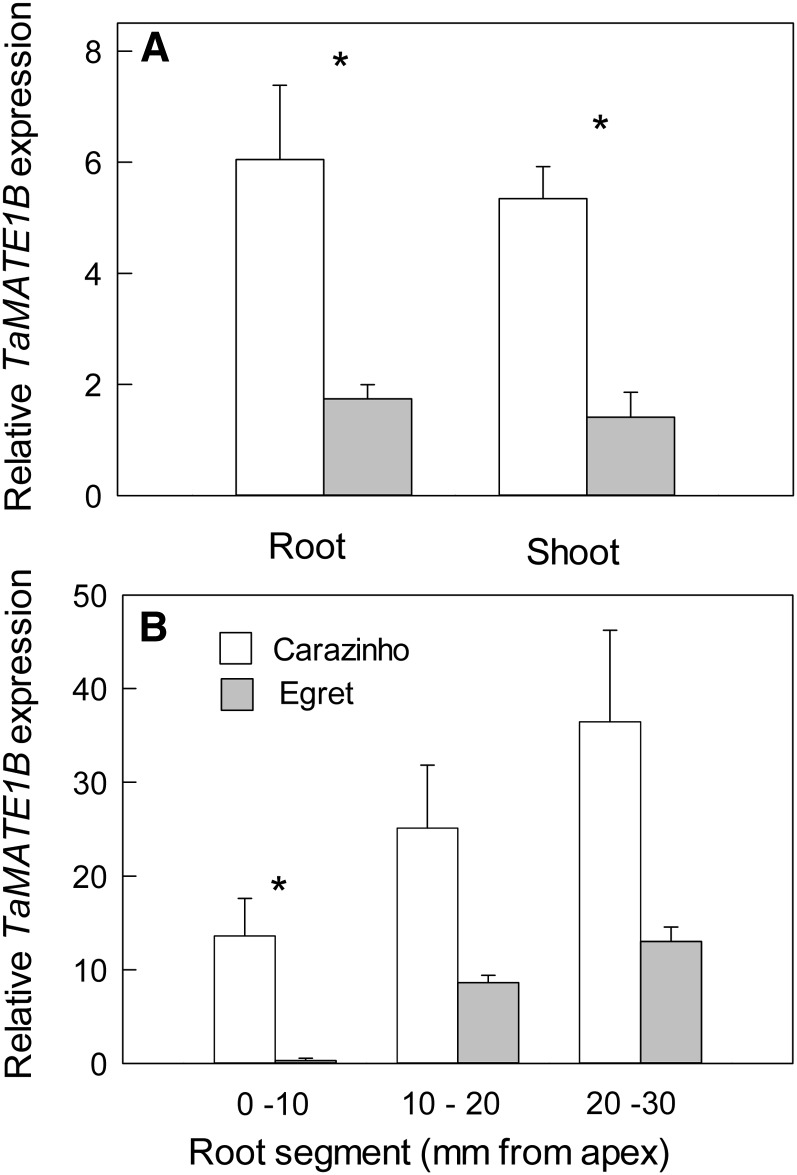
TaMATE1B expression in whole roots and whole shoots of seedlings was 3-fold greater in cv Carazinho than in cv Egret (Fig. 6A). However, a more detailed analysis of roots showed that cv Carazinho had over 50-fold greater expression than cv Egret in the apical 10-mm region (Fig. 6B). We attempted to analyze the expression of the other TaMATE1 homeologs in the roots using allele-specific primers, but the expression in both cv Carazinho and Egret was too low to obtain reliable qRT-PCR data (data not shown).
Figure 6.
Expression of TaMATE1B in wheat ‘Carazinho’ and ‘Egret’. Expression of TaMATE1B was measured by qRT-PCR in whole roots and whole shoots of 12-d-old seedlings (A) and in excised root segments of 4-d-old seedlings (B). Data show mean relative expression and se from three biological replicates using GAPDH as a reference gene. When cv Carazinho was compared with cv Egret for the various tissues, Student’s t test (P < 0.05) identified significant differences (asterisks).
Fe Deficiency Induces TaMATE1B Expression
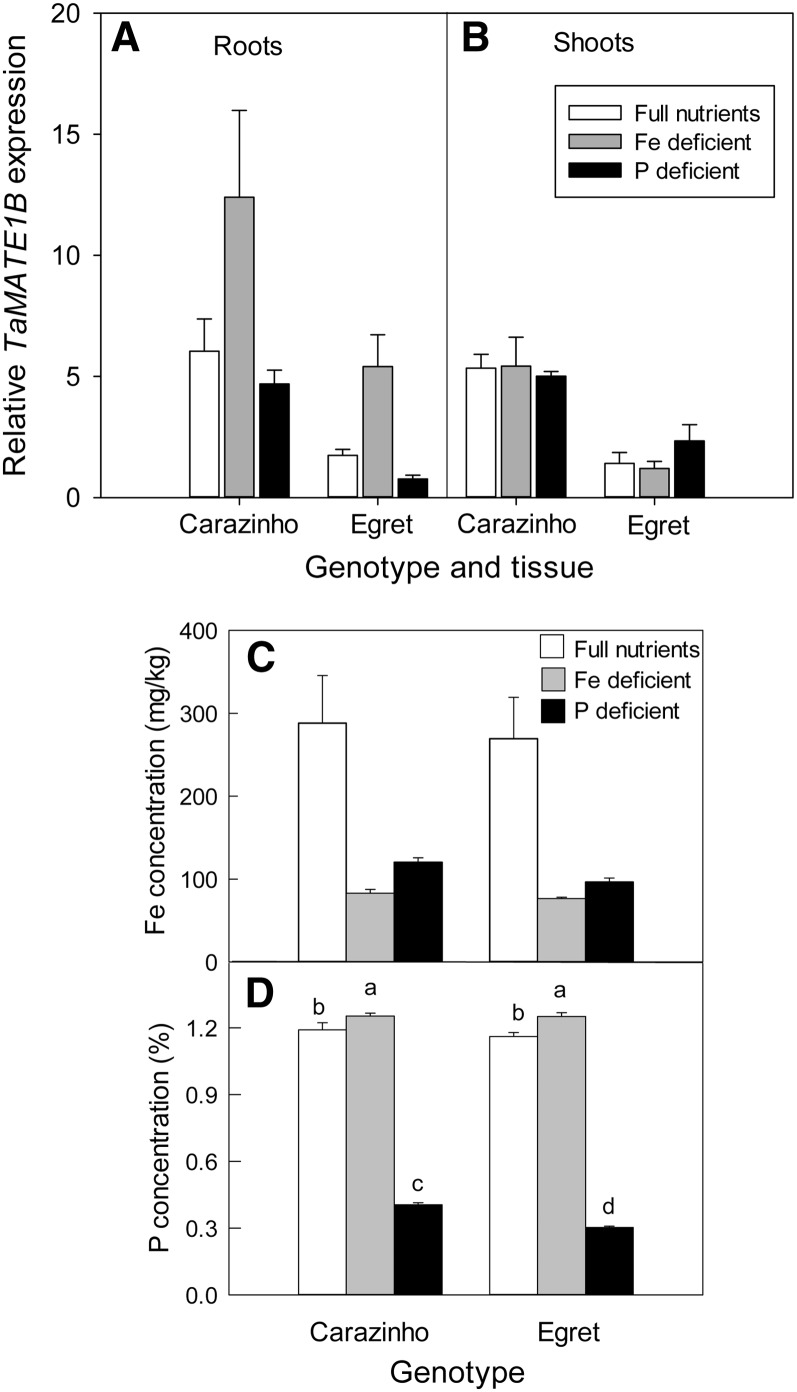
Several members of the MATE gene family have been implicated in the transport and distribution of Fe within plants, Al3+ tolerance mechanisms, or the acquisition of phosphorus from soil. TaMATE1B expression, therefore, was measured in the roots and shoots of seedlings after being deprived of Fe or phosphorus for 12 d (Fig. 7A). Elemental analysis of shoots established that the plants were deficient in Fe and phosphorus following these treatments. Interestingly, withdrawal of either Fe or phosphorus from the nutrient solution reduced the shoot Fe concentration to approximately 30% of the full-nutrient controls, and cv Carazinho and Egret responded similarly (Fig. 7C). Phosphorus starvation reduced the phosphorus concentration of shoots by a similar amount for both genotypes (Fig. 7C). Fe deficiency induced a 2- to 3-fold increase in TaMATE1B expression in roots of both cv Carazinho and Egret but not in the shoots (Fig. 7A). There was no consistent effect of phosphorus starvation on TaMATE1B expression in roots and shoots of either cultivar (Fig. 7A). Al3+ treatment did not induce TaMATE1B expression in either root apices or whole roots of cv Carazinho (50 µm AlCl3 for 24 h; data not shown).
Figure 7.
Expression of TaMATE1B under Fe and phosphorus (P) deficiency. A and B, Expression of TaMATE1B in roots (A) and shoots (B) as measured by qRT-PCR in wheat ‘Carazinho’ and ‘Egret’ grown for 12 d in hydroponics with full nutrients, full nutrients without Fe, and full nutrients without phosphorus. Data show mean relative expression and se from three biological replicates using GAPDH as a reference gene. For roots, two-way ANOVA identified significant treatment and genotype effects, but the interaction of treatment by genotype was not significant. The minus-Fe treatment was significantly different from both phosphorus deficiency and full nutrient treatments, and cv Carazinho was significantly different from cv Egret (P < 0.05). For shoots, two-way ANOVA only identified a significant genotype effect with cv Carazinho significantly different from cv Egret (P < 0.05). C and D, Shoot Fe (C) and phosphorus (D) concentrations measured by inductively coupled plasma-mass spectrometry. Data for Fe concentrations were not normally distributed, and analysis with a Mann-Whitney U test showed significant treatment effects (full nutrients differing from both Fe-deficient and phosphorus-deficient treatments) but no genotype differences. Two-way ANOVA identified a significant genotype-by-treatment interaction for shoot phosphorus concentrations (P < 0.05), with the different letters indicating values that are significantly different from one another.
The Transposable Element in the cv Carazinho Promoter Alters the Level and Pattern of Expression
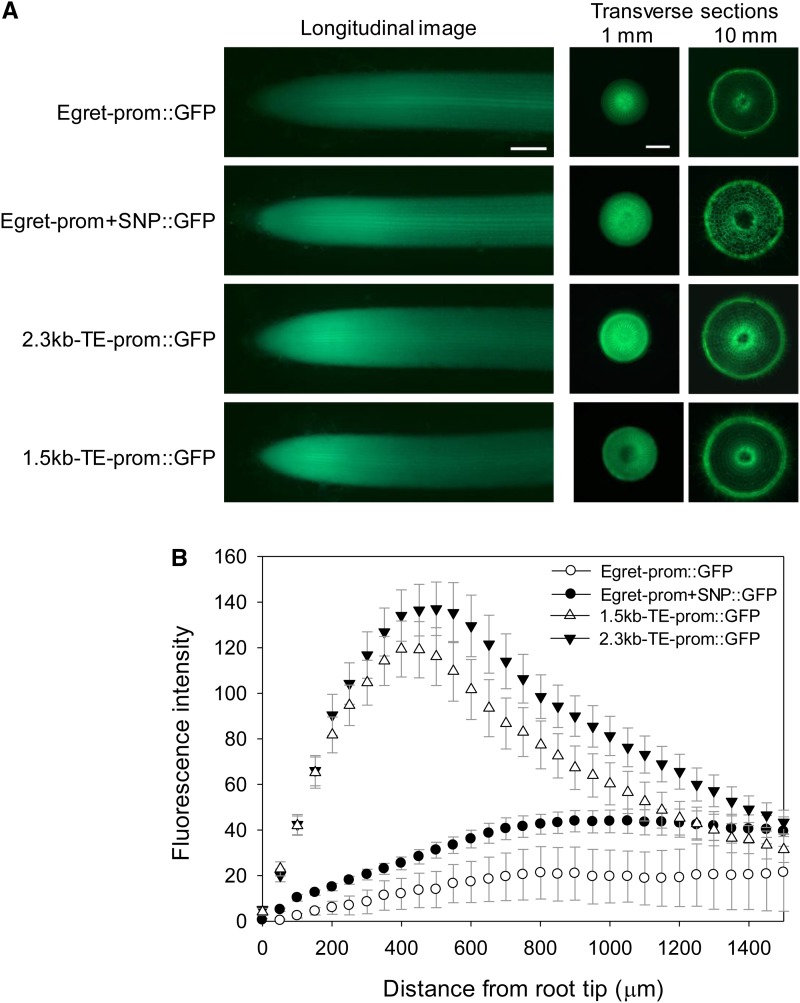
To analyze the influence of the TE and SNP on TaMATE1B expression, different promoter fragments were fused to GFP for expression analysis in transgenic rice. The four constructs were (1) approximately 3 kb of the promoter derived from cv Egret (Egret-prom::GFP); (2) the same fragment incorporating the SNP present in cv Carazinho (Egret-prom+SNP::GFP); (3) 2.3 kb of the cv Carazinho promoter that includes part of the TE (2.3kb-TE-prom::GFP); and (4) 1.5 kb of the cv Carazinho promoter that includes a shorter part of the TE (1.5kb-TE-prom::GFP). Apart from the SNP and TE fragments, the constructs were identical, and specifically, the 25 bp immediately upstream of the coding region was identical for all promoter constructs.
Eight to 10 independently transformed lines were randomly selected and analyzed for each promoter construct, and quantitative measurements were made for GFP fluorescence and GFP transcripts in roots. Fluorescence was first assessed along a longitudinal transect of seminal roots using a fluorescence binocular microscope. Figure 8A shows representative images of GFP expression, with enhanced expression in the apical region of the roots for the TE and SNP constructs. Differences between promoters were further evident in transverse sections taken at 1 mm but not at 10 mm from the root apex (Fig. 8A). In the 1-mm section, the Egret-prom::GFP promoter construct differed from the other three in having less intense expression at the root periphery compared with the central region of the root, where vasculature would be developing. The presence of the SNP alone was sufficient to alter this pattern. For example, the ratio of fluorescence in the periphery to the central region of these images was 0.46 ± 0.04 (se; n = 3) for the native cv Egret promoter (Egret-prom::GFP) and 0.81 ± 0.07 (se; n = 3) for the promoter with the SNP (Egret-prom+SNP::GFP).
Figure 8.
Promoter regions (TE and SNP) that enhance the level of gene expression. A, Rice plants were transformed with 3 kb of the cv Egret promoter (Egret-prom::GFP), 3 kb of the cv Egret promoter incorporating the SNP present in cv Carazinho (Egret-prom+SNP::GFP), 2.3 kb of the cv Carazinho promoter that includes part of the TE (2.3kb-TE prom::GFP), and 1.5 kb of the cv Carazinho promoter that includes a shorter part of the TE (1.5kb-TE prom::GFP). The regions of the TaMATE1B promoters used to prepare the various constructs are shown in Figure 1. Shown are fluorescence images of representative longitudinal root sections (approximately 1.5 mm) and transverse root sections taken at 1 and 10 mm from the root apex as indicated. Due to large differences in absolute fluorescence, images were not taken with the same exposure settings. Bars = 200 µm. B, Quantification of fluorescence from longitudinal root images (1.5 mm) for the four promoter constructs. The symbols represent average levels of GFP intensity (arbitrary units) from eight to nine independently generated transgenic plants, and the error bars denote se.
Fluorescence intensity in longitudinal transects (Fig. 8A) was quantified over the 1.5-mm apical region and averaged over all replicates (Fig. 8B). Promoters containing the TE (1.5kb-TE-prom::GFP and 2.3kb-TE-prom::GFP) had up to 20-fold greater fluorescence at the root apex than the Egret-prom::GFP promoter (Fig. 8B). The two TE promoters showed distinct peaks in fluorescence intensity approximately 0.5 mm from the root apex. By contrast, the two non-TE promoters showed a gradual increase in fluorescence intensity over the entire 1.5-mm region. The TaMATE1B promoter that included the SNP (Egret-prom+SNP::GFP) consistently conferred about a 2-fold increase in GFP fluorescence compared with the native cv Egret promoter (Egret-prom::GFP).
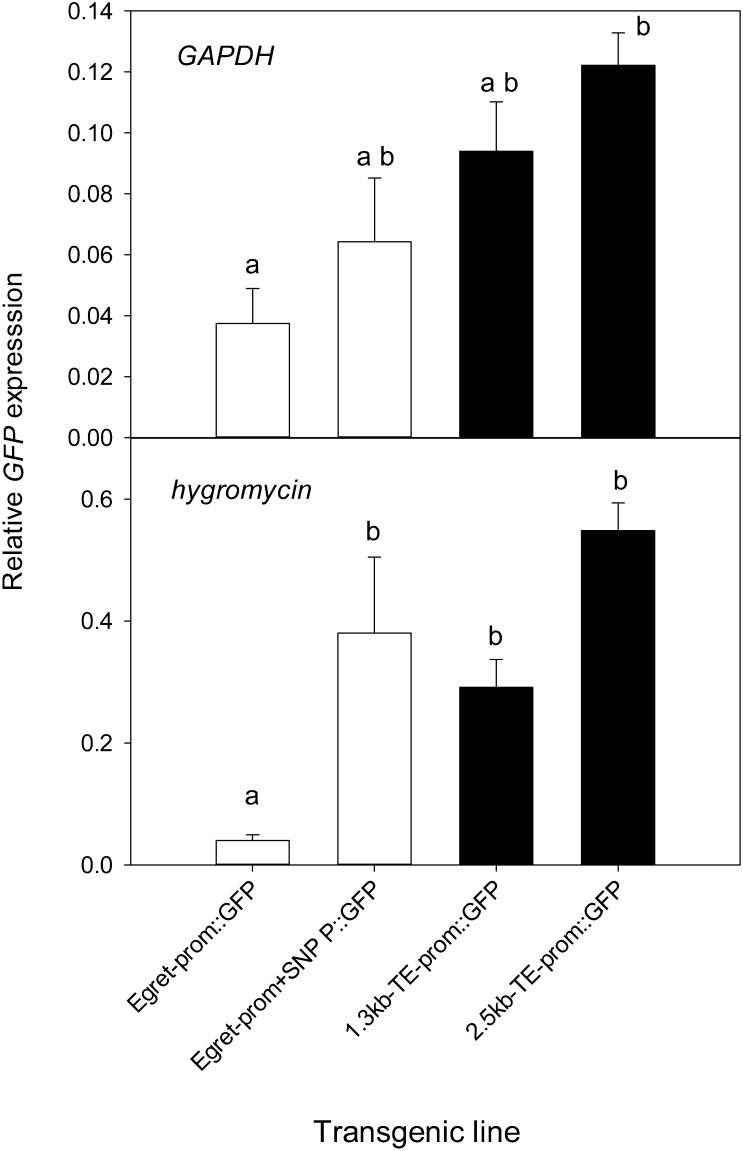
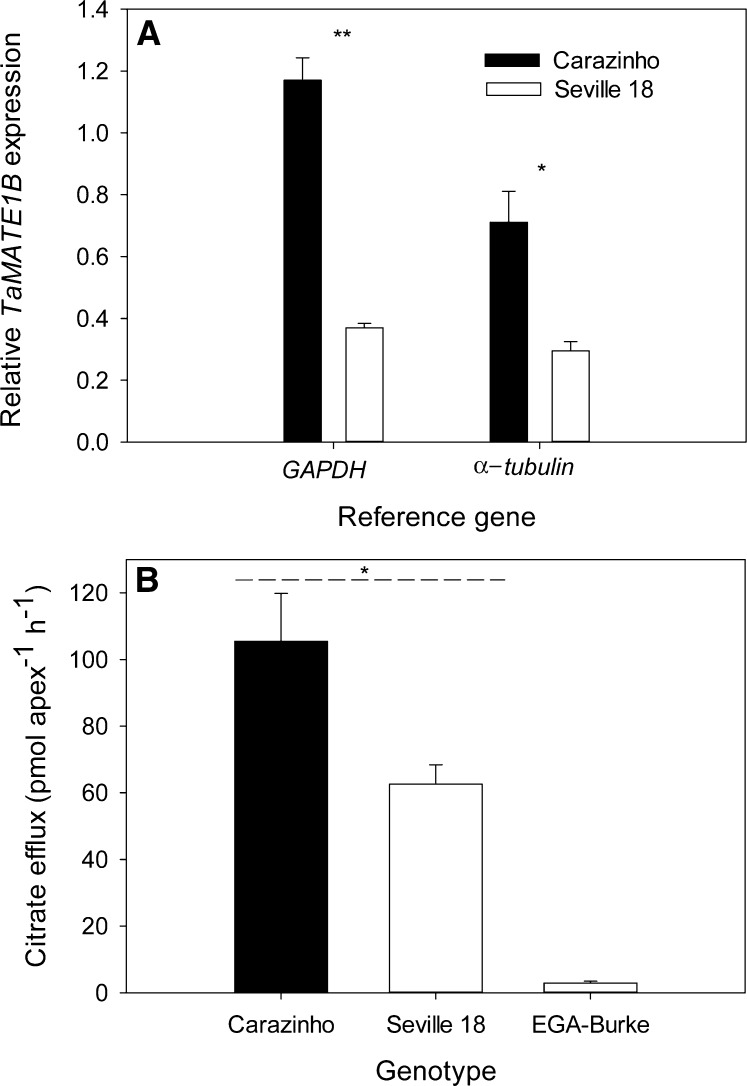
GFP transcript levels in root apices generally followed the pattern of GFP fluorescence. An endogenous gene (Glutamate Dehydrogenase [GAPDH]) and a gene on the plasmid vector (Hygromycin) were used as reference genes in the qRT-PCR. The Hygromycin gene was included to reduce the variation that site of transgene insertion can have on gene expression. For example, if the construct inserts in a region of the genome that provides enhancer activity, then both the promoter construct and the Hygromycin gene are likely to be similarly affected. When the Hygromycin gene was used to normalize the data, GFP expression from the TE promoters as well as the Egret-prom+SNP::GFP construct were all greater than the Egret-prom::GFP construct (Fig. 9). With GAPDH as a reference gene, the only significant difference was between the 2.3kb-TE-prom::GFP and Egret-prom::GFP constructs. The influence of the SNP was further investigated by comparing the expression of TaMATE1B in cv Carazinho and the landrace Seville 18. Both genotypes contain the TE but had different alleles for the SNP. Expression of TaMATE1B in cv Carazinho was 2- to 3-fold greater than in Seville 18, and this was associated with an approximately 2-fold difference in citrate efflux (Fig. 10). Citrate efflux from both genotypes was significantly greater than that for EGA-Burke, which contains the cv Egret-like TaMATE1B promoter.
Figure 9.
Analysis of GFP expression in root apices (approximately 5 mm) of transgenic rice by qRT-PCR with GAPDH and Hygromycin as reference genes. Data show mean relative expression and se (n = 6–10). Since the data were either not normally distributed or had unequal variances, a Kruskal-Wallis ANOVA on ranks was undertaken and a pairwise comparison using Dunn’s method was applied. Constructs with different letters denote statistically significant differences (P < 0.05).
Figure 10.
Comparison of TaMATE1B expression and citrate efflux from the wheat landrace Seville 18 and cv Carazinho. A, TaMATE1B expression was measured by qRT-PCR in root apices (approximately 5 mm) excised from seedling grown for 3 d in hydroponics with all nutrients supplied. Shown are mean relative expression levels of TaMATE1B and se (n = 4 or 5) using GAPDH and α-Tubulin as reference genes. B, Citrate efflux from excised root apices from seedlings grown for 6 d in sterile flasks containing 0.5 mm CaCl2 (pH 5.0). EGA-Burke is a wheat cultivar that possesses the same TaMATE1B allele as cv Egret. Asterisks denote significant differences as identified by paired Student’s t tests (*P < 0.05, **P < 0.001).
DISCUSSION
TaMATE1 was previously identified as a candidate gene encoding the constitutive citrate efflux observed in several Brazilian wheat cultivars (Ryan et al., 2009). In this study, we describe the isolation of three related TaMATE1 genes from wheat and identify TaMATE1B on chromosome 4BL as the homeolog that encodes constitutive citrate efflux in cv Carazinho. This conclusion is supported by the findings that TaMATE1B is located on the plasma membrane and that transgenic plants expressing TaMATE1B constitutively release citrate from their roots. The function of TaMATE1B as a citrate transporter was further supported by its expression in X. laevis oocytes. Inward and outward currents were observed in oocytes expressing TaMATE1B that were absent from control oocytes, but only when citrate was added to the bathing solution. This is consistent with TaMATE1B facilitating the efflux of citrate, or other anions, at negative membrane potentials and citrate uptake at positive membrane potentials. It is unclear why citrate is required in the bathing solution to generate these large currents in oocytes, whereas citrate efflux is constitutive in plants. It could indicate that transactivation occurs in oocytes where substrate is required on both sides of the membrane to trigger function. TaMATE1B-dependent currents were observed whether or not oocytes were loaded with citrate prior to the measurements, but the magnitude tended to be greater when oocytes were loaded with citrate. These results support the conclusion that citrate is a substrate of TaMATE1B but also indicate that other endogenous substrates in oocytes are also transported by TaMATE1B. The inward currents were increased at lower pH values, which is consistent with TaMATE1B functioning as an antiporter with protons, but this result and other aspects of TaMATE function in oocytes require further investigation.
The high expression of TaMATE1B in root apices of cv Carazinho compared with cv Egret can be largely attributed to the presence of the 11.1-kb TE inserted 25 bp upstream of the start codon in cv Carazinho. The TE insertion contains a 3.9-kb Sukkula-like element at its 3′ end. Sukkula elements were first described in barley and possess long terminal repeats typical of retrotransposons, but they are nonautonomous and have been described as large retrotransposon derivatives (Kalendar et al., 2004). TEs can have promoter activity. For example, the long terminal repeats of barley BARE1 retrotransposons are capable of driving the expression of transgenes in barley (Suoniemi et al., 1996). Here, we demonstrated that promoter::GFP constructs that included 3′ fragments of the TE (1.5 or 2.3 kb) conferred stronger expression in root apices than the TaMATE1B promoter of cv Egret. In addition to the TE, a SNP located 2 kb upstream of the start codon in the TaMATE1B promoter also increased overall expression, but to a smaller degree, and extended GFP expression toward the periphery of the root apex. Although the SNP increased promoter activity in transgenic plants with Hygromycin as the reference gene, its ability to affect expression in vivo was uncertain because it is positioned more than 13 kb upstream from the coding region in the native cv Carazinho promoter. However, the landrace Seville 18, which like cv Carazinho has the TE in its promoter but possesses the cv Egret allele for the SNP, had lower TaMATE1B expression and less citrate efflux from root apices than cv Carazinho. Although this result is consistent with the SNP influencing promoter activity in vivo, it is possible that the difference in TaMATE1B expression between Seville 18 and cv Carazinho is caused by other regulatory loci. Similarly, if the Fe-responsive element approximately 1 kb from the ATG start codon in cv Egret were responsible for the induction of TaMATE1B expression in roots when Fe deficient, then the same Fe-responsive element would need to have regulated expression when located about 12 kb from the coding region in cv Carazinho. Alternatively, a different Fe-responsive element not identified by a motif search could have been located downstream of the TE, such as 25 bp upstream of the TaMATE1B coding region or within the intron/exon region of the gene. Ryan and Delhaize (2010) proposed that some genes encoding organic anion transport have been coopted from other functions into Al3+ tolerance by mutations that altered their expression level or pattern. The most convincing evidence in support of this hypothesis was provided by Fujii et al. (2012), who recently described a 1-kb insertion upstream of the HvAACT1 coding region that occurs only in Al3+-tolerant barley cultivars. The 1-kb insertion functions as a promoter that enhances HvAACT1 expression and extends its distribution from the pericycle to the root apices, thus expanding HvAACT1 function from a role in Fe nutrition to Al3+ tolerance by releasing citrate from root apices. This study provides evidence that the expression and function of TaMATE1B in wheat have similarly been extended to include a role in Al3+ tolerance. TaMATE1B expression in cv Egret is induced by Fe deficiency and has a similar expression pattern to OsFRDL1, a MATE gene from rice with a known role in Fe nutrition (Yokosho et al., 2009). The presence of the TE upstream of the TaMATE1B coding region has extended its expression to the root apices of cv Carazinho, resulting in constitutive citrate efflux from those tissues.
In addition to influencing promoter activity, insertion of the TE changed the TSS of TaMATE1B. The 5′ UTR sequences in the roots of cv Carazinho and Egret are different, and the encoded protein is predicted to be shorter in cv Egret. Whether the TaMATE1B protein is functional in the roots of cv Egret is not known, but the other two homeologs identified in this study could also transport citrate due to their strong similarity with TaMATE1B. This illustrates the flexibility of a polyploid species such as wheat in recruiting existing genes for new functions. As one gene incurs a mutation and changes function, any loss of original function may be compensated for by the homeologs. Further work to investigate the expression and localization of the TaMATE1 homeologs and their possible function in citrate transport in different tissues of wheat is warranted.
TEs constitute a significant proportion of genomes, and in the case of wheat, about 80% of the genome consists of various types of TEs (Choulet et al., 2010). In the past, TEs were viewed as nonfunctional DNA sequences that were potentially deleterious to an individual if they inactivated gene function by inserting into coding regions. It is now becoming apparent that TEs can generate variation within genomes, including changes in the expression of specific genes (Morgante et al., 2007; Muotri et al., 2007). In some instances, these changes in expression may benefit plants by generating favorable phenotypes. In the case of TaMATE1B and HvAACT1, the altered expression caused by unrelated TEs have enhanced Al3+ tolerance and conferred a phenotype that benefits plant growth on acid soils. Despite the parallels in barley and wheat, the insertion of TEs near these MATE genes occurred independently. The approximately 1-kb TE near HvAACT1 in barley is unrelated to the 11.1-kb TE upstream of TaMATE1B in wheat and has homology with the CACTA transposon family of rice (Fujii et al., 2012). Furthermore, the transposon is located 25 bp immediately upstream of the TaMATE1B coding region in wheat, whereas the transposon is located approximately 4.8 kb upstream of the HVAACT1 coding region in barley. In both cases, the transposons have generated new TSSs that enhance expression within root apices and increase citrate efflux. Curiously, HvAACT1 requires Al3+ to activate citrate efflux, whereas TaMATE1B functions constitutively. Presumably, HvAACT1 has motifs that interact with Al3+ despite its original location on the xylem parenchyma, where it is unlikely to interact with soluble Al3+. Nevertheless, certain MATE genes appear to be predisposed for recruitment into Al3+ tolerance mechanisms if mutations extend their expression to the root apices, where citrate efflux can protect this very susceptible tissue from Al3+ toxicity.
MATERIALS AND METHODS
Plant Growth and Elemental Analysis
Seeds of wheat (Triticum aestivum) and rice (Oryza sativa) were obtained from the collections at the Commonwealth Scientific and Industrial Research Organization Plant Industry in Canberra, Australia. Wheat seeds were germinated for 2 d on moist filter paper and then planted over 20 L of aerated nutrient solution (Delhaize et al., 2004). Plants were maintained in a glasshouse, where they were grown for 4 d (gene expression in root segments) or 12 d (gene expression in whole roots and shoots). To generate seedlings that were Fe or phosphorus deficient, germinated seedlings were grown for 12 d in the same nutrient solutions except that Fe:EDTA (Fe deficiency) or KH2PO4 (phosphorus deficiency) were omitted from the nutrient solutions.
Rice plants (T0 transformants) were grown in hydroponic culture as described previously (Schünmann et al., 2004).
For elemental analysis, shoots were dried at 65°C, digested in nitric acid, and analyzed for elements by inductively coupled plasma-mass spectrometry.
Isolation of TaMATE1 cDNA and Genes
A BLAST search of the EST databases with barley (Hordeum vulgare) HvAACT1 cDNA identified homologous wheat ESTs that included GenBank accessions BE498331, BJ279544, BJ258950, BJ245240, BE605049, CK206051, and CA663871. The wheat EST BE498331 is a partial sequence from the TaMATE1 gene previously identified as a candidate encoding the citrate efflux phenotype in cv Carazinho (Ryan et al., 2009). The seven ESTs were used to construct a composite TaMATE cDNA, and primers (primer pairs 1 and 2; all primers are listed in Supplemental Table S1) were then designed to amplify two overlapping fragments of this composite cDNA. PCR products from cv Carazinho were cloned and sequenced to yield the full coding region of a cDNA named TaMATE1B. Primers based on the TaMATE1B sequence were then used to amplify TaMATE1 fragments from genomic DNA of both cv Carazinho and Egret (primer pairs 3–7). The PCR products were cloned, and up to 20 individual clones were sequenced for each genotype. Three related sequences were identified from each amplification, and subsequent PCR amplifications used primers that incorporated identified sequence polymorphisms in overlapping regions to allow contigs of the three sequences to be constructed (TaMATE1 homeologs). To obtain sequences upstream and downstream of the contigs, BAC libraries were screened with primers (primer pair 8) designed to amplify all three homeologs. Two BAC libraries held by the Centre National de Resources Genomiques Vegetales in Toulouse, France, were screened with the primer pair. Four positive clones were identified from a library prepared from cv Chinese Spring (BAC clones 139A1, 321B14, 384M22, and 408P21), and six positive clones were identified from a library prepared from cv Renan (BAC clones 242H9, 566A11, 2110P14, 1835D2, 2471A16, and 2472O24). Partial sequences obtained from the BAC clones verified that all three homeologs were represented, and the TaMATE1 gene of at least one BAC clone for each homeolog was sequenced.
The sequence of TaMATE1B obtained from BAC clones 2110P14 and 321B14 was used to design primers that amplified regions upstream and downstream of TaMATE1B from genomic DNA of cv Carazinho and Egret (primer pairs 9–17). The PCR products were sequenced, and it became apparent that a region immediately upstream of the cv Carazinho coding region was recalcitrant to amplification. Long-range PCR (primer pair 18 with a Takara Long Range Amplification kit) amplified a large product (greater than 10 kb) only from cv Carazinho, and this fragment was subsequently cloned and sequenced. An SNP between cv Carazinho and Egret in the TaMATE1B locus located approximately 2 kb upstream of the first exon in cv Egret was used to develop a CAPS marker. Primer pair 19 amplified a 161-bp fragment, and only the cv Egret allele could be digested with MnlI. Another marker using primer pair 20 amplified a product only from the cv Carazinho allele and included part of a large TE insertion present in cv Carazinho. An Invitrogen GeneRacer Kit was used to identify the TSS and 3′ UTRs of the TaMATE1B genes. Primer pairs 21 and 22, composed of TaMATE1B-specific primers along with the 5′ primers provided by the kit, were used to amplify the 5′ UTR. Similarly, primer pairs 23 and 24 were used as TaMATE1B-specific primers along with the 3′ primers provided by the kit to amplify the 3′ UTR.
The four programs used to predict transmembrane regions of the predicted TaMATE1B protein were SOSUI (bp.nuap.nagoya-u.ac.jp/sosui/), DAS (www.sbc.su.se/~miklos/DAS/), TMpred (www.ch.embnet.org/software/TMPRED_form.html), and TMHMM (www.cbs.dtu.dk/services/TMHMM-2.0/).
Subcellular Localization of TaMATE1B
Subcellular localization of the TaMATE1B protein was determined by transiently expressing chimeric proteins using constitutive promoters in leek (Allium ampeloprasum) and Nicotiana benthamiana leaves. GFP was fused to the N- and C-terminal ends of the TaMATE cDNA. For leek transformation, the fusion construct was inserted into the pWUbi plasmid vector under the control of the ubiquitin promoter. The plasmid (1 µg) was precipitated on gold particles (0.6 µm diameter; Bio-Rad) and bombarded into leek tissue as described previously (Delhaize et al., 2007). In brief, the procedure used the Bio-Rad Biolistic Particle Delivery System model PDS-1000/He (600 mm Hg vacuum; 6,000 kP pressure). The tissues were viewed 16 h after bombardment with a Leica SP2 confocal microscope. For N. benthamiana transformation, the cDNA::GFP fusions were inserted into the pPLEX502 plasmid vector under the control of the cauliflower mosaic virus 35S promoter. The vector was transformed into Agrobacterium tumefaciens (strain AG1) by electroporation, and N. benthamiana leaves were transformed by an infiltration method (Wood et al., 2009). Three days after inoculation, epidermal strips were peeled from the abaxial side of the leaves and viewed with the confocal microscope. A control construct consisting of GFP alone was also transformed into cells.
Characterization of TaMATE1B in Xenopus laevis Oocytes
The plasmid pGEMHE-DEST containing TaMATE1B cDNA was linearized and used as the template for capped cRNA using a Message Machine T7 kit (Ambion). Oocytes were harvested and maintained using a modification of a previously described method (Virkki et al., 2006). The oocytes were injected with 46 nL of cRNA (0.6 μg μL−1) or RNase-free water using a microinjector (Nanoject II automatic nanoliter injector; Drummond Scientific) and incubated for 2 d at 18οC in ND88 solution (Sasaki et al., 2004), which was replaced daily. Recordings were performed on both control oocytes (water injected) and oocytes expressing TaMATE1B that were either injected with 46 nL of 0.1 m sodium citrate 2 h prior to TEVC or not injected with sodium citrate. Bathing solutions consisted of basal solution: 1 mm MgCl2, 1.8 mm CaCl2, and 5 mm MES with or without 10 mm sodium citrate. pH was adjusted to 4.5, 5.5, or 6.5 with 1 m Tris, and osmolality was adjusted to 200 mosmol with sorbitol. Whole-cell currents were recorded under constant perfusion and temperature (22°C) with a GeneClamp 500 amplifier (Axon Instruments) using conventional TEVC. From a holding potential of −40 mV, the voltage was stepped from 60 to −160 mV in 20-mV increments. The duration of each voltage pulse was 0.5 s, with a 0.5-s resting phase between successive voltage steps. The output was digitized and analyzed using a Digidata 1322A-pCLAMP 8 data acquisition system (Axon Instruments). Recording electrodes filled with 3.0 m KCl had resistances between 0.5 and 1.2 MΩ. Data analysis was performed using Clampfit software (version 8.2; Molecular Devices) and GraphPad (GraphPad Software) to construct current-voltage curves.
Analysis of Gene Expression by qRT-PCR
Plant tissues were ground in liquid nitrogen, and RNA was extracted with the RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized with the SuperScript III First-Strand Synthesis System (Invitrogen) following methods provided with the kit. Gene expression was determined by qRT-PCR using the SYBR Green Supermix (Bio-Rad) kit on a Bio-Rad CFX96 Real Time System. Primer pairs were designed to amplify all three TaMATE1 homeologs of wheat (primer pair 25) or specifically TaMATE1B (primer pair 26), TaMATE1A (primer pair 27), or TaMATE1D (primer pair 28). GAPDH (GenBank accession no. EF592180; primer pair 29) and α-Tubulin (primer pair 30) were used as reference genes. For transgenic rice expressing GFP as a reporter gene, primer pair 31 amplified the GFP transcripts, while primer pairs 32 and 33 amplified GAPDH and the Hygromycin transcripts, respectively, as reference genes. PCR products were sequenced to confirm their identities. Data were analyzed with the Bio-Rad CFX Manager software, and TaMATE genes were expressed relative to the reference genes.
Transgenic Plants Expressing TaMATE1B
To express TaMATE1B cDNA in rice, the full coding sequence was inserted into the pVec8 vector under the control of the ubiquitin promoter (Schünmann et al., 2004). Rice ‘Nipponbare’ was transformed with the vector using the A. tumefaciens method (Toki et al., 2006). Transgenic controls were generated at the same time by transforming plants with the empty vector. Transgenic (T0) rice lines expressing TaMATE1B were selected to generate T1 seed. Tobacco (Nicotiana tabacum ‘WI 38’) was transformed with a pPLEX502 vector (Schünmann et al., 2003) that expressed TaMATE1B under the control of the cauliflower mosaic virus 35S promoter. Transgenic tobacco lines were generated by A. tumefaciens-mediated transformation, and selected T0 plants identified to be expressing TaMATE1B were grown to produce the T1 generation. Relative expression of TaMATE1B in transgenic rice (root apices) and tobacco (whole seedlings) was measured by qRT-PCR. The forward and reverse primers targeting TaMATE1B expression in transgenic plants were primer pair 26. Reference genes were GAPDH (primer pair 32) in rice and Malate Dehydrogenase (primer pair 34) in tobacco.
TaMATE1B Promoter Constructs
To assess the effects of polymorphisms on promoter activity, regions upstream of the TaMATE1B gene were amplified from genomic DNA (cv Carazinho) or from BAC clone 2110P14 with identical TaMATE1B sequence as cv Egret. PCR products were cloned between the PacI and AscI sites (located upstream of the AdhI intron and GFP gene) in a derivative of the plasmid vector pWBvec8 as described by Schünmann et al. (2004). Primers were designed to include PacI and AscI sites and to amplify regions immediately upstream of the TaMATE1B coding region. A 3.013-kb region upstream of the cv Egret TaMATE1B and the same approximately 3-kb sequence that included a SNP specific to the cv Carazinho allele were amplified with primer pair 35 (Supplemental Table S1). Fragments of 1.471 kb (primer pair 36) and 2.321 kb (primer pair 37) that included part of a TE were amplified from cv Carazinho genomic DNA. Plasmid constructs were transferred to A. tumefaciens strain AGL1 by heat shock, and rice was transformed as described above.
Assay of Citrate Efflux
Transgenic rice lines with the greatest expression were used to generate T1 seed for the assay of citrate efflux. T1 seeds were surface sterilized with 10% bleach, rinsed thoroughly with deionized water, and then germinated and grown in flasks containing 0.5 mm CaCl2 (pH 5.2) for 8 d. The seedlings were transferred to tanks that contained 12 L of nutrient solution and grown under artificial lighting in a constant-temperature room (28°C) for 18 d. Nutrient solution was replaced every 5 to 8 d. Root apices (5 mm) were excised from the plants and collected in 8-mL glass sample tubes (12 apices per tube) containing 1 mL of sterile 0.5 mm CaCl2 (pH 5.2). After rinsing and adding 1.0 mL of fresh solution, the top of the tube was sealed with Parafilm and the tube was placed horizontally on a shaker for 2 h. The solution was collected and dried under vacuum before being resuspended in buffer for citrate assays using an enzyme assay (Wang et al., 2007).
To assay for citrate efflux from tobacco, 40 seeds were surface sterilized with chlorine gas and placed onto sterile 12-well tissue culture plates. Nutrient solution (2 mL) was added to each well and comprised the following components: 4 mm KNO3, 4 mm Ca(NO3)2, 1.5 mm MgSO4, 3 mm NH4Cl, 40 µm FeNa2EDTA, 1 mm KH2PO4, micronutrients, and 10 g L−1 Suc, pH 5.6); seedlings were grown on a shaker (approximately 60 rpm) for 10 to 12 d at 24°C with a 16-h/8-h light/dark period. After the growth period, the nutrient solution was completely removed and replaced with 1 mL of fresh nutrient solution that had FeNa2EDTA and Suc omitted and the KH2PO4 concentration reduced to 0.1 mm. After 30 h of further growth, 250 µL of the nutrient solution was collected for citrate assay.
Microscopy and Image Processing of Promoter::GFP-Expressing Plants
Gene expression in plants transformed with constructs that expressed promoter::GFP constructs was assessed using a Zeiss Axioimager fluorescence microscope. Images from roots of eight to nine independent transgenic lines were obtained from seminal roots for each construct. ImageJ software (Schneider et al., 2012) was used to quantify GFP fluorescence levels along a longitudinal transect of the root for each of the images. ImageJ was used to split color images into the three primary colors, and the green channel was converted into a grayscale. Intensity values for pixels along a line starting at the apex and drawn down the middle of the root were obtained. Data were exported to Microsoft Excel for further calculations that included subtraction of background fluorescence as measured in rice roots transformed with an empty plasmid. A similar procedure was taken to measure GFP fluorescence intensity in transverse root sections, except that values obtained from the center and edge of the root were used to calculate a ratio.
Sequences submitted to GenBank include sequences of the TaMATE1 loci of BAC clones 2110P14 (TaMATE1B; GenBank accession no. KC152454), 321B14 (TaMATE1B; KC152455), 384M22 (TaMATE1D; KC152456), and 242H9 (TaMATE1A; KC152453), TaMATE1B cDNA (KC152457), the TaMATE1B genomic locus of wheat ‘Egret’ (KC152458), and the TaMATE1B genomic locus of wheat ‘Carazinho’ (KC152459).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the TaMATE1 homeologs from hexaploid wheat.
Supplemental Table S1. Primer pairs used for PCR.
Glossary
- SNP
single-nucleotide polymorphism
- Al3+
aluminum
- Fe
iron
- UTR
untranslated region
- BAC
bacterial artificial chromosome
- TSD
target site duplication
- TE
transposon-like element
- CAPS
cleaved-amplified polymorphic sequence
- TSS
transcription start site
- qRT
quantitative real-time
- cRNA
copy RNA
- cDNA
complementary DNA
- TEVC
two-electron voltage clamping
- T0
transgenic
References
- Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, Schlub S, Le Paslier MC, Magdelenat G, Gonthier C, et al. (2010) Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22: 1686–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE. (2007) A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J 51: 198–210 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ma JF, Ryan PR. (2012) Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci 17: 341–348 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H. (2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA 101: 15249–15254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ. (1993) Aluminum tolerance in wheat (Triticum aestivum L.). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE. (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famoso AN, Zhao K, Clark RT, Tung C-W, Wright MH, Bustamante C, Kochian LV, McCouch SR. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7: e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Yokosho K, Yamaji N, Saisho D, Yamane M, Takahashi H, Sato K, Nakazono M, Ma JF. (2012) Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun 3: 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, Katsuhara M, Takeda K, Ma JF. (2007) An aluminum-activated citrate transporter in barley. Plant Cell Physiol 48: 1081–1091 [DOI] [PubMed] [Google Scholar]
- Gruber BD, Ryan PR, Richardson AE, Tyerman SD, Ramesh S, Hebb DM, Howitt SM, Delhaize E. (2010) HvALMT1 from barley is involved in the transport of organic anions. J Exp Bot 61: 1455–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou X-F, Saier MH. (2003) The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur J Biochem 270: 799–813 [DOI] [PubMed] [Google Scholar]
- Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH. (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166: 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovermann P, Meyer S, Hörtensteiner S, Picco C, Scholz-Starke J, Ravera S, Lee Y, Martinoia E. (2007) The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J 52: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Krill AM, Kirst M, Kochian LV, Buckler ES, Hoekenga OA. (2010) Association and linkage analysis of aluminum tolerance genes in maize. PLoS ONE 5: e9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Pauluzzi I, Gunning BES. (2000) A plasmolytic cycle: the fate of cytoskeletal elements. Protoplasma 212: 174–185 [Google Scholar]
- Liu J, Magalhaes JV, Shaff J, Kochian LV. (2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J 57: 389–399 [DOI] [PubMed] [Google Scholar]
- Ma JF, Nagao S, Sato K, Ito H, Furukawa J, Takeda K. (2004) Molecular mapping of a gene responsible for Al-activated secretion of citrate in barley. J Exp Bot 55: 1335–1341 [DOI] [PubMed] [Google Scholar]
- Magalhaes JV. (2010) How a microbial drug transporter became essential for crop cultivation on acid soils: aluminium tolerance conferred by the multidrug and toxic compound extrusion (MATE) family. Ann Bot (Lond) 106: 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes JV, Liu J, Guimarães CT, Lana UG, Alves VM, Wang YH, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE, et al. (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39: 1156–1161 [DOI] [PubMed] [Google Scholar]
- Maron LG, Piñeros MA, Guimarães CT, Magalhaes JV, Pleiman JK, Mao C, Shaff J, Belicuas SN, Kochian LV. (2010) Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J 61: 728–740 [DOI] [PubMed] [Google Scholar]
- Meyer S, Mumm P, Imes D, Endler A, Weder B, Al-Rasheid KA, Geiger D, Marten I, Martinoia E, Hedrich R. (2010) AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J 63: 1054–1062 [DOI] [PubMed] [Google Scholar]
- Meyer S, Scholz-Starke J, De Angeli A, Kovermann P, Burla B, Gambale F, Martinoia E. (2011) Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J 67: 247–257 [DOI] [PubMed] [Google Scholar]
- Morgante M, De Paoli E, Radovic S. (2007) Transposable elements and the plant pan-genomes. Curr Opin Plant Biol 10: 149–155 [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Gage FH. (2007) The necessary junk: new functions for transposable elements. Hum Mol Genet 16: R159–R167 [DOI] [PubMed] [Google Scholar]
- Piñeros MA, Cançado GM, Kochian LV. (2008) Novel properties of the wheat aluminum tolerance organic acid transporter (TaALMT1) revealed by electrophysiological characterization in Xenopus oocytes: functional and structural implications. Plant Physiol 147: 2131–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman H, Ryan PR, Raman R, Stodart BJ, Zhang K, Martin P, Wood R, Sasaki T, Yamamoto Y, Mackay M, et al. (2008) Analysis of TaALMT1 traces the transmission of aluminum resistance in cultivated common wheat (Triticum aestivum L.). Theor Appl Genet 116: 343–354 [DOI] [PubMed] [Google Scholar]
- Raman H, Zhang KR, Cakir M, Appels R, Garvin DF, Maron LG, Kochian LV, Moroni JS, Raman R, Imtiaz M, et al. (2005) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48: 781–791 [DOI] [PubMed] [Google Scholar]
- Ryan P, Delhaize E, Jones D. (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52: 527–560 [DOI] [PubMed] [Google Scholar]
- Ryan P, Delhaize E, Randall P. (1995) Malate efflux from root apices and tolerance to aluminium are highly correlated in wheat. Funct Plant Biol 22: 531–536 [Google Scholar]
- Ryan PR, Delhaize E. (2010) The convergent evolution of aluminium resistance in plants exploits a convenient currency. Funct Plant Biol 37: 275–284 [Google Scholar]
- Ryan PR, Raman H, Gupta S, Horst WJ, Delhaize E. (2009) A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiol 149: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PR, Tyerman SD, Sasaki T, Furuichi T, Yamamoto Y, Zhang WH, Delhaize E. (2011) The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J Exp Bot 62: 9–20 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Mori IC, Furuichi T, Munemasa S, Toyooka K, Matsuoka K, Murata Y, Yamamoto Y. (2010) Closing plant stomata requires a homolog of an aluminum-activated malate transporter. Plant Cell Physiol 51: 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37: 645–653 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünmann PH, Richardson AE, Smith FW, Delhaize E. (2004) Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J Exp Bot 55: 855–865 [DOI] [PubMed] [Google Scholar]
- Schünmann PHD, Surin B, Llewllyn DJ, Surin B, Boevink P, DeFeyter RC, Waterhouse PM. (2003) A suite of novel promoters and terminators for plant biotechnology. Funct Plant Biol 30: 453–460 [DOI] [PubMed] [Google Scholar]
- Suoniemi A, Narvanto A, Schulman AH. (1996) The BARE-1 retrotransposon is transcribed in barley from an LTR promoter active in transient assays. Plant Mol Biol 31: 295–306 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Virkki LV, Murer H, Forster IC. (2006) Voltage clamp fluorometric measurements on a type II Na+-coupled Pi cotransporter: shedding light on substrate binding order. J Gen Physiol 127: 539–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Raman H, Zhou MX, Ryan PR, Delhaize E, Hebb DM, Coombes N, Mendham N. (2007) High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 115: 265–276 [DOI] [PubMed] [Google Scholar]
- Wood CC, Petrie JR, Shrestha P, Mansour MP, Nichols PD, Green AG, Singh SP. (2009) A leaf-based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnol J 7: 914–924 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Sasaki T, Sivaguru M, Yamamoto Y, Osawa H, Ahn SJ, Matsumoto H. (2005) Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1). Plant Cell Physiol 46: 812–816 [DOI] [PubMed] [Google Scholar]
- Yang XY, Yang JL, Zhou Y, Piñeros MA, Kochian LV, Li GX, Zheng SJ. (2011) A de novo synthesis citrate transporter, Vigna umbellata multidrug and toxic compound extrusion, implicates in Al-activated citrate efflux in rice bean (Vigna umbellata) root apex. Plant Cell Environ 34: 2138–2148 [DOI] [PubMed] [Google Scholar]
- Yokosho K, Yamaji N, Ueno D, Mitani N, Ma JF. (2009) OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol 149: 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W-H, Ryan PR, Sasaki T, Yamamoto Y, Sullivan W, Tyerman SD. (2008) Characterization of the TaALMT1 protein as an Al3+-activated anion channel in transformed tobacco (Nicotiana tabacum L.) cells. Plant Cell Physiol 49: 1316–1330 [DOI] [PubMed] [Google Scholar]
- Zhou LL, Bai GH, Ma HX, Carver BF. (2007) Quantitative trait loci for aluminum resistance in wheat. Mol Breed 19: 153–161 [Google Scholar]