Summary: Description of a novel feedback loop between the circadian clock and the iron homeostasis network in which the circadian clock regulates the transcription of multiple iron homeostasis genes and iron nutritional status feeds back on the clock to modulate circadian period.
Abstract
In plants, iron (Fe) uptake and homeostasis are critical for survival, and these processes are tightly regulated at the transcriptional and posttranscriptional levels. Circadian clocks are endogenous oscillating mechanisms that allow an organism to anticipate environmental changes to coordinate biological processes both with one another and with the environmental day/night cycle. The plant circadian clock controls many physiological processes through rhythmic expression of transcripts. In this study, we examined the expression of three Fe homeostasis genes (IRON REGULATED TRANSPORTER1 [IRT1], BASIC HELIX LOOP HELIX39, and FERRITIN1) in Arabidopsis (Arabidopsis thaliana) using promoter:LUCIFERASE transgenic lines. Each of these promoters showed circadian regulation of transcription. The circadian clock monitors a number of clock outputs and uses these outputs as inputs to modulate clock function. We show that this is also true for Fe status. Fe deficiency results in a lengthened circadian period. We interrogated mutants impaired in the Fe homeostasis response, including irt1-1, which lacks the major high-affinity Fe transporter, and fit-2, which lacks Fe deficiency-induced TRANSCRIPTION FACTOR1, a basic helix-loop-helix transcription factor necessary for induction of the Fe deficiency response. Both mutants exhibit symptoms of Fe deficiency, including lengthened circadian period. To determine which components are involved in this cross talk between the circadian and Fe homeostasis networks, we tested clock- or Fe homeostasis-related mutants. Mutants defective in specific clock gene components were resistant to the change in period length under different Fe conditions observed in the wild type, suggesting that these mutants are impaired in cross talk between Fe homeostasis and the circadian clock.
The rotation of the earth on its axis means that life has evolved on a world characterized by dramatic, recurrent, and rhythmic environmental change. Considerable evidence has accumulated in support of the hypothesis that the ability to measure and use time to coordinate biology with the environment in anticipation of coming change confers a fitness advantage (Ouyang et al., 1998; Resco et al., 2009; Yerushalmi and Green, 2009). Thus, circadian rhythms have been described in organisms from all domains of life, including bacteria (Mackey et al., 2011) and archaea (Whitehead et al., 2009; Edgar et al., 2012), as well as eucarya, including plants and animals (Lowrey and Takahashi, 2011; McClung, 2011; Zhang et al., 2011).
Circadian rhythms, the subset of rhythms with a period of approximately 24 h, are generated by an endogenous circadian clock. The fitness advantage conferred by the circadian clock emerges from its regulation of many aspects of biology, including basic metabolism, hormone signaling, and responses to biotic and abiotic stress (Doherty and Kay, 2010; Wang et al., 2011; Sahar and Sassone-Corsi, 2012). In plants, the circadian clock is emerging as a key player in the coordination of metabolism and growth (Dodd et al., 2005; Nozue et al., 2007; Gutiérrez et al., 2008; Michael et al., 2008a; Fukushima et al., 2009; Graf et al., 2010; Kerwin et al., 2011; Kunihiro et al., 2011; Nozue et al., 2011). One major mechanism by which the clock coordinates so many pathways and processes is via pervasive control of gene expression at the levels of transcription, transcript processing, and transcript abundance (Covington et al., 2008; Doherty and Kay, 2010; Filichkin et al., 2010; Sanchez et al., 2010). It is becoming increasingly clear that in plants, clock function not only regulates many aspects of cellular metabolism and physiology (Harmer, 2009; McClung and Gutiérrez, 2010; Pruneda-Paz and Kay, 2010; McClung, 2011), including solute transport (Haydon et al., 2011), but is, in turn, modulated by the cellular metabolic state. Plant clock function has been shown to respond to the uptake and homeostasis of Suc (Bläsing et al., 2005; Knight et al., 2008; Dalchau et al., 2011) and other nutrients, including magnesium (Mg; Hermans et al., 2010), copper (Cu; Andrés-Colás et al., 2010; Peñarrubia et al., 2010), and nitrogen (N; Gutiérrez et al., 2008). Similarly, clock function is highly responsive to metabolic state, including redox status, in cyanobacteria (Ivleva et al., 2005; Rust et al., 2011) and mammals (Rutter et al., 2002; Asher and Schibler, 2011; Bass, 2012).
The Arabidopsis (Arabidopsis thaliana) circadian clock is a complex network of interlocked feedback loops. Central to the clock is a feedback loop consisting of two Myb transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), with a transcriptional repressor, TIMING OF CAB EXPRESSION1 (TOC1), of the PSEUDO-RESPONSE REGULATOR (PRR) family (McClung, 2011; Nagel and Kay, 2012). Much of the regulation of clock components is transcriptional, but this is augmented with considerable posttranscriptional (Staiger and Green, 2011) and posttranslational (McClung, 2011; Nagel and Kay, 2012) regulation. Many clock components, including CCA1, LHY, and TOC1, are differentially phosphorylated throughout the circadian cycle, and this posttranslational modification alters protein activity and, in some cases, protein stability (Kusakina and Dodd, 2012). In particular, an F-box protein, ZEITLUPE (ZTL), targets TOC1 and a second PRR protein, PRR5, for ubiquitylation and subsequent proteasomal degradation (Más et al., 2003; Kiba et al., 2007; Fujiwara et al., 2008).
Plants carry out oxygenic photosynthesis and aerobic respiration, both of which require iron (Fe) and generate toxic reactive oxygen species (ROS). The circadian clock regulates photosynthetic activity at the cellular, organismal, and ecosystem levels (Hennessey and Field, 1991; Salomé et al., 2002; Resco de Dios et al., 2012). Not surprisingly, the production of ROS varies throughout the circadian day, and the circadian clock is emerging as a key regulator of cellular ROS-responsive pathways (Edgar et al., 2012; Lai et al., 2012). Principal sources of ROS include the electron transport chains associated with both photosynthesis and respiration. Fe-containing proteins are key components of these electron transport chains, because Fe can exist in multiple redox states, serving as either electron acceptor or donor. The redox-active nature of Fe allows it to generate ROS via the Fenton reaction, in which Fe(II) reacts with hydrogen peroxide to yield hydroxyl free radicals (Halliwell and Gutteridge, 1992). Thus, Fe is a necessary micronutrient, yet its levels must be tightly controlled to prevent the accumulation of damaging levels of ROS.
Fe homeostasis is imposed at multiple levels. First, Fe acquisition is typically limiting because Fe is only sparingly soluble at neutral pH in aerobic conditions (Palmer and Guerinot, 2009). Dicots and some monocots, although not the grasses, apply a tripartite response to assimilate Fe, and each of these three responses is induced in response to Fe limitation. These plants acidify the soil by proton extrusion to increase Fe solubility, reduce Fe from the ferric [Fe(III)] to the ferrous [Fe(II)] form, and then take up ferrous Fe via high-affinity Fe(II) transporters. In Arabidopsis, H+-ATPases of the AHA (for Arabidopsis H+ ATPase) family are likely responsible for the proton extrusion (Santi and Schmidt, 2009), the ferric chelate reductase FERRIC REDUCTION OXYGENASE2 (FRO2) reduces Fe(III) to Fe(II) (Robinson et al., 1999), and the high-affinity Fe(II) transporter IRON-REGULATED TRANSPORTER1 (IRT1) takes up Fe into the root epidermis (Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002). mRNAs for several AHA genes, FRO2, and IRT1 accumulate only under Fe limitation (Palmer and Guerinot, 2009). Second, within the plant, Fe is sequestered in the vacuole (Kim et al., 2006) or stored in ferritin nanocages (Palmer and Guerinot, 2009). FERRITIN (FER) gene expression is induced by Fe sufficiency (Gaymard et al., 1996; Petit et al., 2001).
In plants, the diurnal rhythm in photosynthetic activity confers a rhythm in the generation of ROS (Lai et al., 2012). Because photosynthetic activity is under circadian control and because the circadian clock is emerging as a central player in the control of levels of ROS (Edgar et al., 2012; Lai et al., 2012), we wished to determine whether the circadian clock might regulate the expression of Fe homeostasis genes. Such a link was suggested by the identification of TIME FOR COFFEE (TIC), which encodes a nuclear circadian clock component (Ding et al., 2007), as a regulator of Fe overload-responsive genes, including FER1 (Duc et al., 2009). We examined the transcription and mRNA accumulation of three key Fe homeostasis genes, IRT1, the basic helix-loop-helix transcription factor gene basic Helix-Loop-Helix39 (bHLH39; Vorwieger et al., 2007; Wang et al., 2007, 2013), and FER1. IRT1 and bHLH39 are critical elements of the Fe acquisition mechanism and are both induced by Fe deficiency, whereas FER1 encodes a key Fe storage protein and is induced by Fe sufficiency. We observed that the circadian clock regulates the transcription and mRNA accumulation of each of these three Fe homeostasis genes. In addition, we showed that mutants known to affect clock function also affected the circadian period length of expression of these Fe homeostasis genes. Finally, we also showed that the Fe status of the plants affected circadian period length, indicating that the Fe homeostasis network is not only an output of the circadian clock but that Fe status is a nutritional input that modulates the pace of the clock.
RESULTS
The Plant Circadian Clock Regulates the Expression of Fe Homeostasis Genes
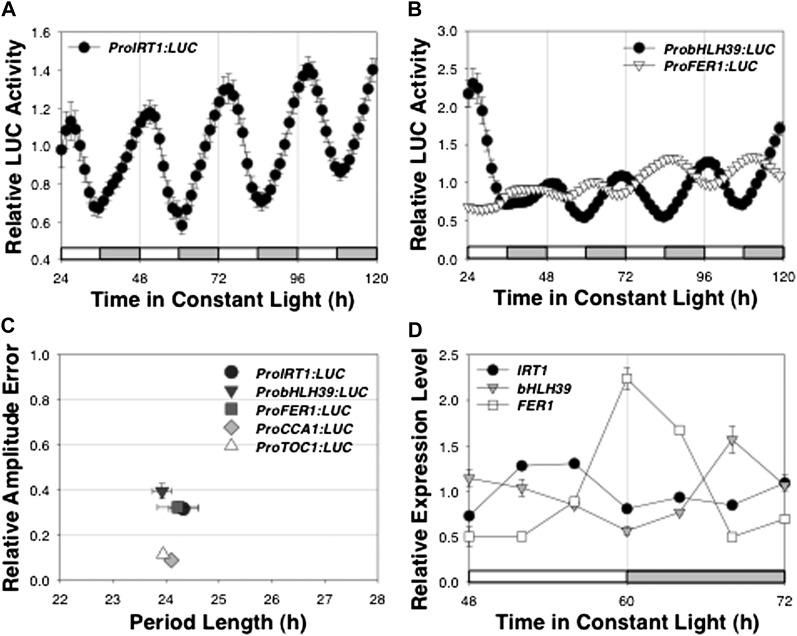
We wished to determine whether key elements of the Fe homeostasis network were under the control of the circadian clock. Although mRNA abundance of IRT1 and FRO2 had earlier been shown to cycle in plants growing in light/dark (LD) cycles (Vert et al., 2003), this cycling had not been shown to persist in continuous conditions and so had not been established to be under circadian clock control. The transcript abundance of FER1 has been shown to cycle in both LD and continuous light (LL; Duc et al., 2009). We first interrogated the DIURNAL database (Mockler et al., 2007) and confirmed that steady-state mRNA abundance for three representative Fe homeostasis genes, IRT1, bHLH39, and FER1, oscillated in LD or warm/cold environmental cycles (Supplemental Fig. S1A). We generated transgenic Arabidopsis lines in the Columbia-0 (Col-0) background carrying gene fusions in which the promoters of IRT1, bHLH39, or FER1 were used to drive the expression of the firefly LUCIFERASE (LUC) gene. Expression of each promoter:LUC (Pro:LUC) fusion transgene recapitulated the spatial patterns previously described on the basis of transcript accumulation (Supplemental Fig. S2). Expression of ProIRT1:LUC was detected only in roots of Fe-deficient but not in Fe-replete roots or in shoots regardless of Fe status (Eide et al., 1996). Expression of ProbHLH39:LUC was detected in both shoots and roots (Vorwieger et al., 2007; Wang et al., 2007). Expression of ProFER1:LUC was detected primarily in shoots but weakly in roots (Gaymard et al., 1996). Expression of ProIRT1:LUC and of ProbHLH39:LUC was greater at Zeitgeber time (ZT)48 than at ZT36 (where ZT0 is dawn), whereas expression of ProFER1:LUC showed the opposite pattern (Supplemental Fig. S2), consistent with the temporal expression patterns shown in Supplemental Figure S1A. We measured LUC activity in seedlings growing in LD cycles and observed robust daily cycling of promoter activity (Supplemental Fig. S1B). To test whether the transcription of these three genes was independent of environmental cycles and, hence, under circadian control, we measured LUC activity in seedlings entrained in LD and released into continuous conditions (LL and constant temperature). Transcription of each Pro:LUC fusion cycled in LL with periods that were statistically indistinguishable from those of ProCCA1:LUC and ProTOC1:LUC (Fig. 1, A–C) and, hence, was clock regulated. We confirmed circadian oscillation of the abundance for each of the IRT1, bHLH39, and FER1 transcripts in LL by quantitative reverse transcription (qRT)-PCR (Fig. 1D).
Figure 1.
Circadian regulation of Fe homeostasis gene expression. A and B, Seedlings growing on minimal medium were entrained to photocycles (LD 12/12 h) for 6 d before release in LL. Average traces (mean ± se, n = 24) are shown for luciferase activity of ProIRT1:LUC (A) and ProbHLH39:LUC and ProFER1:LUC (B) expression, normalized to the average activity over the duration of the experiment. C, Period versus relative amplitude error (RAE) for Fe homeostasis (IRT1, bHLH39, and FER1) and clock (CCA1 and TOC1) gene expression. RAE is a measure of the strength of the oscillation, with RAE = 0 corresponding to a perfect sine wave and RAE = 1 defining the lower limit of statistically significant rhythmicity. D, Transcript levels (from two independent experiments) of IRT1, bHLH39, and FER1 were estimated by qRT-PCR and normalized to tubulin expression. White and gray bars indicate subjective day and night, respectively.
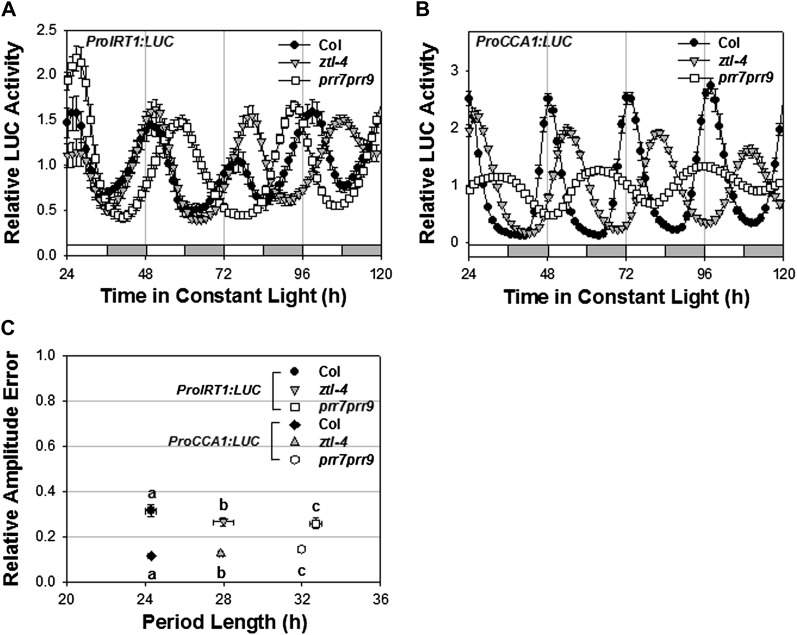
To confirm that the circadian clock regulates the expression of IRT1, we asked whether the period of IRT1 expression was lengthened in mutant backgrounds in which the circadian period is lengthened. Indeed, the period of IRT1 expression lengthened to the same extent as that of the known clock gene, CCA1, in two mutant backgrounds, ztl-4 and prr7 prr9, known to alter period length (Fig. 2; Somers et al., 2000; Farré et al., 2005; Salomé and McClung, 2005), consistent with the clock regulation of IRT1 expression. Therefore, we conclude that the circadian clock regulates the expression of multiple genes involved in the Fe deficiency response (IRT1 and bHLH39) and in Fe storage (FER1).
Figure 2.
Period length of IRT1 promoter activity is lengthened in long-period clock mutants. A and B, Seedlings of the indicated genotypes growing on minimal medium were entrained to photocycles (LD 12/12 h) for 6 d before release in LL. Average traces (mean ± se, n = 12–24) are shown, normalized to the average activity over the duration of the experiment, of ProIRT1:LUC (A) and ProCCA1:LUC (B) expression. White and gray bars indicate subjective day and night, respectively. C, Period versus relative amplitude error (RAE) of ProIRT1:LUC and ProCCA1:LUC expression. RAE is a measure of the strength of the oscillation, with RAE = 0 corresponding to a perfect sine wave and RAE = 1 defining the lower limit of statistically significant rhythmicity. Error bars represent se. Different letters indicate significant differences (P < 0.0001) as determined by ANOVA.
Fe Nutrition Status Regulates the Expression of Fe Homeostasis Gene Transcription
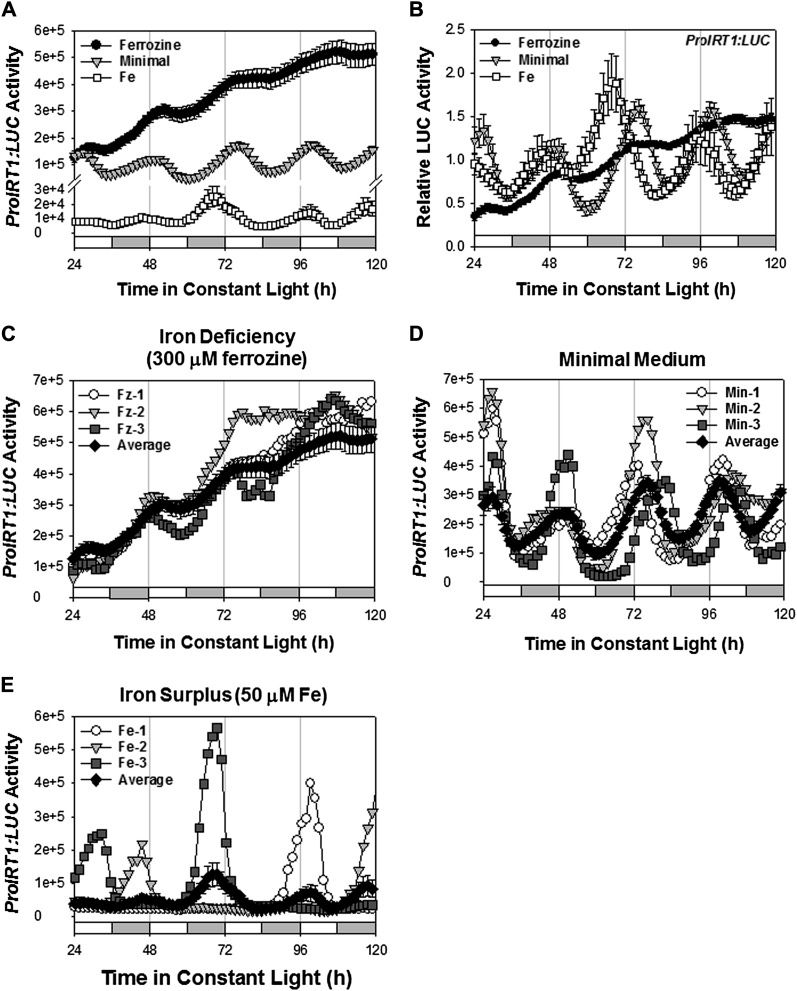
The expression of many Fe homeostasis genes responds to Fe nutrition status. For example, genes associated with Fe acquisition (e.g. IRT1 and bHLH39) are commonly induced during Fe starvation (Vert et al., 2002; Colangelo and Guerinot, 2004; Long et al., 2010), whereas genes associated with Fe storage (e.g. FER1) are induced during Fe sufficiency (Gaymard et al., 1996; Petit et al., 2001). We observed that expression of ProIRT1:LUC was greatly increased during conditions of Fe limitation induced by addition of the Fe chelator, ferrozine (300 µm; Fig. 3, A–C) and was robust in seedlings grown on minimal medium with no Fe supplementation (Fig. 3, A, B, and D). We also detected low-level cycling expression of ProIRT1:LUC during growth in conditions of Fe excess (50 µm Fe; Fig. 3, A, B, and E). Interestingly, although a population of seedlings exhibited daily peaks of expression in Fe-replete conditions, individual seedlings did not express ProIRT1:LUC every day. Rather, an individual seedling expressed ProIRT1:LUC every 2 or 3 d (Fig. 3E). We interpret this to mean that expression on a single day allows sufficient IRT1 activity to take up enough Fe to support growth for more than 1 d, and only when Fe is again limiting is IRT1 expression renewed. Because IRT1 expression is gated by the circadian clock, expression always peaks at the same circadian phase, so the population average, which aggregates expression patterns of multiple seedlings, shows daily expression (Fig. 3, A, B, and E).
Figure 3.
IRT1 promoter activity under different Fe conditions. A and B, Seedlings growing on minimal medium were entrained to photocycles (LD 12/12 h) for 6 d, transferred to three different Fe conditions (300 µm ferrozine, minimal medium, and 50 µm Fe), and released in LL. Absolute (A) and relative (B) traces, normalized to the average activity over the duration of the experiment, of ProIRT1:LUC expression are shown. Data are presented as means ± se (n = 24). C to E, Traces of three individual ProIRT1:LUC seedlings and their average values under the indicated Fe conditions. White and gray bars indicate subjective day and night, respectively.
Fe Nutrition Status Feeds Back to Regulate the Plant Circadian Clock
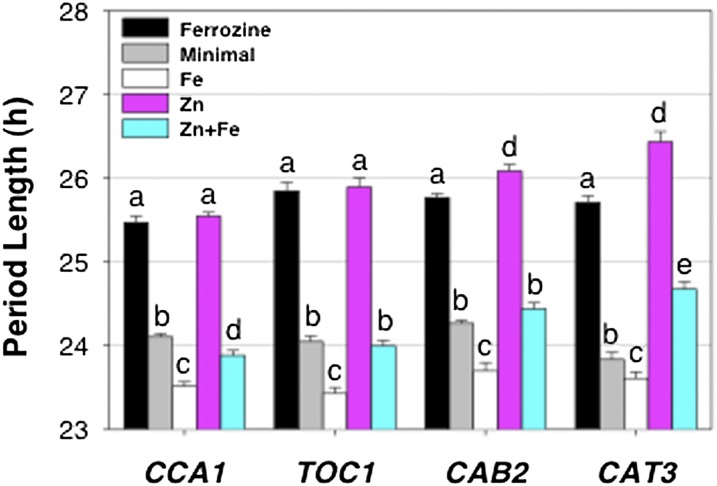
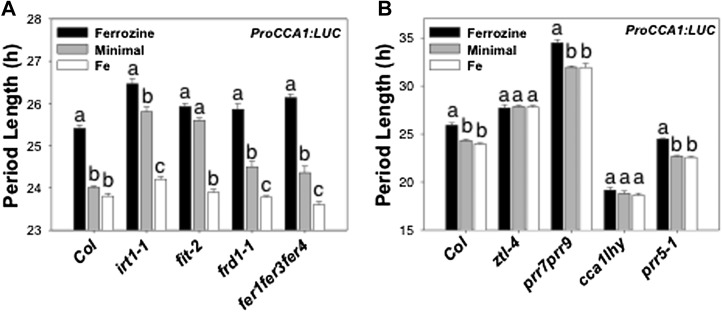
There is considerable interplay between the clock and the uptake and homeostasis of nutrients, including Mg, Cu, and N (Gutiérrez et al., 2008; Andrés-Colás et al., 2010; Hermans et al., 2010; Peñarrubia et al., 2010). The period of multiple clock (CCA1 and TOC1) and clock-controlled (CHLOROPHYLL a/b BINDING PROTEIN2 [CAB2] and CATALASE3 [CAT3]) genes lengthens in response to Fe limitation induced by either chelation of Fe in the growth medium with ferrozine (300 µm) or by the inclusion of excess zinc (Zn; 50 µm), which induces Fe deficiency through competition for the high-affinity Fe uptake system (Fig. 4; Supplemental Fig. S3; Shanmugam et al., 2011). The effect of Zn is confirmed to be through competition for Fe uptake, because the period lengthening in response to excess Zn is blocked by the provision of excess Fe. Furthermore, the period of these clock and clock-controlled genes shortens relative to the period in minimal medium in the presence of excess (50 µm) Fe. Thus, we conclude that the circadian clock responds to Fe status.
Figure 4.
Fe status specifically changes the period length of clock gene promoter activity. Seedlings (n = 24) were grown on minimal medium and entrained to photocycles (LD 12/12 h) for 6 d, transferred to different metal ion conditions (300 µm ferrozine, minimal medium, 50 µm Fe, 50 µm Zn, or 50 µm Zn plus 50 µm Fe), and released in LL. Period data of ProCCA1:LUC, ProTOC1:LUC, ProCAB2:LUC, and ProCAT3:LUC are presented as means ± se. Different letters indicate significant differences (P < 0.004) as determined by ANOVA (comparisons are of the effect of growth conditions on period length as measured within each transgene, and not among different genes; thus, periods measured on different growth media with CCA1 are compared with one another and periods measured on different growth media with TOC1 are compared with one another, but periods measured with CCA1 are not compared with those measured with TOC1).
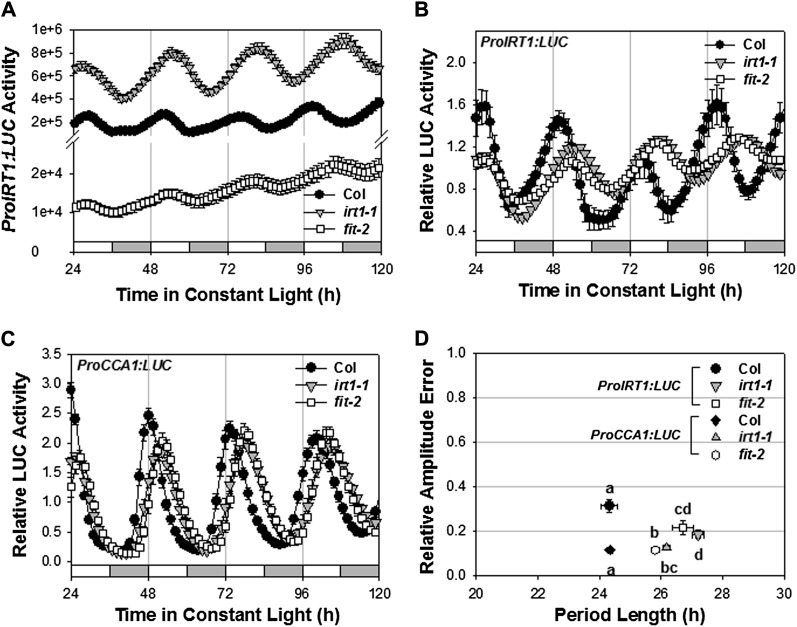
Mutants defective in Fe uptake exhibit Fe deficiency symptoms when grown in minimal medium without Fe supplementation. For example, mutants defective in IRT1, which encodes the high-affinity Fe(II) transporter, are chlorotic and die prior to seed set, but the chlorosis and lethality are rescued by Fe supplementation (Vert et al., 2002). Consistent with these mutants being Fe deficient on minimal medium, ProIRT1:LUC expression was greatly elevated in the irt1-1 mutant (Fig. 5A). FIT encodes an essential bHLH transcription factor necessary for IRT1 induction (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Bauer et al., 2007). Accordingly, IRT1 expression was low in the fit-2 mutant, even though these mutant seedlings are Fe deficient (Fig. 5A). Consistent with irt1-1 and fit-2 mutants being Fe deficient, the period length of both IRT1 and CCA1 expression was lengthened in both irt1-1 and fit-2 mutants (Figs. 5, B–D, and 6A). Similarly, frd1-1 seedlings, which are defective in FRO2 ferric chelate reductase activity and Fe deficient on minimal medium (Robinson et al., 1999), also showed a lengthened period on minimal medium relative to Fe-supplemented medium (Fig. 6A; Supplemental Figs. S4 and S5). The period lengthening in each of the Fe homeostasis mutant backgrounds was seen in minimal medium and exacerbated by the addition of the Fe chelator, ferrozine (Supplemental Fig. S4). In each genotype, the long-period phenotype was rescued by the addition of Fe (50 µm) to the medium, confirming that the period lengthening was in response to Fe limitation. Thus, Fe deficiency resulting from either Fe limitation in the medium or from a genetically imposed impairment of Fe accumulation from the environment resulted in a lengthened circadian period.
Figure 5.
Circadian period is lengthened in Fe deficiency mutants. A to C, Seedlings of the indicated genotypes growing on minimal medium were entrained to photocycles (LD 12/12 h) for 6 d before release in LL. Absolute (A) and relative (B) traces, normalized to the average activity over the duration of the experiment, of ProIRT1:LUC expression (n = 24) and relative traces of ProCCA1:LUC expression (C; n = 12) are presented as means ± se. White and gray bars indicate subjective day and night, respectively. D, Period versus relative amplitude error (RAE) of ProIRT1:LUC and ProCCA1:LUC. RAE is a measure of the strength of the oscillation, with RAE = 0 corresponding to a perfect sine wave and RAE = 1 defining the lower limit of statistically significant rhythmicity. Different letters indicate significant differences in period length (P < 0.025) as determined by ANOVA.
Figure 6.
The response in circadian period to Fe status is altered by mutations affecting Fe homeostasis or circadian clock function. Seedlings (n = 12) growing on minimal medium were entrained to photocycles (LD 12/12 h) for 6 d, transferred to different Fe conditions (300 µm ferrozine, minimal medium, and 50 µm Fe), and released in LL. The period of ProCCA1:LUC in Fe homeostasis (A) or clock-related mutants (B) is shown. Data are presented as means ± se. Different letters indicate significant differences (P < 0.013) as determined by ANOVA (comparisons are of the effect of growth conditions on period length as measured within each mutant, and not among different mutants; thus, periods measured on different growth media with irt1-1 are compared with one another and periods measured on different growth media with fit-2 are compared with one another, but periods measured with irt1-1 are not compared with those measured with fit-2).
We also examined the effects of perturbation of Fe homeostasis via the loss of leaf ferritin in the fer1 fer3 fer4 triple mutant (Ravet et al., 2009). The period of ProCCA1:LUC expression was lengthened in the triple mutant relative to the wild type both in seedlings grown on minimal medium and in Fe-deficient (ferrozine-treated) seedlings (Fig. 6A; Supplemental Figs. S4 and S5). We also note that the period in Fe-replete seedlings was shortened in the triple fer1 fer3 fer4 mutant (Fig. 6A; Supplemental Figs. S4 and S5). The mutant lacks ferritin in the shoot and, as a consequence, mutant plants have higher levels of ROS (Ravet et al., 2009). ROS has been shown to alter the expression of the evening-expressed FLAVIN-BINDING, KELCH REPEAT, F BOX1 (FKF1) clock-controlled gene, although not the midday-expressed CAB2 gene (Lai et al., 2012), and we speculate that the period shortening seen in fer1 fer3 fer4 in Fe-supplemented conditions may result from a feedback of elevated ROS on clock function. However, the mechanistic details and target(s) of this feedback remain mysterious.
To determine which circadian clock components are required for the period-lengthening response to Fe deficiency, we examined a number of single and double mutants with defective clocks (Fig. 6B; Supplemental Fig. S6). The long-period ztl-4 mutant (Michael et al., 2003) did not respond to Fe status and exhibited statistically indistinguishable, albeit long, periods in minimal, Fe-deficient (+ferrozine) and Fe-sufficient (+Fe) media. In contrast, the long-period prr7 prr9 double mutant (Farré et al., 2005; Salomé and McClung, 2005) responded to Fe deficiency with lengthened period. Similarly, the prr5-1 mutant, which exhibits only a slight shortening of circadian period length, responded to Fe deficiency with a lengthened period. The short-period cca1 lhy double mutant did not respond to Fe status and exhibited statistically indistinguishable, albeit short, periods in minimal, Fe-deficient (+ferrozine) and Fe-sufficient (+Fe) media. This loss of sensitivity to Fe in circadian period was seen both in the Wassilewskija (Ws) background with the cca1-11 lhy-21 double mutant (Hall et al., 2003; data not shown) and in the Col-0 background, in which the cca1-1 allele, originally isolated in the Ws background (Green and Tobin, 1999), had been introgressed into the lhy-20 mutant in Col-0 through five generations of backcrossing. These data implicate CCA1, LHY, and ZTL as circadian clock targets of the Fe deficiency signal.
DISCUSSION
The circadian clock exerts pervasive control of gene expression (Michael et al., 2008b) and thereby regulates many aspects of physiology and metabolism, including photosynthetic carbon assimilation (Hennessey and Field, 1991; Dodd et al., 2004), utilization of stored photosynthate (Graf and Smith, 2011; Stitt and Zeeman, 2012), and growth (Nozue et al., 2007; Nusinow et al., 2011; Yazdanbakhsh et al., 2011). Circadian regulation of growth suggests that there should be concomitant oscillations in water and solute fluxes (Haydon et al., 2011). Data have accumulated supporting circadian oscillations in fluxes of several molecules, including carbon into sugar and from starch (Dodd et al., 2004; Stitt and Zeeman, 2012), inorganic macronutrients (e.g. N, sulfur, K+; Gutiérrez et al., 2008; Haydon et al., 2011) and micronutrients (e.g. Mg and Cu; Andrés-Colás et al., 2010; Hermans et al., 2010; Peñarrubia et al., 2010), and the key signaling molecule Ca2+ (Dodd et al., 2006; Xu et al., 2007; Haydon et al., 2011). Moreover, some of these cycling solutes have been shown to serve as feedback regulators of clock function. For example, levels of sugars (Bläsing et al., 2005; James et al., 2008; Dalchau et al., 2011), Cu (Andrés-Colás et al., 2010; Peñarrubia et al., 2010), Mg (Hermans et al., 2010), and N (Gutiérrez et al., 2008) each affect clock function.
We have uncovered a reciprocal relationship between the micronutrient Fe and the circadian clock. The circadian clock regulates the transcription and transcript accumulation of the IRT1 gene encoding the high-affinity Fe(II) transporter responsible for Fe uptake from the soil (Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002) as well as of a gene, bHLH39, encoding a key transcription factor in the Fe deficiency response (Wang et al., 2013). In addition, we describe circadian regulation of FER1, which encodes a key Fe storage protein, ferritin (Gaymard et al., 1996). This establishes broad circadian control of the Fe homeostasis gene network. Moreover, we show that Fe status feeds back to regulate clock function, because circadian period lengthens during Fe deficiency. Such feedback regulation in which the clock regulates Fe homeostasis while Fe status feeds back to modulate clock function has also been observed in Drosophila melanogaster (Mandilaras and Missirlis, 2012). Down-regulation of clock-neuron expression of an Fe storage ferritin gene or of an Fe-carrying transferrin gene disrupted circadian rhythmicity in D. melanogaster (Mandilaras and Missirlis, 2012). In mice, there is a reciprocal regulation of the biosynthesis of heme, the important Fe-binding porphyrin, and the circadian clock, although Fe status has not been directly implicated (Kaasik and Lee, 2004). Heme levels in mice cycle with circadian period (Kaasik and Lee, 2004), and heme binding to the orphan nuclear receptor Rev-erbα, a critical negative component of the mammalian circadian clock, regulates its interaction with a nuclear receptor corepressor complex, thereby affecting broad patterns of gene expression (Yin et al., 2007). However, this interaction of the clock with heme in mice is distinct from the interaction of the clock with Fe homeostasis in fruit flies. Although the expression of heme biosynthetic and degradative genes is under robust circadian control in D. melanogaster heads (Ceriani et al., 2002), down-regulation of multiple D. melanogaster heme biosynthetic genes did not disrupt circadian rhythmicity (Mandilaras and Missirlis, 2012).
In plants, CCA1 is emerging as a critical hub in the circadian network that is the target for nutrient status input to the circadian clock. For example, the amplitude of CCA1 and LHY transcript cycling increased in response to the addition of Cu and decreased in response to Cu chelation (Andrés-Colás et al., 2010). Mg chelation increased CCA1, LHY, and PRR9 expression (Hermans et al., 2010). Systems analysis established CCA1 as a critical hub in the N-responsive gene network, and N metabolites were shown to shift circadian phase as monitored by CCA1 transcription (Gutiérrez et al., 2008). In this study, we have shown that the period lengthening in response to Fe deficiency was abolished in the cca1 lhy double mutant, establishing a requirement for either CCA1 or LHY, or both, in the response of the clock to Fe status. A second clock mutant, ztl-4, also was defective in lengthening period in response to Fe deficiency, which implicates ZTL in the clock response to Fe deficiency. ZTL is not believed to directly regulate either CCA1 or LHY, which may suggest a second route of Fe signaling to the clock, independent of these two key transcription factors. ZTL is an F-box protein that regulates the proteasomal degradation of two important clock proteins, TOC1 and PRR5 (Más et al., 2003; Kiba et al., 2007; Fujiwara et al., 2008), both of which are regulators of CCA1 and LHY expression (Nakamichi et al., 2010; Gendron et al., 2012; Huang et al., 2012; Pokhilko et al., 2012). Thus, it is possible that the expression of CCA1 and LHY is the ultimate target of Fe status signaling via ZTL.
MATERIALS AND METHODS
Plant Mutant Genotypes
The following Arabidopsis (Arabidopsis thaliana) mutant genotypes were used: fer1 fer3 fer4 (Ravet et al., 2009), fit-2 (Colangelo and Guerinot, 2004), frd1-1 (Yi and Guerinot, 1996), irt1-1 (Vert et al., 2002), prr7-3 prr9-1 (Salomé and McClung, 2005), prr5-1, and ztl-4 (Michael et al., 2003) in the Col-0 background; cca1-1 lhy-20, in which the cca1-1 allele was originally isolated in the Ws background (Green and Tobin, 1999), was introgressed into a lhy-20 mutant in the Col-0 background (Michael et al., 2003) through five sequential backcrosses; and cca1-11 lhy-21 (Hall et al., 2003) in the Ws background.
Generation of Constructs and Transgenic Plants
Firefly luciferase was driven from Arabidopsis clock and clock-controlled gene promoters, including ProCCA1:LUC and ProLHY:LUC (Salomé and McClung, 2005), ProTOC1:LUC (Michael and McClung, 2002), ProCAB2:LUC (Millar et al., 1992), and ProCAT3:LUC (Michael and McClung, 2002). Promoters of IRT1 (−1,086 to −63, where the A of the ATG start codon = +1), of bHLH39 (−755 to −1), and of FER1 (−1,394 to −5) were amplified from Col-0 genomic DNA using gene-specific primers (Supplemental Table S1). The amplified products were cloned into pCR8/GW/TOPO (Invitrogen) and subcloned into pZPΩLUC+ (Schultz et al., 2001). The resulting binary vectors were introduced into Agrobacterium tumefaciens strain AGL1 by electroporation, and ProbHLH39:LUC and ProFER1:LUC were transformed into Col-0 (Bechtold et al., 1993). ProIRT1:LUC was initially transformed into Col-gl1 and moved into the Col-0 wild-type background by genetic crossing. With the exception of fer1 fer3 fer4, Pro:LUC transgenes were introduced into the mutants via genetic crossing and selection of F2 plants with the appropriate morphological and/or circadian phenotype in LL after LD entrainment. fer1 fer3 fer4 plants were transformed by infiltration via A. tumefaciens (Bechtold et al., 1993).
Bioluminescence Assay
Rhythm assays were performed as described (Salomé and McClung, 2005) except that seeds were sterilized, stratified in the dark at 4°C for 3 d, and sown on minimal medium consisting of 2 mm Ca(NO3)2, 0.75 mm K2SO4, 0.65 mm MgSO4, 0.1 mm KH2PO4, 10 µm H3BO3, 0.1 µm MnSO4, 0.05 µm CuSO4, 0.05 µm ZnSO4, 0.005 µm (NH3)6Mo7O24, 0.5 g of MES, and 0.5% Suc (w/v), adjusted to pH 6.0 and solidified with 0.7% type M agar (Marschner et al., 1982; Yi and Guerinot, 1996). Seedlings were entrained for 6 d in photocycles (LD 12/12 h) before transfer to LL for LUC activity measurement using a Packard TopCount Luminometer. Fe deficiency was imposed by the addition of the Fe chelator ferrozine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate (HACH Chemical)] to the minimal medium at 300 µm (Yi and Guerinot, 1996). Fe repletion was imposed through the addition of 50 µm Fe(III)-EDTA to the minimal medium (Yi and Guerinot, 1996). Rhythms were analyzed by fast Fourier transform nonlinear least-squares (Plautz et al., 1997). Whole-seedling LUC imaging was performed using an ORCA II ER CCD camera (C4742-98 ERG; Hamamatsu Photonics; http://www.hamamatsu.com), with data collected at the time of peak expression for each transgene (Fig. 1, A and B) with 60-min exposure times. Images were analyzed with MetaMorph software (Molecular Devices; http://www.moleculardevices.com/Products/Software/Meta-Imaging-Series.html).
Expression Analysis by qRT-PCR
Seedlings were entrained for 10 d in photocycles (LD 12/12 h) and transferred to LL. Samples were collected every 4 h for the following 3 d. RNA was extracted using the Qiagen RNeasy Plant Mini Kit. First-strand complementary DNA synthesis used 2 μg of total RNA with the SuperScript III first-strand synthesis system (Invitrogen). The complementary DNA was diluted 10 times with water, and 1 μL was used for PCR amplification using a SYBR Premix Ex Taq II (Takara) with gene-specific primers (Supplemental Table S1). mRNA abundances were calculated using the comparative cycle threshold method, with TUB3 (At5g62700) as the normalization control.
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this study are as follows: bHLH39 (At3g56980), CAB2 (At1g29920), CAT3 (At1g20620), CCA1 (At2g46830), CCR2 (At2g21660), CHE (At5g08330), FER1 (At5g01600), FER3 (At3g56090), FER4 (At2g40300), FIT (At2g28160), FRD1/FRO2 (At1g01580), GI (At1g22770), IRT1 (At4g19690), LHY (At1g01060), PRR5 (At5g24470), PRR7 (At5g02810), PRR9 (At2g46790), TIC (At3g22380), TOC1 (At5g61380), and ZTL (At5g57360).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Diurnal and circadian regulation of Fe homeostasis gene expression.
Supplemental Figure S2. Temporal and spatial expression of Fe homeostasis gene expression.
Supplemental Figure S3. Fe status specifically changes period length of clock gene promoter activity.
Supplemental Figure S4. Circadian period is lengthened in Fe homeostasis mutants.
Supplemental Figure S5. Circadian period is lengthened in Fe homeostasis mutants.
Supplemental Figure S6. The response in circadian period to Fe status is altered by mutations affecting Fe homeostasis or circadian clock function.
Supplemental Table S1. Oligonucleotide primers used in this study.
Acknowledgments
We thank the members of our laboratories for helpful discussions. We thank J. Hayes, Perkin-Elmer support engineer, for keeping the TopCount running.
Glossary
- Mg
magnesium
- Cu
copper
- Fe
iron
- N
nitrogen
- ROS
reactive oxygen species
- LD
light/dark
- Col-0
Columbia-0
- LL
constant light and temperature
- qRT
quantitative reverse transcription
- Zn
zinc
- Ws
Wassilewskija
References
- Andrés-Colás N, Perea-García A, Puig S, Peñarrubia L. (2010) Deregulated copper transport affects Arabidopsis development especially in the absence of environmental cycles. Plant Physiol 153: 170–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Schibler U. (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13: 125–137 [DOI] [PubMed] [Google Scholar]
- Bass J. (2012) Circadian topology of metabolism. Nature 491: 348–356 [DOI] [PubMed] [Google Scholar]
- Bauer P, Ling HQ, Guerinot ML. (2007) FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem 45: 260–261 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199 [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible W-R, Stitt M. (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. (2002) Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci 22: 9305–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, Stancombe MA, Haydon MJ, Stan G-B, Gonçalves JM, et al. (2011) The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci USA 108: 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Millar AJ, Davis AM, Davis SJ. (2007) TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19: 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Jakobsen MK, Baker AJ, Telzerow A, Hou S-W, Laplaze L, Barrot L, Poethig RS, Haseloff J, Webb AAR. (2006) Time of day modulates low-temperature Ca2+ signals in Arabidopsis Plant J 48: 962–973 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Parkinson K, Webb AAR. (2004) Independent circadian regulation of assimilation and stomatal conductance in the ztl-1 mutant of Arabidopsis. New Phytol 162: 63–70 [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Kay SA. (2010) Circadian control of global gene expression patterns. Annu Rev Genet 44: 419–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc C, Cellier F, Lobréaux S, Briat J-F, Gaymard F. (2009) Regulation of iron homeostasis in Arabidopsis thaliana by the clock regulator time for coffee. J Biol Chem 284: 36271–36281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong W-K, Mockler TC. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE. (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K. (2009) Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci USA 106: 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F, Boucherez J, Briat JF. (1996) Characterization of a ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem J 318: 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM. (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Smith AM. (2011) Starch and the clock: the dark side of plant productivity. Trends Plant Sci 16: 169–175 [DOI] [PubMed] [Google Scholar]
- Green RM, Tobin EM. (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96: 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, et al. (2003) The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. (1992) Biologically relevant metal ion-dependent hydroxyl radical generation: an update. FEBS Lett 307: 108–112 [DOI] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Bell LJ, Webb AAR. (2011) Interactions between plant circadian clocks and solute transport. J Exp Bot 62: 2333–2348 [DOI] [PubMed] [Google Scholar]
- Hennessey TL, Field CB. (1991) Circadian rhythms in photosynthesis: oscillations in carbon assimilation and stomatal conductance under constant conditions. Plant Physiol 96: 831–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jásik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C. (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50: 587–597 [DOI] [PubMed] [Google Scholar]
- Hermans C, Vuylsteke M, Coppens F, Craciun A, Inzé D, Verbruggen N. (2010) Early transcriptomic changes induced by magnesium deficiency in Arabidopsis thaliana reveal the alteration of circadian clock gene expression in roots and the triggering of abscisic acid-responsive genes. New Phytol 187: 119–131 [DOI] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P. (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. (2005) LdpA: a component of the circadian clock senses redox state of the cell. EMBO J 24: 1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Wang H-Y, Reidt W, Weisshaar B, Bauer P. (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG. (2008) The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322: 1832–1835 [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. (2004) Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430: 467–471 [DOI] [PubMed] [Google Scholar]
- Kerwin RE, Jimenez-Gomez JM, Fulop D, Harmer SL, Maloof JN, Kliebenstein DJ. (2011) Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua N-H. (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li LT, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Knight H, Thomson AJW, McWatters HG. (2008) SENSITIVE TO FREEZING6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol 148: 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, Mizuno T. (2011) Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol 52: 1315–1329 [DOI] [PubMed] [Google Scholar]
- Kusakina J, Dodd AN. (2012) Phosphorylation in the plant circadian system. Trends Plant Sci 17: 575–583 [DOI] [PubMed] [Google Scholar]
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JHM, Dijkwel PP. (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci USA 109: 17129–17134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. (2011) Genetics of circadian rhythms in mammalian model organisms. Adv Genet 74: 175–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey SR, Golden SS, Ditty JL. (2011) The itty-bitty time machine genetics of the cyanobacterial circadian clock. Adv Genet 74: 13–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandilaras K, Missirlis F. (2012) Genes for iron metabolism influence circadian rhythms in Drosophila melanogaster. Metallomics 4: 928–936 [DOI] [PubMed] [Google Scholar]
- Marschner H, Römheld V, Ossenberg-Neuhaus H. (1982) Rapid method for measuring changes in pH and reducing processes along roots of intact plants. Z Pflanzenphysiol 105: 407–416 [Google Scholar]
- Más P, Kim W-Y, Somers DE, Kay SA. (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- McClung CR. (2011) The genetics of plant clocks. Adv Genet 74: 105–139 [DOI] [PubMed] [Google Scholar]
- McClung CR, Gutiérrez RA. (2010) Network news: prime time for systems biology of the plant circadian clock. Curr Opin Genet Dev 20: 588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J. (2008a) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, McClung CR. (2002) Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol 130: 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008b) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua N-H, Kay SA. (1992) A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J. (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Nagel DH, Kay SA. (2012) Complexity in the wiring and regulation of plant circadian networks. Curr Biol 22: R648–R657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H. (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nozue K, Harmer SL, Maloof JN. (2011) Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 156: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA 95: 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Guerinot ML. (2009) Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol 5: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñarrubia L, Andrés-Colás N, Moreno J, Puig S. (2010) Regulation of copper transport in Arabidopsis thaliana: a biochemical oscillator? J Biol Inorg Chem 15: 29–36 [DOI] [PubMed] [Google Scholar]
- Petit J-M, van Wuytswinkel O, Briat J-F, Lobréaux S. (2001) Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. J Biol Chem 276: 5584–5590 [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ. (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Kay SA. (2010) An expanding universe of circadian networks in higher plants. Trends Plant Sci 15: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57: 400–412 [DOI] [PubMed] [Google Scholar]
- Resco V, Hartwell J, Hall A. (2009) Ecological implications of plants ability to tell the time. Ecol Lett 12: 583–592 [DOI] [PubMed] [Google Scholar]
- Resco de Dios V, Goulden ML, Ogle K, Richardson AD, Hollinger DY, Davidson EA, Alday JG, Barron-Gafford GA, Carrara A, Kowalski AS, et al. (2012) Endogenous circadian regulation of carbon dioxide exchange in terrestrial ecosystems. Glob Change Biol 18: 1956–1970 [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rust MJ, Golden SS, O’Shea EK. (2011) Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science 331: 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight SL. (2002) Metabolism and the control of circadian rhythms. Annu Rev Biochem 71: 307–331 [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. (2012) Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab 23: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, McClung CR. (2005) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Michael TP, Kearns EV, Fett-Neto AG, Sharrock RA, McClung CR. (2002) The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol 129: 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, et al. (2010) A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116 [DOI] [PubMed] [Google Scholar]
- Santi S, Schmidt W. (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam V, Lo J-C, Wu C-L, Wang S-L, Lai C-C, Connolly EL, Huang J-L, Yeh K-C. (2011) Differential expression and regulation of iron-regulated metal transporters in Arabidopsis halleri and Arabidopsis thaliana: the role in zinc tolerance. New Phytol 190: 125–137 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Staiger D, Green R. (2011) RNA-based regulation in the plant circadian clock. Trends Plant Sci 16: 517–523 [DOI] [PubMed] [Google Scholar]
- Stitt M, Zeeman SC. (2012) Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol 15: 282–292 [DOI] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert GA, Briat J-F, Curie C. (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol 132: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwieger A, Gryczka C, Czihal A, Douchkov D, Tiedemann J, Mock H-P, Jakoby M, Weisshaar B, Saalbach I, Bäumlein H. (2007) Iron assimilation and transcription factor controlled synthesis of riboflavin in plants. Planta 226: 147–158 [DOI] [PubMed] [Google Scholar]
- Wang H-Y, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P. (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling H-Q. (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant (in press) [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, Lee DU, Fu X-D, Dong X. (2011) Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead K, Pan M, Masumura K-i, Bonneau R, Baliga NS. (2009) Diurnally entrained anticipatory behavior in archaea. PLoS ONE 4: e5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hotta CT, Dodd AN, Love J, Sharrock R, Lee YW, Xie Q, Johnson CH, Webb AAR. (2007) Distinct light and clock modulation of cytosolic free Ca2+ oscillations and rhythmic CHLOROPHYLL A/B BINDING PROTEIN2 promoter activity in Arabidopsis. Plant Cell 19: 3474–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanbakhsh N, Sulpice R, Graf A, Stitt M, Fisahn J. (2011) Circadian control of root elongation and C partitioning in Arabidopsis thaliana. Plant Cell Environ 34: 877–894 [DOI] [PubMed] [Google Scholar]
- Yerushalmi S, Green RM. (2009) Evidence for the adaptive significance of circadian rhythms. Ecol Lett 12: 970–981 [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML. (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. (2007) Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ. (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zhang L, Jones CR, Ptacek LJ, Fu Y-H. (2011) The genetics of the human circadian clock. Adv Genet 74: 231–247 [DOI] [PubMed] [Google Scholar]